Abstract
RNA silencing in plants is a natural defense system against foreign genetic elements including viruses. This natural antiviral mechanism has been adopted to develop virus-resistant plants through expression of virus-derived double-stranded RNAs or hairpin RNAs, which in turn are processed into small interfering RNAs (siRNAs) by the host's RNA silencing machinery. While these virus-specific siRNAs were shown to be a hallmark of the acquired virus resistance, the functionality of another set of the RNA silencing-related small RNAs, microRNAs (miRNAs), in engineering plant virus resistance has not been extensively explored. Here we show that expression of an artificial miRNA, targeting sequences encoding the silencing suppressor 2b of Cucumber mosaic virus (CMV), can efficiently inhibit 2b gene expression and protein suppressor function in transient expression assays and confer on transgenic tobacco plants effective resistance to CMV infection. Moreover, the resistance level conferred by the transgenic miRNA is well correlated to the miRNA expression level. Comparison of the anti-CMV effect of the artificial miRNA to that of a short hairpin RNA-derived small RNA targeting the same site revealed that the miRNA approach is superior to the approach using short hairpin RNA both in transient assays and in transgenic plants. Together, our data demonstrate that expression of virus-specific artificial miRNAs is an effective and predictable new approach to engineering resistance to CMV and, possibly, to other plant viruses as well.
RNA silencing is a natural regulatory mechanism of eukaryotes acting against invasive nucleic acids or modulating endogenous gene expression (4, 41). Silencing is initiated when double-stranded RNAs (dsRNAs) or hairpin RNAs (hpRNAs) are processed into 21- to 24-nucleotide (nt) small interfering RNA (siRNA) or microRNA (miRNA) duplexes by dsRNA-specific RNase III-type Dicer enzymes. These small RNAs (sRNAs) are then incorporated into the RNA-induced silencing complex to guide the degradation or translation repression of cRNA targets (4, 39). Unlike animals, higher plants have evolved diversified RNA silencing pathways to generate different sRNA classes with specialized functions (9, 40). For example, Arabidopsis thaliana encodes four Dicer-like (DCL) proteins (DCL1 to DCL4). While DCL1 mainly processes imperfectly base-paired fold-back precursors to produce miRNAs (3), the other three DCL proteins are responsible for generating various endogenous siRNAs from perfect dsRNAs, such as stress-related 24-nt natural-antisense-transcript siRNAs by DCL2 (6), 24-nt repeat-associated siRNAs that guide heterochromatin formation by DCL3 (46), and 21-nt trans-acting (ta-) siRNAs that control some aspects of plant development by DCL4 (17, 45). However, there is some functional redundancy among the four Arabidopsis DCL proteins. For instance, DCL1 is also capable of producing 21-nt ta-siRNAs in the dcl2 dcl3 dcl4 triple mutant (7), and DCL2 can generate 22-nt ta-siRNAs when the DCL4 activity is compromised (17, 45). In addition, plant DCL enzymes can also generate exogenous siRNAs that are derived from invading viruses or hpRNA-forming transgenes. siRNAs that mediate antiviral defense consist of primarily 21-nt and 22-nt species which are produced by DCL4 and DCL2, respectively (1, 7, 13, 16, 29, 46). Specifically, the 21-nt siRNAs appear to form the first line of antiviral defense, whereas the DCL2-dependent 22-nt siRNAs become an alternative arsenal when DCL4 is inactivated (7, 13). Although miRNAs have been shown to be involved in developmental regulation and many other cellular processes (3), the possibility that miRNA-guided RNA silencing could play a role in host defense against virus infection has been proposed (25). Indeed, the antiviral function of natural miRNAs has been described in animal systems (23), and more recently the plant miRNA pathway was also shown to be implicated in impairing the replication of an engineered potyvirus bearing an miRNA target (36).
Engineered antiviral strategies in both animals and plants mimicked natural RNA silencing mechanisms. In mammalian cells, two plasmid vector-based RNA interference approaches have been used to induce specific inhibition of virus replication. The first approach was to use RNA polymerase III (Pol III) promoter-driven short hairpin RNA (shRNA) expression vectors to direct siRNA expression (10, 27). The second relied on a pre-miRNA backbone to produce artificial miRNA, transcribed by Pol II or Pol III promoters (5, 47). The latter approach was shown to have more reliable and profound effects on knock-down of viral genes (5). In plants, RNA silencing-based antiviral strategies involved expression of long pathogen-derived hpRNAs (37) which in turn would be processed into siRNAs. It became clear recently that the hpRNA-derived siRNA classes that are responsible for directing target RNA degradation are predominantly the DCL4-processed 21-nt species and the 22-nt species produced by DCL2 when DCL4 is absent (16), very similar to the siRNA size profile observed in antiviral silencing (7, 13, 16). In most of the reported long viral hpRNA-transgenic studies, the observed virus resistance level was correlated to the abundance of virus-specific siRNAs in plants (12, 22, 28), reminiscent of the earlier findings that siRNAs serve as molecular markers for active RNA silencing (19). It is therefore interesting to see whether directly expressing virus-related sRNAs in plants through the miRNA precursor or the shRNA structure can achieve virus resistance, as in animal systems. Relevant techniques for generating RNA silencing-related sRNAs in plants are already available. Arabidopsis 7SL RNA Pol III promoter-driven shRNA vector was used to produce glucuronidase (GUS)-specific 21-nt siRNAs which could down-regulate GUS expression in transgenic plants (26). More recently, miRNA precursor vectors under the control of an RNA Pol II promoter have been exploited as carriers to express artificial miRNAs which can specifically inhibit the expression of targeted host genes in plants (2, 31, 35).
In this research, we designed two plant expression vectors, one harboring a Cauliflower mosaic virus (CaMV) 35S promoter-driven miRNA precursor sequence and the other harboring a plant Pol III promoter-driven shRNA construct, to generate an artificial miRNA and an sRNA tentatively named shRNA-derived sRNA (sh-sRNA), respectively, both targeting the same region of the gene coding for the viral suppressor 2b of Cucumber mosaic virus (CMV). CMV 2b is one of the first examined viral suppressors of RNA silencing (8), and earlier studies have revealed that it can block the movement of the systemic silencing signal, disrupt DNA methylation-induced silencing (18), and interfere with the host's salicylic acid-mediated antiviral pathway (21). Recently, CMV 2b was found to directly interact with Arabidopsis Argonaute1 and block its slicer activity (48). Through this research we attempted to address whether expression of artificial sRNAs targeting the suppressor 2b sequences can counteract the suppressor function of 2b and confer plants with CMV resistance. Here, we show that expression of 2b-specific miRNA and sh-sRNA can inhibit the expression of the 2b gene and 2b suppressor function in transient expression assays. Transgenic tobacco plants expressing the 2b-specific miRNA or sh-sRNA inhibited multiplication of the CMV inoculum and exhibited resistance to CMV. In both systems, expression of miRNA was more effective than expression of sh-sRNA in inhibiting viral functions. There was a strong correlation between virus resistance and the expression level of the miRNA but not of the sh-sRNA. We conclude that the miRNA-mediated viral silencing is an effective approach against CMV infection, and in light of a recent report showing high resistance in Arabidopsis to two other plant RNA viruses by expressing virus-specific miRNAs (30), we believe that this miRNA-mediated approach would be useful to engineer crop plants resistant to a wide range of viruses.
MATERIALS AND METHODS
Construction of DNA vectors.
The −319 to −7 region of the Arabidopsis thaliana U6-29 promoter (42) was PCR-amplified from A. thaliana (ecotype Columbia) genomic DNA. To facilitate cloning into pUC19, the original −6 to −1 sequence of the U6-29 promoter was changed to a BamHI site. The modified promoter was cloned into pUC19 to generate pUC-U6. The sh-sRNA target site in the 2b gene of CMV strain SD (SD-CMV) (GenBank accession no. D86330) was identified using the siRNA Target Finder program (http://www. ambion.com/techlib/misc/siRNA_finder.html). Accordingly, two cDNA oligonucleotides, 5′-GATCCGTAGATGGTTCGGAACTGATTCAAGAGATCAGTTCCGAACCATCTACTTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAAGTAGATGGTTCGGAACTGATCTCTTGAATCAGTTCCGAACCATCTACG-3′ (the sense and antisense sequences of the sh-sRNA target site are underlined), were synthesized and annealed. The resulting DNA duplex (called sh2b) with overhangs of the BamHI and HindIII sites was first cloned into pUC-U6, and then the U6-sh2b cassette was transferred into the plant binary vector pCAMBIA 1300 to generate pU6-sh2b to express a 21-nt 2b-specific sh-sRNA (shsR2b) in planta.
