Abstract
The interaction of simian virus 40 (SV40) T antigen (T-ag) with the viral origin has served as a model for studies of site-specific recognition of a eukaryotic replication origin and the mechanism of DNA unwinding. These studies have revealed that a motif termed the “beta-hairpin” is necessary for assembly of T-ag on the SV40 origin. Herein it is demonstrated that residues at the tip of the “beta-hairpin” are needed to melt the origin-flanking regions and that the T-ag helicase domain selectively assembles around one of the newly generated single strands in a manner that accounts for its 3′-to-5′ helicase activity. Furthermore, T-ags mutated at the tip of the “beta-hairpin” are defective for oligomerization on duplex DNA; however, they can assemble on hybrid duplex DNA or single-stranded DNA (ssDNA) substrates provided the strand containing the 3′ extension is present. Collectively, these experiments indicate that residues at the tip of the beta-hairpin generate ssDNA in the core origin and that the ssDNA is essential for subsequent oligomerization events.
Replication of DNA is a fundamental biological process that has been highly conserved (36). However, at the molecular level, many steps during DNA replication are not well understood. For example, our understanding of initiation of replication in higher eukaryotes is limited owing to the failure to identify sequences that serve as origins of replication (28). An additional complexity is that while a number of factors needed to initiate eukaryotic DNA replication have been identified, the functions of many of these proteins have yet to be established (36, 86).
Therefore, fundamental issues related to the initiation of DNA replication in eukaryotes are being addressed using the relatively simple DNA tumor viruses (78). The DNA sequences that serve as viral origins of replication have been characterized extensively (reviewed in references 10 and 26). For instance, the simian virus 40 (SV40) core origin is 64 bp long and contains three regions, including a central region containing four GAGGC pentanucleotides arranged as inverted repeats and two flanking regions termed the early palindrome (EP) and the AT-rich region. Viral origins are recognized by virally encoded proteins termed initiators; the SV40-encoded initiator is a multidomain, multifunctional protein termed the large T antigen (T-ag) (reviewed in references 14, 26, and 73). T-ag belongs to helicase superfamily III and is also a member of the AAA+ (ATPase associated with cellular activities) family of proteins (31). It contains an N-terminal J domain, a central origin binding domain (T-ag-obd), and a C-terminal helicase domain (reviewed in references 14, 26, and 73). The determination of much of the structure of T-ag, including the J domain (40), T-ag-obd (49, 54), and the helicase domain (27, 42), has greatly expanded our understanding of T-ag and its assembly on the origin. For example, the T-ag-obd has been shown to form an open ring structure in the crystal, with a positively charged central channel that is sufficiently large to accommodate duplex DNA (54). The T-ag helicase domain oligomerizes to form hexameric rings containing a central channel whose diameter ranges from ∼7 Å (ATP bound) to ∼15 Å (apoenzyme form) (27, 42). This structural information and related electron microscopy studies (e.g., see references 83-85) have greatly enhanced our understanding of the hexamers and double hexamers that form on the SV40 origin (reviewed in reference 10).
The interaction of T-ag with the viral origin is an ideal model system to establish fundamental mechanisms needed for initiation of DNA replication, including origin recognition, cell cycle-dependent assembly into a functional double-hexamer helicase, and origin-specific DNA unwinding. Regarding site-specific binding to the origin, previous studies demonstrated that the GAGGC repeats in the central region of the origin are recognized by residues within the T-ag-obd (49, 74). The recent determination of the costructures of T-ag-obd131-260 bound to GAGGC repeats has greatly expanded our understanding of this interaction (9, 55). However, origin recognition also depends upon a poorly understood interaction between a motif in the helicase domain termed the “beta-hairpin” (35, 65, 71) (around residues 507 to 517) and either of the origin “flanking regions.”
The “beta-hairpin” is present in a number of viral initiators (1, 25, 27, 65, 69, 93) and in the archaeal minichromosome maintenance (MCM) complex (52); therefore, its interactions with DNA are of general interest. In addition to origin recognition, it has been proposed that the “beta-hairpin” plays a direct role in the initial melting of the origin “flanking sequences” (65, 71). Moreover, recent studies of bovine papillomavirus E1 protein demonstrated that within the helicase domain of this protein, the “beta-hairpins” surround single-stranded DNA (ssDNA) (25). Furthermore, movements of the “beta-hairpins” within bovine papillomavirus E1 (25) and T-ag (27) are coupled to ATP hydrolysis. Although these studies clearly implicate “beta-hairpins” in the ATP-dependent helicase activities of superfamily III helicases, the precise role(s) played by the “beta-hairpin” during origin recognition and subsequent DNA melting events has not been established. Therefore, we have further investigated the role(s) played by the “beta-hairpin” during T-ag-dependent initiation of viral DNA replication.
MATERIALS AND METHODS
Protein purification.
Both wild-type (wt) T-ag and a mutant designed to inactivate the tip of the “beta-hairpin,” the KH512/513AA double mutant, were expressed in insect (Sf9) cells via infection with baculovirus expression vectors. The wt vector was previously described (59). The expression vector encoding the T-ag KH512A/513AA double mutant was prepared as follows. A QuikChange kit (Stratagene) was used to generate the double mutant, employing the pCVM-T-ag vector (17) as the substrate. After cleavage of the plasmid vector with BamHI and isolation of the fragment encoding the mutant T-ag, the fragment was ligated into the pVL1392 expression vector (Pharmingen) at the BamHI site. The pVL1392 vector encoding the mutant T-ag was cotransfected into Sf9 cells along with linearized baculovirus DNA and then amplified to produce a viral stock (∼108 PFU/ml) that was used to produce the mutant T-ag protein. Proteins were purified using immunoaffinity techniques with the Pab419 monoclonal antibody (23, 72). Purified proteins were stored in T-ag storage buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride, 0.2 μg of leupeptin per ml, 0.2 μg of antipain per ml, 10% glycerol) (90) and frozen at −80°C until needed.
