Abstract
The human cytomegalovirus (HCMV) major immediate-early (MIE) enhancer contains five functional cyclic AMP (cAMP) response elements (CRE). Because the CRE in their native context do not contribute appreciably to MIE enhancer/promoter activity in lytically infected human fibroblasts and NTera2 (NT2)-derived neurons, we postulated that they might have a role in MIE enhancer/promoter reactivation in quiescently infected cells. Here, we show that stimulation of the cAMP signaling pathway by treatment with forskolin (FSK), an adenylyl cyclase activator, greatly alleviates MIE enhancer/promoter silencing in quiescently infected NT2 neuronal precursors. The effect is immediate, independent of de novo protein synthesis, associated with the phosphorylation of ATF-1 serine 63 and CREB serine 133, dependent on protein kinase A (PKA) and the enhancer's CRE, and linked to viral-lytic-cycle advancement. Coupling of FSK treatment with the inhibition of either histone deacetylases or protein synthesis synergistically activates MIE gene expression in a manner suggesting that MIE enhancer/promoter silencing is optimally relieved by an interplay of multiple regulatory mechanisms. In contrast, MIE enhancer/promoter silence is not overcome by stimulation of the gamma interferon (IFN-γ) signaling pathway, despite the enhancer having two IFN-γ-activated-site-like elements. We conclude that stimulation of the cAMP/PKA signaling pathway drives CRE-dependent MIE enhancer/promoter activation in quiescently infected cells, thus exposing a potential mode of regulation in HCMV reactivation.
The reactivation of latent cytomegalovirus (CMV) becomes more apparent when host cellular immunity weakens. The CMV major immediate-early (MIE) enhancer/promoter is a sentinel checkpoint in the regulation of viral reactivation (26, 51, 59). Tumor necrosis factor alpha (TNF-α), lipopolysaccharide, interleukin-1β, or allogeneic stimulation induces murine CMV MIE enhancer/promoter reactivation in the lungs or kidneys of latently infected mice (8, 16, 51), possibly by way of signal transduction-mediated activation of NF-κB and AP-1, which bind the enhancer (15). Latent human CMV (HCMV) in blood monocytes of healthy donors is reported to reactivate in culture after allogeneic stimulation with unrelated blood mononuclear cells (55). This reactivation is tied to macrophage-dendritic cell differentiation and exposure to specific cytokines, including gamma interferon (IFN-γ) (54). HCMV also reactivates from blood monocytes and CD34+ hematopoietic progenitor cells cultured in cytokine mixtures favoring dendritic cell differentiation (46). In contrast, the culturing of latently infected monocyte-derived macrophages in granulocyte-macrophage colony-stimulating factor plus hydrocortisone reactivates the MIE enhancer/promoter but not subsequent lytic-cycle events (59). Induction of HCMV reactivation by cellular differentiation or by the cytokine TNF-α or IFN-γ is reproduced in human monocyte-dendritic cell precursors that are quiescently infected in vitro (14, 24, 45, 66). Either allogeneic stimulation (18, 52) or treatment with TNF-α or IFN-γ (53) also enhances HCMV replication in productively infected monocyte-derived macrophages. Taken together, CMV lytic-cycle reactivation can be viewed as a deliberate response to specific cellular changes imposed by distinct cellular-differentiation programs and external stimuli.
Murine CMV latency is not limited to cells of myeloid lineage (23, 60), and the same might be true for HCMV. The periventricular location of neuronal-cell precursors from which murine CMV reactivates upon culturing of latently infected mouse brain explants (60) is reminiscent of HCMV's proclivity for subependymal neuronal cells in the ventriculoencephalitis that results from HCMV reactivation in persons with advanced AIDS (5, 64). The mechanisms that underlie HCMV reactivation disease in the central nervous system are unknown, though terminal differentiation of the neuronal lineage is linked to permissiveness for HCMV replication (43).
Human NTera2/D1 cells (NT2) are recognized as a useful model in which to study the regulatory mechanisms behind MIE enhancer/promoter silencing during quiescent HCMV infection (13, 27, 35, 39, 50). NT2 closely resemble early neuronal precursors and gradually differentiate into dopaminergic central nervous system neurons after exposure to retinoic acid (RA) (1, 21, 42). Unlike with NT2, HCMV infection of differentiated NT2 neuronal derivatives gives rise to MIE enhancer/promoter activation and viral replication (13, 27, 35, 39, 50). However, a delay in the initiation of RA-induced cellular differentiation until the time of infection fails to alleviate MIE enhancer/promoter silencing (32), despite RA's ability to activate MIE enhancer/promoter segments in transient-transfection assays via multiple copies of the enhancer's RA response element (3, 4, 11). Trichostatin A (TSA), an inhibitor of histone deacetylases, brings about MIE enhancer/promoter reactivation in quiescently infected NT2 (32) in a manner independent of cellular differentiation (38). This treatment disrupts heterochromatin nucleation at the MIE enhancer/promoter (38), akin to the chromatin disruption that accompanies HCMV reactivation in endogenously infected dendritic cells (45).
The MIE enhancer/promoter is dynamically regulated to control the expression of the IE1 p72 and IE2 p86 proteins, which are vital for advancing the viral lytic cycle but are likely detrimental to viral latency (36). This regulation entails the integration of variable input provided by the cell, the virus, and external surroundings. The 540-bp enhancer is composed of structurally dissimilar distal and proximal halves that operationally harmonize to efficiently activate MIE promoter-dependent transcription in acutely infected cells (19, 34).
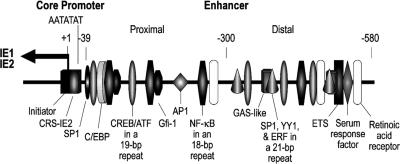
A complex interplay of the enhancer's multiple cis-acting elements and the specific transcription factors that act on them is believed to control enhancer performance (Fig. 1). Little is known about the regulatory mechanisms involved in triggering HCMV MIE enhancer/promoter reactivation in quiescently infected cells. The two IFN-γ-activated-site (GAS)-like elements recently discovered in the distal enhancer (Fig. 1) augment MIE enhancer/promoter-dependent transcription in acutely infected human foreskin fibroblasts (HFF), in a fashion further stimulated by IFN-γ (40). Whether these cis-acting elements are linked to IFN-γ's putative involvement in HCMV reactivation from cells of myeloid lineage is unknown.
FIG. 1.
HCMV MIE promoter/enhancer and its cis-acting elements. The enhancer is divided into proximal (−39 to −299) and distal (−300 to −579) portions. Positions of binding sites for the initiator complex, C/EBP, CREB/ATF, Gfi-1, AP1, NF-κB, SP1, YY1, ETS2 repressor factor (ERF), ETS, serum response factor, RA receptor, and the GAS-like complex (36, 40) are shown relative to the +1 RNA start site (leftward arrow) of the core promoter. Binding sites for NF-κB, CREB/ATF, and the combination of SP1, YY1, and ERF are nested in the 18-, 19-, and 21-bp repeats, respectively. The SP1 site depicted in the proximal enhancer exists as two closely spaced sites that are bound in vitro by SP1 or SP3 (20). IE2 p86 binds to the cis repression signal (CRS-IE2).
The five cyclic AMP (cAMP) response elements (CRE) dispersed throughout the enhancer are among an assortment of cis-acting elements with the capacity for carrying out stimulus-induced transcriptional activation (Fig. 1). The CRE motif is bound in vitro by CREB and ATF-1 (44, 47, 56, 57), which are expressed in most cell types (31, 49). Stimulation of the cAMP signaling pathway drives CRE-dependent transcription from MIE enhancer/promoter segments positioned exogenously in gene delivery vectors in diverse cell types (17, 44, 57, 61). However, the CRE in the context of all other enhancer elements add little to transcription from HCMV's native MIE enhancer/promoter when analyzed in either stimulated or unstimulated acutely infected HFF and differentiated NT2 (22). We therefore postulated that the CRE might have a greater role in MIE enhancer/promoter reactivation (22).
In this report, we show that the stimulation of the cAMP pathway with forskolin (FSK), an adenylyl cyclase activator, greatly alleviates MIE enhancer/promoter silencing in quiescently infected NT2. The mechanism behind the induction of this activation and its role in subsequent viral lytic events are characterized. This outcome is distinct from that of stimulation of the IFN-γ signaling pathway, which fails to reverse MIE enhancer/promoter silencing.
MATERIALS AND METHODS
Cells and viruses.
NT2 (13) were grown in Dulbecco's modified Eagle's medium supplemented with 4 μM glutamine, 4.5 g/liter glucose, 10% knockout serum replacement (Life Technologies, Rockville, MD), and 3% charcoal-treated fetal bovine serum (FBS), as described previously (32). FBS was treated with charcoal (5 g per 100 ml) overnight at 4°C. Cells were fed daily. Attention to the culture method, such as avoidance of trypsin, cell aggregation, and low-density seeding (6, 12), is important to minimize background levels of cellular differentiation and MIE expression. Where indicated below, knockout serum replacement was removed from the medium for a short period to further minimize the background NT2 subpopulation, permitting MIE expression. The differentiation of NT2 was achieved by the addition of 10 μM RA (Sigma, St. Louis, MO) to the growth medium for 5 days. HFF were isolated and grown as described previously (35).