The CaMV 35S promoter was chosen to direct the expression of 2b-specific miRNA (miR2b) from the A. thaliana pre-miR171a backbone (31). Two primers, 5′-CGGGATCCATGAGAGAGTCCCTTTGTAGATGGTACGGAACTTATAGATCTTACCTGACCACACACG-3′ and 5′-GCGAGCTCACGAGAGAGTACTGAGTAGATGGTTCGGAACTGATAGATAATCTAGAGAGAATAATG-3′, corresponding to the 5′ (sense) and 3′ (antisense) portion of pre-miR171a, respectively, and each harboring a 21-nt 2b-specific sequence (underlined), were annealed to the DNA template present in plasmid T-miR171prec (31) and extended by PCR. The resulting amplified product pre-miR171a/CMV 2b chimeric DNA (miR2bprec) was cloned into the plasmid pBI221 via the BamHI and SacI sites to generate pBI-35S-miR2bprec. The 35S-miR2bprec expression cassette was transferred into pCAMBIA 1300 to generate p35S-miR2bprec.
The binary vector pCAMBIA 1302 containing the 35S green fluorescent protein (GFP) cassette (p35S-GFP) has been described before (GenBank accession no. AF234298). All the pCAMBIA 1300 derivatives contain a hygromycin phosphotransferase (hpt)-selectable marker gene under the control of a CaMV 35S promoter.
Agroinfiltration, plant transformation, and virus challenge.
The DNA vectors were separately moved into Agrobacterium tumefaciens strain EH105. Agrobacterium-mediated transient expression in Nicotiana benthamiana leaves was conducted by pressure infiltration essentially as described previously (24) with minor modifications. The bacterial cultures were adjusted to a final optical density at 600 nm (OD600) of 1.0 prior to infiltration. For coinfiltrations, the final OD ratio of the sRNA-expressing strain to the target-expressing strain was 3:1. The Agrobacterium strain harboring p35S-miRGFP which can express a gfp-specific miRNA, miRGFP, was used as a control for shsR2b- and miR2b-expressing strains in coinfiltrations with 2b transcript-expressing strains. The test sample and the control sample were separately infiltrated into the opposite halves of the same leaf for comparison.
Tobacco (Nicotiana tabacum cv SR1) plants were transformed with pU6-sh2b, p35S-miR2bprec, or pCAMBIA 1300 control vector by the Agrobacterium-mediated leaf disc method (20). The primary transformants (T0) were first screened by PCR for the presence of the respective transgenes. PCR-positive plants were allowed to self-pollinate, and T1 seeds were selected on the germination medium containing hygromycin. SD-CMV-infected fresh tobacco leaves were ground with phosphate-buffered saline buffer at the ratio of 1:10 (wt/vol), and the homogenate was used to inoculate two fully expanded upper leaves of T1 progeny plants at the 5- to 6-leaf stage. A five-grade disease index system was adopted to describe the symptoms of CMV at certain days postinoculation (dpi): 0, no symptoms; 1, mild mosaic on one top leaf; 2, mild mosaic on more than two leaves; 3, pronounced mosaic on older leaves and mild deformation on two top leaves; 4, similar symptoms as for grade 3 but plants also became stunted; and 5, pronounced leaf deformation and plants severely stunted. The overall disease severity for a certain group of inoculated plants was represented by the disease severity index (DSI) calculated as follows: DSI = [Σ (grade of each plants)/5 × number of plants scored] ×100. T1 lines of shsR2b- and miR2b-expressing plants exhibiting virus resistance were harvested for seeds. Hygromycin-resistant T2 progeny of CMV-resistant lines were subsequently tested for resistance by inoculation with 50 μg of SD-CMV/ml.
GFP imaging and quantitative fluorescence analysis.
To visually detect GFP fluorescence on leaf patches and whole plants, a hand-held 100 W, long-wave UV lamp (UV Products, Upland, CA) was used, and fluorescence images were taken using a Nikon Coolpix 995 digital camera (Tokyo, Japan) mounted with UV and Kenko yellow lens (Tokyo, Japan).
The BMG FLUOstar OPTIMA multidetection reader (Offenburg, Germany) was used in quantitative analysis of GFP fluorescence according to the manufacturer's protocol. The excitation and emission wavelengths are 485 nm and 520 nm, respectively. Protein concentrations were determined using protein assay dye reagent concentrate (Bio-Rad) and a spectrophotometer (model 550; Bio-Rad).
Antibodies and protein gel blot analysis.
His6-tagged GFP and glutathione S-transferase-2b fusion proteins were purified by Ni-nitrilotriacetic acid agarose (QIAGEN) and glutathione Sepharose (Pharmacia) affinity chromotographies, respectively, and rabbit polyclonal antibodies against each of the proteins were prepared. The rabbit antibody to CMV coat protein (CP) was previously prepared by our laboratory. Total plant proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G was used as a secondary antibody. Immunoreactive bands were visualized using 5-bromo-4-chioro-3-indolylphosphate/nitroblue tetrazolium as substrates (Promega).
RNA analysis.
sRNA enrichment, probe construction, and sRNA detection were performed by using a mirVana miRNA Isolation Kit, Probe Construction Kit, and Detection Kit (Ambion), respectively, according to the supplier's protocols. Briefly, 250 mg of leaf materials was disrupted in a lysis buffer and sRNAs were purified over glass fiber filters. For the probe preparation, an oligonucleotide (5′-TCAGTTCCGAACCATCTACCCTGTCTC-3′) was synthesized to yield a 5′ 19-nt sequence identical to nt 3 to 21 of the miR2b strand and nt 1 to 19 of the antisense strand of shsR2b and a 3′ 8-nt sequence (underlined) complementary to the 3′ end of the T7 Promoter Primer included in the kit. This target-specific template was annealed to the T7 Promoter Primer and filled in with the Klenow enzyme. The formed double-stranded DNA template was in vitro transcribed with T7 RNA polymerase in the presence of [α-32P]UTP to generate a 19-nt RNA probe for the detection of miR2b and shsR2b. For solution hybridizations, 1 μg of enriched-plant sRNA was mixed with the 32P-labeled probe in the hybridization buffer. After heat denaturation, each mixture was incubated at 42°C overnight, followed by RNase A and T1 digestion to remove unhybridized RNAs and excess probe. Protected 19-nt RNA fragments were resolved by electrophoresis in a 15% denaturing polyacrylamide gel and detected by autoradiography.
For analysis of CMV RNAs by conventional RNA gel blot hybridization, total plant RNA was extracted using Trizol reagent (Invitrogen). Twenty micrograms of total RNA for each sample was electrophoresed in a 1.2% agarose gel containing 1% formaldehyde and transferred onto Hybond-N+ membrane (Amersham). RNA bands on the membrane were visualized by methylene blue staining, and 18S rRNA was used to monitor equal RNA loading. A 32P-labeled RNA probe complementary to the 3′ terminal 200 nt of SD-CMV RNA 2 was synthesized by in vitro transcription with T7 RNA polymerase and employed to detect CMV RNAs. Hybridization, washing, and autoradiography were performed using standard methods (34).
Quantitative real-time RT-PCR.
For quantitative analysis of the RNA transcript produced from the CaMV 35S promoter-driven 2b cDNA encompassing the entire open reading frame (ORF) in agroinfiltrated leaves, 2 μg of total plant RNA pretreated with RQ1 DNase I was used in a reverse transcription (RT) reaction performed according to Promega's protocol. One microliter of the reaction mixture was added to 25 μl of PCR mixture containing 12.5 μl of Realtime PCR Master Mix (TOYOBO), a 0.3 μM concentration of the forward primer (5′-ACAAAAGTCCCAGCGAGAG-3′; nt 113 to 131 of the 2b ORF) and the reverse primer (5′-CGTTACCAGCGAACCAATC-3′; nt 295 to 313 of the 2b ORF). The N. benthamiana EF1α mRNA (nt 124 to 283; GenBank accession no. CN744397) served as an internal control using PCR primers EF1U (5′-TGGTGTCCTCAAGCCTGGTATGGTTG-3′) and EF1D (5′-ACGCTTGAGATCCTTAACCGCAACATTCTT-3′). Real-time PCR was carried out in Opticom II (Bio-Rad) using the following thermal cycling profile: 95°C for 1 min, followed by 40 cycles of amplification (95°C for 30 s, 60°C for 30 s, and 72°C for 30 s). All samples were run in triplicates, and the cycle threshold values were compared to a standard curve generated from the known copy number of plasmid pBI221-2b or T-EF1α (the EF1α gene cloned in pGEM-T vector).