DNA purification and labeling.
Oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer, purified by electrophoresis through 10% polyacrylamide-urea gels, and isolated as described previously (67, 75). Upon hybridization, double-stranded oligonucleotides were 32P labeled at their 5′ termini and isolated by standard procedures (67, 75). The SV40 origin-containing plasmid used in the replication assays, pSV01ΔEP, was previously described (90) and was purified using conventional methods (67).
Biochemical assays. (i) DNA replication assays.
SV40 DNA replication reactions (43, 80, 90) were conducted as previously described (15, 16). The replication reaction mixtures (60 μl) contained 7 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 40 mM creatine phosphate (di-Tris salt [pH 7.6]), 1.4 μg of creatine phosphate kinase, the deoxynucleoside triphosphates dATP, dGTP, and dTTP (100 μM [each]), the nucleoside triphosphates CTP, GTP, and UTP (200 μM [each]), [α-32P]dCTP (20 μM; ∼5 cpm/fmol), 750 ng (∼0.4 pmol) of the SV40 origin-containing plasmid pSV01ΔEP, 2 μg (∼24 pmol) of T-ag or the KH512/513AA mutant, and 30 μl of HeLa cell extract (∼12 mg/ml). The reaction mixtures were incubated at 37°C for the indicated lengths of time, and then aliquots were removed and acid-insoluble radioactivity determined by trichloroacetic acid precipitation.
(ii) EMSAs.
Electrophoretic mobility shift assays (EMSAs) with T-ag and the T-ag KH512/513AA double mutant were conducted under SV40 in vitro replication conditions (90) in the presence of 4 mM AMP-PNP (a nonhydrolyzable analog of ATP), as previously described (21, 65). 32P-labeled DNA substrates (∼25 fmol) included a 64-bp double-stranded oligonucleotide containing the core origin and smaller derivatives, some of which contained ssDNA (see below). After a 20-min incubation at 37°C, glutaraldehyde was added (0.1% final concentration) for 5 min. The samples were then applied to a 4 to 12% gradient polyacrylamide gel and electrophoresed in 0.5% Tris-borate-EDTA (pH 8.4) for ∼1.5 h (10 W). The gels were dried on Whatman 3MM paper and subjected to autoradiography.
(iii) KMnO4 footprinting.
The KMnO4 footprinting technique was conducted using standard methods (12). The 30-μl reaction mixtures were incubated under replication conditions (90), using 4 mM of the indicated nucleotide, 1 μg of T-ag (∼12 pmol), and 0.5 μg of the SV40 core origin-containing plasmid pSV01ΔEP (∼0.27 pmol). Oligonucleotide 2 (5′ TATCACGAGGCCCTTTCG 3′), which was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase (67), was used in the primer extension reactions. The samples were electrophoresed for ∼4 h at 1,500 V and 40 mA in a 7% polyacrylamide gel containing 8 M urea. The locations of the oxidized residues were determined by using a dideoxy sequencing ladder (68), with oligonucleotide 2 as the primer.
(iv) Nitrocellulose filter binding assays.
Previously described nitrocellulose filter binding assays were used to measure binding of T-ag to the SV40 origin (11, 51). Reaction mixtures (20 μl) contained 7 mM MgCl2, 0.5 mM DTT, 40 mM creatine phosphate (di-Tris salt [pH 7.6]), 0.48 μg of creatine phosphate kinase, 4 mM AMP-PNP, 0.2 mg of bovine serum albumin per ml, 0.8 μg of HaeIII-digested pBR322 DNA, 25 fmol (∼0.4 × 106 cpm/pmol) of the 64-bp core origin or a 64-bp control oligonucleotide derived from the SV40 enhancer (41), and 490 ng (6 pmol) of T-ag or the KH512/513AA double mutant. After incubation for 20 min at 37°C, the mixtures were filtered under suction through alkali-treated nitrocellulose filters (Millipore type HAWP; pore size, 0.45 μm; stored in 100 mM Tris-HCl [pH 7.5]). The filters were then washed with 5 ml of 100 mM Tris-HCl (pH 7.5), dried, and counted in a Beckman LS 3801 scintillation counter.
Molecular modeling.
The coordinates of the hexameric T-ag helicase domain (PDB accession code 1N25 [apo form]) were generated from the reported dimer (42), using crystallographic symmetry operators. Visualization of residues within the C terminus of T-ag was performed using the computer program VMD (NIH resource for macromolecular modeling and bioinformatics). The electrostatic potential of the surface of the helicase domain was calculated using the program APBS (6), and the results were displayed using the program PyMOL (22).
RESULTS
Formation of hexamers and double hexamers of T-ag on the SV40 origin requires the interaction of the “beta-hairpin” with regions of the flanking sequences that undergo structural distortions (65, 71), suggesting that this interaction is necessary for the initial melting of the origin. However, the precise role(s) played by the “beta-hairpin” during assembly of T-ag on the origin has yet to be established. Therefore, the function(s) of the T-ag “beta-hairpin” during origin recognition and subsequent melting events was further examined.
Residues at the tip of the “beta-hairpin” are essential for initiation of DNA replication.
