Abstract
Many ichneumonid and braconid endoparasitoids inject a polydnavirus (PDV) into their caterpillar hosts during oviposition. The viral entities carried by wasps of these families are referred to as “ichnoviruses” (IVs) and “bracoviruses” (BVs), respectively. All IV genomes characterized to date are found in wasps of the subfamily Campopleginae; consequently, little is known about PDVs found in wasps of the subfamily Banchinae, the only other ichneumonid taxon thus far shown to carry these viruses. Here we report on the genome sequence and virion morphology of a PDV carried by the banchine parasitoid Glypta fumiferanae. With an aggregate genome size of ∼290 kb and 105 genome segments, this virus displays a degree of genome segmentation far greater than that reported for BVs or IVs. The size range of its genome segments is also lower than those in the latter two groups. As reported for other PDVs, the predicted open reading frames of this virus cluster into gene families, including the protein tyrosine phosphatase (PTP) and viral ankyrin (ank) families, but phylogenetic analysis indicates that ank genes of the G. fumiferanae virus are not embedded within the IV lineage, while its PTPs and those of BVs form distinct clusters. The banchine PDV genome also encodes a novel family of NTPase-like proteins displaying a pox-D5 domain. The unique genomic features of the first banchine virus examined, along with the morphological singularities of its virions (IV-like nucleocapsids, but enveloped in groups like some of the BVs), suggest that they could have an origin distinct from those of IVs and BVs.
The family Polydnaviridae comprises double-stranded DNA (dsDNA) viruses transmitted by hymenopteran endoparasitoids to their caterpillar hosts during oviposition. Although they cause no apparent pathology in the wasp carrier, polydnaviruses (PDVs) induce physiological perturbations in their lepidopteran hosts, in the absence of which survival of the developing wasp egg or larva would be severely compromised. The observed dysfunctions include a suppression of the host cellular immune response and/or an inhibition of metamorphosis, both of which are brought about by PDV gene expression (32). Viral replication, however, does not take place in the lepidopteran host; rather, this is confined to calyx cells of the wasp ovary, where chromosomally integrated copies of the segmented viral genome are excised, circularized, packaged, and then released into the lumen of the oviduct, where the virions form the particulate fraction of a so-called “calyx fluid” bathing the mature eggs.
The finding that PDVs are transmitted through the wasp germ line (22, 40) led to predictions about their obligate association with certain wasp taxa. Indeed, early (16, 24, 27) and more recent (34) surveys of ichneumonid and braconid species belonging to several subfamilies made it possible to identify those taxa displaying a stable association with PDVs. Within the family Braconidae, a relatively homogenous group of viral entities, the bracoviruses (BVs), were found to be distributed among four subfamilies, the Cardiochilinae, the Microgastrinae, the Miracinae, and the Cheloninae. The BVs carried by members of these four wasp subfamilies are now believed to have a common origin (35) and indeed share several characteristics, including cylindrical nucleocapsids of various lengths, the envelopment of single or multiple nucleocapsids by a single unit membrane, and their release into the lumen of the oviduct by lysis of calyx epithelial cells. Within the family Ichneumonidae, two subfamilies have been identified as containing wasps carrying PDVs: the Campopleginae and the Banchinae. Two studies (16, 24), however, suggested that the viral entities found in these two taxa may not be as similar to one another as those found among the braconid subfamilies. Perhaps for practical reasons, all subsequent work on ichneumonid PDVs focused entirely on viruses associated with campoplegine wasps, from which the defining features of the ichnovirus (IV) taxon emerged: lenticular nucleocapsids enveloped singly by two unit membranes, the outer one being acquired from the plasma membrane of calyx cells during exocytosis into the lumen of the oviduct.
Until recently, evidence for the existence of PDVs among banchine wasps was based upon electron micrographs of sections taken through calyx tissue obtained from field-collected individuals of Glypta spp. (24), as well as on agarose gel fractionation of viral genome segments extracted from the ovaries of Glypta sp. (16; D. Stoltz, unpublished data). Early reports (16, 24) indicated that the size range of genome segments packaged within banchine PDVs appeared to be significantly smaller than that of typical IVs and BVs.
In this paper, we provide the first thorough analysis of a banchine PDV, including the integration of its genome within wasp chromosomes, the sequencing and annotation of its genome segments, and the morphological features of its virions. For the purpose of the present analysis, we chose the banchine wasp Glypta fumiferanae, a common parasitoid of the spruce budworm, Choristoneura fumiferana, in eastern Canada (5). Recent efforts at sequencing packaged the genomes of selected IVs and BVs (9, 10, 29, 33) have led to the recognition that (i) these genomes have a remarkably low coding density; (ii) the genes they carry do not encode viral structural proteins or proteins required for replication; (iii) several putative viral genes are most closely related to wasp cellular genes; and (iv) IV and BV genes, with the exception of those encoding ankyrin proteins, are apparently unrelated. The latter observation is in support of the hypothesis that IVs and BVs have arisen independently (27, 35). The data presented here show that the virus of G. fumiferanae (referred to here as GfV) displays morphological features that are more similar to those of IVs than BVs but that the gene content of its genome has more in common with BVs than IVs. Altogether, our results indicate that the banchine virus may form a PDV subgroup distinct from IVs and BVs.
MATERIALS AND METHODS
Insects.
The banchine parasitoid Glypta fumiferanae was obtained from C. fumiferana larvae collected in a small stand of mixed balsam fir and white spruce near Trois-Rivières, Quebec, Canada, in May 2003. Host larvae were transferred from foliage to artificial diet poured into a small glass vial to monitor parasitoid egression and cocoon spinning. Upon emergence, male and female wasps were held separately in petri dishes and fed honey and water; their identification as G. fumiferanae was confirmed by G. Pelletier (Canadian Forest Service, Quebec City, Canada). Dissection of ovaries, virion purification, and DNA extraction were carried out as described previously (17, 23).
Southern blot analysis.
To confirm the presence of the GfV genome within G. fumiferanae chromosomes, DNA extracted from male wasps was digested with either EcoRI or HindIII, fractionated by agarose gel electrophoresis (AGE), and assessed by Southern hybridization as described previously (3), with the exception that the 32P labeling of the probes was carried out using the Megaprime DNA labeling kit (GE Healthcare). We used two different probes: total GfV DNA and cloned GfV genome segment B43, from which the TN transposon (see “Construction of EZ::TN library” below) was first removed by digestion with HindIII; this genome segment contains 0 and 1 restriction sites for HindIII and EcoRI, respectively. For comparative purposes, we also conducted a hybridization using the same B43 probe on undigested GfV DNA.
