Abstract
White spot syndrome virus (WSSV) is a virulent pathogen known to infect various crustaceans. It has bacilliform morphology with a tail-like appendage at one end. The envelope consists of four major proteins. Envelope structural proteins play a crucial role in viral infection and are believed to be the first molecules to interact with the host. Here, we report the localization and crystal structure of major envelope proteins VP26 and VP28 from WSSV at resolutions of 2.2 and 2.0 Å, respectively. These two proteins alone account for approximately 60% of the envelope, and their structures represent the first two structural envelope proteins of WSSV. Structural comparisons among VP26, VP28, and other viral proteins reveal an evolutionary relationship between WSSV envelope proteins and structural proteins from other viruses. Both proteins adopt β-barrel architecture with a protruding N-terminal region. We have investigated the localization of VP26 and VP28 using immunoelectron microscopy. This study suggests that VP26 and VP28 are located on the outer surface of the virus and are observed as a surface protrusion in the WSSV envelope, and this is the first convincing observation for VP26. Based on our studies combined with the literature, we speculate that the predicted N-terminal transmembrane region of VP26 and VP28 may anchor on the viral envelope membrane, making the core β-barrel protrude outside the envelope, possibly to interact with the host receptor or to fuse with the host cell membrane for effective transfer of the viral infection. Furthermore, it is tempting to extend this host interaction mode to other structural viral proteins of similar structures. Our finding has the potential to extend further toward drug and vaccine development against WSSV.
In the shrimp aquaculture industry, more than 20 viruses have been reported to date. Of these, white spot syndrome virus (WSSV) is the major and most serious pathogen (4, 5, 24). The virulence of WSSV is very high, resulting in mortality rates from 90 to 100% within 3 to 7 days of infection and leading to complete devastation of the shrimp culture industry. This virus has a wide range of hosts among crustaceans and induces distinctive clinical signs of white spots on the carapace, appendages, and the inner surface of the body. WSSV belongs to a new virus family, Nimaviridae, under a new genus Whispovirus (www.ncbi.nih.gov/ICTVdb/Ictv/index.htm), which shares a low homology with other known DNA viruses (25, 41). WSSV is an enveloped virus with a 305-kb double-stranded circular DNA genome (41, 46) and approximately 180 open reading frames. However, most of its presumptive open reading frames do not have any homology with known proteins and are functionally unknown.
In order to understand the mechanism of WSSV infection and morphogenesis, we focused on the characterization of structural proteins. Among these structural proteins, the envelope proteins are extremely important because they are believed to be the first molecules to interact with the host and, consequently, play critical roles in cell targeting as well as in triggering host defenses (38, 39). Using proteomic methods, almost 40 structural proteins of WSSV have been identified (16, 17, 38), of which 22 are envelope proteins. VP28 and VP26 are the two most abundant structural proteins observed in the envelope, accounting for approximately two-thirds of the envelope, in addition to the two other major envelope proteins of WSSV, VP24 and VP19. Using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) profiles of WSSV envelope fractions, VP28, VP26, VP24, and VP19 were shown to constitute the major content of the entire envelope (44). Preliminary studies show that VP28 is crucial for virus entry and vaccine-induced protection (42, 43, 47). Neither VP28 nor VP26 shares significant homology with other known structural proteins from other viruses. However, these two proteins share a sequence similarity of 41%. Further, VP24 shares significant sequence similarities of 46% and 41% with VP28 and VP26, respectively, whereas VP19 has no sequence similarity with either WSSV proteins or other known viral proteins. In addition, the length of VP19 is approximately 85 amino acids [aa] shorter than the rest of the major WSSV envelope proteins. VP24, VP26, and VP28 have a predicted transmembrane region, approximately 30 aa in length at the N terminus, whereas in VP19, the transmembrane region is predicted to be in the middle of the sequence.
As a continuation of our efforts to understand the structure and function of WSSV proteins (23), here we report the localization and crystal structure of two major envelope proteins, VP26 and VP28. This is the first report of envelope protein structures from WSSV. Both proteins adopt similar β-barrel architectures with a protruding N-terminal region. Structural comparison reveals the evolutionary relationship between WSSV envelope proteins and other viral structural proteins. Our immunoelectron microscopy investigation indicates that VP26 and VP28 protrude into the outer surface of the virus envelope. Combining our results with what is known from the literature, we propose a possible role for VP26 and VP28 in host-pathogen interactions.
MATERIALS AND METHODS
Cloning, expression, and purification.
The gene coding for the truncated constructs of VP26 and VP28 was amplified from the WSSV genome and inserted into vector pGEX6P-1 (Amersham). The recombinant plasmid was transformed into Escherichia coli BL21(DE3) Star (Novagen). Overexpressed VP26 and VP28 proteins with a glutathione S-transferase tag were purified by glutathione-Sepharose 4B (Amersham). After the removal of the glutathione S-transferase tag by PreScission protease (Amersham) cleavage, the eluted untagged protein was further purified by Superdex 75 size exclusion chromatography (Amersham) with a buffer solution consisting of 20 mM Tris, 150 mM NaCl, 10 mM dithiothreitol, and 5% (vol/vol) glycerol, pH 7.4. Purified protein fractions were collected and concentrated to a final concentration of 8 to 10 mg/ml using a Vivaspin 20 instrument (Vivascience). The selenomethionine (SeMet)-substituted VP26 and VP28 were expressed in defined LeMaster medium (13) using the methionine auxotrophic strain (DL41). Purification and concentration steps are the same as for the native protein.