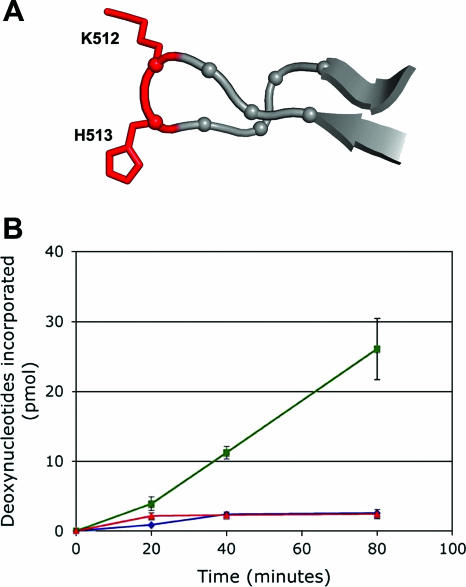
To address the role(s) of the “beta-hairpin” during origin recognition and assembly, a T-ag molecule containing a double mutation at the tip of the beta-hairpin was isolated, giving the KH512/513AA mutant (see Materials and Methods). The relative positions of the two mutated residues at the tip of the “beta-hairpin” are shown in Fig. 1A. The mutant and wt T-ags had nearly identical ATPase activities (data not shown), indicating that the C-terminal domain of the mutant molecule was folded correctly. Structure-based studies have also established that mutations in the “beta-hairpin” do not disrupt the fold of the helicase domain (71). Therefore, we tested whether the residues at the tip of the beta-hairpin are essential for T-ag-dependent DNA replication in vitro (Fig. 1B). It is apparent from these data that the KH512/513AA T-ag mutant is unable to support an in vitro SV40 DNA replication reaction.
FIG. 1.
Testing the ability of the KH512/513AA double mutant to catalyze initiation of DNA replication. (A) View of the “beta-hairpin” (residues 507 to 519) showing the positions of terminal residues K512 and H513 (in red), which were mutated to alanine. (B) SV40 in vitro replication assays (43, 80, 90) conducted for the indicated amounts of time with wt T-ag (green line) and the KH512/513AA double mutant (red line). As a negative control, the amount of T-ag-independent nonspecific incorporation was also determined (blue line).
Using full-length T-ag isolated from baculovirus, it has been demonstrated that the interaction of the beta-hairpin with the origin flanking sequences is essential for T-ag assembly (65). Therefore, it was likely that the replication defect of the KH512/513AA mutant reflected an assembly defect. To test this prediction, a series of EMSA experiments were conducted to monitor oligomerization on the core origin, which is a key step during initiation of replication. A representative experiment is presented in Fig. 2; it is apparent that the KH512/513AA mutant was unable to form either single or double hexamers on a 64-bp duplex oligonucleotide containing the core origin. Based on these studies, it was concluded that the residues at the tip of the beta-hairpin are essential for the oligomerization of T-ag on duplex DNA containing the core origin.
FIG. 2.
Comparison of the abilities of T-ag and the KH512/513AA double mutant to oligomerize on the SV40 core origin. Band shift experiments, conducted with a 32P-labeled 64-bp duplex oligonucleotide containing the SV40 core origin and either 3, 6, or 12 pmol of T-ag, are shown in lanes 2, 4, and 6, respectively. Reactions conducted with the same amounts of the KH512/513AA double mutant are shown in lanes 3, 5, and 7. The control reaction in lane 1 was conducted in the absence of protein. The positions of T-ag hexamers (H), double hexamers (DH), and free DNA (F) are indicated.
Determining the stage in the initiation process that is blocked in the KH512/513AA mutant.
The next series of experiments was conducted to establish the exact stage in the assembly process that was defective in the double mutant. Initially, we tested the possibility that the failure of the double mutant to oligomerize was due to a defect in DNA binding.
(i) Testing the DNA binding ability of the KH512/513AA mutant.
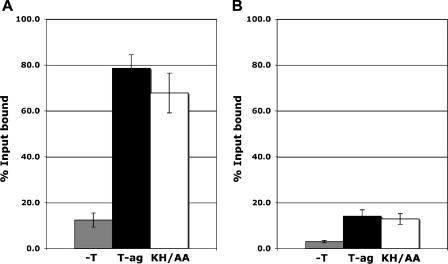
Whether the KH512/513AA mutation disrupted DNA binding was investigated by a series of filter binding assays. Inspection of Fig. 3A reveals that as with wt T-ag, the double mutant bound to an oligonucleotide containing the SV40 core origin. A similar result was obtained when these experiments were repeated with a 64-bp non-sequence-specific “control oligonucleotide” (Fig. 3B), although the overall level of DNA binding was reduced relative to that for the core origin. Consistent with these findings, T-ags containing several different substitutions at residue H513 were competent for binding to a duplex subfragment of the origin when assayed in the presence of ATP (71). Collectively, these studies indicate that DNA binding can occur in the absence of the residues at the tip of the beta-hairpin. However, under replication conditions (90), binding levels of the KH512/513AA mutant were reproducibly reduced relative to those of wt T-ag. Moreover, fluorescence anisotropy DNA binding assays, conducted as previously described (82) with a DNA substrate containing a single pentanucleotide, established that the affinity of the KH512/513AA mutant for this substrate was reduced as much as threefold relative to that of the wt (data not presented). Finally, since the KH512/513AA mutant binds fragments of DNA containing the SV40 origin, one may question why binding was not detected in the EMSA experiments (Fig. 2). Note that the KH512/513AA mutation is in the T-ag helicase domain. Therefore, the T-ag-obd should still be able to bind to the GAGGC sequences, an event presumably detected by the filter binding and fluorescence anisotropy experiments. In contrast, our hypothesis is that the KH512/513AA mutant, which fails to properly oligomerize, is dislodged from the DNA substrate during the relatively harsh conditions encountered during the EMSA experiments.