Agarose gel electrophoresis.
To compare the size range of genome segments in the banchine virus with that of genome segments in a typical IV, DNA (500 ng) extracted from the Tranosema rostrale ichnovirus (TrIV) was fractionated by AGE (0.8% gel) along with DNA (500 ng) from the G. fumiferanae virus and stained with ethidium bromide after electrophoresis.
Construction of EZ::TN library.
Virions were purified and their DNA was extracted as described previously (17, 23). Individual GfV genome segments were isolated, replicated, and stably maintained in Escherichia coli using the EZ::TN transposon system, following the manufacturer's directions (Epicenter). In this process, bacterial origins of replication and a Kanr gene were inserted into the circular genome segments, which permitted the cloning of individual genome segments in their entirety. The resulting plasmid DNAs were purified from bacterial cultures using the QIAprep Spin Miniprep kit (QIAGEN).
Sequencing and assembly.
To sequence the GfV genome, we processed individual EZ::TN clones, using both vector- and virus-specific primers and the ABI Prism BigDye terminator cycle sequencing ready reaction kit (PE Applied Biosystems), on an ABI model 310 Prism DNA sequencer (PE Applied Biosystems) at the University of Kentucky. The DNA sequences were first edited to remove vector sequences and then assembled using either SeqMan II (Lasergene DNAStar) or Genchek v. 2061 (Ocimum Biosolutions). Contigs were built using a minimum of a 50-nucleotide overlap, and sequence variations below 0.5% were regarded as polymorphism. The gaps between contigs on the same genome segment were closed by primer walking. Genome coverage is estimated at threefold.
ORF predictions and sequence similarity searches.
The GfV genome was scanned for putative open reading frames (ORFs) using NCBI's ORF Finder (www.ncbi.nlm.nih.gov/gorf/gorf.html), the Gene Construction Kit 2 program (Texco Inc.), Genchek v. 2061 (Ocimum Biosolutions), and FGENESH, with the Apis mellifera settings (www.softberry.com). Initially, only ORFs encoding ≥100 amino acids (aa) with an initiator methionine were considered as putative genes; however, a few smaller ones (75 to 99 aa) were also counted as putative genes on the basis of the significant similarity of their protein products to other GfV putative proteins (see reference 29). Database searches were performed using NCBI's BLASTN, BLASTX, and BLASTP (www.ncbi.nlm.nih.gov/BLAST/). The amino acid sequences were also checked for alignments with protein families at InterProScan (www.ebi.ac.uk/InterProScan/) and for protein families and domains at Prosite (au.expasy.org/prosite/) and Pfam (www.sanger.ac.uk/Software/Pfam/). Putative promoter elements, transcription initiation sites, and polyadenylation signals were identified as described previously (29).
Phylogenetic analyses.
For the phylogenetic analysis of ank and PTP genes, all of the sequences were aligned using Clustal X version 1.81 and the Gonnet 250 protein matrix (4), with the gap penalties recommended by Hall (14). Portions of the sequences that did not correspond among gene copies were then pruned. Data sets thus treated were then analyzed using MrBayes version 3.1 (21; MrBayes v3.1, J. P. Huelsenbeck and F. Ronquist [http://mrbayes.csit.fsu.edu/]), which uses Markov chain Monte Carlo sampling to explore the “space” of possible alternative trees while resampling from the model parameters. The protein model employed for each was the matrix of Jones et al. (15), which is similar to the Gonnet matrix mentioned above and is available in MrBayes. A total of 8,000,000 generations of four chains were run in two parallel analyses and then checked for convergence of the two runs using Tracer version 1.2.1 (A. Rambaut and A. Drummond [http://evolve.zoo.ox.ac.uk/beast/]). The majority rule consensus tree from each two-run analysis was visualized using TreeView (19), after deleting the first 25% of the samples as burn in.
The “backbone phylogeny” of the wasps was obtained from analysis of partial DNA sequences of seven genes (16S rRNA, cytochrome oxidase 1, 28S rRNA, elongation factor 1α, wingless, arginine kinase, and longwave rhodopsin genes as detailed in reference 2). Modeltest version 3.06 (20; Modeltest version 3.06, D. Posada [http://darwin.uvigo.es/software/modeltest.html]) was used to select the appropriate maximum likelihood model for anaysis of each gene; the general time-reversible model with the proportion of invariant sites and a gamma distribution of four rate classes estimated (GTR + I + G) model was selected in each case. MrBayes 3.1 was then used to estimate the phylogeny employing 8,000,000 generations, four chains and a 25% burn in, in two parallel runs. Tracer 1.2.1 was used to check for convergence of the two runs. Raw sequences were deposited in GenBank.
Electron microscopy.
Ovaries to be processed for thin sectioning were dissected out into the primary fixative (3% glutaraldehyde in 50 mM sodium cacodylate buffer containing 250 mM sucrose) and left at room temperature for 2 h. Postfixation was in 2% OsO4 in the same buffer, followed by overnight staining in 0.1% aqueous uranyl acetate. Embedment was in epon/araldite resin, following dehydration in acetone.
Attempts were made to disrupt virions such that enveloped nucleocapsids could be visualized in situ by negative staining. To this end, calyx fluid was dissected out into TASS buffer (50 mM ammonium sulfate, 30 mM Tris, 5% [wt/vol] sucrose [pH 7.5]) and adsorbed onto carbon-coated Formvar films. Grids were subsequently floated onto a variety of detergents at 0.1% (wt/vol) concentration for 30 to 60 s, in order to partially disrupt the viral envelopes, prior to negative staining using 0.1% aqueous uranyl acetate.
RESULTS
Linear copies of GfV genome within G. fumiferanae chromosomal DNA.