HCMV (Towne strain) was propagated on HFF. The green fluorescent protein (GFP)-expressing HCMVs rΔ−640/−1108gfp (29) and rΔ−582/−1108Egfp (34) were derived from the Towne strain and are herein denoted HCMV-GFP1 and HCMV-GFP2, respectively. The GFP is expressed with early/late kinetics during the viral lytic cycle from virus-native UL127 and heterologous adenovirus E1b promoters, respectively (29, 32, 34). The wild type (WT) and rCREM5 (Towne strain) were characterized previously in acutely infected permissive cells (22).
HCMV was prepared by passing supernatants of infected HFF through a 0.45-μm filter. Partially purified HCMV represents filtered virus that was centrifuged through a 20% sorbitol cushion in phosphate-buffered saline (PBS), followed by resuspension of the virion pellet in fresh NT2 growth medium lacking knockout serum replacement. Virus absorption was carried out for 1 to 1.5 h, and cells were subsequently washed twice or thrice with Hanks' balanced salt solution without calcium and magnesium.
FSK (10 mM), H-89 (10 mM), calphostin C (1 mM), and staurosporin (100 μM) stocks (Sigma, St. Louis, MO) were prepared in dimethyl sulfoxide. Protein kinase inhibitor (14-24) amide (Alexis Corporation) and recombinant human IFN-γ (10,000 units/μg; R&D Systems, Inc., Minneapolis, MN) were dissolved in water.
Quantification of HCMV DNA and RNA.
HCMV DNA was quantified by real-time PCR using ABI PRISM 7700 and 7000 sequence detection systems (Applied Biosystems, Foster City, CA). Cell-associated DNA was prepared using methods described previously (35). The MIE enhancer of HCMV genomes was targeted for amplification with primers 5′-CATGGTGATGCGGTTTTGG-3′ and 5′-TGGAAATCCCCGTGAGTCA-3′ and detected with the reporter probe 5′-6-carboxyfluorescein-ACCGCTATCCACGCCCATTGATGT-nonfluorescent minor groove binder-3′, which were obtained from Integrated DNA Technologies (Coralville, IA). The primers and probe used for amplification and detection of cellular ribosomal 18S RNA, respectively, were obtained from Applied Biosystems. The standard-curve method was applied to determine the relative concentration of target DNA in relationship to the threshold cycle (CT) value.
Whole-cell RNA was isolated according to the method of Chomczynski and Sacchi (7). cDNA was made using Superscript II RNase H− reverse transcriptase (RT; Life Technologies, Gaithersburg, MD) and random hexamers, as described previously (33). The MIE RNA-derived cDNA was quantified by real-time PCR using the primers, probe, and amplification conditions reported previously (33). The primers target MIE exons 1 and 2 to produce an MIE amplicon spanning intron A. Spliced UL89 RNA was quantified in the same way, but the primers and probe that were described by White et al. (62) were used. The cDNAs derived from ribosomal 18S, 2′-5′-oligo(A) synthetase-2 (OAS-2), and indoleamine 2,3-dioxygenase RNAs were amplified and detected with reagents supplied by Applied Biosystems. The standard-curve method was applied to determine the concentration of target cDNA relative to the CT value. CT values of samples not treated with RT but analyzed in parallel with the RT-treated samples did not differ from baseline.
Flow cytometry analysis and microscopy.
Samples of live cells were analyzed on a FACScan flow cytometer using CellQuest software (Becton Dickinson, Franklin Lakes, NJ). Cells were resuspended in PBS containing propidium iodide (50 μg/ml) and 1% FBS. Ten thousand live-cell events (as determined by forward and side scatter) were acquired for analysis. Flow sorting of live NT2 was performed with a Coulter EPICS 753 cytometer (Beckman Coulter, Inc.). Ten thousand live cells either emitting or lacking GFP fluorescence were collected in growth medium. These cells were cocultivated with subconfluent adherent HFF in a 12-well dish (400 NT2 per 105 HFF in each well) containing Dulbecco's modified Eagle's medium supplemented with 10% FBS.
An indirect immunofluorescence assay was applied for the detection of HCMV pp65 and MIE proteins. Cells were fixed in ice-cold methanol for 5 min, permeablilzed with 0.3% Triton X-100 in PBS for 5 min, and blocked with 10% goat serum in PBS for 30 min at room temperature. Monoclonal murine antibodies against CMV pp65 tegument protein (C9100-25; U.S. Biologicals, Swampscott, MA) or IE1 and IE2 proteins (MAB810; Chemicon International, Charlottesville, VA) at a 1:200 dilution in 10% goat serum in PBS were applied for 1 h at 37°C for the detection of pp65 or MIE protein, respectively. Secondary goat anti-mouse antibody conjugated to Alexa Fluor 555 (Molecular Probes Invitrogen, Eugene, OR) was applied at a 1:1,000 dilution for 1 h at room temperature. Cells were counterstained with 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI).
Cytopathic effect and fluorescence were assessed and images were captured using an inverted Olympus IX 51 fluorescence microscope equipped with an X-Cite 120 fluorescence illumination system.
Immunoblotting.
Cells were lysed in a buffer (10% glycerol, 1% Triton X-100, 20 mM Tris [pH 7.9], 137 mM NaCl, 1 mM Na3VO4, 5 mM EDTA, 1 mM EGTA, 10 mM NaF, 1 mM Na pyrophosphate, 1 mM β-glycerophosphate) containing Complete Mini, EDTA-free protease inhibitor mixture (Roche Diagnostics GmbH, Penzberg, Germany) and phenylmethlysulfonyl fluoride (0.5 mM). Lysates were sonicated on ice for 10 s and boiled in sodium dodecyl sulfate (SDS) sample buffer for 5 min. Protein extracts (50 μg/lane) were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), using either 10% or 4 to 20% Tris-glycine gels (Invitrogen, Carlsbad, CA), and transferred to Protran BA85 nitrocellulose membranes (Schleicher & Schuell BioScience, Keene, NH). Blots were incubated overnight at 4°C with primary antibody in 1× PBS containing 5% dried milk, incubated with ImmunoPure peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (1:5,000 dilution) at room temperature for 90 min in PBS containing 5% dried milk, treated with SuperSignal West Femto maximum-sensitivity substrate (Pierce Biotechnology, Inc., Rockford, IL), and exposed to autoradiography film. The primary antibodies were used at 1:1,000 to 1:2,000 dilutions. They include anti-CREB5-24, an immunoaffinity-purified polyclonal rabbit antibody raised against amino-terminal residues 5 to 24 of human CREB (Upstate Biotechnology, Lake Placid, NY); anti-CREB260-280, a monoclonal rabbit antibody raised against residues 260 to 280 (Epitomics, Burlingame, CA); anti-pCREB122-136, an affinity-purified polyclonal rabbit antibody raised against a synthetic phosphopeptide corresponding to residues 126 to 136 (phospho-Ser133) of rat CREB (Upstate Biotechnology); and anti-ATF-139-271 (25C10G), a monoclonal murine antibody raised against residues 39 to 271 of human ATF-1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). HCMV IE1 p72 and IE2 p86 were detected using monoclonal murine antibody MAB810 (Chemicon International), which reacts to an epitope in both proteins. Blots were reprobed with murine monoclonal anti-beta tubulin (E7) antibody (1:500 dilution; University of Iowa Hybridoma Bank, Iowa City, IA). Rabbit monoclonal antibody against human Oct4 (Epitomics) was applied at a 1:1,000 dilution. Densitometry analysis was performed using the NIH ImageJ 1.34s program.
RESULTS
FSK, but not IFN-γ, activates HCMV lytic-cycle gene expression.
The roles of FSK and IFN-γ in activating viral-lytic-cycle gene expression from quiescent HCMV genomes were examined in the NT2 neuronal-precursor model, in which both the CRE (9, 47) and IFN-γ (2) signaling pathways function prominently.
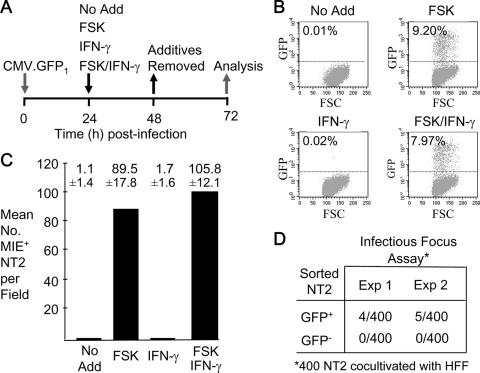
NT2 were infected with partially purified HCMV virions (multiplicity of infection [MOI] of 3 to 5 PFU per cell) to obviate possible confounding effects of factors unassociated with viral particles (see Materials and Methods). An HCMV (Towne strain) modified to express GFP from the viral UL127 early/late kinetic-class promoter (28, 29, 32), designated HCMV-GFP1, enabled the use of fluorescence emission to mark infected cells that permit viral-lytic-cycle activation. Over 98% of NT2 nuclei contain HCMV pp65 at 1 h postinfection (p.i.) (data not shown), corroborating an earlier report of NT2 nuclei containing at least three HCMV genome equivalents per nucleus at an MOI of 10 (35). Using an experimental strategy outlined in Fig. 2A, quiescently infected NT2 were exposed at 24 h p.i. to FSK, IFN-γ, or a combination of FSK and IFN-γ for a duration of 24 h at concentrations of these agents found previously to stimulate HCMV lytic-cycle events in other cell types (22, 53).