FIG. 3.
Analysis of the relative abilities of T-ag and the KH512/513AA mutant to bind to duplex DNA substrates. Filter binding assays (11, 51) were used to establish the relative affinities of T-ag (black bars) and the KH512/513AA mutant (KH/AA; white bars) for either (A) a 64-bp oligonucleotide containing the SV40 core origin or (B) a 64-bp control oligonucleotide. As a control, filter binding assays were conducted in the absence of protein (−T; gray bars). The reactions were conducted under replication conditions (see Materials and Methods) in the presence of AMP-PNP and 6 pmol of either T-ag or the KH512/513AA mutant. The percentage of oligonucleotide bound was determined by nitrocellulose filter binding assays and scintillation counting.
(ii) Are the residues at the tip of the “beta-hairpin” necessary for the structural distortions within the origin flanking sequences?
The experiments presented in Fig. 2 and 3 established that the KH512/513AA mutant binds to the SV40 origin but fails to oligomerize. In view of these observations, it was of interest to determine whether the tip of the beta-hairpin is needed to catalyze the previously described distortions in the flanking sequences (reviewed in reference 10).
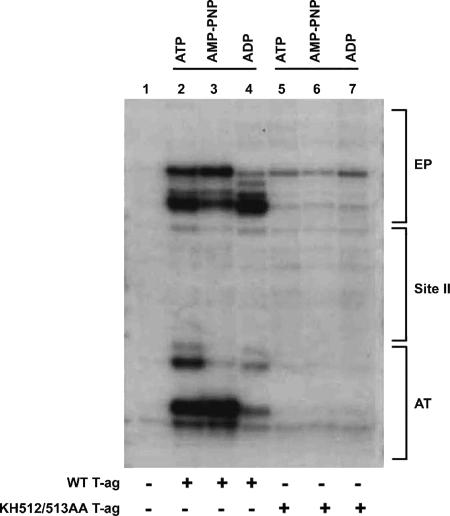
KMnO4 oxidizes thymine rings that are not properly base paired, and this compound has been employed in a technique designed to detect structural distortions in DNA. Therefore, standard KMnO4 assays were conducted with either T-ag or the KH512/513AA mutant and a plasmid that contains the SV40 core origin (12). Results of a representative study are presented in Fig. 4. A control reaction was conducted in the absence of T-ag and loaded in lane 1. The products of reactions conducted with T-ag in the presence of ATP, AMP-PNP, and ADP are presented in lanes 2 to 4, respectively. The previously described alterations in the AT-rich region and EP regions are indicated. Identical reactions were conducted with the KH512/513AA double mutant (lanes 5 to 7). It is apparent from these studies that the residues at the tip of the beta-hairpin are needed to promote the structural distortions in the origin flanking sequences.
FIG. 4.
Structural distortions in the SV40 origin are not catalyzed by the KH512/513AA mutant. T-ag (12 pmol) was incubated under replication conditions with pSV01ΔEP (0.27 pmol) in the presence of ATP (lane 2), AMP-PNP (lane 3), or ADP (lane 4). Additional reactions were conducted with 12 pmol of the KH512/513AA double mutant (lanes 5 to 7) in the presence of the identical nucleotides. As a control, the reaction in lane 1 was conducted in the absence of T-ag. After treatment with KMnO4, the locations of the oxidized bases were determined via primer extension reactions with a 32P-labeled oligonucleotide (see Materials and Methods). The products of the primer extension reactions were analyzed by electrophoresis on a 7% polyacrylamide gel containing 8 M urea. The locations of regions of the SV40 core origin, including the EP, site II, and the AT-rich region, are indicated to the right of the gel.
T-ag assembly on DNA substrates designed to mimic origins containing melted flanking regions.
It was previously reported that the structural distortions in the EP are associated with the generation of ssDNA (12). Therefore, it was of interest that the KH512/513AA double mutant was able to bind to duplex DNA containing the core origin but was unable to distort the EP (Fig. 4). These observations raised the possibility that oligomerization of T-ag on the origin is dependent upon the presence of ssDNA in the EP and that the “beta-hairpins” play a direct role in the generation of ssDNA in this region. In view of these considerations, we analyzed whether hybrid duplex/ssDNA substrates would support the assembly of both wt T-ag and the KH512/513AA double mutant.
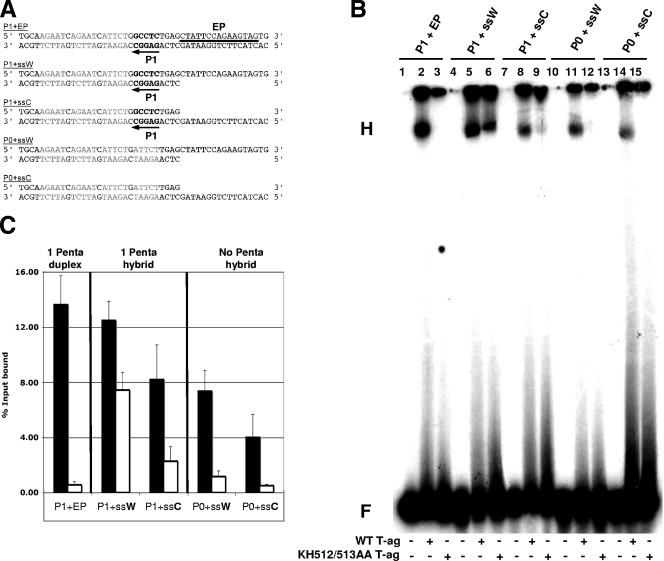
The origin-based substrates used in the EMSA experiments are presented in Fig. 5A. The first three oligonucleotides contain a single GAGGC pentanucleotide (penta 1) and were designed to limit T-ag assembly to a single hexamer. Oligonucleotides with ssDNA on the top, or “Watson,” strand are termed ssW, and those with ssDNA on the bottom, or “Crick,” strand are termed ssC. A representative EMSA experiment conducted with these substrates is presented in Fig. 5B. As a positive control, a reaction was conducted with the entirely duplex “penta 1+EP” oligonucleotide; as previously reported (37, 65, 76), T-ag readily formed hexamers on a subfragment of the origin containing a single pentanucleotide (lane 2). However, consistent with the EMSA data presented in Fig. 2, T-ag containing the KH512/513AA double mutation failed to oligomerize on the duplex “penta 1+EP” oligonucleotide (lane 3).