Immunogold labeling for VP26, VP28, and WSSV particles.
WSSV was purified from hemolymph of artificially WSSV-infected red claw crayfish, Cherax quadricarinatus, by sucrose gradient centrifugation as previously described (15). Purified viruses suspended in TNE buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 5 mM EDTA) were subsequently treated with 0.1% Tween 20 (Bio-Rad) or Triton X-100 (Bio-Rad) at room temperature for 30 s. After being washed one time with 0.2 M phosphate buffer (pH 7.3) to remove the detergent, the virus suspension was then absorbed to carbon-coated nickel grids. Rabbit anti-VP26 and -VP28 antibodies, raised from truncated recombinant proteins, respectively, were used to recognize the VP26 and VP28 in the viral particles. Preimmune rabbit serum was used to bind virus particles in parallel as a negative control, and subsequent immunogold labeling was performed according to Leu et al. (21) and examined under a Jeol JEM 2010F or Philips CM 10 electron microscope.
Oligomerization state analysis by Western blotting and MS.
The purified WSSV was treated with radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride) followed by nonreducing 12% SDS-PAGE electrophoresis. Proteins separated by SDS-PAGE were transferred by electroblotting to a nitrocellulose membrane, and Western blotting was carried out using antibodies against VP26 and VP28. Based on the Western blotting result, the protein bands in the corresponding SDS-PAGE gel were excised and analyzed by matrix-assisted laser desorption ionization—tandem time of flight mass spectrometry (MALDI-TOF/TOF MS).
Crystallization and data collection.
Crystals of VP26 and VP28 were obtained using the hanging-drop method at 20°C. The initial screens were carried out using Hampton Research crystallization screen kits (crystal screens 1 and 2). The initial conditions were further optimized to obtain diffraction-quality crystals. The best crystals of VP26 were found with a reservoir solution of 0.1 M citric acid, pH 3.5, 3.0 M sodium chloride, and 1% (wt/vol) polyethylene glycol 3350. For VP28, the reservoir solution was 25% polyethylene glycol 8000, 0.2 M calcium acetate, 0.1 M Na-HEPES, pH 7.5, and 1.5% (wt/vol) 1,2,3-heptanetriol. Prior to data collection, crystals were briefly soaked for 10 s in a cryo-protectant solution consisting of 30% glycerol, picked up in a nylon loop, and flash-cooled at 100 K in a nitrogen gas cold stream (Oxford Cryosystems, Oxford, United Kingdom). A complete multiwavelength anomalous diffraction (MAD) data set was collected for VP26 and VP28 at three wavelengths in the synchrotron beam lines X12C and X29 at the National Synchrotron Light Source, Brookhaven National Laboratory, using a charge-coupled device detector (Area Detector Systems Corp., Poway, CA). Data sets were processed and scaled using the HKL2000 package (30). The data collection and refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics of VP26 and VP28
| Data set | VP26
|
VP28
|
||||
|---|---|---|---|---|---|---|
| Peak | Inflection | Remote | Peak | Inflection | Remote | |
| Cell parameters (Å) | a = b = 73.915; c = 199.313 | a = 105.33; b = 106.71; c = 200.37 | ||||
| Space group | R32 | P212121 | ||||
| Data collection | ||||||
| Resolution range (Å) | 50-2.2 | 50-2.6 | 50-2.7 | 50-2.0 | 50-2.1 | 50-2.1 |
| Wavelength (Å) | 0.9782 | 0.9787 | 0.9600 | 0.9796 | 0.9799 | 0.9600 |
| Observed reflections >1 | 187787 | 188815 | 120101 | 1818612 | 1608124 | 774774 |
| Unique reflections | 10798 | 10841 | 11519 | 152547 | 132247 | 131772 |
| Redundancy | 17.4 | 17.4 | 10.4 | 11.9 | 12.2 | 5.9 |
| Completeness (%) | 99.9 | 99.7 | 99.8 | 100 | 100 | 99.7 |
| Overall (I/σI) | 23 | 19.1 | 12.3 | 15.3 | 15.9 | 15.5 |
| Rsyma (%) | 0.090 | 0.088 | 0.082 | 0.080 | 0.072 | 0.073 |
| Refinement | ||||||
| Resolution range (Å) | 45.0-2.2 | 45.0-2.0 | ||||
| No. of reflections | 19998 | 294116 | ||||
| Rworkb | 21.68 | 24.82 | ||||
| Rfreec | 27.85 | 28.12 | ||||
| RMSD bond lengths (Å) | 0.0074 | 0.0075 | ||||
| RMSD bond angles (°) | 1.22 | 1.24 | ||||
| Avg B-factors (Å2) | 73.74 | 60.00 | ||||
| Ramachandran plot | ||||||
| Most favored region (%) | 85.3 | 89.7 | ||||
| Additional allowed regions (%) | 14.0 | 9.7 | ||||
| Generously allowed regions (%) | 0.7 | 0.6 | ||||
| Disallowed regions (%) | 0.0 | 0.0 | ||||
Rsym=Σ|Ii − <I>/Σ|Ii where Ii is the intensity of the ith measurement, and <I> is the mean intensity for that reflection.