Although the virus found in G. fumiferanae ovaries displays several of the hallmark features of PDVs (see below), the integration of its genomic DNA within the chromosomal DNA of the wasp host remained to be ascertained. To this end, we carried out Southern hybridizations on digested male G. fumiferanae DNA, using either total GfV DNA or cloned genome segment B43 (see Fig. 1) as a probe. Hybridization with radiolabeled GfV DNA yielded the expected smear for both HindIII and EcoRI digests, strongly suggestive of integration (Fig. 1A). To further confirm the presence of GfV genome segments within G. fumiferanae genomic DNA, we hybridized both undigested GfV DNA and parasitoid DNA with radiolabeled genome segment B43, which contains 0 and 1 restriction sites for HindIII and EcoRI, respectively. The GfV DNA blot yielded a strong signal corresponding to a DNA fragment of the expected size (2,641 bp); there were, however, additional, fainter signals corresponding to cross-hybridizing genome segments (Fig. 1B). Upon hybridization with HindIII-digested wasp DNA, the B43 probe yielded one off-size fragment of about 7 kb (Fig. 1C). Because digestion of genome segment B43 with EcoRI is predicted to yield fragments of 2,484 and 157 bp, the strongest signal (∼2.8 kb) observed upon hybridization of B43 with EcoRI-digested host DNA is assumed to be the off-size fragment corresponding to the larger B43 fragment. The fastest-migrating signal (<1 kb) is assumed to be the off-size fragment corresponding to the 157-bp B43 fragment; the signal of intermediate mobility (∼2.6 kb) may have been generated by integrated, cross-hybridizing genome segments or may result from incomplete digestion of wasp DNA (Fig. 1C).
FIG. 1.
Southern hybridization strongly suggests that linear copies of GfV genome segments are present within the genomic DNA of G. fumiferanae wasps. (A) Male G. fumiferanae genomic DNA digested with either HindIII (H) or EcoRI (E), fractionated by AGE, blotted, and probed with 32P-labeled GfV DNA. (B) AGE-fractionated, undigested GfV DNA probed with 32P-labeled genome segment B43 (2641 bp). Note that there are cross-hybridizing genome segments; the two upper, fainter signals are presumed to have been generated by DNAs in their open circular topology. (C) Same blot as in panel A but hybridized with 32P-labeled genome segment B43, which has 0 and 1 restriction sites for HindIII and EcoRI, respectively. Note that the positions of only a sample of bands of the 1-kb DNA ladder (kb) and of the supercoiled DNA ladder [kb (ccc)] are shown, so as to not overload the illustration.
Agarose gel electrophoresis of GfV DNA.
Fractionation of both GfV and TrIV DNA by AGE showed the size range of GfV genome segments to be significantly lower than that of the ichnovirus TrIV (Fig. 2). Although there appears to be no overlap in the size range of visually detectable genome segments, cloning and sequencing of individual TrIV (29) and GfV (this study) genome segments indicated that the size of the smallest TrIV genome segment (4.1 kb) is smaller than that of the largest in GfV (5.2 kb; see below). The majority of GfV DNA molecules detected on this gel are believed to be in their superhelical topology, although low-abundance open circular species are likely present as well, as suggested by the detection of larger-size DNAs by Southern blot analysis (Fig. 1B).
FIG. 2.
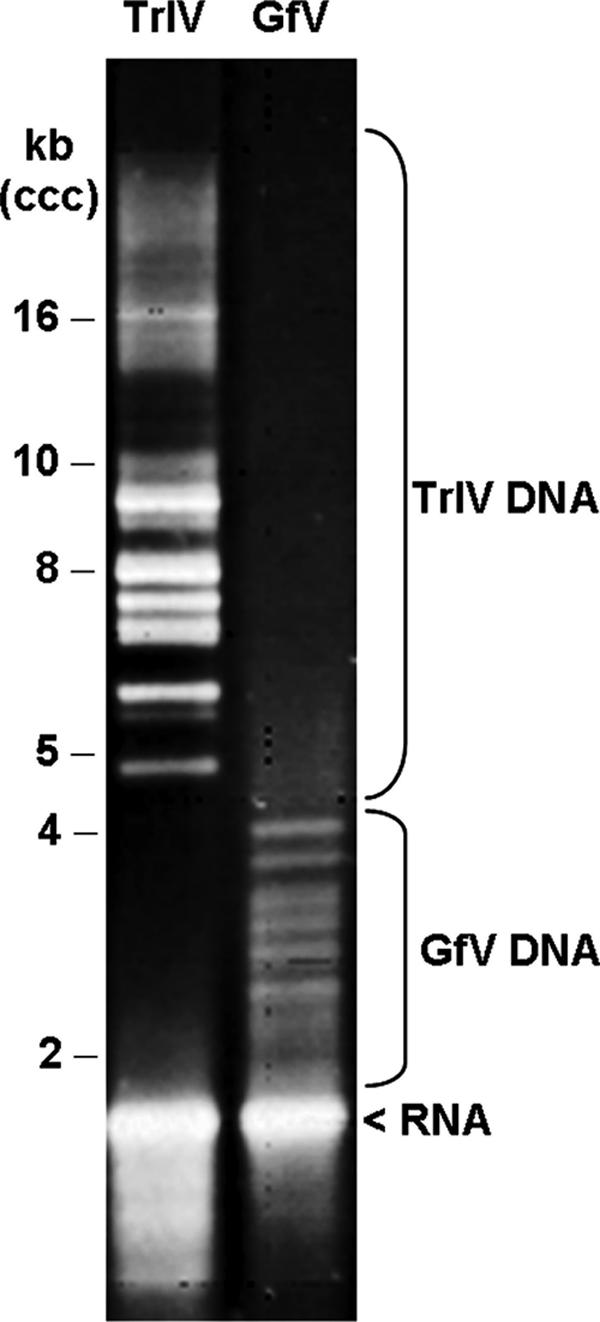
Electrophoretic fractionation of undigested GfV and TrIV DNA on a 0.8% agarose gel. Note the difference in the size ranges of the GfV and TrIV genome segments.
General features of the GfV genome.