FIG. 2.
FSK, but not IFN-γ, activates the HCMV lytic cycle. (A) Diagram of the experimental design. At 24 h p.i., FSK (10 μM), IFN-γ (1,000 U/ml), or both FSK (10 μM) and IFN-γ (1,000 U/ml) were added to or omitted from (No Add) the growth medium of NT2 infected with partially purified HCMV-GFP1 (MOI of 3 to 5) and then removed at 48 h p.i. (B) Activation of the viral UL127 promoter expressing GFP. Percentages of GFP+ NT2 were determined by fluorescence-activated cell sorting (FACS) at 72 h p.i. for 10,000 live cells per experimental group. Uninfected NT2 gave a background of 0.01% GFP+ cells. The proportions of treated and untreated (No Add) infected cells excluding propidium iodide varied by less than 10%. FSC, forward scatter. (C) Quantification of NT2 expressing MIE proteins at 72 h p.i., using indirect immunofluorescence microscopy analysis. Cells were reacted with MA810 antibody against MIE (IE1 p71 and IE2 p86) proteins and secondary antibody conjugated with Alexa Fluor 555, followed by counterstaining with DAPI. MIE+ NT2 were counted in 10 random fields (original magnification, ×20). Means ± standard deviations are depicted. (D) FSK-induced production of infectious virus. GFP+ and GFP− NT2 from two independent experiments (Exp) were separated by FACS at 72 h p.i. GFP+ NT2 represent the very bright fluorescent cells in the FSK-treated population. An infectious-focus assay was performed on each of the groups by cocultivation of 400 NT2 with subconfluent HFF.
Several experiments were analyzed by flow cytometry to assess the size of the GFP+ NT2 population at 72 h p.i. in relation to treatment condition. One such experiment depicted in Fig. 2B reveals that virally produced GFP is evident in 0.01% of the untreated NT2 population. Treatment with IFN-γ does not significantly change this outcome. In contrast, the proportion of NT2 expressing GFP increases to 9.2% in response to FSK but does not increase further by the addition of IFN-γ. The groups did not differ in degree of cell viability as measured by propidium iodide exclusion. While FSK reproducibly caused a large expansion in the GFP+ NT2 population in all experiments (data not shown), the absolute sizes of GFP+ NT2 populations before and after stimulation varied between experiments for reasons that partly reflect variations in growth conditions (e.g., growth medium components and method of culture) that have an impact on the heterogeneity of the starting NT2 population.
The number of infected NT2 expressing HCMV MIE proteins in relation to treatment condition was determined in situ by indirect immunofluorescence microscopy with antibody against IE1 p72 and IE2 p86. Results of the experiment depicted in Fig. 2C indicate that FSK stimulation increases the size of the MIE+ NT2 population by 81-fold, compared to that of untreated NT2. IFN-γ does not significantly expand this population. Ratios of MIE+ to GFP+ cells were additionally determined by fluorescence microscopy to be 4.3 and 5.3 for NT2 receiving FSK alone and NT2 receiving FSK plus IFN-γ, respectively, whereas the ratio was 9 or 11 for NT2 receiving no treatment or IFN-γ alone, respectively (data not shown). In separate studies of colocalization of GFP and MIE proteins, GFP fluorescence was observed only in NT2 emitting MIE protein-specific Alexa Fluor 555 fluorescence, while a subset of MIE+ NT2 lacked GFP fluorescence (data not shown).
The question of whether FSK-induced activation of viral-lytic-gene expression yields infectious progeny was addressed by the method of assaying the infectious foci of GFP+ cells that had been isolated by flow cytometry (32). The findings reveal that approximately 1% of GFP+ cells are able to transmit virus to HFF but that their GFP− counterparts are noninfectious (Fig. 2D). It is unclear why such low frequencies of GFP+ cells produce or release infectious virus. The expression of viral lytic genes may be extinguished in a subset of these cells, because they cease to emit fluorescence and resume cellular division (data not shown).
Thus, HCMV replication appears to be restricted at multiple levels. FSK stimulation lifts constraints to viral MIE gene expression and other early lytic-cycle events in a sizable subpopulation of quiescently infected NT2.
FSK, but not IFN-γ, alleviates HCMV MIE enhancer/promoter silencing.
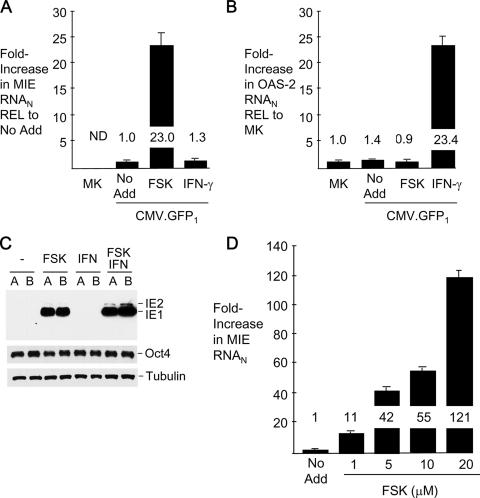
We next examined the relative abilities of FSK and IFN-γ to activate MIE enhancer/promoter-dependent transcription from the quiescent viral genomes. Spliced MIE message for IE1 p71 and IE2 p86 was quantified at 24 h after the addition of these agents. As shown in Fig. 3A, FSK increases MIE RNA expression by 23-fold in comparison to its expression after mock treatment. In contrast, IFN-γ does not significantly increase MIE RNA production yet appropriately elevates by 23.4-fold the expression of RNA from a characteristic cellular-IFN-stimulated gene, the OAS-2 gene (Fig. 3B). OAS-2 gene expression is up-regulated neither by FSK nor by viral particles at 48 h p.i. A comparable set of findings was observed for another IFN-stimulated gene, the indoleamine 2,3-dioxygenase gene (data not shown).
FIG. 3.
FSK, but not IFN-γ, activates HCMV MIE gene transcription. At 24 h p.i., FSK (10 μM), IFN-γ (1,000 U/ml), or both FSK (10 μM) and IFN-γ (1,000 U/ml) were added to or omitted from (No Add) the growth medium of NT2 infected with partially purified CMV-GFP1 (MOI of 3). Viral MIE (A) and cellular OAS-2 (B) RNAs were quantified by real-time RT-PCR at 48 h p.i. after normalization to the cellular 18S RNA abundance (RNAN). Depicted are the means ± standard deviations of changes (n-fold) in levels of MIE and OAS-2 RNAN relative to (REL) levels in infected and mock-infected (MK) NT2, respectively, receiving No Add. ND, not detected. (C) In a separate experiment of identical design, the expression of the IE1 p71 and IE2 p86 proteins was analyzed. Whole-cell extracts from experimental groups in duplicate (A and B samples) were fractionated by SDS-PAGE and subjected to immunoblotting. Proteins were visualized using an MA810 antibody against an epitope in both IE1 p71 and IE2 p86. The blot was reprobed with antibody against β-tubulin and again with antibody against human Oct4. (D) The dose-response curve of MIE RNA expression was determined after FSK was added at the designated concentrations to quiescently infected NT2 at 24 h p.i. The amount of the dimethyl sulfoxide vehicle was kept constant. MIE RNA was quantified at 6 h p.i., using the standard-curve method and normalization to the 18S RNA amount.
The FSK-induced rise in MIE RNA expression results in the production of both the IE1 p71 and IE2 p86 proteins (Fig. 3C). These proteins are not detected by the Western blot method in whole-cell extracts of untreated and IFN-γ-treated infected NT2. IFN-γ adds minimally to FSK's effect on MIE protein production. Neither FSK nor IFN-γ results in the down-regulation of cellular Oct4 expression, which is required to maintain the undifferentiated NT2 state and causes differentiation when down-regulated (30).
FSK raises levels of MIE RNA expression in relation to its concentration (Fig. 3C). FSK concentrations producing a submaximal response are applied throughout this work to lessen the potential contribution of CRE-independent effects that can mount with higher FSK doses (22). Notably, the 121-fold increase in MIE RNA expression induced by 20 μM FSK for 6 h greatly exceeds the reported 4.4-fold rise in MIE RNA production in acutely infected HFF receiving 50 μM FSK for the initial 6 h p.i. (22).
Hence, FSK stimulation alleviates MIE enhancer/promoter silencing. In contrast, stimulation of the IFN-γ signaling pathway does not reverse MIE enhancer/promoter silencing, an outcome like the previous finding of RA's inability to reverse the silencing despite the presence of multiple RA response elements in the enhancer (32).