Quantification of sRNAs in agroinfiltrated N. benthamiana leaves or transgenic tobacco plants was performed essentially following the method of Chen et al. (11) except that the Taqman probe was replaced with SYBR Green I. For miR2b, the RT primer is 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTAGAT-3′, and the forward primer is 5′-TGCGGTATCAGTTCCGAACC-3′. For the antisense strand of shsR2b, the RT primer and the forward primer are 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGTAG-3′ and 5′-GCGGTCAGTTCCGAACCAT-3′, respectively. Sequences in italics confer specificity to miR2b and the antisense strand of shsR2b. The universal reverse primer (underlined in sequences) is 5′-GTGCAGGGTCCGAGGT-3′. The EF1α gene was used as an internal control, as mentioned before. The RT-PCR products of sRNAs were cloned into pGEM T-vector (Promega), sequenced, and used for generation of a standard curve. The amount of 2b-specific sRNAs is expressed as the copy number of the sRNAs present in an RNA sample containing 1 × 106 copies of the EF1α transcript.
ELISA.
Enzyme-linked immunosorbent assay (ELISA) plate wells were coated with crude extracts from systemic leaves of various CMV-inoculated transgenic T1 plants in sodium carbonate buffer (pH 9.6) and incubated at 37°C for 4 h. The primary antibody was rabbit polyclonal antibodies against SD-CMV CP. Goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Promega) was used as the secondary antibody. 3,3′,5,5′-Tetramethylbenzidine was used as a substrate for color development. The absorbance was recorded by a Bio-Rad ELISA reader at 450 nm.
RESULTS
Two strategies to produce sRNA in plant cells.
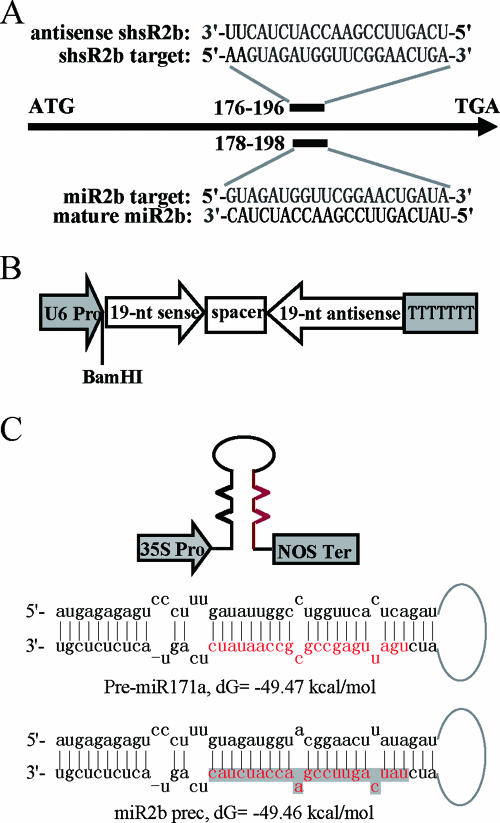
To produce CMV 2b transcript-targeting artificial sRNAs in plants, we constructed two expression vectors, pU6-sh2b and p35S-miR2bprec, essentially following the design of DNA vectors used in animal systems to express shRNAs or chimeric pre-miRNAs. In pU6-sh2b, an A. thaliana RNA Pol III promoter, the U6-29 promoter was used to drive the expression of 2b-specific shRNA. This promoter has extragenic regulatory elements and needs only a stretch of uridine residues as a transcription termination signal (42). The upstream elements of the promoter comprise a −30 TATA-like box and a −65 motif ATCCCACATCG, called the upstream sequence element, separated by 25 nt capable of forming three helical DNA turns. Moreover, its transcription start site is T/G (−1/+1) which fits the consensus dinucleotide Y/R (−1/+1) of RNA Pol III-transcribed plant U-snRNA genes (15). The −319 to −1 region of the U6-29 promoter was cloned and modified by changing the −6 to −1 sequence GAGATT to a BamHI site GGATCC to keep the spacing between the upstream sequence element and the transcription start site and retain the nucleotide at the −1 site as a pyrimidine. By using the siRNA Target Finder program of Ambion and to the criteria mentioned in Reynolds et al. (33), a 21-nt sequence (5′-AAGUAGAUGGUUCGGAACUGA-3′) corresponding to nt 176 to 196 of the 2b RNA (numbering from the translation initiation codon) was selected as a preferential target (Fig. 1A). We thus synthesized a composite sequence containing the sense and antisense sequences of the target linked by a 9-nt spacer and connected it to the U6-29 promoter via a BamHI site (Fig. 1B). The resulting transcript was predicted to form a short hairpin structure which would be processed in plants to produce a 19-bp RNA duplex with 3′ UU overhangs (shsR2b). The 21-nt antisense strand of shsR2b would act as the guide molecule to mediate degradation of the 2b RNA.
FIG. 1.
Two strategies to produce 2b-specific sRNAs (shsR2b and miR2b) in plant cells. (A) Locations of shsR2b- and miR2b-targeting sites in the 2b gene. The predicted base pairings between sRNAs and their target sequences are shown. (B) A schematic diagram of the shsR2b expression cassette. The cassette contains the Arabidopsis U6-29 promoter engineered with a BamHI site at positions −6 to −1, followed by the shRNA-coding sequence and a 7-thymidine termination signal. (C) A schematic diagram of the 35S promoter (35SPro)-driven miR2b expression cassette with a termination sequence from the nopaline synthase gene (NOS Ter). The miR171 (in red) and miR171* sequences in the stem of the Arabidopsis pre-miR171a (the upper hairpin) were replaced with 21-nt miR2b (in red and shadowed) and miR2b* sequences, resulting in miR2bprec (the lower hairpin). Note that two mismatches between miR171 and miR171* are maintained between miR2b and miR2b*. The free energy of pre-miR171a and that of miR2bprec are indicated.
It has been shown that the fold-back structure of pre-miR171a, the precursor of the Arabidopsis miR171, was sufficient for synthesis of the mature miRNA in vivo (31). We thus exploited the pre-miR171a backbone to produce an artificial miRNA as an alternative approach to specifically cleave the 2b RNA. We replaced the miR171 sequence in pre-miR171a with a 21-nt sequence complementary to nt 178 to 198 of 2b RNA (5′-GUAGAUGGUUCGGAACUGAUA-3′) (Fig. 1A), which covers the target of shsR2b except its 5′ AA. The PCR-amplified DNA duplex corresponding to the modified miRNA precursor, termed miR2bprec, was placed downstream of a 35S promoter to form p35S-miR2bprec (Fig. 1C). The in vivo produced 21-nt artificial miRNA (miR2b) started with a U and ended with a C residue, like miR171. The secondary structure of miR2bprec was exactly the same as that of the original pre-miR171a, and the free energy was also preserved (−49.47 and −49.46 kcal/mol, respectively).
Artificial sRNAs down-regulated the expression of the 2b gene with sequence specificity.