Rwork=Σ|Fobs − Fcalc/Σ|Fobs where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree is calculated using the same equation as that for Rwork but 10% for VP26 and 3% for VP28 of reflections were chosen randomly and omitted from the refinement.
Structure determination and refinement.
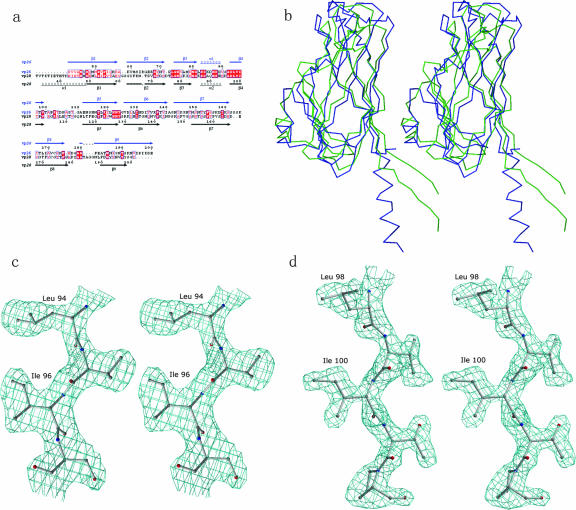
For VP26, the SOLVE program was used to locate the Se sites, whereas the BNP program was used for VP28. The phases were further developed using RESOLVE, which improved the overall figure of merit to 0.67 and 0.75 for VP26 and VP28, respectively (36, 37). The initial models were built automatically by RESOLVE containing ∼50% of the total residues for VP26 and VP28. The rest of the model was manually built using program O (18) and refined using the program CNS (2), interspersed with several rounds of manual refitting. All the measured reflections were used in the refinement. The asymmetric unit of VP26 consists of 1 molecule whereas there are 12 molecules in the asymmetric unit of VP28. Throughout the refinement, noncrystallographic symmetry restraints were used for VP28. The final R factors for VP26 and VP28 are 0.21 (Rfree, 0.27) and 0.24 (Rfree, 0.28) at resolutions of 2.2 and 2.0 Å, respectively. Statistics for the Ramachandran plot from an analysis using PROCHECK (20) showed no outliers (Table 1).
Protein structure accession numbers.
Coordinates and structures factors for VP26 and VP28 have been deposited in the RCSB Protein Data Bank (PDB) under accession numbers 2EDM and 2ED6, respectively.
RESULTS AND DISCUSSION
Localization of VP26 and VP28.
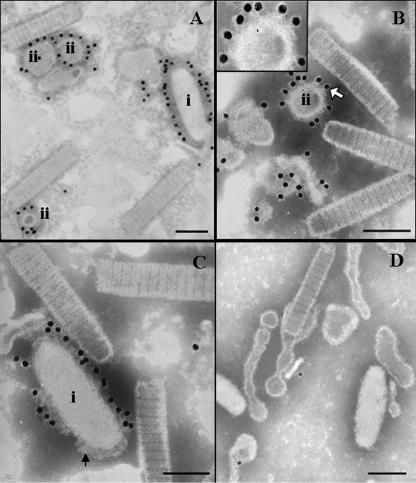
VP26 was first identified as a nucleocapsid protein (40) and later reported to be an envelope protein (49). Our previous immunogold labeling attempt to localize VP26 in the intact virion had shown only a few gold particles. However, recent studies show that VP26 should be a linker protein (45) or a tegument protein (39). To clarify the location of VP26 in the virion, virus treatment with Tween 20 gave a mixture of partial and complete separation of the viral envelope from the nucleocapsid to reveal the location of the bound gold particles. Interestingly, some of the completely separated envelope formed vesicles of different sizes and shapes. For the first time, visible surface protrusions were observed after the treatment, but these surface protrusions were not found on the envelope of an intact virion. This indicates that some inner structures become exposed as the envelope becomes loosely packed. Figure 1A and B show immunogold-labeled VP26, and Fig. 1C shows immunogold-labeled VP28. All the gold particles were bound to the outer surface of the envelope and not to the inner surface of the envelope, nucleocapsid, or the space between them. Some gold particles were clearly localized on the surface protrusions when anti-VP26 immunogold labeling was used (Fig. 1B, inset).
FIG. 1.
Purified WSSV virion was treated with 0.1% Tween 20 and then was immunogold labeled with VP26 and VP28 antibodies. Panels A and B show VP26 antibody specifically binding to the outer surface with the gold particles localized on the outer surface of virions (A-i) and separated envelope (envelope vacuoles, A-ii and B-ii). Panel C shows that immunogold-labeled VP28 is also localized on the outer surface of the envelope. Surface-protruding structures (arrow) clearly appeared on some of the envelope and vesicles. The inset in panel B shows a large view of the surface protrusions. Panel D shows a very low level of nonspecific binding when a preimmune rabbit antibody or gold-conjugated secondary antibody was used. The image possibly also shows the process of vesicle formation. Bar, 100 nm.