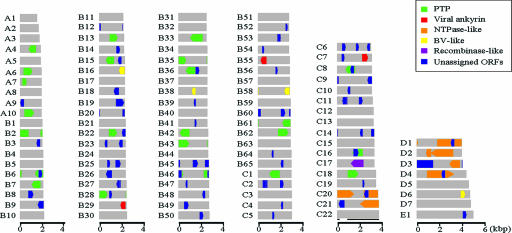
With an estimated genome size of 291.4 kb and 105 genome segments (GenBank accession no. AB289903 to AB290007 and AB295392 for the polymorphic variant GfV-B55′), GfV has the most highly segmented PDV genome examined to date. As a consequence, the size range of its genome segments (1,533 to 5,156 bp; see Fig. 2 and 3) is also the smallest of all characterized PDVs. As shown for both IVs and BVs, the GfV genome displays a strong A+T bias (63.3%) and a low coding density (20.2%), although these values are somewhat closer to those reported for Microplitis demolitor bracovirus (MdBV) (66% A+T; 17% coding density) (33) than to those of IVs (57 to 59% A+T; 22 to 30% coding density) (29, 33). Strikingly, 31 of the 105 annotated GfV genome segments (30%) contained no detectable ORF (see Fig. 3); the coding density of the subset of genome segments in which ORFs were detected was 31.8%, ranging from 6.3 to 77.8% for individual segments. While genome segment nesting (6, 29, 41) was clearly observed in only one instance (C22; see Fig. 3), we found large portions of perfect sequence identity within each of three genome segment pairs (B10 and B34, B54 and C19, and D1 and D4); however, other portions displayed high levels of mismatch (17 to 34% in the low-identity portions, corresponding to 3 to 9% over the length of the smaller DNA), precluding the assignment of the smaller genome segment as a daughter segment or as a polymorphic variant.
FIG. 3.
Graphic representation of GfV genome sequences and annotations, with the 105 nonredundant circular genome segments shown as linear molecules. Individual genome segments are displayed from smallest (A1 = 1.5 kb) to largest (E1 = 5.2 kb). Colored boxes show the sizes and locations of ORFs, with orientation indicated by the arrowhead on each box. Refer to the legend for color coding of gene families. Noncoding sequences are shown in gray. Genome segment B55 has a polymorphic variant (B55′) not shown here; this variant encodes an additional ankyrin protein (B55′-ORF1) that is not present in B55 due to a deletion-induced frameshift. One of the two unassigned ORFs found on genome segment C7 is not shown here because it is of the same size and is in the same location (but on opposite strand) as the ankyrin ORF. The black line shown under genome segment C22 indicates the size and position of the daughter segment C22′.
Predicted GfV ORFs.
The different algorithms employed for ORF identification yielded essentially the same list of putative genes, except that FGENESH, used with the A. mellifera settings, occasionally predicted spliced transcripts in cases where the other algorithms did not. However, all of the ORFs presented here are reported as being devoid of introns, based on observations made on other PDVs; indeed, demonstrated cases of gene splicing are thus far limited to secreted products (33), none of which are predicted from the present genome sequence.
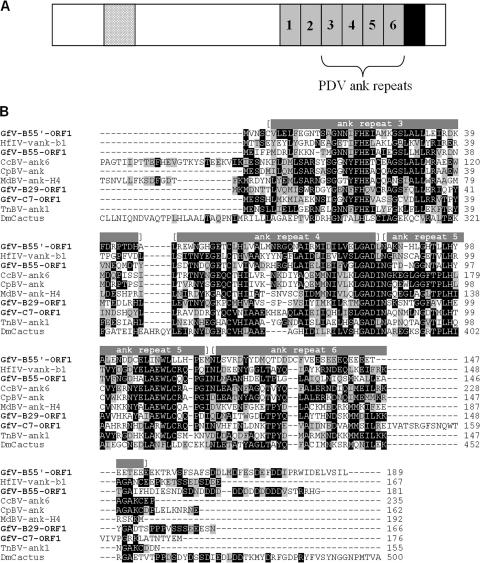
The GfV genome contains members of two gene families identified earlier in other PDVs (Table 1 and Fig. 3). With 23 members, protein tyrosine phosphatases (PTPs) constitute the largest family of identifiable protein-coding genes in this virus; PTPs are also abundant in BV genomes but have not yet been detected in those of IVs (9, 10, 29, 33). The GfV genome was also found to contain four viral ankyrin (ank) genes, a ubiquitous family with members found in both IVs and BVs (18, 30). The four putative ankyrins of GfV, like those of IVs and BVs, contain only the last four of the six ankyrin repeats that form the ankyrin repeat domain (ARD) identified in the Drosophila melanogaster Iκβ protein, Cactus (see 30), although the portion downstream from ankyrin repeat 5 in GfV-B55′-ORF1 shows little or no similarity to ankyrin repeat 6 in other PDV ankyrin proteins (see Fig. 4B). As described for IV and BV ankyrins, the GfV ankyrins also lack the N-terminal signal-receiving domain and PEST domain involved in protein turnover (see reference 30). The C termini of three of them, however, do contain the proline, glutamic acid, serine, and threonine residues characteristic of PEST domains (see Fig. 4), but yield subthreshold PEST scores upon scanning with the PESTfind algorithm (https://bioweb.pasteur.fr/seqanal/interfaces/pestfind.html), suggesting that these sequences have recently lost their PEST domain functionality.
TABLE 1.
Family distribution of ORFs identified within the GfV genome
| Family | No. of ORFs |
|---|---|
| PTP | 23 |
| Viral ankyrin | 4 |
| NTPase-like | 7 |
| BV-like | 4 |
| Recombinase-like | 1 |
| Unassigned | 62 |
FIG. 4.
(A) Diagram showing the structure of the D. melanogaster Iκβ protein, Cactus (AAA85908). Gray boxes, individual ankyrin repeats within the ankyrin repeat domain (ARD); black box, PEST domain; dotted box, signal-receiving domain. Of these, the PDV ankyrins have retained only the last four ankyrin repeats of the ARD. (B) Amino acid alignment (ClustalW) of the four deduced GfV ankyrin proteins, along with selected representatives of IV and BV ankyrins [CcBV-ank6, CAG17493; TnBV-ank1, CAE47441; CpBV-ank, AAZ04266; MdBV-ank(H4), YP_239384; HfIV-vank-b1, AAX24120] and Dm-Cactus. Within a column, identical and similar residues are shaded in black and gray, respectively. Positions of ankyrin repeats 3 to 6 (see reference 30) are shown above the aligned sequences.