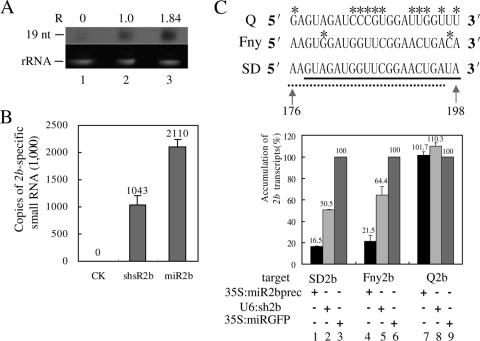
To examine the ability of the plant expression vectors to produce 2b-specific sRNAs in vivo, an Agrobacterium strain harboring pU6-sh2b (U6:sh2b), p35S-miR2bprec (35S:miR2bprec), or the control pCAMBIA 1300 vector (CK) was infiltrated into N. benthamiana leaves. Total RNA was extracted at 3 dpi, and artificial sRNAs could be detected by solution hybridization-based Northern analysis in both U6:sh2b- and 35S:miR2bprec-infiltrated leaves but not in control leaves (Fig. 2A). The expression level of miR2b was 80% higher than that of the antisense strand of shsR2b (Fig. 2A). Real-time quantification of the 2b-specific sRNAs by the newly developed stem-loop RT-PCR method (11) also revealed that the arbitrary copy number of miR2b in infiltrated leaves was twice that of shsR2b or that of NbEF1a mRNA, which served as a normalization control (Fig. 2B). EF1α mRNA is an abundant RNA species, accounting for 0.1% to 1.0% of the poly(A)+ RNA in Lycopersicon esculentum tissues (32). So both pU6-sh2b and p35S-miR2bprec can produce artificial sRNAs in planta at high levels.
FIG. 2.
Artificial 2b-specific small RNAs down-regulated the expression of the 2b gene with sequence specificity. (A) Solution hybridization analysis of artificial sRNAs generated in agroinfiltrated N. benthamiana leaves. sRNAs hybridized with a 32P-labeled miR2b- and shsR2b-specific probe and protected from RNase A/T1 digestion were resolved by 15% polyacrylamide gel electrophoresis and detected by autoradiography (top). The migration position of a 19-nt RNA oligonucleotide is indicated on the left. The relative intensity (R) of the hybridizing bands determined by density scanning is shown above the panel. Ethidium bromide-stained 5.8S rRNA, indicating an equal amount of RNA (1 μg), was used in solution hybridization (bottom). Lane 1, control strain-infiltrated leaves; lane 2, U6:sh2b-infiltrated leaves; lane 3, 35S:miR2bprec-infiltrated leaves. (B) Real-time RT-PCR quantification of the expression levels of artificial sRNAs. Total RNA extracted from the agroinfiltrated area of N. benthamiana leaves was used as the template in the RT reaction. The arbitrary copy number of 2b-specific sRNAs is presented relative to 1 × 106 copies of the EF1α transcript. Error bars represent standard deviations of three determinations, and similar results were obtained in two independent RT-PCR experiments. CK, control strain-infiltrated leaves; shsR2b, U6:sh2b-infiltrated leaves; miR2b, 35S:miR2bprec-infiltrated leaves. (C) Artificial sRNAs inhibited the expression of the 2b genes with sequence specificity. The target regions in 2b genes of three CMV strains (Q, Fny, and SD) are shown (top). The nucleotides unpaired with the sequence of miR2b (thick line) or shsR2b (broken line) are marked with asterisks. The accumulation levels of the 35S promoter-produced 2b transcripts (SD 2b, Fny 2b, and Q 2b) in N. benthamiana leaves coinfiltrated with the miR2b-producing (35S:miR2bprec) or the shsR2b-producing (U6:sh2b) Agrobacterium strain were determined by real-time RT-PCR and compared to levels when a miRGFP-producing strain (35S:miRGFP) was coinfiltrated (bottom). Error bars represent standard deviations of three determinations, and similar results were obtained in three independent RT-PCR experiments.
We next examined whether the two artificial sRNAs can inhibit the target expression with sequence specificity. 2b-encoding ORFs of three CMV strains belonging to different subgroups (Fny from subgroup IA, SD from subgroup IB, and Q from subgroup II) were separately constructed into pCAMBIA 1300 under the control of a 35S promoter to express the target sequences. The three 2b RNA sequences differ in the shsR2b- and miR2b-targeted regions (nt 176 to 198) with variations at two positions between SD and Fny and as many as 11 changes between SD and Q (Fig. 2C). pU6-sh2b or p35S-miR2bprec was codelivered into the N. benthamiana leaves with each of the three target-expressing vectors by agroinfiltration; the final OD ratio of the sRNA-expressing strain to the 2b-expressing strain was 3:1. The accumulation level of the target transcripts at 4 dpi was quantified by real-time RT-PCR after normalization against EF1α transcript. The expression level of the target transcript in the presence of miRGFP, an irrelevant sRNA control which by itself did not show any effect on accumulation of each of the 2b transcripts (data not shown), was used as a null-silencing control. As shown in Fig. 2C, shsR2b and miR2b reduced the levels of their cognate target SD 2b RNA and the closely resembled Fny 2b target (lanes 1 to 6), but had no effect on Q 2b RNA (lanes 7 to 9). Moreover, miR2b was more efficient than shsR2b to knock down the target expression. Compared with the miRGFP control, miR2b reduced the SD 2b and Fny 2b RNA levels to 16.5% and 21.5% (lanes 1 and 4), whereas action of shsR2b still retained 50.5% of SD 2b RNA and 64.4% of Fny 2b RNA (lanes 2 and 5). It is obvious that degradation of 2b transcript by artificial sRNAs is sequence specific and that the miR2b-mediated approach is effective in silencing the expression of the target gene coding for the silencing suppressor 2b.
Artificial miRNA can efficiently interfere with the suppressor activity of 2b both on local and systemic silencing.
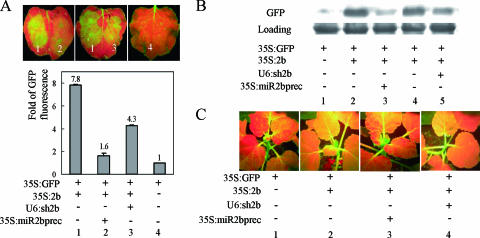
It has been shown that CMV 2b, when coexpressed with the GFP gene in the leaf of the gfp-transgenic N. benthamiana line 16c, could suppress local silencing of GFP expression in the infiltrated leaf and later on inhibit systemic GFP silencing in the upper leaves (18). To test if miR2b and shsR2b can interfere with the 2b silencing suppressor activity, we used GFP as a reporter and coinfiltrated three Agrobacterium strains each harboring p35S-GFP (35S:GFP), p35S-SD 2b (35S:2b), or one of the sRNA-expressing vectors (U6:sh2b or 35S:miR2bprec) into 16c or wild-type (WT) N. benthamiana leaves. As shown in Fig. 3A, at 4 dpi GFP fluorescence on 16c leaves infiltrated with 35S:GFP plus 35S:2b (lane 1) was enhanced by 7.8-fold compared to leaves infiltrated with 35S:GFP alone (lane 4). Adding 35S:miR2bprec (lane 2) or U6:sh2b (lane 3) reduced the enhancement of GFP fluorescence to 1.6-fold or 4.3-fold. The interfering function of the sRNAs on 2b's suppressor activity was also demonstrated in WT N. benthamiana leaves as shown by GFP-specific immunoblotting experiments (Fig. 3B). These data showed that sRNAs can interfere with the activity of 2b on suppression of local silencing and that miR2b is more efficient than shsR2b in this regard, consistent with the results of reduction of the 2b RNA level shown in Fig. 2C.
FIG. 3.
Artificial sRNAs can interfere with the silencing suppressor activity of 2b. (A) Coexpression of miR2b or shsR2b with the 2b protein reduced the activity of 2b in suppression of silencing of GFP expression in N. benthamiana 16c leaves. The GFP fluorescence images of the agroinfiltrated leaves were taken at 4 dpi under UV illumination. The bottom graph shows results of quantitative GFP fluorescence analysis averaged from two leaves for each infiltration treatment. (B) Effects of miR2b and shsR2b on the activity of 2b in enhancing the expression level of the GFP protein in WT N. benthamiana leaves. Total protein was extracted from the agroinfiltrated leaf area for each infiltration treatment at 4 dpi and subjected to Western blot analysis probed with rabbit anti-GFP antibodies. Coomassie Bright Blue staining of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase indicates equal loading. (C) miR2b and shsR2b have differential effects on the activity of 2b in inhibiting systemic silencing. Images show different patterns of agroinfiltration into 16c plants and their effects on systemic silencing of GFP expression at 14 dpi. A representative plant coinfiltrated with 35S:GFP/35S:2b/35S:miR2bprec showed systemic silencing of GFP expression (frame 3) as the plant infiltrated with 35S:GFP alone (frame 1), and a plant coinfiltrated with 35S:GFP/35S:2b/U6:sh2b exhibited GFP expression in systemic leaves (frame 4), similar to that observed in the 35S:GFP/35S:2b coinfiltrated plant (frame 2).