FSK's action is immediate, does not require de novo protein synthesis, and corresponds to heightened levels of pCREB and pATF-1.
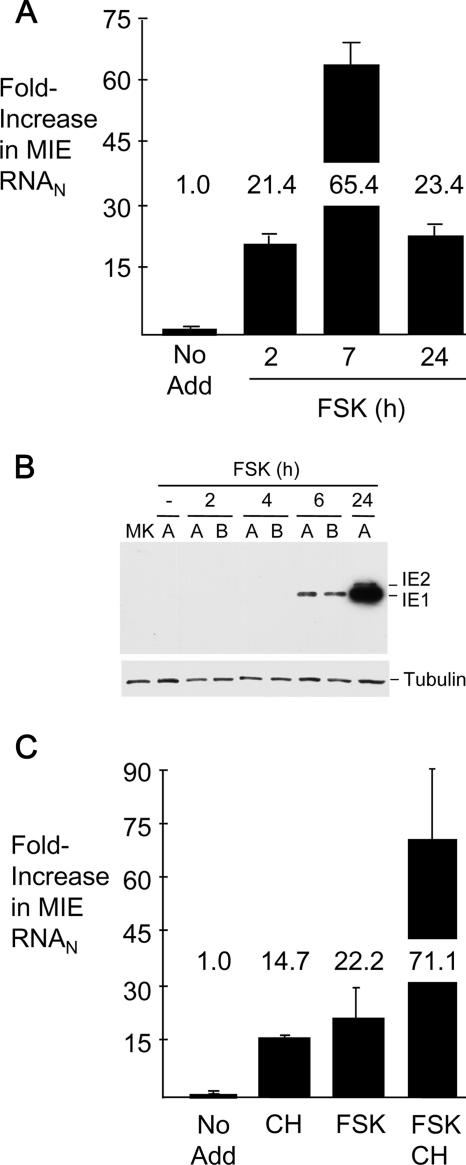
FSK stimulation is known to activate the cAMP/CRE signaling pathway to quickly exert an effect on genes directly regulated by this pathway (49). We investigated whether FSK functions in this manner when activating MIE gene expression in quiescently infected NT2. The findings shown in Fig. 4A reveal that FSK increases the MIE RNA amount by 21-fold as early as 2 h after the beginning of the treatment. This prompt escalation in expression parallels the immediate-early activation kinetics of MIE gene expression in productively infected cells (58). The increase in MIE RNA abundance is greatest at 7 h posttreatment (65.4-fold) and wanes modestly at 24 h (23.4-fold), possibly in response to negative autoregulation provided by the viral IE2 p86 protein. The IE1 p72 protein becomes detectable by the Western blot method at 6 h after FSK treatment, and the abundance of both the IE1 p72 and IE2 p86 proteins increases substantially by 24 h (Fig. 4B).
FIG. 4.
FSK's action is immediate and does not require de novo protein synthesis. Time course for FSK-induced expression of MIE RNA (A) and protein (B). FSK (10 μM) was added to or omitted from (No Add) quiescently infected NT2 at 24 h p.i. RNA and protein were isolated at the designated times thereafter. MIE RNA was quantified by real-time RT-PCR, using the standard-curve method and normalization to the 18S RNA amount (RNAN). The IE1 p71 and IE2 p86 proteins were analyzed by the immunoblotting method specified in the legend of Fig. 3C. MK, mock-infected NT2. (C) FSK-induced MIE RNA expression in the absence of protein synthesis. FSK (10 μM), CH (200 μg/ml), or both FSK and CH were added to or omitted from (No Add) quiescently infected NT2 at 24 h p.i., except that CH was added 30 min prior to the addition of FSK. MIE RNA was quantified at 4 h after FSK addition, using methods described above for panel A. Error bars represent standard errors of the means from two experiments.
Because FSK can enhance the production of c-Fos (49) (and possibly other cellular proteins) with the potential for activating the MIE enhancer/promoter, we examined whether blocking protein synthesis for 4 h abrogates FSK's effect on MIE gene expression. Several experiments consistently showed that the cessation of de novo protein synthesis, by itself, increases MIE RNA abundance, as exemplified in Fig. 4C. However, the combination of protein synthesis inhibitor and FSK acts synergistically to increase MIE RNA production (71-fold), exceeding the sum of the individual effects of protein synthesis inhibitor (12-fold) and FSK (22-fold). GFP RNA expression from the upstream UL127 promoter in HCMV-GFP1 was not increased appreciably by cycloheximide (CH) and FSK treatment, as assessed by quantitative RT-PCR (data not shown). This experimental approach was not suitable for assessing FSK's effect beyond the immediate time frame because of overwhelming toxicity produced by the prolonged inhibition of protein synthesis (data not shown).
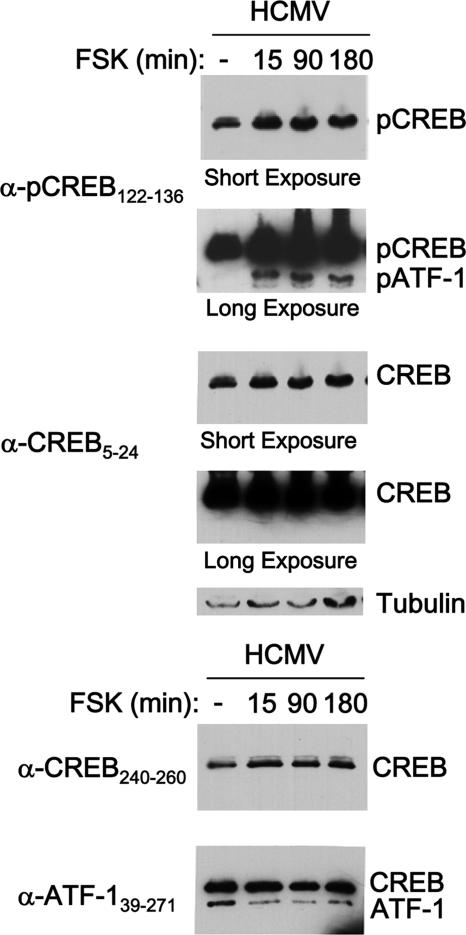
FSK stimulation is known to activate CRE-dependent transcription through the signaling-mediated phosphorylation of Ser133 and Ser63 in CREB and ATF-1, respectively (37, 49). The phosphorylation at these locations is either transient or sustained during FSK exposure, depending on cell type (37, 49). We addressed the question of whether FSK stimulation modulates the phosphorylation and total concentration of CREB and ATF-1 in quiescently infected NT2. Because the amino acid residues surrounding Ser133 of CREB (43 kDa) and Ser63 of ATF-1 (38 kDa) are nearly identical, the same phospho-specific antibody is used in Western blot assays to monitor the phosphorylation of both proteins (63). As shown in Fig. 5, the level of pCREB is substantial at baseline in unstimulated cells. FSK increases the baseline pCREB amount by 2.0- to 2.2-fold. The basal level of phosphorylation at Ser63 of ATF-1 (pATF-1) in unstimulated cells is below the limit of detection. FSK dramatically elevates the pATF-1 level as early as 15 min, and this change is sustained throughout the 180 min of examination. The FSK-induced pattern of change in pCREB and pATF-1 levels cannot be ascribed to changes in total amounts of CREB and ATF-1, as assessed with a panel of antibodies that selectively target various segments of these proteins (Fig. 5 and data not shown).
FIG. 5.
FSK increases the phosphorylation of ATF-1 Ser63 and CREB Ser133. FSK (10 μM) was added to or omitted from quiescently infected NT2 at 24 h p.i. (partially purified CMV-GFP1 at an MOI of 3). Cells were lysed at 15, 90, and 180 min after FSK addition. Whole-cell extracts were fractionated by SDS-PAGE and transferred to a membrane for immunoblotting. Proteins were visualized using the primary antibodies against the indicated regions of CREB or ATF-1, secondary antibodies conjugated to horseradish peroxidase, and a chemiluminescence detection system. The anti-pCREB122-136 (α-pCREB122-136) cross-reacts with phospho-Ser63 in corresponding residues of ATF-1. The blot was reprobed with anti-CREB5-24 (α-CREB5-24) and again reprobed for β-tubulin. Autoradiographs were subjected to long or short exposures. Densitometry analysis revealed that 15 min of FSK increased the pCREB abundance by 2.0- or 2.2-fold after normalization to total CREB or tubulin levels, respectively. The abundance of pATF-1 in unstimulated cells was too low to permit a determination of the relative increase of pATF-1 by FSK treatment. Another blot of the same protein extracts was probed with anti-ATF-139-271, which cross-reacts with CREB, and reprobed with anti-CREB240-260.
Thus, FSK's action is immediate, does not absolutely require de novo protein synthesis, and correlates with increased levels of phosphorylation of CREB Ser133 and ATF-1 Ser63. This constellation of findings is consistent with the view that FSK likely acts directly on the MIE enhancer/promoter through the CRE signaling pathway.