Our result showed that 0.1% Tween 20 could gently separate the envelope from the nucleocapsid, but 0.1% Triton X-100 was too harsh, resulting in a complete loss of envelope during subsequent steps of immunogold labeling (data not shown). More interestingly, surface protrusions were visible in the outer surface of the envelope after the Tween 20 treatment, possibly indicating that as the envelope structure becomes loosely packed, some inner structures and binding sites for both VP26 and VP28 are well exposed, giving high labeling efficiency. Tsai et al. (39) also reported that both VP28 and VP664 (a major capsid protein) could be digested when the intact virion was exposed to trypsin, while VP26 could not be digested. Taking these results together, we speculate that VP26 itself is an envelope protein located on the outer surface, but, somehow, it is not accessible to antibodies or proteases. Nevertheless, when treated with Tween 20, VP26 may become accessible.
Similar to our previous report, here we have reconfirmed the localization of VP28 by immunogold labeling experiments using the intact WSSV virion (48). The purified virus was first treated with Tween 20 before the immunogold labeling by anti-VP28. Figure 1C shows immunogold-labeled VP28. All the gold particles were observed on only the surface-protruding structures on the outer surface of the envelope, thus confirming the localization of VP26 and VP28 in the envelope.
Structural study. (i) Identification of constructs.
The expression and purification trials on full-length VP26 and VP28 resulted in very low expression and insoluble proteins. An extensive detergent screen to solubilize the protein and to crystallize it was unsuccessful. Sequence analysis using the DAS server (Stockholm University) indicated the presence of a transmembrane region at the N-terminal region of both proteins. We have postulated that the removal of the predicted transmembrane region would increase the solubility and yield crystals. Subsequently, the transmembrane truncated constructs of VP26 (Asn35 to Lys204) and VP28 (Asn31 to Glu204) readily gave diffraction quality crystals.
(ii) Quality of the models.
The structure of recombinant SeMet VP26 was determined by the MAD method using the synchrotron data and refined to a final R factor of 0.21 (Rfree, 0.27) up to a resolution of 2.2 Å. Similarly, the recombinant SeMet VP28 was solved using a MAD data set collected from the synchrotron beam line at the Brookhaven National Laboratories. The structure was refined at a resolution of 2.0 Å to a final R factor of 0.24 (Rfree, 0.28). Both models have been refined with good stereochemical parameters (Table 1). Statistics for the Ramachandran plot from an analysis using PROCHECK (20) for VP26 and VP28 models gave over 85.3% and 89.7%, respectively, of nonglycine residues in the most favored region and no residues in the disallowed region.
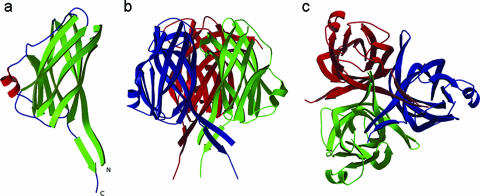
(iii) VP26.
Fig. 2a shows the crystal structure of VP26. The asymmetric unit consists of a VP26 molecule comprising 161 residues from Ser41 to Ile201 and a total of 153 water molecules. The residues Asn35 to Arg40 and Lys202 to Lys204, part of the N and C termini had no interpretable density and were not modeled. The organization of the molecules in the crystal is consistent with its being a monomer in solution. The full-length VP26 consists of a nine-stranded β-barrel with mostly antiparallel β-strands (β1↓β2↑β3↓β4↓β5↑β6↓β7↑β8↓β9↑). In addition, a two-stranded β-sheet (β1↓β9↑) protrudes out of the β-barrel almost parallel to the barrel axis. The extreme N- and C-terminal β-strands come together to form this protruding β-sheet. The N-terminal β-strand (β1) is the longest in the molecule, approximately 50 Å in length. Interestingly, this β-strand is part of the β-barrel as well as the protruding β-sheet. Additionally, there is a two-turn α-helix hanging outside the β-barrel structure. The pore of the β-barrel is highly hydrophobic in nature with side chains from three Phe, seven Ile, two Leu, five Met, and four Val residues, for a total of 21 hydrophobic side chains lining the inner pore surface. The approximate dimensions of the β-barrel are 38.5Å (height) by 15.5Å (diameter).
FIG. 2.
Structure of VP26. (a) Ribbon diagram of the VP26 monomer. The N and C termini are labeled. (b) Ribbon diagram of the VP26 trimer (crystallographic symmetry-related molecules). (c) Top view of the VP26 trimer. Diagrams were prepared using the programs MOLSCRIPT (19) and Raster3D (27).
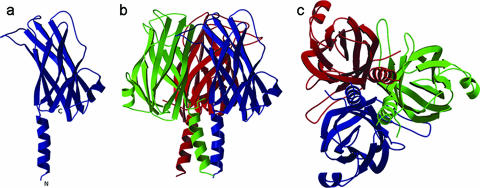
(iv) VP28.
The full-length VP28 exhibits a single β-barrel and an α-helix protruding from the β-barrel architecture (Fig. 3a). The extended N-terminal α-helix protrudes approximately 20 Å from the surface of the β-barrel. This protruding α-helix (15 residues, Thr32 to Asn47) is linked by a 2-aa coil to the core β-barrel. The overall dimensions of the β-barrel are approximately 35 Å (height) by 15 Å (diameter). A total of nine β-strands (β1↓β2↑β3↓β4↓β5↑β6↓β7↑β8↓β9↑), mostly antiparallel, form the β-barrel. In addition, similar to VP26, one α-helix hangs outside the β-barrel, and the pore of the β-barrel is highly hydrophobic in nature. There are 25 hydrophobic side chains lining the inner surface of the pore, contributed by the side chains from seven Phe, eight Ile, one Leu, four Met, and five Val residues.