FIG. 5.
EMSA experiments conducted with the pentanucleotide 1-based set of oligonucleotides, T-ag, and the KH512/513AA mutant. (A) Pentanucleotide 1-based set of oligonucleotides used for EMSA experiments. The first three SV40 origin-derived oligonucleotides contained pentanucleotide 1 (P1; arrow). The 48-bp “P1+EP” oligonucleotide was entirely duplex; pentanucleotide 1 is indicated, the transition mutations that replaced the other pentanucleotides are shown in light gray, and the location of the EP region is indicated by a horizontal bar. The hybrid duplex/single-stranded “P1+ssW” and “P1+ssC” oligonucleotides contained pentanucleotide 1 and either a 3′ extension (Watson [W]) or a 5′ extension (Crick [C]) of the EP. The two control oligonucleotides, “P0+ssW” and “P0+ssC,” lacked pentanucleotide 1 but contained the same single-stranded extensions of the EP found in the “P1+ssW” and “P1+ssC” oligonucleotides. (B) Representative band shift experiment used to compare the abilities of T-ag (lanes 2, 5, 8, 11, and 14) and the KH512/513AA mutant (lanes 3, 6, 9, 12, and 15) to assemble on the indicated substrates (i.e., P1+EP, P1+ssW, P1+ssC, P0+ssW, and P0+ssC). The positions of the hexamers (H) and free DNA (F) are indicated. As controls, the reactions in lanes 1, 4, 7, 10, and 13 were conducted in the absence of protein. (C) Three separate EMSA experiments were quantitated with a Molecular Dynamics PhosphorImager and used to determine the percentages of input DNA present in the hexamers. Black bars symbolize experiments conducted with wt T-ag, while white bars symbolize those performed with the KH512/513AA mutant. It was apparent that the KH512/513AA mutant assembled at appreciable levels only on the P1+ssW substrate. Moreover, for the hybrid duplex/ssDNA substrates and either T-ag or the KH512/513AA mutant, the Watson strand was preferred over the Crick strand. Finally, for wt T-ag, the presence of a pentanucleotide in the hybrid duplex/ssDNA substrates led to a relatively minor increase (∼2-fold) in the level of assembly.
The next two sets of reactions were conducted with oligonucleotides that were designed to mimic the viral origin after “beta-hairpin”-dependent melting occurred. T-ag formed hexamers on the “penta 1+ssW” oligonucleotide at levels comparable to those with the duplex substrate (Fig. 5B, lane 5). It was of considerable interest that the KH512/513AA double mutant, which was defective for hexamerization on the duplex substrate (lane 3), formed hexamers on this hybrid substrate (lane 6). The next series of reactions were conducted with the “penta 1+ssC” oligonucleotide. T-ag formed hexamers on this substrate (lane 8), although at reduced levels relative to its assembly on the “penta 1+ssW” oligonucleotide. In addition, little assembly of the KH512/513AA double mutant was detected on the “penta 1+ssC” oligonucleotide. Finally, T-ag is known to assemble on “forked substrates” in a pentanucleotide-independent manner (70, 77, 89). Therefore, additional control reactions were conducted to establish the extent to which pentanucleotide 1 was needed for assembly. T-ag formed relatively high levels of hexamers on the “0 penta+ssW” oligonucleotide (lane 11), but the KH512/513AA mutant did not (lane 12). A similar conclusion was derived from studies of assembly on the “0 penta+ssC” oligonucleotide (lanes 14 and 15). However, comparison of lanes 14 and 11 indicates that T-ag preferentially assembled on the molecule that contained a 3′ single-stranded extension (the Watson strand). The reactions in lanes 1, 4, 7, 10, and 13 were conducted in the absence of protein.
The reaction products from Fig. 5B and two additional sets of experiments were quantified using a phosphorimager, and the results are presented in Fig. 5C. The substrates used in a given experiment are indicated. The black bars represent data derived from experiments conducted with T-ag, while data derived using the KH512/513AA mutant are represented by white bars. Collectively, these experiments demonstrate that T-ag preferentially oligomerizes on hybrid duplex/ssDNA substrates containing a 3′ extension of the EP (Watson [W] strand). They also establish that on substrates containing hybrid duplex/ssDNA, assembly takes place in a manner that is relatively independent of pentanucleotide 1 (Fig. 5C, compare the middle and right panels). Moreover, they demonstrate that a mutation that renders the tip of the “beta-hairpin” defective for assembly on duplex DNA is “rescued” by DNA containing pentanucleotide 1 and ssDNA derived from the Watson strand in the EP region. In contrast, the other hybrid duplex/ssDNA substrates support little or no assembly of the KH512/513AA mutant.