The activity of 2b in suppressing systemic silencing was also dampened by artificial sRNAs. At 14 dpi, all 32 16c plants that were infiltrated with only 35S:GFP developed severe systemic silencing of GFP expression in upper leaves (Fig. 3C, frame 1), whereas upper leaves of the other 30 plants coinfiltrated with 35S:GFP and 35S:2b together with a strain harboring the empty vector CK did not show any GFP silencing (Fig. 3C, frame 2) because of 2b's suppressor activity. When the CK strain was replaced by 35S:miR2bprec, significant systemic silencing could be observed in upper leaves of 10 of 30 infiltrated plants (Fig. 3C, frame 3) although it developed at a slower rate. Again, the efficacy of shsR2b was lower than that of miR2b: 28 of 30 plants infiltrated with 35S:GFP, 35S:2b, and U6:sh2b still showed systemic GFP expression (Fig. 3C, frame 4).
Taken together, these results indicate that artificial 2b-specific sRNAs, especially miR2b, can inhibit 2b function to suppress both local and systemic silencing, and they suggest that transgenic expression of the sRNAs in plants may be an effective way to combat CMV infection.
Artificial sRNAs conferred virus resistance on transgenic plants.
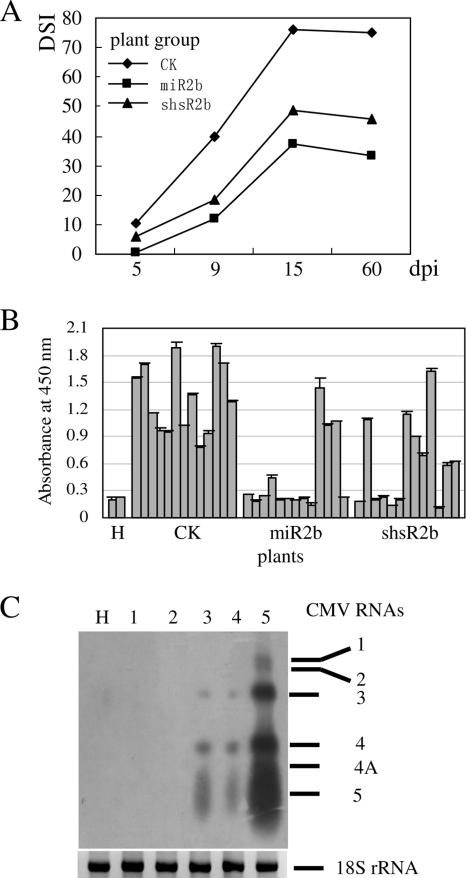
The majority of the tobacco plants derived from transformation with pU6-sh2b or p35S-miR2bprec did not show abnormal developmental phenotypes, and the expression level of the endogenous miR171 in several checked shsR2b- and miR2b-expressing transgenic lines was unaltered compared to that in the control vector-transformed tobacco plants (data not shown), suggesting that the in vivo expressed 2b-specific sRNAs have little effect on host gene expression. Primary transformants which had given positive results in the PCR tests for checking the presence of the sRNA expression cassettes were selected, and their T1 progeny plants generated from hygromycin-containing medium were used in an initial viral resistance test. In total, 55 T1 plants (named miR2b plants) from 16 p35S-miR2bprec lines, 43 T1 plants (named shsR2b plants) from 14 pU6-sh2b lines, 62 T1 plants from 18 control vector-transformed lines, and 3 to 5 plants from each T0 line were mechanically inoculated with CMV. The appearance of disease symptoms on each inoculated plant was monitored at 5, 9, 15, and 60 dpi, and the disease severity was scored using a five-grade disease index system (0 to 5) and used to calculate the DSI which represents the level of CMV resistance for each entire group of transgenic plants. As illustrated in Fig. 4A, at 5 dpi, 18 of the control plants showed obvious symptoms (DSI, 10.3), while only 1 of the miR2b plants and 6 of the shsR2b plants showed symptoms, giving a DSI of 0.36 and 4.2, respectively. In the period from 5 to 15 dpi, disease symptoms developed rapidly, and the highest severity level was recorded at 15 dpi. Control plants showed severe symptoms, and the DSI reached 76.12. At the same time, miR2b plants and shsR2b plants had significantly lower DSIs of 39.07 and 48.84, respectively, indicating that a considerable fraction of these transgenic plants were resistant or tolerant to CMV infection.
FIG. 4.
Transgenic plants expressing miR2b or shsR2b exhibited resistance to CMV infection. (A) Symptom development in T1 progeny of 16 p35S-miR2bprec-, 14 pU6-sh2b-, and 18 control vector-transformed plant lines after CMV inoculation. Three to five T1 plants, each from independent T0 lines, were used in the challenge experiments. DSIs of each group of transgenic plants (abbreviated as miR2b, shsR2b and CK, respectively) were calculated based on the disease index grade of all individual plants within each group scored at 5, 9, 15, and 60 dpi. (B) Accumulation of CMV CP in CMV-inoculated transgenic plants. From each transgenic group, 13 representative T1 plants, each from an individual T0 line, were selected at 9 dpi, and the level of CMV CP in upper uninoculated leaves of each plant was determined by ELISA. Two uninoculated tobacco plants (H) were used as a blank control in the ELISA. Error bars represent the standard deviations of two repeat assays. (C) Northern blot analysis of CMV RNAs accumulated in CMV-inoculated transgenic plants. Total RNA from upper uninoculated leaves at 15 dpi was used in the experiments. Lanes 1 and 2, an miR2b-plant (m-1-2) and an shsR2b-plant (s-4-13), respectively, showing no symptoms; lanes 3 and 4, an miR2b-plant (m-19-33) and a shsR2b-plant (s-25-3), respectively, showing mild symptoms; lane 5, control vector-transformed plant; lane H, uninoculated tobacco plant. On the right, the migration positions of CMV RNAs 1, 2, 3, 4, 4A, and 5 are indicated. Ethidium bromide-stained 18S rRNA was used as an RNA loading control (bottom).
More specifically, the numbers of plants showing different responses to CMV challenge were separately counted in each transgenic group. In our experiments, plant responses to CMV infection could be divided into four major categories: “resistant,” “recovery,” “delayed,” and “susceptible” (see explanations of the categories in the footnote of Table 1). As can be seen in Table 1, 21.82% and 16.36% of the miR2b plants belonged to resistant and recovery categories, these percentages are higher than the corresponding values of 13.95% and 11.63%, respectively, for the shsR2b plants. The percentage of delayed miR2b plants was slightly higher than that of delayed shsR2b plants (25.45% versus 23.26%). Collectively, a significant portion of the miR2b plants (63.63%) and the shsR2b plants (48.84%) showed considerable resistance against CMV infection, compared to 8.07% for the control plants. On the other hand, 69.35% of the control plants showed severe symptoms at 60 dpi (a severe-subtype of the category susceptible), whereas only 23.64% of the miR2b plants and 32.56% of the shsR2b plants exhibited similarly severe symptoms.
TABLE 1.
Classification of T1 plants of different transgenic groups according to responses to CMV infection
| Transgenic groupa | Plant response to infection (no. of plants in group/total no. of plants challenged [%])b
|
||||
|---|---|---|---|---|---|
| Resistant | Recovered | Delayed | Susceptible
|
||
| Moderately | Severely | ||||
| CK | 0/62 (0) | 3/62 (4.84) | 2/62 (3.23) | 14/62 (22.58) | 43/62 (69.35) |
| miR2b | 12/55 (21.82) | 9/55 (16.36) | 14/55 (25.45) | 7/55 (12.73) | 13/55 (23.64) |
| shsR2b | 6/43 (13.95) | 5/43 (11.63) | 10/43 (23.26) | 8/43 (18.60) | 14/43 (32.56) |
CK, control vector-transformed plants; miR2b, p35S-miR2bprec-transformed plants; shsR2b, pU6-sh2b-transformed plants.