FIG. 3.
Ribbon diagrams of VP28. (a) Monomer of VP28. The N and C termini are labeled. (b) One asymmetric unit consists of 12 molecules consisting of four trimers. This figure shows one trimer of the asymmetric unit. Each monomer of the trimer is shown in a different color. (c) Top view of the trimer. Figures were prepared using the programs MOLSCRIPT (19) and Raster3D (27).
Considering the size and the hydrophobic nature of the VP26 and VP28 pores, the pores may recognize small, specific, hydrophobic ligands or ions which could potentially establish hydrophobic or Van der Waals interactions with the side chains of the pore residues. However, the exact role of these pores for the function of VP26 and VP28 is yet to be established.
(v) Oligomerization of VP26 and VP28.
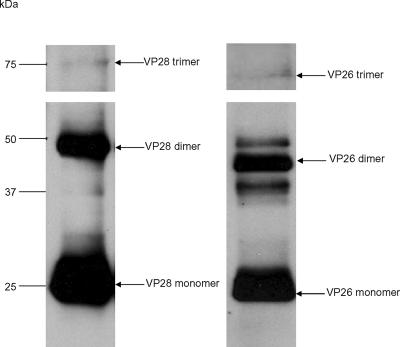
The oligomerization state of recombinant VP26 was investigated by gel filtration chromatography and SDS-PAGE analysis. All the experiments showed an apparent molecular size of 20 kDa that corresponded to that of a monomer. SDS-PAGE analysis (nonreduced) on the purified WSSV showed the presence of trimer (∼66 kDa), dimer (∼44 kDa), and monomer (∼22 kDa, the most abundant species) forms of VP26 (Fig. 4). This suggests that VP26 favors trimers in the viral envelope but that the trimer species may become unstable during virus purification and lysis treatment and becomes a monomer in solution. However, VP26 crystallized in the R32 space group, and its crystal packing showed a trimeric arrangement within the crystal lattices from the symmetry-related molecules (Fig. 2b and c). There was no other evidence to support the formation of a trimer of VP26. We speculated that the trimer formation may be the desirable aggregated form of this protein in the viral envelope. Monomers of the trimer may be held together by weak interactions but not sufficiently strong to retain its oligomerization in solution.
FIG. 4.
Western blotting analysis of WSSV proteins using VP28 and VP26 antibodies. The bands in the corresponding SDS-PAGE gel were analyzed by MALDI-TOF/TOF MS. The arrow indicates the match to VP28 and VP26.
In the case of VP28, the gel filtration and SDS-PAGE analysis showed a monomer in solution. However, in the crystal, an asymmetric unit consists of 12 copies of VP28 molecules with 170 residues from Thr32 to Thr201 assembled into four trimers (Fig. 3b and c). There are 10 hydrogen bonding contacts (<3.2 Å) between two monomers of the trimer and a few hydrophobic interactions among the monomers of the trimer. 1,170 Å2 of the monomer surface (or 13%) is buried at the monomer interface of the trimer. The purified WSSV was analyzed by nonreduced SDS-PAGE, Western blotting, and MALDI-TOF/TOF MS. VP28 forms, corresponding to a trimer (∼75 kDa), dimer (∼50 kDa), and monomer (∼25 kDa, the most abundant species), were detected (Fig. 4). This suggests that VP28 naturally forms trimers in the viral envelope, similar to VP26, but the trimer species may be fragile during virus purification and lysis treatment; hence, it becomes a monomer in solution. However, during crystal formation the monomers tend to aggregate as trimers, the most natural oligomerization state for VP28.
In both cases, a channel is formed by the protruding N-terminal region and the wall of the β-barrel from each monomer of VP26 and VP28 of the trimer. The height and the diameter of VP26 and VP28 channels are approximately 58 by 12 Å. The channel may allow the passage of ions or small molecules, but the exact role of these channels remains to be identified.
It has been shown that viral structural proteins can exhibit multiple functions by adopting different oligomeric states (12). For instance, Ebola virus matrix protein VP40 was shown to form monomers when targeted to cellular membranes but to change its oligomerization state to form hexamers via specific interactions of the N-terminal domain that plays an important role in viral assembly and budding (28). Here, we propose that trimers of VP26 and VP28 may have important functional roles, and possibly the anchoring to viral envelope membrane allows the trimer to interact with the host during virus infection.
It is worth mentioning here that the enveloped viruses always gain entry into the cytoplasm by fusion of their lipid envelope with the host cell membrane (6). Though VP26 and VP28 are not highly homologous to any of the viral envelope fusion proteins, they retain the overall β-barrel as well as trimer architecture similar to the structure of other viral envelope fusion proteins such as the E1 structural protein of Semliki Forest virus (9). Furthermore, Yi et al. (47) showed that only in low-pH environments can VP28 bind to shrimp cells as an attachment protein to help the virus enter the cytoplasm. It has been well established that some of the enveloped viruses enter the cells by an endocytic pathway, and this fusion depends on the acidification of the endosomal compartment (7). Considering this, we suggest that the fusion of WSSV viral envelope and the host membrane through VP26 and VP28 is triggered by the exposure to low-pH conditions in the endocytic pathway or the combination of receptor binding and low pH.