We identified a third, seven-member family of distinctly related protein-coding genes, four of which encode products displaying similarity to the pox-D5 family of NTPases (11). With two exceptions (D2-ORF2 and D3-ORF3), these putative GfV genes encode large proteins (574 to 972 aa) that are found on some of the largest GfV genome segments (Fig. 5A); they are designated here as NTPase-like proteins (Fig. 3). Interestingly, most BLASTP hits corresponded to proteins of viral origin (Mimiviridae, Iridoviridae, Poxviridae, and Ascoviridae), including one from the Spodoptera frugiperda ascovirus 1a (YP_762453).
FIG. 5.
(A) Amino acid alignment (ClustalW) of portions of the seven GfV NTPase-like proteins identified in this study. The arrowheads indicate the boundaries of the region in which pox-D5 domains were identified by Motif Scan. (B) Amino acid alignment (ClustalW) of the four BV-like GfV proteins identified in this study, along with that of CpBV ORF1103 (AAZ04291). Within a column, identical and similar residues are shaded in black and gray, respectively.
We also detected four putative genes whose protein products display similarity to a hypothetical protein encoded by ORF1103 of the Cotesia plutellae BV (AAZ04291). The two larger ones, GfV-B16-ORF1 (179 aa) and GfV-B58-ORF1 (178 aa), are very similar to one another and show higher levels of similarity (BLASTP expect values of 2e-12 and 2e-07, respectively) to the CpBV hypothetical protein than do the two smaller ones, GfV-D6-ORF1 (131 aa) and GfV-B38-ORF1 (106 aa; BLASTP expect values of 3e-06 and 5.0, respectively; see Fig. 5B). The function of these putative proteins (referred to here as “BV-like”; see Fig. 3), is unknown but one of them (GfV-B16-ORF1) displays a bipartite nuclear targeting sequence, as determined by Prosite.
Finally, we identified a gene whose conceptual product shows similarity to two Chelonus inanitus bracovirus (CiBV) putative proteins (CAA91234 and CAC82011, BLASTP expect values of 2e-10 and 2e-07, respectively), one of which is considered a putative recombinase. In fact, all three PDV proteins contain a recombinase domain (as determined by Pfam) also seen in three related dipteran putative recombinases (CAC37683, CAC37681, and CAC37684).
The remaining ORFs that we detected in the GfV genome could not be associated with reasonable confidence with any known genes. Among these ORFs, however, some were found to encode putative proteins that are clearly related to one another, forming families of two to five proteins (see supplemental material for sequence alignments). Given that known PDV genes tend to cluster into families, the present observation suggests that these unassigned ORFs are likely to represent functional genes. For further details on genome annotation, please refer to the supplemental material.
Phylogenetic analyses.
To examine the relationships among IVs, BVs and the PDV of G. fumiferanae, we conducted three types of phylogenetic analyses: (i) a “backbone” phylogeny based on seven wasp genes; (ii) a PTP phylogeny based on all known PTPs from BVs and GfV; and (iii) an ankyrin phylogeny based on all known ankyrin proteins from IVs, BVs, and GfV.
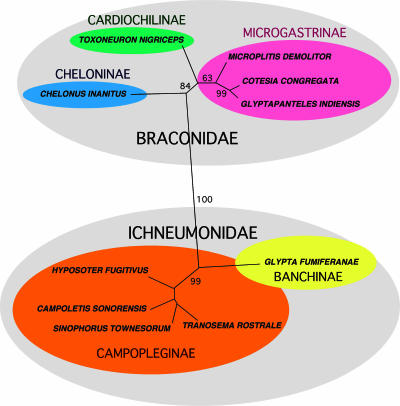
The relationships found among the wasps from the analysis of seven genes are as expected from previous analyses of microgastroid wasp subfamilies and genera (2, 8, 35-37) and from current broader ichneumonoid classification (Fig. 6). Thus, if PDV relationships mirror those of their wasp carriers, we expect that the BVs in Cheloninae will be most genetically distant from those in Microgastrinae and PDVs in Banchinae to be distant from those in Campopleginae. In the latter case, it is of interest to note that not all members of the Campopleginae have IVs (D. Stoltz, unpublished data) and that other ichneumonid subfamilies not possessing PDVs are more closely related to this taxon than Banchinae, thus suggesting that the Campopleginae and Banchinae viruses could have separate origins.
FIG. 6.
Unrooted Bayesian phylogeny of wasps from which PDV genomes are being sequenced, based on analysis of DNA sequences from seven genes. Values on branches represent bootstrap support, and branch lengths are proportional to corrected distances. See Materials and Methods for genes and analysis details.
Phylogenetic analysis of all PTPs from the GfV and the five BV genomes for which partial or complete genomic sequences are available (Fig. 7) revealed that all PTPs from the former virus form a lineage separate from the BV PTPs. This result suggests that the two wasp groups acquired the viruses independently or that only a single PTP copy was present ancestrally in the PDV genome. It is not currently possible to distinguish these two scenarios phylogenetically.
FIG. 7.
Unrooted Bayesian phylogeny of PTPs from the five available BV genomes and the GfV genome. Gene copies are color coded by wasp. Thick branches are supported by posterior probabilities above 0.90; dotted internal branches are supported by lower values. Glypta virus PTPs are colored red. See the supplemental material for a complete list of GenBank accession numbers corresponding to the gene labels shown on the tree.
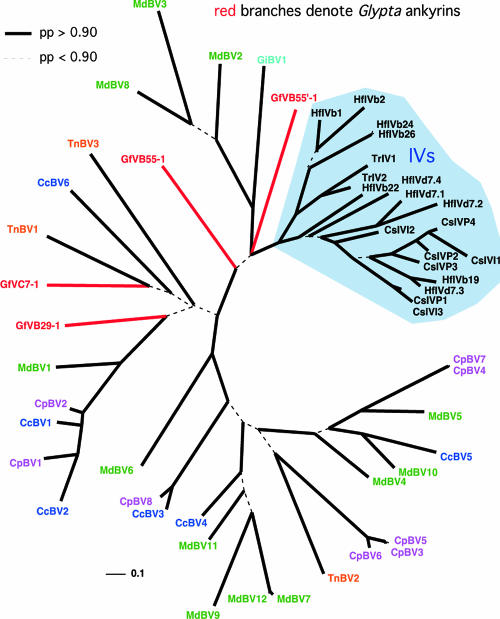
Phylogenetic analysis of the ankyrins from the same BV and three IV genomes as well as from that of GfV (Fig. 8) showed that the GfV ankyrins are not embedded within the IV ankyrin lineage but instead generally fall near the base of the BV lineage. While they do not appear to form a coherent lineage themselves, it would be premature to conclude that they do not, since the data are limited in resolution due to high sequence divergence between the IV and BV copies. Nevertheless, the evidence strongly suggests that GfV is not closely related to the campoplegine IVs.