Resistant, no disease symptoms throughout the testing period; recovered, initially showed symptoms of variable severity but no symptoms on young leaves formed at a later stage; delayed, plants with no symptoms until 15 dpi but with symptoms at 60 dpi and plants with no symptoms at 9 dpi but with mild or moderate symptoms at 15 and 60 dpi; susceptible, showed disease symptoms similar to those in CMV-infected WT tobacco plants, with a division into moderate and severe subgroups according to the symptoms at 60 dpi.
To demonstrate the inhibition of CMV multiplication by 2b-specific sRNAs, 13 representative plant lines from each group of challenged plants, which had a calculated DSI equal to that of the entire group, were selected and subjected to ELISA of CMV CP to measure the accumulation level of CMV in systemic leaves at 9 dpi. The OD450 values are shown in Fig. 4B. Nine of the miR2b plant lines (69.2%) and six of the shsR2b plant lines (46.1%) had values similar to those of uninoculated tobacco plants (OD450 of <0.3). The remaining plants contained virus at titers similar to those determined in inoculated control vector plants.
Furthermore, we have determined the levels of CMV RNAs in challenged transgenic plants at 15 dpi to correlate the inhibition of viral RNA replication with the observed virus resistance. Northern blot analysis showed that in T1 plants m-1-2 and s-4-13, which had a disease index grade of 0, CMV genomic RNAs (RNA 1, 2, and 3) and subgenomic RNAs (RNA 4 and RNA 4A) were undetectable (Fig. 4C, lanes 1 and 2). The levels of CMV RNAs in plants m-19-33 and s-25-3 which showed mild symptoms (disease index grade of 1) were much lower (Fig. 4C, lanes 3 and 4) compared to those in the inoculated control vector plant (Fig. 4C, lane 5).
Taken together, the above results proved that the strategy of transgenic production of sRNAs targeting the 2b RNA is useful to generate CMV-resistant plants, and in this respect expression of miR2b is more effective than expression of shsR2b.
Virus resistance correlates with the expression level of miR2b but not always with the level of shsR2b.
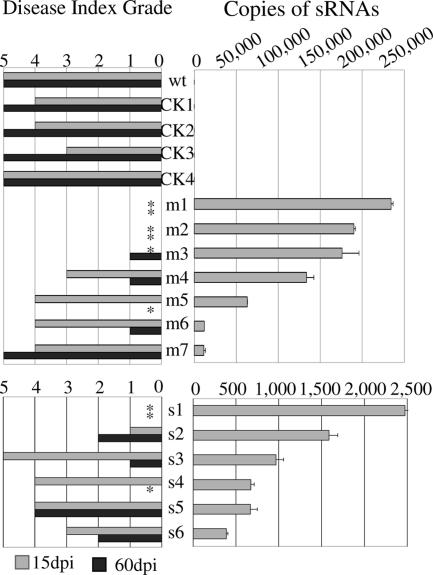
Various degrees of resistance to CMV challenge were observed among different lines of miR2b plants and shsR2b plants. To determine if the resistance level is correlated with the level of in vivo expressed sRNAs, four control vector transgenic T1 plants (CK1 to CK4), seven T1 miR2b plants (m1 to m7) and six T1 shsR2b plants (s1 to s6)—all hygromycin-resistant and each from a different T0 line—and one nontransformed tobacco plant (WT) were subjected to real-time RT-PCR analysis to quantify the copy numbers of sRNAs using the leaves collected before CMV challenge. Figure 5 gives the arbitrary copy numbers of sRNAs and the disease index grades recorded at 15 and 60 dpi. There was a general correlation between the expression level of 2b-specific sRNAs and virus resistance. This correlation was obvious in the miR2b plants. The plant m1 which had been shown to accumulate no CMV RNAs after virus challenge (Fig. 4C, lane 1) contained the highest level of miR2b and showed no disease symptoms throughout the whole life cycle (disease index grade of 0), whereas plant m7, whose miR2b level was only 1/20 of that of m1, was highly susceptible to CMV infection. One plausible explanation is that, in plant m7, the number of the miR2b molecules was not high enough to activate the host RNA silencing process to fully suppress the invading virus. Consequently, the copy number dependence of CMV resistance was less stringent when miR2b levels were low (m5 to m7). On the other hand, the shsR2b level is not well correlated with the degree of virus resistance. Surprisingly, the copy numbers of shsR2b in transgenic plants were 400 to 2,000 times lower than those determined in agroinfiltrated leaf tissues (Fig. 2B). The reason for this discrepancy in the expression level of shsR2b is unclear. Despite the low copy number, expression of shsR2b still conferred CMV resistance on some plants. For instance, in plant s1, although the copy number of shsR2b was 1/100 that of the miR2b in plant m1, the plant showed full resistance to CMV. This suggests that RNA interference induced by artificial shRNA may be controlled by other factors beyond sRNA-mRNA stoichiometry.
FIG. 5.
Correlation between the expression level of miR2b and virus resistance in T1 progeny plants. The disease index grades of individual plants scored at 15 and 60 dpi are given. An asterisk indicates that there were no symptoms (grade 0) in the challenged plant at the time point. The copy numbers of 2b-specific sRNAs determined by real-time RT-PCR analysis and defined as described in the legend of Fig. 2B are given. Error bars represent the standard deviations of three determinations, and similar results were obtained in two independent RT-PCR experiments.
Virus resistance in T2 generation plants.
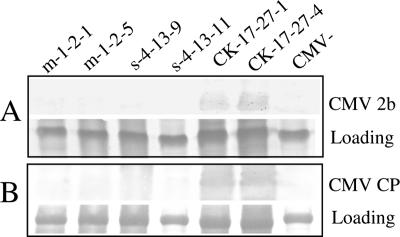
Two T1 plant lines, m-1-2 and s-4-13, which expressed high levels of sRNAs and exhibited high CMV resistance, were chosen to test virus resistance in T2 progeny plants. Eleven T2 plants of m-1-2 and 12 T2 plants of s-4-13, together with 10 T2 plants of a control vector-transformed line CK-17-27, all generated from hygromycin-containing medium, were inoculated with a high concentration of SD-CMV virions (50 μg/ml). Most of the m-1-2 T2 plants (9 plants, or 82%) and half of the s-4-13 T2 plants were free of symptoms at 15 dpi, when all the control plants showed typical CMV symptoms. The expression levels of sRNAs in the resistant T2 plants were similar to those of their parental lines (data not shown), indicating that the expressed sRNAs are able to protect these T2 plants from high-dose virus infection. Western blot analyses revealed that the 2b protein (Fig. 6A) and the CP (Fig. 6B) were below the detection limit in systemic leaves of the resistant T2 plants but accumulated at high levels in susceptible control plants, confirming the complete inhibition of 2b synthesis and virus propagation in these sRNA-expressing T2 plants. The residual two m-1-2 T2 plants developed mild symptoms (disease index grade of 1) and also expressed lower levels of miR2b (20% to 30% of that of the parental line). The other half of the challenged s-4-13 T2 plants reacted with symptoms of variable severity: two plants with mild symptoms and four with moderate severity. The shsR2b levels in these plants are 15% to 45% lower than the level of the parental T1 line and are not directly related to the severity of symptoms, as previously observed in T1 shsR2b plants (Fig. 5). Variations of expression levels of the sRNAs and hence the degree of virus resistance in T2 siblings might be caused by some host regulatory mechanisms or some environmental factors imposed on the production or stability of the sRNAs. Nevertheless, our results indicated that the 2b-specific sRNA-induced CMV resistance, especially that triggered by miR2b, is inherited in a large population of the T2 generation plants.
FIG. 6.
Western blot analyses of CMV 2b and CP in CMV-inoculated T2 generation plants. Total plant proteins were extracted from upper uninoculated leaves of two m-1-2 T2 plants (m-1-2-1 and m-1-2-5), two s-4-13 T2 plants (s-4-13-9 and s-4-13-11), and two T2 plants of the control vector-transformed plant line CK-17-27 (CK-17-27-1 and CK-17-27-4) at 15 dpi. Total protein extracted from an uninoculated tobacco plant (CMV−) was used as a negative control. These four sRNA-transgenic T2 plants showed no CMV symptoms, whereas the two control plants exhibited typical mosaic symptoms. The plant proteins were subjected to immunoblot analysis using antibodies against glutathione S-transferase-2b fusion protein (A) or CMV CP (B). Coomassie Bright Blue staining of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase indicates equal loading.