The VP26 and VP28 immunoelectron microscopic images indicated that the surface protrusion favors the trimer. Considering the presence of the transmembrane region at the N terminus and the similarity of the trimer architecture of VP26 and VP28 to their structural homologs, its role in membrane fusion at the viral envelope and interaction of the trimer with the host are supported.
(vi) Homology of VP26 and VP28.
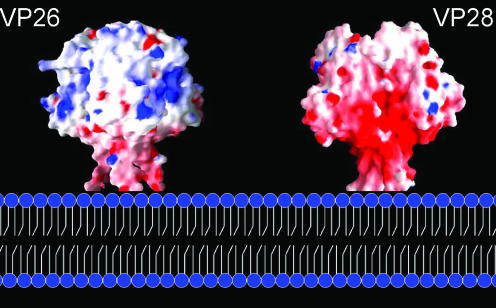
VP26 and VP28 display significant structural and sequence homology (Fig. 5a and b). The structure-based sequence alignment revealed that most of the structurally invariant residues are located at β1, β3, α1 (equivalent of α2 in VP28), β4, and β5. VP26 and VP28 monomers possess a similar hydrophobic pore. The hydrophobic side chain residues lining the inside of the pore are quite conserved between these two proteins. Though both proteins have an N-terminal protruding region, the most significant difference between them is an extended β-strand in VP26 and an α-helix in VP28. In the case of VP26, the N-terminal part and the C-terminal strand come together to form a two-stranded β-sheet (β1↓β9↑) protruding out of the β-barrel parallel to the barrel axis. In VP28, however, at the N terminus an α-helix protrudes outside the β-barrel. Further, an interesting feature of these two proteins is their extreme opposite charges (Fig. 6), while the inner pore region of the β-barrel maintains a strong hydrophobic environment with highly conserved residues lining the pore. The calculated isoelectric focusing point for VP26 is 9.9 whereas for VP28 it is 4.7. More recently Xie et al., using far-Western and coimmunoprecipitation experiments, have shown that VP28 interacts with both VP26 and VP24 to form a complex (44). Similar to VP26 and VP28, the presence of a predicted N-terminal transmembrane region of VP24 was suspected because of its insolubility. Brief expression and purification trials of full-length VP24 resulted in an insoluble protein. Our attempt to improve the solubility by truncating the N-terminal transmembrane region was not successful. However, from the sequence analysis of VP26, VP28, and VP24, combined with the known structural data, we can speculate that VP24 may adopt a β-barrel structure with a hydrophobic pore. An example of the electron density from a simulated annealing Fo-Fc omit map showing the conserved region of VP26 and VP28 is shown Fig. 5c and d.
FIG. 5.
(a)Structure-based sequence alignment was performed using the program O (18). The secondary structural elements and the sequence numbering for VP26 and VP28 are shown at the top and the bottom, respectively. The conserved residues are highlighted in red boxes outlined in blue. This figure was created by using the program ESPript (10). (b) Stereo Cα superposition of VP26 (green) and VP28 (blue). The superposition was generated using the program O (18), using the conserved residues as the starting point. The RMSD between VP26 and VP28 is 2.6 Å for 144 Cα atoms. This figure was prepared using the program MOLSCRIPT (19). (c) Simulated annealing Fo-Fc omit map showing the conserved region of VP26. (d) The same map as used in panel c applied to VP28. All the residues shown in the figure as well as all the atoms within 3 Å of these residues were omitted prior to refinement and map calculation. The map was contoured at a level of 2σ for VP26 and 2.5σ for VP28. This figure was prepared using the program BOBSCRIPT (8).
FIG. 6.
Molecular surface representation for VP26 and VP28 along with the proposed membrane fusion. VP26 trimer (crystallographic symmetry-related molecules) depicting the patches of positively charged surfaces and VP28 trimer (part of one asymmetric unit) depicting the negatively charged regions are shown. These figures were prepared using the program GRASP (29).
(vii) Gene duplication.
The superposition of VP26 and VP28 shows that their core β-barrels are highly similar, with 144 Cα atoms having an root mean square deviation (RMSD) of 2.6 Å. The structure-based sequence alignment shows 18% identical residues and 36% similar residues between the two proteins, with the preservation of the hydrophobic character of the residues forming the pore of these β-barrels. These hydrophobic residues are also found conserved in VP24. Sequence homology of VP26, VP28, and VP24 indicates that these three proteins are highly related. The structural similarity of VP26 and VP28 combined with the model prediction of VP24 suggests that the three envelope proteins may have evolved from a common ancestral gene via duplication (Fig. 5a). A recent study by Merckel et al. (26) showed that the similarities between the tertiary structure markers are indicators of gene duplication. As shown in Fig. 5b, the β-strands of the β-barrel of both VP26 and VP28 superimpose very well, indicating similar distances and similar angles between them. This structure is likely to have been conserved upon gene duplication within the viral genome.
(vii) Comparison with other viral proteins.