FIG. 8.
Unrooted Bayesian phylogeny of ankyrins from the five available BV genomes, three available IV genomes, plus the GfV genome. Glypta virus copies are colored red; BV copies are color coded by wasp. Thick branches are supported by posterior probabilities above 0.90; dotted internal branches are supported by lower values. See the supplemental material for a complete list of GenBank accession numbers corresponding to the gene labels shown on the tree.
Virion morphology and morphogenesis.
As has been shown for both IVs (26, 31) and BVs (7, 25, 39), GfV morphogenesis takes place within the nuclei of calyx epithelial cells. Like the ichnoviruses (26), the mature GfV particle possesses two unit membrane envelopes. The inner one appears to form de novo on the surfaces of an intranuclear virogenic stroma (Fig. 9A), after which the enveloped particle exits the nucleus by budding through the nuclear envelope, thereby acquiring two additional membranes (Fig. 9B), which are subsequently lost within the cytosol, as also occurs in the case of typical ichnoviruses. Virions finally exit the cell in a nonlytic manner by budding through the plasma membrane (Fig. 9C); thus, the mature extracellular particle is surrounded by two envelopes.
FIG. 9.
(A) GfV morphogenesis occurring in the ovarian calyx epithelium. Envelopes (arrows) appear to assemble de novo on the surfaces of an intranuclear virogenic stroma (VS); several nucleocapsids (NC) are packaged within each envelope. (B) Particles exit the nucleus (N) through the nuclear envelope (NE) into the cytosol (C), where the two membranes derived from the nuclear envelope are, in some as-yet-undetermined manner, lost, such that only the original envelope (IM) is retained. (C) Virus release by budding through the plasma membrane (PM) into the lumen (L) of the calyx; mature, extracellular virions thus have two envelopes: IM, inner membrane; OM, outer membrane. Bar, 200 nm.
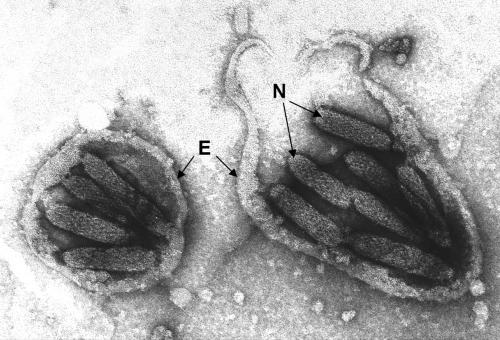
Interestingly, and unlike what has been described for the ichnoviruses, each GfV virion appears to contain several nucleocapsids (Fig. 9 and 10). GfV nucleocapsids appear much smaller than those of typical IVs and are also less obviously lenticular in shape, although not blunt ended like BV nucleocapsids (1, 7, 25, 39); by negative staining (Fig. 10), GfV nucleocapsid surface substructure is more readily apparent than is the case for either BV or IV nucleocapsids.
FIG. 10.
Negatively stained GfV nucleocapsids (N). Virions in calyx fluid were adsorbed to carbon-coated grids, which were then floated briefly on 0.1% TX114 in order to disrupt the envelopes, remnants of which (E) remain surrounding groups of nucleocapsids derived from two virions.
DISCUSSION
Early studies (16, 24, 27; D. Stoltz, unpublished data) had indicated that females of field-collected banchine wasps, namely Glypta sp. and Lissonota sp., carried ovarian particles whose apparent site of replication (calyx epithelial cells), general morphology, and segmented DNA genomes strongly suggested that they were PDVs. Here we confirm these observations and in addition provide Southern hybridization evidence to the effect that the genome of GfV is integrated within the chromosomal DNA of its wasp host (Fig. 1); thus, GfV possesses all the structural and life cycle features that define PDVs. Given that the BVs associated with the braconid subfamilies Cardiochilinae, Microgastrinae, Miricinae, and Cheloninae are believed to have a common origin (35), the question arose as to whether the PDVs observed in campoplegine (i.e., IVs) and banchine wasps also share a common origin. We believe that the data presented here suggest that campoplegine and banchine PDVs could have arisen independently.
First, while GfV morphology (quasicylindrical nucleocapsids surrounded by two envelopes) and morphogenesis (noncytolytic exit by budding through both the nuclear envelope and the plasma membrane) parallel those of typical IVs, there are at least two notable differences. For example, nucleocapsids in the case of banchine PDVs are much smaller than those of typical IVs; this can be seen by comparing Fig. 1a and b of an earlier publication (24). More importantly, the mature extracellular virion contains not one, but several nucleocapsids, unlike any IV thus far described. Taken together, our electron microscopy observations lend support to the idea that the banchine PDVs could represent a separate lineage.
Second, the level of genome segmentation is higher and the size range of genome segments is lower in GfV (Fig. 2 and 3) than in the IV and BV genomes characterized to date; we recognize that the values of these two variables do vary among the sequenced IV and BV genomes (9, 10, 29, 33) but clearly not to the same extent as shown here. Third, although the A+T bias and coding density of the GfV genome are roughly similar to those calculated for other PDVs, the values of these two variables are closer to those of BVs than to those of IVs.
Perhaps most importantly, GfV gene composition is strikingly different from that of characterized IVs (29, 33). In fact, PTPs, which have not yet been observed in IVs, constitute the dominant gene family of the GfV genome (Table 1 and Fig. 3), a feature shared with BVs (9, 10, 13). Our phylogenetic analysis, however, suggests that GfV PTPs form a different lineage from those of BVs (Fig. 7), thus supporting the hypothesis that banchine PDVs could have an origin distinct from those of either IVs or BVs. Similarly, GfV ankyrin genes, a family which has members in both IVs and BVs, appear phylogenetically more removed from those of IVs than those of BVs (Fig. 8), despite the fact that the members of the Banchinae are more closely related to the Campopleginae than to the BV-carrying braconid wasps (Fig. 6). Interestingly, two other groups of genes identified here in the GfV genome show some similarity to putative ORFs found in the genomes of BVs (i.e., BV-like and recombinase-like proteins; Fig. 3), as opposed to those of IVs. Clearly, gene composition of the GfV genome has little in common with that of characterized IVs.