Support derived from T-ag mutations for the selective assembly of the helicase domain around one strand of DNA.
The previous observations suggest a model whereby residues at the tip of the beta-hairpin play a critical role in melting the EP, followed by selective assembly of the helicase domain around the strand containing the 3′ extension. Within the T-ag helicase domain, the positions of the beta-hairpins vary as a function of the nucleotides (27); however, they all project into the center of the channel (Fig. 6A). Thus, it is likely that the strand containing the 3′ extension transits past the beta-hairpins in the central channel. However, little is known regarding the path taken by the second strand of DNA through hexamers or double hexamers, the products of the assembly process. Therefore, the following analyses were performed.
FIG. 6.
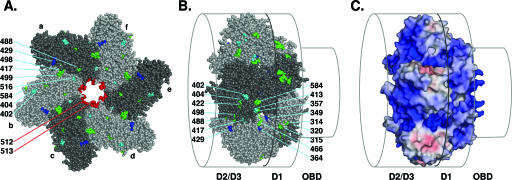
Support for the hypothesis that there exists an external path for ssDNA over the helicase domain. (A) Distribution of residues on the C-terminal surface of the helicase domain whose mutation selectively disrupted binding to ssDNA (blue residues, i.e., P429), helicase activity (green residues, i.e., G488, R498, D499, and P584), or both activities (cyan residues, i.e., D402, V404, P417, and K516). The six T-ag monomers in the T-ag hexamer (a to f) are indicated. Residues K512 and H513 are situated at the tip of the beta-hairpin, and their positions within the central channel are indicated (shown in red). (B) Residues on the lateral surface of the helicase domain which are defective, when mutated, for binding to ssDNA (blue residues, i.e., R349, R364, and D429), helicase activity (green residues, i.e., Y314, K315, H320, R357, W422, D466, G488, R498, and P584), or both activities (cyan residues, i.e., D402, V404, V413, and P417). The relative positions of the T-ag-obd (OBD) and the helicase subdomains (D2/D3 and D1) are indicated. (C) Electrostatic potential of the surface of the helicase domain. Blue surfaces indicate positive potential, along which negative ssDNA could transit, and red surfaces indicate negative potential (±10 kT/e). The relative positions of the T-ag domains are indicated as in panel B.
Initially, all of the previously described replication-defective mutations in the helicase domain were tabulated, along with information regarding the step(s) in the initiation process that was defective (Table 1). The mutations were mapped onto the structure of the helicase domain and then analyzed for information regarding the transit of ssDNA through the helicase domain. A C-terminal view of a T-ag251-627 hexamer is presented in Fig. 6A, with the individual T-ag monomers colored in two alternating shades of gray. The residues in blue represent the subset of mutations that are defective for binding to ssDNA. The distribution of mutations that are defective for helicase activity (green residues) is also informative in terms of potential paths for ssDNA. It is apparent that the C-terminal face of the helicase domain contains several residues that are defective, when mutated, for binding to ssDNA (i.e., P429) and for helicase activity (i.e., G488, R498, D499, and P584). Residues defective for both activities are colored cyan (i.e., D402, V404, P417, and K516). (Note that the mutations defective for binding to ssDNA and for helicase activity include all of the mutations that are defective for DNA unwinding [Table 1]; thus, the unwinding mutations were not considered separately.) In summary, the C-terminal surface of the helicase domain contains a number of residues that are defective for DNA replication when mutated, many of which play important roles in binding to ssDNA and in DNA helicase and unwinding activities. Moreover, it is apparent that replication-defective mutations also cluster in the central channel (beta-hairpin residues K512 and H513 are colored red).
TABLE 1.
Compilation of mutations within the T-ag helicase domain causing defects in DNA replicationa
| Mutation(s) in helicase domain | Defect caused by mutation
|
Reference(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| DNA binding | ssDNA binding | Oligomerization | DNA distortions | DNA unwinding | Helicase activity | ATPase activity | Replication | ||
| C302S | O, I, II | 3, 33, 46 | |||||||
| C305S | O, I, II | 3, 33, 46 | |||||||
| K308A | • | 91 | |||||||
| H313L | • | 46 | |||||||
| H313R | • | 46 | |||||||
| Y314S | • | 46 | |||||||
| K315E | • | 46 | |||||||
| Y314G/K315E | • | • | • | 47 | |||||
| H317Q | • | 46 | |||||||
| H320L | • | • | • | 46, 47 | |||||
| R349A | • | 91 | |||||||
| R357K | O | • | • | L | • | • | 48, 63, 66 | ||
| R364A | • | • | 91 | ||||||
| R371A | • | 91 | |||||||
| G377A | • | 91 | |||||||
| W393C | • | 48 | |||||||
| L398V | • | 45 | |||||||
| P399L | • | 45 | |||||||
| D402N/V404M/V413M | • | L | • | • | 19, 60 | ||||
| P417S | • | • | L | • | • | 19, 60, 62 | |||
| W422C | O | • | • | • | • | 48, 63 | |||
| G426A | • | 62 | |||||||
| P427L | O | • | • | • | 48, 63 | ||||
| D429A | • | L | 91 | ||||||
| G431ALE | • | • | • | L | • | • | 19, 60 | ||
| A438V | O | • | • | 48, 63, 66 | |||||
| P453S | • | 48 | |||||||
| R456A | NS | • | L | • | 35 | ||||
| F459Ab | 1,2+EP | • | • | 71 | |||||
| E460D | NS | • | L | • | 35 | ||||
| E460A | • | L | 91 | ||||||
| G462A | NS | • | L | • | 35 | ||||
| D466E | L | • | • | 35 | |||||
| G488A | L | • | • | 35 | |||||
| R498K | L | • | • | 35 | |||||
| D499A | NS | • | L | • | 35 | ||||
| K512A | 1,2+EP | • | • | 71 | |||||
| K513Ab | 1,2+EP | • | • | 71 | |||||
| K516A | 1,2+EP | • | • | 71 | |||||
| K516R | O | 50, 58, 79 | |||||||
| P522S | • | C | • | • | 50, 58 | ||||
| P522S/P549R | • | 5, 50, 79 | |||||||
| P549R | • | 50 | |||||||
| F562S | • | 48 | |||||||
| P584L | NS | • | • | • | 20, 44, 61, 81 | ||||
In many instances, the step(s) in the initiation process that is defective is known. For example, mutants defective for binding to double-stranded DNA substrates (i.e., with defects in binding to the core origin [O], site I [I], site II [II], or an origin subfragment containing pentanucleotides 1 and 2 and the EP [1,2+EP] or in non-sequence-specific [NS] binding to duplex DNA) have been reported. Additional mutants are defective for binding to ssDNA, oligomerization on origin-containing DNA, DNA distortions of the origin flanking regions, the ability to unwind covalently closed circle (C) or linear (L) DNA, helicase activities, or ATPase activities. However, in several instances, the stage at which the mutants are defective has not been determined. Therefore, these mutants were simply classified as “replication defective.”