The β-barrel architecture is one of the most commonly found structural features of viral proteins (1, 11, 31-33, 35). Neither VP28 nor VP26 shares any sequence or structural homology with any other known structural protein from other viruses. A DALI search for structural similarities (14) shows the highest structural homology with the gamma COP appendage domain of an intracellular trafficking pathway protein (PDB accession code 1pzd; RMSD is 3.3 Å for 89 Cα atoms of VP26 and for 87 Cα atoms of VP28). The only viral protein structures showing a partial structural similarity are bluetongue virus VP7 (PDB accession code 1bvp; RMSD of 3.6 Å for only 82 Cα atoms of VP26) and human coxsackievirus A21 fragment with VP28 (PDB accession code 1z7z; RMSD of 3.6 Å for only 72 Cα atoms). Structurally, the most common feature of these proteins is the presence of an overall β-barrel architecture. However, unlike the predominant eight-stranded β-barrels, VP26 and VP28 have a nine-stranded β-barrel, a feature not commonly observed in other viral proteins. This unique feature of WSSV proteins is consistent with the phylogenetic tree of the WSSV genes’ DNA polymerase and shows that this virus is not related to any of the larger double-stranded DNA virus family (3).
(viii) Functional implications.
We have investigated the localization of VP26 and VP28 and determined the crystal structures of these two major envelope proteins which make up approximately two-thirds of the WSSV envelope. We speculate that the observed trimer shapes of the VP26 and VP28 crystal structures could be the surface protrusions found in our immunoelectron microscopy images. Further, we suggest that VP26 and VP28 might anchor as trimers on the envelope membrane through the N-terminal transmembrane regions (Fig. 6). It is worth mentioning here that VP28 has been shown to play a crucial role in systemic WSSV infection in shrimp (22). VP28 can bind to shrimp cells in a low-pH environment and aid viral entry into the cytoplasm (47). More recently it has been shown that the interaction between the host and WSSV is mediated through the host protein PmRab7 and VP28 (34). In addition, here we speculate that, only under certain circumstances, this mushroom-like structure (trimer) will be projected out for the interaction with the host cell for an effective transfer of viral infection. Furthermore, it is tempting to propose that the possible role of envelope protein trimers in the virus-host interaction may represent a common feature in other homologous structural viral proteins. This study can be further extended toward drug design and vaccine development against WSSV.
Acknowledgments
We acknowledge the use of the X12C and X29 beam lines, Brookhaven National Laboratory, National Synchrotron Light Source, for data collection, and we thank Anand Saxena for help during data collection. We thank Shashikant Joshi for valuable discussions. We thank Sunita Subramanian for the structure-based sequence analysis. We thank Jing Chen for her help with Western blotting analysis. We also acknowledge the facilities provided by the Protein and Proteomic Center, Department of Biological Sciences, National University of Singapore.
J.S. acknowledges research support from the Academic Research Fund, National University of Singapore. We are grateful for funding support from the Ministry of Education of Singapore to C.L.H.
Footnotes
Published ahead of print on 4 April 2007.
REFERENCES
- 1.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Chen, L. L., H. C. Wang, C. J. Huang, S. E. Peng, Y. G. Chen, S. J. Lin, W. Y. Chen, C. F. Dai, H. T. Yu, C. H. Wang, C. F. Lo, and G. H. Kou. 2002. Transcriptional analysis of the DNA polymerase gene of shrimp white spot syndrome virus. Virology 301:136-147. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X. F., C. Chen, D. H. Wu, H. Huai, and X. C. Chi. 1997. A new baculovirus of cultured shrimp. Sci. China Ser. 40:630-635. [DOI] [PubMed] [Google Scholar]
- 5.Chou, H. Y., C. Y. Huang, C. H. Wang, H. C. Chiang, and C. F. Lo. 1995. Pathogenicity of a baculovirus infection causing white spot syndrome in cultured penaeid shrimp in Taiwan. Dis. Aquat. Org. 23:165-173. [Google Scholar]
- 6.Da Poian, A. T., F. A. Carneiro, and F. Stauffer. 2005. Viral membrane fusion: is glycoprotein G of rhabdoviruses a representative of a new class of viral fusion proteins? Braz. J. Med. Biol. Res. 38:813-823. [DOI] [PubMed] [Google Scholar]
- 7.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model 15:132-134, 112-113. [DOI] [PubMed] [Google Scholar]
- 9.Gibbons, D. L., M. C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 10.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 11.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 12.Hartlieb, B., and W. Weissenhorn. 2006. Filovirus assembly and budding. Virology 344:64-70. [DOI] [PubMed] [Google Scholar]
- 13.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478-480. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C., L. Zhang, J. Zhang, L. Xiao, Q. Wu, D. Chen, and J. K. Li. 2001. Purification and characterization of white spot syndrome virus (WSSV) produced in an alternate host: crayfish, Cambarus clarkii. Virus Res. 76:115-125. [DOI] [PubMed] [Google Scholar]
- 16.Huang, C., X. Zhang, Q. Lin, X. Xu, and C. L. Hew. 2002. Characterization of a novel envelope protein (VP281) of shrimp white spot syndrome virus by mass spectrometry. J. Gen. Virol. 83:2385-2392. [DOI] [PubMed] [Google Scholar]