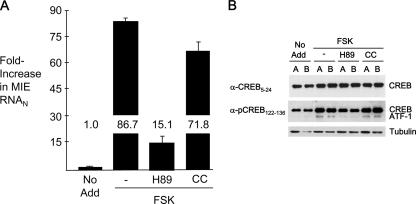
Inhibition of PKA, but not PKC, attenuates FSK-induced MIE gene activation and ATF-1 phosphorylation.
In transient-transfection assays, the stimulation of CRE-dependent MIE enhancer/promoter activation involves the role of either protein kinase A (PKA) (44) or protein kinase C (PKC) (17, 57), depending on cell type. Notably, CMV MIE gene expression is inhibited in lytically infected cells by blocking PKC activity but not by blocking PKA (25, 40).
The roles of PKA and PKC in mediating FSK-induced MIE gene activation in NT2 were explored through a method of selective inhibition of these enzymes. H-89, an inhibitor of PKA, and calphostin C (CC), an inhibitor of classical and novel PKC isoforms, were applied at concentrations used previously for other infected cell types (25). As shown in Fig. 6A, H-89 lowers FSK-induced MIE RNA production by 73%, whereas CC is unsuccessful in blocking FSK's effect. These results accord with those showing that H-89, but not CC, substantially attenuates FSK's ability to elevate levels of pATF-1 (Fig. 6B). In additional studies, the PKA inhibitor PKI (14-24) amide, but not the PKC inhibitor stauroporine, likewise decreased FSK-induced MIE gene activation and ATF-1 Ser63 phosphorylation (data not shown).
FIG. 6.
The inhibition of PKA, but not PKC, lowers FSK-induced MIE transcription and levels of pCREB and pATF-1. At 24 h p.i. of NT2, FSK (10 μM) was added to the growth medium with or without H-89 (10 μM), an inhibitor of PKA, or 1 μM CC, an inhibitor of classical and novel PKC isoforms. Inhibitors were added 30 min prior to the addition of FSK, as described in Materials and Methods. No Add, FSK was not added. (A) MIE RNA was isolated 4 h later and quantified by real-time RT-PCR, using a standard-curve method and normalization to the 18S RNA amount (RNAN). The basal level of MIE RNA expression was not significantly altered by H89 treatment (data not shown). (B) Whole-cell extracts from duplicate samples (A and B) of each experimental group were prepared 30 min after FSK addition, fractionated by SDS-PAGE, and transferred to a membrane for immunoblotting. Proteins were visualized using the indicated primary antibodies, secondary antibodies conjugated to horseradish peroxidase, and a chemiluminescence detection system. The primary antibody anti-pCREB122-136 cross-reacts with phospho-Ser63 in ATF-1. The blot was reprobed with anti-CREB5-24 and again reprobed for β-tubulin. Densitometry analysis revealed that FSK increased pCREB abundance by 2.1-fold in the infected cells after normalization to total CREB.
We surmise that PKA activity is likely involved in mediating FSK's effect on MIE enhancer/promoter activation but that PKC does not play this role in quiescently infected NT2.
FSK and TSA synergistically activate the HCMV lytic cycle.
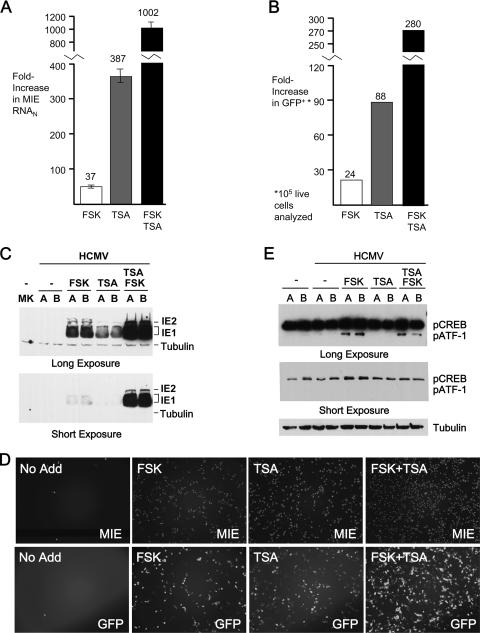
Our findings so far suggest that MIE gene activation likely involves additional regulatory mechanisms that are unaffected by FSK. We further tested this notion by combining FSK (10 μM) with TSA at 100 ng/ml, a concentration reported previously to strongly activate the MIE enhancer/promoter in quiescently infected NT2 (32). The findings in Fig. 7A reveal that the two agents interact synergistically to greatly increase MIE RNA production (1,002-fold), compared to RNA production after treatment with either agent alone (FSK, 37-fold; TSA, 387-fold). Synergy is also reflected in the frequency that NT2 allow GFP expression from the upstream UL127 promoter (Fig. 7B).
FIG. 7.
FSK and TSA synergistically activate the MIE genes. (A) At 24 h p.i., FSK (15 μM), TSA (100 ng/ml), or both FSK (15 μM) and TSA (100 μg/ml) were added to or omitted from (No Add) the growth medium of NT2 infected with purified CMV-GFP1 (MOI of 3) and then removed at 48 h p.i. HCMV MIE RNA was quantified by real-time RT-PCR at 48 h p.i. after normalization to the cellular 18S RNA abundance (RNAN). Means and standard deviations depict changes (n-fold) in MIE RNAN levels relative to the level in infected NT2 receiving No Add. (B) Increases (n-fold) in the proportions of NT2 expressing GFP from the viral UL127 promoter were determined at 72 h p.i. by FACS analysis of 10,000 live cells per experimental group. Results are depicted relative to the result for No Add, with 0.04% GFP+ NT2 present in the No Add group. (C) FSK (15 μM), TSA (10 ng/ml), or both FSK (15 μM) and TSA (10 ng/ml) were added to or omitted from (No Add) the growth medium of NT2 at 24 h p.i. At 48 h p.i., whole-cell extracts were fractionated by SDS-PAGE and subjected to immunoblotting. MIE (IE1 p71 and IE2 p86) proteins were visualized with antibody MA810 and a chemiluminescence detection system. MK, mock-infected cells. (D) Fluorescence microscopy was performed at 72 h p.i. for the experimental conditions specified for panel C. MIE proteins were visualized by indirect immunofluorescence, using MA810 and secondary antibody conjugated with Alexa Fluor 555. The MIE+ cell/GFP+ cell ratios were 5.9, 2.5, 2.4, and 3.0 for NT2 with No Add, FSK, TSA, and both FSK and TSA, respectively. (E) Quiescently infected NT2 at 24 h p.i. were treated with FSK (10 μM), TSA (100 ng/ml), or both FSK (10 μM) and TSA (100 ng/ml) for 15 min. Whole-cell extracts from experimental groups in duplicate (A and B samples) were fractionated by SDS-PAGE and subjected to immunoblotting. Proteins were visualized using the primary antibody anti-pCREB122-136 and a chemiluminescence detection system. Blots were reprobed for β-tubulin. Densitometry analysis revealed that FSK increased pCREB abundance by 1.8-fold after normalization to tubulin levels. The abundance of pATF-1 in unstimulated cells was too low to permit a determination of the relative increase of pATF-1 by FSK treatment.
The magnitude of activation of MIE gene expression is dependent on concentrations of the individual agents. As shown in Fig. 7C, the synergy for the activated expression of the IE1 p71 and IE p86 proteins is maintained when the TSA concentration is lowered to 10 ng/ml and the FSK concentration is kept constant. The extent of expansion of the NT2 population expressing MIE proteins in response to the individual or combined agents exceeds that of the GFP+ NT2 population (Fig. 7D).
TSA alone does not increase pCREB or pATF-1 levels, and the combination of FSK and TSA does not elevate pCREB or pATF-1 levels above those produced by FSK alone (Fig. 7E). None of the treatments alter levels of total CREB and ATF-1 (data not shown).
Hence, the FSK and TSA combination synergistically activates the expression of MIE RNA and protein in quiescently infected NT2. The synergy is accompanied by levels of phosphorylation of CREB Ser133 and ATF-1 Ser63 that are equivalent to those induced by FSK alone.
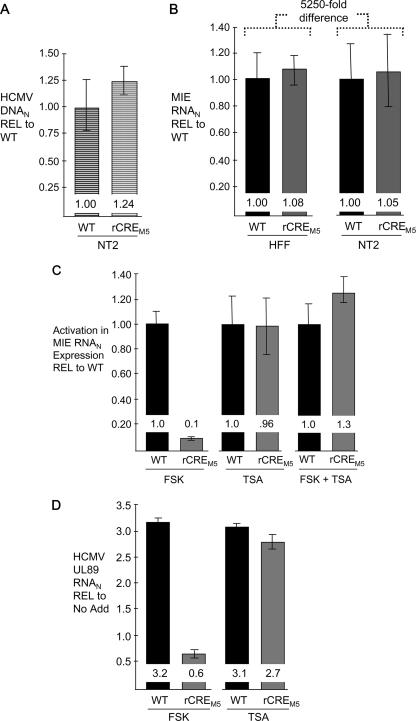
The CRE mediate FSK-induced MIE enhancer/promoter activation.