We also tested whether the CMV resistance conferred by SD 2b-targeting sRNAs is strain specific, as was shown for degradation of 2b transcripts in transient assays (Fig. 2C). For this purpose, T2 generation plants of several SD-CMV-resistant lines, e.g., 10 T2 plants of m-1-2, were challenged with Q-CMV. There was no obvious difference between sRNA-expressing plants and CK plants in either disease phenotype or virus titer level (data not shown), indicating that the miR2b- or shsR2b-mediated virus resistance is strain specific. Since the target selected in our experiments is located in the most variable region of the 2b genes, it is probably necessary to target a conserved viral sequence to achieve cross-strain virus resistance.
DISCUSSION
Most of the recent reports on RNA silencing-based plant virus resistance have relied on expression of long pathogen-related dsRNAs or dsRNAs derived from hpRNAs (>300 base pairs) in plants (12, 14, 22, 28, 37, 43). These long dsRNAs activate the RNA silencing machinery of the host plant and are themselves processed into siRNAs, which are suggested to be responsible for the degradation of invading viral RNA through the preactivated RNA silencing machinery. In several cases, the correlation between the presence or abundance of virus-specific siRNAs and virus resistance was observed (12, 22, 28), reminiscent of the earlier finding that siRNAs serve as molecular markers for active RNA silencing (19). Therefore, the preexistence of a high level of virus-specific siRNAs in plants seems to be a crucial determinant for plants to acquire virus resistance. In addition to siRNAs, miRNAs are another class of RNA silencing effectors that function as negative regulators of gene expression in plants. These two types of sRNAs share some similarities in site-specific cleavage of complementary mRNAs, so it is possible that miRNA-guided silencing might also contribute to plant defense by interfering with viral infection in plants. In fact, a recent report indicated that the plant miRNA pathway might be involved in impairing virus replication (36). One aim of this research was to examine whether directly expressing virus-specific sRNAs in plants, especially the artificial miRNAs, could result in effective virus resistance. In addition, we wanted to see if the expressed sRNAs specially targeting viral suppressors would out-compete the suppressors’ function in inhibition of the host's antiviral silencing processes.
In this report, we used an Arabidopsis U6 promoter-driven shRNA vector (pU6-sh2b) and a 35S promoter-driven miR171 precursor vector (p35S-miR2bprec) to express two kinds of CMV 2b-specific artificial sRNAs in tobacco plants. The sRNA derived from the chimeric pre-miR171 could be regarded as an miRNA (miR2b) as judged from its biogenesis route. We have shown in a temperature shift experiment that the expression level of miR2b in agroinfiltrated N. benthamiana leaves was reduced at 15°C compared to that at 25°C (data not shown). This result is in contrast to results reported earlier that accumulation of miRNAs was temperature independent in Arabidopsis (30, 38). However, the level of N. benthamiana bmiR171, an endogenous control for the artificial sRNAs in our experiments, was also reduced in a temperature-dependent manner similar to miR2b (data not shown), suggesting that the levels of some tobacco miRNAs might be affected by low temperature and that the difference in temperature dependence of the miRNA level between Arabidopsis and tobacco might be due to the fact that Arabidopsis develops normally at 15°C but tobacco does not. On the other hand, the precise nature of the sRNA produced from the sh2b RNA (shsR2b) is unknown, although in an earlier report the 21-nt RNA duplexes derived from the GUS-shRNAs were called siRNAs (26), and sRNAs of different sizes (21 to 24 nt) generated from long hpRNA were regarded as siRNAs (16). It would help to clarify this uncertainty if the plant DCL(s) responsible for processing artificial shRNAs is characterized.
In preliminary transient expression experiments carried out by agroinfiltration on N. benthamiana leaves, the 2b-specific artificial sRNAs were expressed at relatively high levels and could inhibit the accumulation of the 2b transcript. In subsequent experiments with transgenic tobacco plants, a significant portion of 2b-specific sRNA-expressing plants exhibited resistance against CMV challenge. Among them, 38% of miR2b plants and 25% of shsR2b plants were free of disease symptoms either in the whole plants or in newly developed leaves (so-called recovery phenotype). These resistance frequencies are comparable to those observed for transgenic tobacco plants expressing a 747-bp dsRNA of the CMV CP gene (28%) (22) and also to those of transgenic N. benthamiana plants expressing a 490-bp dsRNA of the 3′ terminal of CMV RNA 2 (30%) (12). However, the resistance levels conferred by miR2b and shsR2b are significantly lower than that (75%) induced by a longer dsRNA comprising the 3′ 1,534-nt sequence of CMV RNA 2, which includes the full-length coding sequence of 2b (12). This difference in resistance efficiency might be due to the difference in the number of potentially active virus-specific sRNAs: there is only one from our sh2b or miR2bprec construct but many more could be expected from the long CMV RNA 2 inverted repeat construct, and a fraction of them would specifically target the coding region of 2b. The resistance levels of miR2b plants are also lower than those obtained by a similar virus-specific artificial miRNA approach against two other viruses (turnip yellow mosaic virus and turnip mosaic virus) in Arabidopsis (30). There are several potential reasons for this fact, such as different virus-host systems and different miRNA precursors used to express artificial miRNAs. Although there are some data suggesting that the Arabidopsis miRNA171 could function well in tobacco cells (2, 31), use of miRNA precursors of tobacco origin might be more suitable to express artificial miRNAs in tobacco plants. Nevertheless, our results proved that transgenic production of CMV-specific sRNAs, especially miRNAs, is an effective way to generate CMV resistance in tobacco plants. In addition, shorter virus-origin sequences of the artificial sRNAs will largely reduce the potential biosafety-related risks associated with the developed transgenic plants.
The fact that the 2b ORF is embedded in the CMV RNA 2 sequence precludes us from unambiguously defining the effect of targeting the 2b suppressor on the efficiency of acquired virus resistance. However, miR2b-mediated CMV resistance in tobacco plants indicated that the suppressor activity of 2b of the incoming virus has little effect on processing of the miR171a/CMV 2b precursor and on miR2b-guided antiviral silencing.
We have shown that artificial miR2b expressed from p35S-miR2bprec was more effective than shsR2b from pU6-sh2b in inhibiting viral functions in both the transient expression system and transgenic plants. There are several possible explanations for the results. One might be the higher expression levels of miR2b observed in agroinfiltrated leaves and transgenic plants, partly due to the strong 35S promoter used or because the naturally nonexistent short hairpin structure of sh2b RNA was not readily processed by plant DCLs. Second, miRNAs generally need less extensive base pairings to their target sequences for effective RNA silencing than siRNAs (10, 31), so that variants of the invading virus with microheterogenicity in the target sequence would be more difficult to escape from miRNA-specified silencing. Third, the local folding potential of miR2bprec (ΔG, −49.47 kcal/mol) is higher than that of shsR2b (ΔG, −38.89 kcal/mol). The higher stem-loop stability would make the miR2bprec transcript more rapidly fold into its secondary structure and would make it prone to cleavage by DCL1, resulting in a faster formation of the RNA-induced silencing complex and more potent gene silencing.
We noticed that the copy numbers of miR2b in transgenic tobacco plants were about 10 times lower than those determined in agroinfiltrated N. benthamiana leaves. Similar observations were made previously that the transgene expression levels in agroinfiltrated leaves often exceeded those observed in stably transformed plants (44). However, the copy numbers of shsR2b expressed in transgenic plants were more than two orders of magnitude lower than those determined in agroinfiltrated leaves. One possible cause of this drastic reduction in the level of shsR2b might be the DNA methylation-induced transcriptional gene silencing between the endogenous and transgenic U6 promoter, which is part of a genomic defense mechanism whereby parasitic sequences are inactivated (49). Notably, despite the low expression levels, shsR2b can still confer full resistance to CMV on some plants. Moreover, unlike miR2b, the expression level of shsR2b is not always correlated with the virus resistance phenotype. It seems that the interaction between shsR2b and the 2b RNA is more complicated than we thought.
In conclusion, we demonstrated that transgenic expression of the 2b-specific miRNA is an effective and predictable method to protect hosts from infection by CMV. The correlation between virus resistance and the expression level of the miRNA makes it possible to select for the most promising lines that are highly resistant to the virus. We believe that combined production of multiple virus-specific miRNAs in plants would probably result in better protection against virus infection. By targeting the strain-specific sequences or sequences conserved in different strains, virus resistance with strain specificity or against a broad spectrum of virus strains could be achieved.