Additionally, we identified a new putative seven-member gene family not previously detected in other PDVs. The seven putative proteins are here designated as “NTPase-like” on the basis that four of them display the pox-D5 domain of related NTPases found in several large dsDNA viruses, including the vaccinia virus D5 protein; the latter was the first well-characterized member of this family and was shown to be essential for vaccinia virus DNA replication (11). It is not clear, at this point, whether the similarity of these GfV gene products to viral proteins means anything in terms of the presumed viral origin of PDVs, given that the levels of similarity were not very high (BLASTP expect values of e-07 to e-04). It is interesting, however, that the BLASTP hits included a putative pox-D5 protein from an ascovirus; the Ascoviridae display morphological, genomic, and host-specificity features that have led some workers to suggest that they could be the ancestors of the ichnoviruses (12). Alternatively, it is conceivable that the putative GfV NTPase-like proteins could be derived from the wasp. For example, the GfV proteins also show similarity to a group of related eukaryotic proteins, the SMC2 (structural maintenance of chromosomes 2) proteins, which are large ATPases that are only active as SMC2/SMC4 heterodimeric complexes (28). Since our BLASTP searches identified some insect SMC2-like proteins among the potential homologs, the putative GfV proteins reported here could be derived from wasp SMC2 proteins, none of which, to our knowledge, have been reported yet.
In summary, the data presented here indicate that banchine PDVs have genomic and morphological features that are distinct from those of previously characterized IVs or BVs. Whether this points to an independent origin for the banchine virus needs to be explored further, especially in view of the fact that the genomes of chelonine BVs appear to contain none of the genes identified so far in cardiochiline and microgastrine BVs (38; B. Lanzrein, personal communication), while all BVs are thought to have a common ancestor (35). Irrespective of the outcome of such investigations, we feel that the banchine virus examined here is sufficiently different from IVs and BVs to justify the creation of a distinct PDV taxon. This exercise will require, of course, that the definition of ichnoviruses be revisited. The PDVs of additional banchine species will also need to be examined in order to assess the homogeneity of the features reported here within this wasp subfamily. Finally, the exact role of the banchine viruses in host-parasitoid relationships remains to be ascertained. Interestingly, G. fumiferanae females lay their eggs in pre-diapausing second-instar C. fumiferana hosts in late summer, but their larval progeny complete most of their development in the following spring (5). Whether GfV gene expression is detectable and required for wasp larval development after the winter diapause needs to be examined.
Supplementary Material
Acknowledgments
This research was supported by USDA-NRI-2001-04713 and NSF-MCB-0094403 grants to B.A.W., a grant from Genome Canada through the Ontario Genomics Institute to M.C., and grants from the Natural Sciences and Engineering Research Council of Canada to D.S. and M.C.
We thank G. Pelletier (Canadian Forest Service, Quebec City, Canada) for wasp identification and D. Doucet for constructive comments on the manuscript.
Footnotes
Published ahead of print on 11 April 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
This is publication no. 07-08-045 of the University of Kentucky Agricultural Experiment Station.
REFERENCES
- 1.Albrecht, U., T. Wyler, R. Pfister-Wilhelm, A. Gruber, P. Stettler, P. Heiniger, E. Kurt, D. Schumperli, and B. Lanzrein. 1994. Polydnavirus of the parasitic wasp Chelonus inanitus (Braconidae): characterization, genome organization and time point of replication. J. Gen. Virol. 75:3353-3363. [DOI] [PubMed] [Google Scholar]
- 2.Banks, J. C., and J. B. Whitfield. 2006. Dissecting the ancient rapid radiation of microgastrine wasp genera using additional nuclear genes. Mol. Phylogenet. Evol. 41:690-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Béliveau, C., M. Laforge, M. Cusson, and G. Bellemare. 2000. Expression of a Tranosema rostrale polydnavirus gene in the spruce budworm Choristoneura fumiferana. J. Gen. Virol. 81:1871-1880. [DOI] [PubMed] [Google Scholar]
- 4.Benner, S. A., M. A. Cohen, and G. H. Gonnet. 1994. Amino acid substitution during functionally constrained divergent evolution of protein sequences. Protein Eng. 7:1323-1332. [DOI] [PubMed] [Google Scholar]
- 5.Brown, N. R. 1946. Studies on parasites of the spruce budworm, Archips fumiferana (Clem.). II. Life history of Glypta fumiferanae (Viereck) (Hymenoptera, Ichneumonidae). Can. Entomol. 78:138-147. [Google Scholar]
- 6.Cui, L., and B. A. Webb. 1997. Homologous sequences in the Campoletis sonorensis polydnavirus genome are implicated in replication and nesting of the W segment family. J. Virol. 71:8504-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.deBuron, I., and N. E. Beckage. 1992. Characterization of a polydnavirus (PDV) and virus-like filamentous particle (VLFP) in the braconid wasp Cotesia congregata (Hymenoptera: Braconidae). J. Invertebr. Pathol. 59:315-327. [Google Scholar]
- 8.Dowton, M., and A. D. Austin. 1998. Phylogenetic relationships among the microgastroid wasps (Hymenoptera: Braconidae): combined analysis of 16S and 28S rDNA genes and morphological data. Mol. Phylogenet. Evol. 10:354-366. [DOI] [PubMed] [Google Scholar]
- 9.Dupuy, C., É. Huguet, and J.-M. Drezen. 2006. Unfolding the evolutionary story of polydnaviruses. Virus Res. 117:81-89. [DOI] [PubMed] [Google Scholar]
- 10.Espagne, É., C. Dupuy, É. Huguet, L. Cattolico, B. Provost, N. Martins, M. Poirié, G. Periquet, and J.-M. Drezen. 2004. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science 306:286-289. [DOI] [PubMed] [Google Scholar]
- 11.Evans, E., N. Klemperer, R. Ghosh, and P. Traktman. 1995. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 69:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federici, B. A., and Y. Bigot. 2003. Origin and evolution of polydnaviruses by symbiogenesis of insect DNA viruses in endoparasitic wasps. J. Insect Physiol. 49:419-432. [DOI] [PubMed] [Google Scholar]
- 13.Gundersen-Rindal, D. E., and M. J. Pedroni. 2006. Characterization and transcriptional analysis of protein tyrosine phosphatase genes and an ankyrin repeat gene of the parasitoid Glyptapanteles indiensis polydnavirus in the parasitized host. J. Gen. Virol. 87:311-322. [DOI] [PubMed] [Google Scholar]
- 14.Hall, B. G. 2004. Phylogenetic trees made easy. Sinauer Associates, Sunderland, MA.