Many additional mutations were introduced at these positions (71).
A lateral view of the T-ag helicase domain was obtained by rotating the image in Fig. 6A 90° (Fig. 6B). The set of mutations situated on the surface of the helicase domain that are defective for binding to ssDNA (blue) and for DNA helicase activity (green) are indicated (residues defective for both activities are colored cyan). It is evident that the lateral surface contains a number of residues that are defective, when mutated, for binding to ssDNA (i.e., R349, R364, and D429), helicase activity (i.e., Y314/K315, H320, R357, W422, D466, G488, R498, and P584), or both activities (i.e., D402, V404, V413, and P417). In addition, the electrostatic potential at the surface of the helicase domain, spanning the D2/D3 and D1 domains, was examined (Fig. 6C). The figure demonstrates that the mutations defective for binding to ssDNA and for helicase activity (Fig. 6A and B) cluster in a region that is comprised of a number of basic residues. In summary, the mutagenesis data support the hypothesis that on a given hexamer, one strand of DNA transits past the beta-hairpins and continues on into the central channel, while the second strand is routed over the outer surface of the helicase domain and never enters the central channel.
DISCUSSION
Residues situated at the tip of the beta-hairpin are needed for the structural distortions of the SV40 origin that occur during T-ag assembly. They are also required for T-ag oligomerization on substrates containing a duplex EP region but not for oligomerization on substrates containing a single-stranded 3′ extension of the EP. Based on these findings, it is proposed that the tip of the beta-hairpin plays a critical role in melting the EP and that subsequent oligomerization events are dependent on the ssDNA generated in this process. Furthermore, our studies indicate that the melting of the EP is not simply due to the forcing of the beta-hairpin into duplex DNA. If that were the case, then the mutations at the tip of the hairpin would not have such a dramatic effect on the structural distortions. Therefore, it is proposed that residues at the tip of the beta-hairpin are directly involved in the initial melting process.
Our studies also establish that the T-ag helicase domain preferentially oligomerizes on the strand containing the 3′ extension of the EP. Thus, after assembly on the origin, a given hexamer is positioned such that it can serve as a 3′-to-5′ helicase (29, 89). Studies of synthetic replication forks also established that T-ag binds selectively to the strand that would enable it to travel in a 3′-to-5′ direction (35, 70). The hypothesis that only a single strand of DNA transits through the central channel in T-ag is supported by related structure-based considerations. For example, the diameter of the central channel running between the beta-hairpins ranges from 7 to 15 Å, depending on the nucleotide state (27, 42), and there is little precedent for two strands of DNA passing through such a narrow passage. For instance, the hexameric RuvB helicase, which is specific for duplex DNA, has a 30-Å central channel (56). In contrast, RepA (92) and TrwB (30), two hexameric pumps for ssDNA, have central channels of 17 Å and 20 Å, respectively. Furthermore, in view of the conclusion that only a single strand of DNA transits through the central channel of T-ag, it is of interest that the bovine papillomavirus (BPV) E1 helicase assembles around a single strand of DNA whose 3′ end is directed towards the C terminus (25). Moreover, the steric exclusion model for DnaB and MCM (38, 39) as well as Rho (87) suggests that one DNA strand transits through the central channels, while the second strand passes outside the rings. Similarly, models suggest that the T7 helicase ring surrounds the 5′ strand in its central channel, thus accounting for its 5′-to-3′ polarity, while the 3′ strand is routed over the external surface (2, 24, 32).
The assembly of the helicase domain on one strand of DNA is readily explained if during T-ag assembly, the beta-hairpins coordinate around only a single strand of duplex DNA. Regarding the residues involved in this process, it is apparent that K512 and H513 are needed for the generation of ssDNA. However, since the KH512/513AA mutant still preferentially assembles on one strand of DNA, albeit to a lesser degree than the wt (Fig. 5B, lane 6 versus lanes 9, 12, and 15), additional residues must contribute to strand selection and binding to ssDNA. While the set of residues involved in selection of the 3′ strand remains to be identified, it is of interest that T-ags containing mutations at residues P522 (58) and K516 (71) had markedly reduced abilities to bind to ssDNA. Thus, it is clear that other residues in the beta-hairpin are involved in binding to ssDNA and, perhaps, in strand selection. Why the helicase domain selectively assembles on the 3′ extension of the EP is unresolved. Possibilities include the polarity of the DNA strand, the chemical nonequivalence of the antiparallel strands, and the shape and charge complementarity of the protein for a given strand.