Acknowledgments
We thank Eneida Abreu Parizotto and Olivier Voinnet (Institut de Biologie Moléculaire des Plantes du CNRS, France) for providing A. thaliana pre-miR171a and pmiRGFPprec and David Baulcombe (John Innes Center, United Kingdom) for providing N. benthamiana 16c seeds.
This work was supported by grant 30530500 from the Natural Science Foundation of China.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Akbergenov, R., A. Si-Ammour, T. Blevins, I. Amin, C. Kutter, H. Vanderschuren, P. Zhang, W. Gruissem, F. Meins, Jr., T. Hohn, and M. M. Pooggin. 2006. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 34:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, J. P., I. Pekker, A. Goldshmidt, E. Blum, Z. Amsellem, and Y. Eshed. 2006. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18:1134-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe, D. 2005. RNA silencing. Trends Biochem. Sci. 30:290-293. [DOI] [PubMed] [Google Scholar]
- 5.Boden, D., O. Pusch, R. Silbermann, F. Lee, L. Tucker, and B. Ramratnam. 2004. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 32:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsani, O., J. Zhu, P. E. Verslues, R. Sunkar, and J. K. Zhu. 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123:1279-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouche, N., D. Lauressergues, V. Gasciolli, and H. Vaucheret. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25:3347-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Brodersen, P., and O. Voinnet. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22:268-280. [DOI] [PubMed] [Google Scholar]
- 10.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C., D. A. Ridzon, A. J. Broomer, Z. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar, M. R. Andersen, K. Q. Lao, K. J. Livak, and K. J. Guegler. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Y., D. Lohuis, R. Goldbach, and M. Prins. 2004. High frequency induction of RNA-mediated resistance against Cucumber mosaic virus using inverted repeat constructs. Mol. Breed. 14:215-226. [Google Scholar]
- 13.Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68-71. [DOI] [PubMed] [Google Scholar]
- 14.Di Nicola-Negri, E., A. Brunetti, M. Tavazza, and V. Ilardi. 2005. Hairpin RNA-mediated silencing of Plum pox virus P1 and HC-Pro genes for efficient and predictable resistance to the virus. Transgenic Res. 14:989-994. [DOI] [PubMed] [Google Scholar]
- 15.Edoh, D., T. Kiss, and W. Filipowicz. 1993. Activity of U-snRNA genes with modified placement of promoter elements in transfected protoplasts and stably transformed tobacco. Nucleic Acids Res. 21:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fusaro, A. F., L. Matthew, N. A. Smith, S. J. Curtin, J. Dedic-Hagan, G. A. Ellacott, J. M. Watson, M. B. Wang, C. Brosnan, B. J. Carroll, and P. M. Waterhouse. 2006. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7:1168-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasciolli, V., A. C. Mallory, D. P. Bartel, and H. Vaucheret. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15:1494-1500. [DOI] [PubMed] [Google Scholar]
- 18.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 20.Horsch, R. B., J. E. Fry, N. L. Hoffman, D. Eichholz, S. G. Rogers, and R. T. Fraley. 1985. A simple and general method for transforming genes into plants. Science 227:1229-1231. [DOI] [PubMed] [Google Scholar]
- 21.Ji, L. H., and S. W. Ding. 2001. The suppressor of transgene RNA silencing encoded by Cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol. Plant-Microbe Interact. 14:715-724. [DOI] [PubMed] [Google Scholar]
- 22.Kalantidis, K., S. Psaradakis, M. Tabler, and M. Tsagris. 2002. The occurrence of CMV-specific short RNAs in transgenic tobacco expressing virus-derived double-stranded RNA is indicative of resistance to the virus. Mol. Plant-Microbe Interact. 15:826-833. [DOI] [PubMed] [Google Scholar]
- 23.Lecellier, C. H., P. Dunoyer, K. Arar, J. Lehmann-Che, S. Eyquem, C. Himber, A. Saib, and O. Voinnet. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557-560. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Z. Z., J. L. Wang, X. Huang, W. H. Xu, Z. M. Liu, and R. X. Fang. 2003. The promoter of a rice glycine-rich protein gene, Osgrp-2, confers vascular-specific expression in transgenic plants. Planta 216:824-833. [DOI] [PubMed] [Google Scholar]
- 25.Llave, C. 2004. MicroRNAs: more than a role in plant development? Mol. Plant Pathol. 5:361-366. [DOI] [PubMed] [Google Scholar]
- 26.Lu, S., R. Shi, C. C. Tsao, X. Yi, L. Li, and V. L. Chiang. 2004. RNA silencing in plants by the expression of siRNA duplexes. Nucleic Acids Res. 32:e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaffrey, A. P., H. Nakai, K. Pandey, Z. Huang, F. H. Salazar, H. Xu, S. F. Wieland, P. L. Marion, and M. A. Kay. 2003. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21:639-644. [DOI] [PubMed] [Google Scholar]
- 28.Missiou, A., K. Kalantidis, A. Boutla, S. Tzortzakaki, M. Tabler, and M. Tsagris. 2004. Generation of transgenic potato plants highly resistant to potato virus Y (PVY) through RNA silencing. Mol. Breed. 14:185-197. [Google Scholar]
- 29.Moissiard, G., and O. Voinnet. 2006. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. USA 103:19593-19598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Niu, Q. W., S. S. Lin, J. L. Reyes, K. C. Chen, H. W. Wu, S. D. Yeh, and N. H. Chua. 2006. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24:1420-1428. [DOI] [PubMed] [Google Scholar]
- 31.Parizotto, E. A., P. Dunoyer, N. Rahm, C. Himber, and O. Voinnet. 2004. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18:2237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pokalsky, A. R., W. R. Hiatt, N. Ridge, R. Rasmussen, C. M. Houck, and C. K. Shewmaker. 1989. Structure and expression of elongation factor 1 alpha in tomato. Nucleic Acids Res. 17:4661-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds, A., D. Leake, Q. Boese, S. Scaringe, W. S. Marshall, and A. Khvorova. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22:326-330. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Schwab, R., S. Ossowski, M. Riester, N. Warthmann, and D. Weigel. 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18:1121-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon-Mateo, C., and J. A. Garcia. 2006. MicroRNA-guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J. Virol. 80:2429-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, N. A., S. P. Singh, M. B. Wang, P. A. Stoutjesdijk, A. G. Green, and P. M. Waterhouse. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407:319-320. [DOI] [PubMed] [Google Scholar]
- 38.Szittya, G., D. Silhavy, A. Molnar, Z. Havelda, A. Lovas, L. Lakatos, Z. Banfalvi, and J. Burgyan. 2003. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomari, Y., and P. D. Zamore. 2005. Perspective: machines for RNAi. Genes Dev. 19:517-529. [DOI] [PubMed] [Google Scholar]
- 40.Vaucheret, H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20:759-771. [DOI] [PubMed] [Google Scholar]
- 41.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 42.Waibel, F., and W. Filipowicz. 1990. U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res. 18:3451-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, M.-B., D. C. Abbott, and P. M. Waterhouse. 2000. A single copy of a virus-derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol. Plant Pathol. 1:347-356. [DOI] [PubMed] [Google Scholar]
- 44.Wroblewski, T., A. Tomczak, and R. Michelmore. 2005. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato, and Arabidopsis. Plant Biotechnol. J. 3:259-273. [DOI] [PubMed] [Google Scholar]
- 45.Xie, Z., E. Allen, A. Wilken, and J. C. Carrington. 2005. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102:12984-12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie, Z., L. K. Johansen, A. M. Gustafson, K. D. Kasschau, A. D. Lellis, D. Zilberman, S. E. Jacobsen, and J. C. Carrington. 2004. Genetic and functional diversification of small RNA pathways in plants. PLOS Biol. 2:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X., Y. R. Yuan, Y. Pei, S. S. Lin, T. Tuschl, D. J. Patel, and N. H. Chua. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20:3255-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zilberman, D., and S. Henikoff. 2005. Epigenetic inheritance in Arabidopsis: selective silence. Curr. Opin. Genet. Dev. 15:557-562. [DOI] [PubMed] [Google Scholar]