The CRE's role in FSK- and TSA-induced MIE RNA expression was assessed by a comparison of WT CRE and rCREM5. rCREM5 differs from its parental WT HCMV only in having base substitutions in each of the five CRE in the enhancer to thereby preclude CREB/ATF-mediated transcriptional activation (22, 61). These comparisons required proof that the infectivities and genomic inputs for the two viruses are equivalent. Three different types of controls performed in parallel with the treatment studies demonstrate equivalence in viral DNA uptake in unstimulated NT2 (Fig. 8A), basal levels of MIE RNA in unstimulated NT2 (Fig. 8B), and lytic-cycle MIE RNA production in HFF (Fig. 8B). MIE RNA abundance is vastly greater in acutely infected HFF than in quiescently infected NT2 (>5,250-fold difference) (Fig. 8B), as reported previously (32). The relative difference in levels of activation (n-fold) of MIE RNA expression by FSK, TSA, or both for the WT and rCREM5 is shown in Fig. 8C. Removal of the CRE repetition profoundly weakens FSK's ability to activate MIE gene transcription, lowering its activation ability to approximately 1/10 that for the WT. In contrast, TSA-induced activation is unaffected by the mutations. The CRE repetition is also not required for the synergistic response induced by the combination of FSK and TSA. The impairment in FSK-induced MIE gene activation caused by dismantling the CRE repetition is accompanied by a downstream defect in activation of the viral DNA terminase (UL89) gene, which is normally expressed in the early/late phase of lytic-virus replication (Fig. 8D).
FIG. 8.
The CRE repetition mediates FSK-induced MIE RNA expression. All studies comparing the WT and rCREM5 were performed in parallel, using an MOI of 3. MIE RNA and DNA were quantified by real-time PCR and normalized to the cellular 18S RNA abundance (RNAN). Results are depicted relative to the result with the WT (REL to WT) and reflect means and standard errors from two replicate independent experiments. (A) Relative levels of HCMV MIE DNA in the nuclei of quiescently infected NT2 at 24 h p.i. (B) Relative levels of MIE RNA expression in unstimulated, productively infected HFF and quiescently infected NT2 at 6 and 48 h p.i., respectively. (C) Relative levels of activation of MIE RNA expression in response to stimulation. At 24 h p.i., FSK (15 μM), TSA (100 ng/ml), or both FSK (15 μM) and TSA (100 ng/ml) were added to the growth medium of quiescently infected NT2. MIE RNA levels were quantified 24 h thereafter. The change in MIE RNA production in comparison to the level of MIE RNA in unstimulated quiescently infected NT2 (A) was calculated to determine the activation level. The activation level with the WT is set as 1. (D) Using RNA samples described above for panel C, levels of induced WT and rCREM5 early gene UL89 RNA relative to (REL) the level with no additive (No Add) were determined by real-time RT-PCR at 48 h p.i. after normalization to the abundance of cellular 18S RNA.
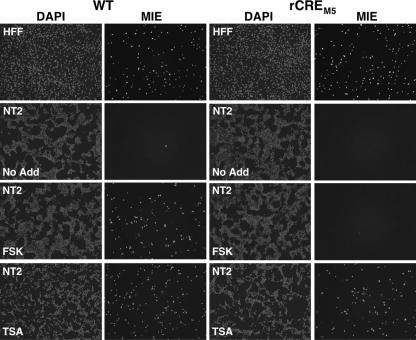
The CRE's role in activating MIE protein expression was assessed in cells in situ by indirect immunofluorescence microscopy (Fig. 9). The WT and rCREM5 are equivalent in their frequencies of MIE+ cells observed in control HFF infections at an MOI of 0.05. In unstimulated NT2 at an MOI of 1.0, the WT and rCREM5 rarely produce MIE+ cells at 48 h p.i. FSK stimulation of WT-infected NT2 greatly increases the size of the MIE+ cell population, corroborating findings produced by the HCMV-GFP1 construct. In striking contrast, FSK stimulation is poorly effective in expanding the MIE+ population of rCREM5-infected NT2. TSA stimulation greatly increases the frequency of MIE+ NT2 for both WT and rCREM5 infections.
FIG. 9.
The CRE repetition mediates FSK-induced MIE protein expression. HFF and NT2 were infected in parallel with the WT and rCREM5 at MOIs of 0.05 and 1.0, respectively. At 24 h p.i. of NT2, FSK (15 μM) or TSA (100 ng/ml) was added to or omitted from (No Add) the growth medium lacking knockout serum replacement. MIE proteins were visualized by indirect immunofluorescence assay, using MA810 and secondary antibody conjugated with Alexa Fluor 555 at 24 and 48 h p.i. of HFF and NT2, respectively. Nuclei were counterstained with DAPI. Images of DAPI and MIE fluorescence for each group are captured from the same cells in the field of magnification. Magnification, ×10.
These findings indicate that FSK activates the enhancer through a CRE-dependent signaling pathway to overcome MIE enhancer/promoter silencing. TSA appears to act mainly through a CRE-independent mechanism.
DISCUSSION
Quiescently infected NT2 are a model with which to investigate the molecular facets underlying HCMV MIE enhancer/promoter reactivation. In this report, we show that MIE enhancer/promoter silencing is alleviated by the stimulation of cAMP/PKA signaling and CRE-dependent transcriptional activation. The implication is that the induction of this regulatory cascade might also contribute to HCMV MIE enhancer/promoter reactivation from latent or smoldering infection in vivo, when MIE gene expression is extremely restricted. Such a scenario would also explain why stimulation of the CRE repetition contributes minimally to MIE enhancer/promoter activity in productively infected HFF and NT2-derived neurons (22). It has been reported that latently infected persons suffering acute myocardial infarction are prone to express HCMV MIE antigen in association with elevated serum levels of catecholamines (norepinephrine and epinephrine), which can stimulate CRE-dependent transcription from transfected MIE enhancer/promoter segments via the cAMP/PKA signaling pathway (44). In the NT2 model, catecholamine exposure does not reverse MIE enhancer/promoter silencing (data not shown), possibly because undifferentiated NT2 do not produce enough functional adrenergic receptors (48).
Several lines of evidence indicate that FSK's effect on MIE gene expression is implausibly the consequence of induction of cellular differentiation. First, the FSK effect is rapid, boosting MIE RNA expression within 2 h after receiving the treatment (Fig. 4A). Second, FSK does not lower the cellular Oct4 level (Fig. 3C), an early step in the direction of NT2 differentiation (30). Third, exposing NT2 to FSK prior to infection and not during the infection fails to activate MIE gene transcription (data not shown). Fourth, Keller et al. (22) have reported that FSK increases CRE-dependent MIE RNA expression by only 2.1- to 2.8-fold in fully differentiated NT2, suggesting that cellular differentiation diminishes the contribution of this activation mechanism. Last, HCMV infection of cultured primary neuronal precursors has been recently shown to inhibit cellular differentiation (41). Notwithstanding these findings, it remains to be determined with the NT2 model whether the induction of cellular differentiation can lift other HCMV lytic-cycle constraints following FSK or TSA stimulation.
The enhancer CRE sequence, 5′-TGACGTCA-3′, is a classical consensus binding site for CREB family members, which include CREB, ATF-1, CREM, and their respective isoforms (31, 49). CREB, ATF-1, and CREM bind to the CRE as either homodimers or heterodimers (31, 49). The protein's transcriptional activation potential is governed by its kinase-inducible domain, which is modulated in function through the phosphorylation of specific serine residues by corresponding kinases (31, 49). By activating adenylyl cyclase, FSK quickly induces cAMP-dependent PKA-mediated phosphorylation of the kinase-inducible domain (e.g., at Ser133 in CREB and Ser63 in ATF-1) to recruit CBP or p300 for leveraging transcriptional activation (49). While CREB and ATF-1 are ubiquitously expressed (49) and bind to the enhancer CRE in vitro (44, 47, 56, 57), whether they or other factors actually bind to this cis-acting element in viral genomes in infected cells is unknown. In the NT2 model, FSK stimulation produces a temporal pattern of ATF-1 Ser63 phosphorylation mirroring that of MIE RNA expression, and these two outcomes are concordantly regulated by PKA (Fig. 4A, 5, and 6). The relationship between MIE RNA expression and CREB Ser133 phosphorylation is less striking, as pCREB is abundant in whole-cell extracts of unstimulated NT2 and increases modestly in response to FSK stimulation (Fig. 5, 6, and 7E). Levels of total CREB and ATF-1 are not appreciably altered by FSK, TSA, or infection. CREM is not detected under these experimental conditions (data not shown). Whether ATF-1 and CREB are the regulatory linchpins in bridging signal transduction with CRE activation in the infected NT2 remains to be determined.