- 15.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biol. Sci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 16.Krell, P. J. 1991. The polydnaviruses: multipartite DNA viruses from parasitic Hymenoptera, p. 141-177. In E. Kurstak (ed.), Viruses of invertebrates (ed.), Marcel Dekker, Inc., New York, NY.
- 17.Krell, P. J., and D. B. Stoltz. 1980. Virus-like particles in the ovary of an ichneumonid wasp: purification and preliminary characterization. Virology 101:408-418. [DOI] [PubMed] [Google Scholar]
- 18.Kroemer, J. A., and B. A. Webb. 2005. Iκβ-related vankyrin genes in the Campoletis sonorensis ichnovirus: temporal and tissue-specific patterns of expression in parasitized Heliothis virescens lepidopteran hosts. J. Virol. 79:7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 20.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 21.Ronquist, F., and J. P. Huelsenback. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 22.Stoltz, D. B. 1990. Evidence for chromosomal transmission of polydnavirus DNA. J. Gen. Virol. 71:1051-1056. [DOI] [PubMed] [Google Scholar]
- 23.Stoltz, D. B., D. Guzo, and D. Cook. 1986. Studies on polydnavirus transmission. Virology 155:120-131. [DOI] [PubMed] [Google Scholar]
- 24.Stoltz, D. B., P. J. Krell, and S. B. Vinson. 1981. Polydisperse viral DNAs in ichneumonid ovaries: a survey. Can. J. Microbiol. 27:123-130. [DOI] [PubMed] [Google Scholar]
- 25.Stoltz, D. B., and S. B. Vinson. 1977. Baculovirus-like particles in the reproductive tracts of female parasitoid wasps. II. The genus Apanteles. Can. J. Microbiol. 23:28-37. [DOI] [PubMed] [Google Scholar]
- 26.Stoltz, D. B., and S. B. Vinson. 1979. Penetration into caterpillar cells of virus-like particles injected during oviposition by parasitoid ichneumonid wasps. Can. J. Microbiol. 25:207-216. [DOI] [PubMed] [Google Scholar]
- 27.Stoltz, D. B., and J. B. Whitfield. 1992. Viruses and virus-like entities in the parasitic Hymenoptera. J. Hymenopt. Res. 1:125-139. [Google Scholar]
- 28.Stray, J. E., and J. E. Lindsley. 2003. Biochemical analysis of the yeast condensin Smc2/4 complex: an ATPase that promotes knotting of circular DNA. J. Biol. Chem. 278:26238-26248. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, K., R. Lapointe, W. Barney, A. Makkay, D. Stoltz, M. Cusson, and B. A. Webb. 14 February 2007. Shared and species-specific features among ichnovirus genomes. Virology doi: 10.1016/j.virol. 2006.11.34. [DOI] [PubMed]
- 30.Thoetkiattikul, H., M. H. Beck, and M. R. Strand. 2005. Inhibitor kappaB-like proteins from a polydnavirus inhibit NF-kappaB activation and suppress the insect immune response. Proc. Natl. Acad. Sci. USA 102:11426-11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkoff, A.-N., M. Ravallec, J.-P. Bossy, P. Cerutti, J. Rocher, M. Cerutti, and G. Devauchelle. 1995. The replication of Hyposoter didymator polydnavirus: cytopathology of the calyx cells in the parasitoid. Biol. Cell 83:1-13. [Google Scholar]
- 32.Webb, B. A. 1998. Polydnavirus biology, genome structure, and evolution, p. 105-139. In L. K. Miller and L. A. Balls (ed.), The insect viruses. Plenum Press, New York, NY.
- 33.Webb, B. A., M. R. Strand, S. E. Dickey, M. H. Beck, R. S. Hilgarth, W. E. Barney, K. Kadash, J. A. Kroemer, K. G. Lindstrom, W. Rattanadechakul, K. S. Shelby, H. Thoetkiattikul, M. W. Turnbull, and R. A. Witherell. 2006. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347:160-174. [DOI] [PubMed] [Google Scholar]
- 34.Wharton, R. A., and H. Sittertz-Bhatkar. 2002. Polydnaviruses in the genus Mirax Haliday (Hymenoptera: Braconidae). J. Hymenopt. Res. 11:358-365. [Google Scholar]
- 35.Whitfield, J. B. 1997. Molecular and morphological data suggest a common origin for the polydnaviruses among braconid wasps. Naturwissenschaften 84:502-507. [Google Scholar]
- 36.Whitfield, J. B. 2002. Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc. Natl. Acad. Sci. USA 99:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitfield, J. B., P. Mardulyn, A. D. Austin, and M. Dowton. 2002. Phylogenetic analysis of relationships among microgastrine braconid wasp genera based on data from the 16S, COI and 28S genes and morphology. Syst. Entomol. 27:337-359. [Google Scholar]
- 38.Wyder, S., A. Tschannen, A. Hochuli, A. Gruber, V. Saladin, S. Zumbach, and B. Lanzrein. 2002. Characterization of Chelonus inanitus polydnavirus segments: sequences and analysis, excision site and demonstration of clustering. J. Gen. Virol. 83:247-256. [DOI] [PubMed] [Google Scholar]
- 39.Wyler, T., and B. Lanzrein. 2003. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. II. Ultrastructural analysis of calyx cell development, virion formation and release. J. Gen. Virol. 84:1151-1163. [DOI] [PubMed] [Google Scholar]
- 40.Xu, D., and D. Stoltz. 1991. Evidence for a chromosomal location of polydnavirus DNA in the ichneumonid parasitoid Hyposoter fugitivus. J. Virol. 65:6693-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, D., and D. Stoltz. 1993. Polydnavirus genome segment families in the ichneumonid parasitoid Hyposoter fugitivus. J. Virol. 67:1340-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.