- 17.Huang, C., X. Zhang, Q. Lin, X. Xu, Z. Hu, and C. L. Hew. 2002. Proteomic analysis of shrimp white spot syndrome viral proteins and characterization of a novel envelope protein VP466. Mol. Cell Proteomics 1:223-231. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 19.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallog. 24:946-950. [Google Scholar]
- 20.Laskowski, R. A., D. S. Moss, and J. M. Thornton. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049-1067. [DOI] [PubMed] [Google Scholar]
- 21.Leu, J. H., J. M. Tsai, H. C. Wang, A. H. Wang, C. H. Wang, G. H. Kous, and C. F. Lo. 2005. The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found. J. Virol. 79:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H. X., S. L. Meng, J. P. Xu, W. Lu, and J. Wang. 2005. Protection of crayfish Cambarus clarkia, from white spot syndrome virus by polyclonal antibodies against a viral envelope fusion protein. J. Fish Dis. 28:285-291. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y., J. Wu, J. Song, J. Sivaraman, and C. L. Hew. 2006. Identification of a novel nonstructural protein VP9 from white spot syndrome virus: its structure reveals a ferredoxin fold with specific metal binding sites. J. Virol. 80:10419-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo, C. F., C. H. Ho, S. E. Peng, C. H. Chen, H. E. Hsu, Y. L. Chiu, C. F. Chang, K. F. Liu, M. S. Su, C. H. Wang, and G. H. Kou. 1996. White spot syndrome associated virus (WSBV) detected in cultured and captured shrimp, crabs and other arthropods. Dis. Aquat. Org. 27:215-225. [Google Scholar]
- 25.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655-1663. [DOI] [PubMed] [Google Scholar]
- 26.Merckel, M. C., J. T. Huiskonen, D. H. Bamfor, A. Goldman, and R. Tuma. 2005. The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture. Mol. Cell 18:161-170. [DOI] [PubMed] [Google Scholar]
- 27.Merrit, E. A., and D. J. Bacon. 1997. Raster3D. Photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, T. L., G. Schoehn, W. Weissenhorn, A. R. Hermone, J. C. Burnett, R. G. Panchal, C. McGrath, D. W. Zaharevitz, M. J. Aman, R. Gussio, and S. Bavari. 2005. An all-atom model of the pore-like structure of hexameric VP40 from Ebola: structural insights into the monomer-hexamer transition. J. Struct. Biol 151:30-40. [DOI] [PubMed] [Google Scholar]
- 29.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 31.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, M. M., J. L. White, M. G. Grutter, and R. M. Burnett. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148-1151. [DOI] [PubMed] [Google Scholar]
- 33.Rossmann, M. G., and J. E. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533-573. [DOI] [PubMed] [Google Scholar]
- 34.Sritunyalucksana, K., W. Wannapapho, C. F. Lo, and T. W. Flegel. 2006. PmRab7 is a VP28-binding protein involved in white spot syndrome virus infection in shrimp. J. Virol. 80:10734-10742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stehle, T., S. J. Gamblin, Y. Yan, and S. C. Harrison. 1996. The structure of simian virus 40 refined at 3.1 Å resolution. Structure 4:165-182. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai, J. M., H. C. Wang, J. H. Leu, H. H. Hsiao, A. H. Wang, G. H. Kou, and C. F. Lo. 2004. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78:11360-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai, J. M., H. C. Wang, J. H. Leu, A. H. Wang, Y. Zhuang, P. J. Walker, G. H. Kou, and C. F. Lo. 2006. Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol. 80:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Hulten, M. C., M. Westenberg, S. D. Goodall, and J. M. Vlak. 2000. Identification of two major virion protein genes of white spot syndrome virus of shrimp. Virology 266:227-236. [DOI] [PubMed] [Google Scholar]
- 41.van Hulten, M. C., J. Witteveldt, S. Peters, N. Kloosterboer, R. Tarchini, M. Fiers, H. Sandbrink, R. K. Lankhorst, and J. M. Vlak. 2001. The white spot syndrome virus DNA genome sequence. Virology 286:7-22. [DOI] [PubMed] [Google Scholar]
- 42.van Hulten, M. C., J. Witteveldt, M. Snippe, and J. M. Vlak. 2001. White spot syndrome virus envelope protein VP28 is involved in the systemic infection of shrimp. Virology 285:228-233. [DOI] [PubMed] [Google Scholar]
- 43.Witteveldt, J., C. C. Cifuentes, J. M. Vlak, and M. C. van Hulten. 2004. Protection of Penaeus monodon against white spot syndrome virus by oral vaccination. J. Virol. 78:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie, X., L. Xu, and F. Yang. 2006. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 80:10615-10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie, X., and F. Yang. 2005. Interaction of white spot syndrome virus VP26 protein with actin. Virology 336:93-99. [DOI] [PubMed] [Google Scholar]
- 46.Yang, F., J. He, X. Lin, Q. Li, D. Pan, X. Zhang, and X. Xu. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi, G., Z. Wang, Y. Qi, L. Yao, J. Qian, and L. Hu. 2004. Vp28 of shrimp white spot syndrome virus is involved in the attachment and penetration into shrimp cells. J. Biochem. Mol. Biol. 37:726-734. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X., C. Huang, X. Xu, and C. L. Hew. 2002. Identification and localization of a prawn white spot syndrome virus gene that encodes an envelope protein. J. Gen. Virol. 83:1069-1074. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, X., C. Huang, X. Xu, and C. L. Hew. 2002. Transcription and identification of an envelope protein gene (p22) from shrimp white spot syndrome virus. J. Gen. Virol. 83:471-477. [DOI] [PubMed] [Google Scholar]