The activation of viral early/late gene expression in lytically infected cells is dependent on the expression of the MIE gene products (reviewed in reference 36). While the primary objective of this work was to elucidate the CRE's role in activating MIE gene expression, the effect of this activation on the advancement of the viral lytic cycle was also explored. We monitored GFP production from the UL127 and adenovirus E1b promoters in HCMV-GFP1 and HCMV-GFP2, respectively, because both promoters are located upstream of the MIE enhancer boundary elements and consequently exhibit early/late transcriptional kinetics in lytically infected HFF (28, 29, 32). We opted to show the results for HCMV-GFP1 in this report. The results of TSA stimulation for HCMV-GFP2 were reported previously (32). HCMV-GFP1 and HCMV-GFP2 behave similarly in NT2 with regard to FSK- and TSA-induced GFP expression (data not shown). The findings linking CRE-dependent MIE gene expression to subsequent activation of the viral early/late DNA terminase (UL89) gene of WT HCMV (Fig. 8D) accord with the surrogate GFP results of HCMV-GFP1 and HCMV-GFP2. The possibility of the CRE in the MIE enhancer modulating GFP expression from the UL127 and E1b promoters at early/late times after FSK stimulation cannot be dismissed. CRE are located in a number of HCMV early/late promoters and function in augmenting transcription (10). Three findings suggest that the UL127 promoter of HCMV-GFP1 in NT2 does not function independently of MIE gene expression: (i) the ratio of MIE+ to GFP+ cells always exceeds 2 regardless of treatment condition (Fig. 2C and 5D), (ii) GFP fluorescence was observed only in NT2 emitting MIE protein-specific fluorescence (data not shown), and (iii) CH blocked FSK-induced GFP RNA expression from the UL127 promoter of HCMV-GFP1 (data not shown).
Only a small fraction of FSK-stimulated GFP+ NT2 release infectious viral progeny (Fig. 2D). The reason for this apparent shortfall is unknown, but additional levels of blockade to HCMV replication likely play a role. A subset of transcriptionally active viral genomes reverts to quiescence after the discontinuation of FSK stimulation (data not shown), a result also observed after TSA was stopped following TSA-induced viral-gene activation (32). Removal of FSK at 24 h of treatment subsequently lessens the level of MIE RNA expression (data not shown), reflecting a need for continual stimulation to optimally sustain MIE gene transcription.
IFN-γ stimulation fails to activate the expression of MIE RNA and protein in quiescently infected NT2, despite the up-regulation of IFN-stimulated genes (Fig. 2C and 3). IFN-γ adds little or nothing to FSK-induced MIE gene expression in this system (Fig. 2C and 3A and C). It is unclear why the enhancer's two GAS-like elements do not respond to IFN-γ signaling to boost transcription, when they reportedly do so in productively infected HFF (40). These sites could serve primarily to support a mechanism of action not involving IFN-γ signaling transduction, as was suggested previously (65). Findings from the NT2 model do not discount the possibility of IFN-γ having a role in MIE enhancer/promoter reactivation in cells of myeloid lineage.
Two separate findings emerging from this work support the idea that the reversal of MIE enhancer/promoter silencing is optimally accomplished through the cooperation of multiple regulatory mechanisms. First, the TSA effect and its amplification by FSK are not dependent on the enhancer CRE (Fig. 8 and 9). TSA was shown previously to disrupt the chromatin-mediated silencing of the MIE enhancer/promoter by increasing histone acetylation (38), although its effects on both nonhistone substrates and the expression of specific cellular genes may also contribute to the reversal of transcriptional silencing. Second, halting the synthesis of new proteins elevates the MIE RNA level (Fig. 4C) in a manner not accompanied by an increase in the level of phosphorylation of ATF-1 Ser63 or CREB Ser133 (data not shown). Coupling the protein synthesis inhibition with FSK works to synergistically increase MIE RNA expression (Fig. 4C) but does not increase ATF-1 Ser63 or CREB Ser133 phosphorylation above levels achieved with FSK alone (data not shown). Why there is need for the combined actions of multiple regulatory mechanisms to overcome MIE enhancer/promoter silencing in some NT2, but not others, is unknown. Heterogeneity in the NT2 population is likely to partly explain this result. Nevertheless, the heterogeneity of the cell populations in vivo is also the reality. It is noteworthy that perceptible HCMV reactivaton events in vivo are infrequent unless amplified by an immunocompromising or inflammatory condition. Teleologically, it might be in the virus's best interest to have multiple requirements to achieve reactivation, so that a lone stimulus cannot inappropriately flush out all dormant viral offspring to risk total annihilation.
This report is the first to show that stimulus-induced cAMP/PKA signaling has the capacity to activate HCMV MIE gene expression in a CRE-dependent manner in quiescently infected cells. Future studies will be needed to determine whether this mechanism applies to HCMV reactivation in vivo and whether neuronal precursors are a cellular site for HCMV latency. Work to identify naturally occurring stimuli that trigger this signaling cascade is under way, though early findings suggest that vasoactive intestinal peptide, an immunosuppressive neuropeptide, is one such stimulus.
Acknowledgments
We are grateful to Mark F. Stinski for critical reading of the manuscript. We thank Xiaoqui Lui for her help with this project. We thank members of the Stinski and Meier laboratories for helpful discussions of this work.
This work was supported by the American Heart Association (Grant-in-Aid Award to J.L.M.), the National Institutes of Health (grant T32 AI007485 to J.I.A.), and the Department of Veterans Affairs (Merit Award to J.L.M.).
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Andrews, P. W. 1984. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev. Biol. 103:285-293. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, P. W., G. Trinchieri, B. Pertussia, and C. Baglioni. 1987. Induction of class I histocompatibility complex antigens in human teratocarcinoma cells by interferon without induction of differentiation, growth inhibition, or resistance to viral infection. Cancer Res. 47:740-746. [PubMed] [Google Scholar]
- 3.Angulo, A., C. Suto, M. F. Boehm, R. A. Heyman, and P. Ghazal. 1995. Retinoid activation of retinoic acid receptors but not of retinoid X receptors promotes cellular differentiation and replication of human cytomegalovirus in embryonal cells. J. Virol. 69:3831-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angulo, A., C. Suto, R. A. Heymen, and P. Ghazal. 1996. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol. Endocrinol. 10:781-793. [DOI] [PubMed] [Google Scholar]
- 5.Arribas, J. R., G. A. Storch, D. B. Clifford, and A. C. Tselis. 1996. Cytomegalovirus encephalitis. Ann. Intern. Med. 125:577-587. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, W. M. W., W. Y. Fu, W. S. Hui, and N. Y. Ip. 1999. Production of human CNS neurons from embryonal carcinoma cells using a cell aggregation method. BioTechniques 26:946-954. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocynate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Cook, C. H., J. Trgovcich, P. D. Zimmerman, Y. Zhang, and D. D. Sedmak. 2006. Lipopolysaccharide, tumor necrosis factor alpha, or interleukin-1b triggers reactivation of latent cytomegalovirus in immunocompetent mice. J. Virol. 80:9151-9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, H., J. Chartier, C. Sodja, A. Desbois, M. Ribecco-Lutkiewicz, R. P. Walker, and M. Sikorska. 2003. Transcriptional activation of the human brain-derived neurotrophic factor gene promoter III by dopamine signaling in NT2/N neurons. J. Biol. Chem. 278:26401-26409. [DOI] [PubMed] [Google Scholar]
- 10.Fortunato, E. A., A. K. McElroy, V. Sanchez, and D. H. Spector. 2000. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Mircobiol. 8:111-119. [DOI] [PubMed] [Google Scholar]
- 11.Ghazal, P., C. DeMattei, E. Giulietti, S. A. Kliewer, K. Umesono, and R. M. Evans. 1992. Retinoic acid receptors initiate induction of the cytomegalovirus enhancer in embryonal cells. Proc. Natl. Acad. Sci. USA 89:7630-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonczol, E., P. W. Andrews, and S. A. Plotkin. 1985. Cytomegalovirus infection of human teratocarcinoma cells in culture. J. Gen. Virol. 66:509-515. [DOI] [PubMed] [Google Scholar]
- 13.Gonczol, E., P. W. Andrews, and S. A. Plotkin. 1984. Cytomegalovirus replicates in differentiated but not in undifferentiated human embryonal carcinoma cells. Science 224:159-161. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 95:3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hummel, M., and M. Abecassis. 2002. A model for reactivation of CMV from latency. J. Clin. Virol. 25:S123-S136. [DOI] [PubMed] [Google Scholar]
- 16.Hummel, M., Z. Zhang, S. Yan, I. Deplaen, P. Golia, T. Varghese, G. Thomas, and M. I. Abecassis. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunninghake, G. W., M. M. Monick, B. Liu, and M. F. Stinski. 1989. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP-response elements. J. Virol. 63:3026-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibanez, C. E., R. Schrier, P. Ghazal, C. Wiley, and J. A. Nelson. 1991. Human cytomegalovirus productively infects primary differentiated macrophages. J. Virol. 65:6581-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isomura, H., and M. F. Stinski. 2003. Effect of substitution of the human cytomegalovirus enhancer or promoter on replication in human fibroblasts. J. Virol. 77:3602-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isomura, H., M. F. Stinski, A. Kudoh, T. Daikoku, N. Shirata, and T. Tsurumi. 2005. Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J. Virol. 79:9597-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones-Villeneuve, E. M. V., M. A. Rudnicki, J. F. Harris, and M. W. McBurney. 1983. Retinoic acid-induced neural differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 3:2271-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller, M. J., D. G. Wheeler, E. Cooper, and J. L. Meier. 2003. Role of the human cytomegalovirus major immediate-early promoter's 19-base-pair-repeat cyclic AMP-response element in acutely infected cells. J. Virol. 77:6666-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koffron, A. J., M. Hummel, B. K. Patterson, S. Yan, D. Kaufman, J. P. Fryer, F. P. Stuart, and M. Abecassis. 1998. Cellular localization of latent murine cytomegalovirus. J. Virol. 72:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, K. J. X., J. Xu, and E. S. Mocarski. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. USA 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kucic, N., H. Mahmutefendic, and P. Lucin. 2005. Inhibition of protein kinases C prevents murine cytomegalovirus replication. J. Gen. Virol. 86:2153-2161. [DOI] [PubMed] [Google Scholar]
- 26.Kurz, S. K., and M. J. Reddehase. 1999. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 73:8612-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaFemina, R., and G. S. Hayward. 1986. Constitutive and retinoic acid-inducible expression of cytomegalovirus immediate-early genes in human teratocarcinoma cells. J. Virol. 58:434-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lashmit, P. E., C. A. Lundquist, J. L. Meier, and M. F. Stinski. 2004. Cellular repressor inhibits human cytomegalovirus transcription from the UL127 promoter. J. Virol. 78:5113-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundquist, C. A., J. L. Meier, and M. F. Stinski. 1999. A strong transcriptional negative regulatory region between the human cytomegalovirus UL127 gene and the major immediate early enhancer. J. Virol. 73:9032-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matin, M. M., J. R. Walsh, P. J. Gokhale, J. S. Draper, A. R. Bahrami, I. Morton, H. D. Moore, and P. W. Andrews. 2004. Specific knockdown of Oct4 and B2-microglobulin expression by RNA interefence in human embryonic stem cells. Stem Cells 22:659-668. [DOI] [PubMed] [Google Scholar]
- 31.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 32.Meier, J. L. 2001. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol. 75:1581-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier, J. L., M. J. Keller, and J. J. McCoy. 2002. Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for activation of viral gene expression and replication. J. Virol. 76:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier, J. L., and J. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier, J. L., and M. F. Stinski. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J. Virol. 71:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meier, J. L., and M. F. Stinski. 2006. Major immediate-early enhancer and its gene products, p. 151-166. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Norfolk, United Kingdom.
- 37.Montminy, M. 1997. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66:807-822. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, J., W. Fischle, E. Verdin, and J. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson, J. A., and M. Groudine. 1986. Transcriptional regulation of the human cytomegalovirus major immediate-early gene is associated with induction of DNase I-hypersensitive sites. Mol. Cell. Biol. 6:452-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Netterwald, J., S. Yang, W. Wang, S. Ghanny, M. Cody, P. Soteropoulas, B. Tian, W. Dunn, F. Liu, and H. Zhu. 2005. Two gamma interferon-activated site-like elements in the human cytomegalovirus major immediate-early promoter/enhancer are important for viral replication. J. Virol. 79:5035-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odeberg, J., N. Wolmer, S. Falci, M. Westgren, A. Seiger, and C. Soderberg-Naucler. 2006. Human cytomegalovirus inhibits neuronal differentiation and induces apoptosis in human neural precursor cells. J. Virol. 80:8929-8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pleasure, S. J., and V. M.-Y. Lee. 1993. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J. Neurosci. 35:585-602. [DOI] [PubMed] [Google Scholar]
- 43.Poland, S. D., L. L. Bambrick, G. A. Dekaban, and G. P. A. Rice. 1994. The extent of human cytomegalovirus replication in primary neurons is dependent on host cell differentiation. J. Infect. Dis. 170:1267-1271. [DOI] [PubMed] [Google Scholar]
- 44.Prosch, S., C. E. C. Wendt, P. Reinke, C. Priemer, M. Oppert, D. H. Kruger, H. D. Volk, and W. D. Docke. 2000. A novel link between stress and human cytomegalovirus (HCMV) infection: sympathetic hyperactivity stimulates HCMV activation. Virology 272:357-365. [DOI] [PubMed] [Google Scholar]
- 45.Reeves, M. B., P. J. Lehner, J. G. P. Sissons, and J. H. Sinclair. 2005. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodeling. J. Gen. Virol. 86:2949-2954. [DOI] [PubMed] [Google Scholar]
- 46.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. P. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rideg, K., G. Hiraka, K. Prakash, L. M. Busher, J. Y. Nothias, R. Weinmann, P. W. Andrews, and E. Gonczol. 1994. DNA-binding proteins that interact with the 19-base pair (CRE-like) element from the HCMV major immediate-early promoter in differentiating human embryonal carcinoma cells. Differentiation 56:119-129. [DOI] [PubMed] [Google Scholar]
- 48.Satoh, J., and Y. Kuroda. 2000. Differential gene expression between human neurons and neuronal progenitor cells in culture: an analysis of arrayed cDNA clones in NTera2 human embryonal carcinoma cell line as a model system. J. Neurosci. Methods 94:155-164. [DOI] [PubMed] [Google Scholar]
- 49.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 50.Shelbourn, S. L., S. K. Kothari, J. G. P. Sissons, and J. H. Sinclair. 1989. Repression of human cytomegalovirus gene expression associated with a novel immediate early regulatory region binding factor. Nucleic Acids Res. 17:9165-9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon, C. O., C. K. Seckert, D. Dreis, M. J. Reddehase, and N. K. A. Grzimek. 2005. Role of tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs. J. Virol. 79:326-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1998. Growth of human cytomegalovirus in primary macrophages. Methods 16:126-138. [DOI] [PubMed] [Google Scholar]
- 53.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Interferon-gamma and tumor necrosis factor-alpha specifically induce formation of cytomegalovirus-permissive monocyte-derived macrophages that are refractory to antiviral activity of these cytokines. J. Clin. Investig. 100:3154-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soderberg-Naucler, C., D. N. Streblow, K. N. Fish, J. Allan-Yorke, P. P. Smith, and J. A. Nelson. 2001. Reactivation of latent human cytomegalovirus in CD14+ monocytes is differentiation dependent. J. Virol. 75:7543-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soderberg-Naucler, N. C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogenic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 56.Staak, K., S. Prosch, J. Stein, C. Priemer, R. Ewert, W. D. Docke, D. H. Kruger, H. D. Volk, and P. Reinke. 1997. Pentoxifylline promotes replication of human cytomegalovirus in vivo and in vitro. Blood 89:3682-3690. [PubMed] [Google Scholar]
- 57.Stamminger, T., H. Fickenscher, and B. Fleckenstein. 1990. Cell type-specific induction of the major immediate-early enhancer of human cytomegalovirus by cAMP. J. Gen. Virol. 71:105-113. [DOI] [PubMed] [Google Scholar]
- 58.Stinski, M. F., D. R. Thomsen, R. M. Stenberg, and L. C. Goldstein. 1983. Organization and expression of the immediate early genes of human cytomegalovirus. J. Virol. 46:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor-Wiedeman, J. A., J. G. P. Sissons, and J. H. Sinclair. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 68:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsutsui, Y., H. Kawasaki, and I. Kosugi. 2002. Reactivation of latent cytomegalovirus infection in mouse brain cells detected after transfer to brain slice cultures. J. Virol. 76:7247-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheeler, D. G., and E. Cooper. 2001. Depolarization strongly induces human cytomegalovirus major immediate-early promoter/enhancer activity in neurons. J. Biol. Chem. 276:31978-31985. [DOI] [PubMed] [Google Scholar]
- 62.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiggin, G. R., A. Soloaga, J. M. Foster, V. Murray-Trait, P. Cohen, and J. S. C. Arthur. 2002. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 22:2871-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiley, C. A., R. D. Schrier, F. J. Denaro, J. A. Nelson, P. W. Lampert, and M. B. Oldstone. 1986. Localization of cytomegalovirus proteins and genome during fulminant central nervous system infection in an AIDS patient. J. Neuropathol. Exp. Neurol. 45:127-139. [DOI] [PubMed] [Google Scholar]
- 65.Yang, S., J. Netterwald, W. Wang, and H. Zhu. 2005. Characterization of the elements and proteins responsible for interferon-stimulated gene induction by human cytomegalovirus. J. Virol. 79:5027-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhuravskaya, T., J. Maciejewski, D. M. Netski, E. Bruening, F. R. Mackintosh, and S. St. Joer. 1997. Spread of human cytomegalovirus (HCMV) after infection of human hematopoietic progenitor cells: model of HCMV latency. Blood 90:2482-2491. [PubMed] [Google Scholar]