It is interesting that the presence or absence of a pentanucleotide in our hybrid duplex/ssDNA substrates did not dramatically influence the level of hexamer assembly (Fig. 5B and C). Following the interaction between the T-ag helicase domain and ssDNA, it is likely that the T-ag-obds are positioned such that they are in the center of the complex, where they maintain certain interactions with dsDNA. Nevertheless, the pentanucleotide-independent assembly of the helicase domain on ssDNA raises the possibility that following beta-hairpin-dependent melting of the origin, there is no longer a requirement for site-specific binding of the T-ag-obd to pentanucleotide 1. Consistent with this hypothesis, it was recently reported that within the “lock-washer” hexameric conformation of the T-ag-obd, residues known to be required for site-specific binding to DNA (i.e., V150, F151, and N153) were instead involved in protein-protein interactions (54). Thus, assembly of the helicase domain around ssDNA may trigger the conversion of the T-ag-obd from a domain needed for site-specific binding to duplex DNA to one engaged in other interactions (e.g., nonspecific binding to duplex DNA and ssDNA [64] and to proteins such as RPA [4, 34]).
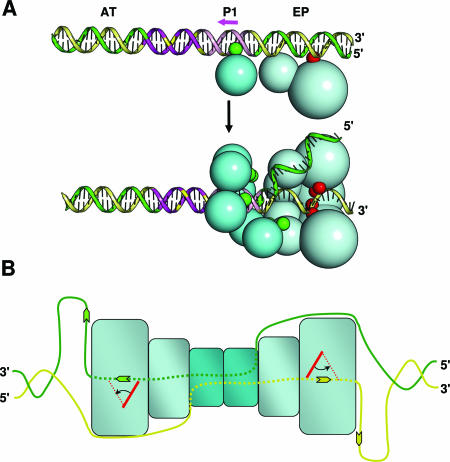
A model for the assembly of T-ag on the SV40 origin is presented in Fig. 7. Important determinants of origin recognition and assembly include the interactions of the A1 and B2 loops with the pentanucleotides and those between the beta-hairpins and the EP (Fig. 7A, top diagram). The present analyses indicate that following beta-hairpin-dependent melting of the EP and subsequent assembly events, only one strand of DNA transits through the central channel of the helicase domain (Fig. 7A, bottom diagram). The same conclusion was reached following the determination of the costructure of BPV E1 bound to ssDNA (25). It follows that if only one strand of DNA travels through the central channel, the second strand must be excluded from this region. In support of this hypothesis, our analysis of T-ag mutations in the helicase domain suggests that the external surface of this domain contains a path for ssDNA. It is likely that the second hexamer forms in much the same way as the first. However, for unknown reasons, assembly of the second hexamer, particularly on single assembly units (41, 76), is promoted by phosphorylation of Thr124 (7, 53, 57, 88).
FIG. 7.
Models for beta-hairpin-dependent unwinding of the core origin and subsequent helicase activity. (A) The two images in panel A present a model for hexamer formation around a single strand of the SV40 origin. As shown in the upper figure, the SV40 core origin consists of a central region, containing four GAGGC pentanucleotides, and two flanking regions, the AT-rich region and the EP. Recognition of the SV40 origin by a T-ag monomer depends upon two interactions, namely, those of the A1 and B2 loops (green spheres) with a pentanucleotide (reviewed in reference 14) (pentanucleotide 1 is depicted by the pink arrow) and those between the beta-hairpin (shown in red) and the flanking sequences (65, 71). The lower figure depicts the subsequent assembly of the helicase domain (light blue spheres) on the 3′ extension of the EP, which is proposed to depend upon the selective coordination of the beta-hairpins around a single strand of origin DNA. The displaced strand is shown transiting the external surface of the helicase domain and then entering the T-ag-obd hexamer (dark blue spheres) via the recently described gap (54). For ease of viewing, two helicase domains have been removed. (B) Upon assembly of the double hexamer, it is proposed that DNA is routed through the complex as depicted (DNA is symbolized by green and yellow strands; DNA situated within the complex is depicted by dotted lines). The present studies indicate that the central channel of each T-ag hexamer contains the 3′ extension of the flanking sequence. Based on recent studies by Enemark and Joshua-Tor (25), ssDNA in the central channel is proposed to be pumped out of the helicase domain owing to beta-hairpin- and ATP-dependent interactions with the sugar-phosphate backbone. (The beta-hairpin is symbolized by red rectangles, and its position following ATP hydrolysis is depicted by red dotted lines). Finally, recent studies established that ssDNA selectively interacts with the T-ag-obd on the helicase proximal surface (64), one indication that upon entering the gap, ssDNA interacts with this surface.
How DNA is currently thought to be routed through the newly formed double-hexamer complex is summarized in Fig. 7B. According to this model, the 3′ strand of ssDNA transits through the central channel of a given hexamer. In contrast, the complementary strand transits over the surface of the helicase domain and is then routed into the central portion of the T-ag-obd via the gap (54, 64). The strands then switch their routes through the second hexamer. Support for this model, particularly for the internal routing of DNA through the T-ag-obd, was obtained from footprinting studies of double hexamers performed with 1,10-phenanthroline-copper ions (37). These studies revealed that within a double hexamer, the DNA comprising site II was protected from cleavage, while the flanking sequences were degraded by the oxygen radicals generated by this reagent. Finally, based on observations derived from the costructure of BPV E1 bound to DNA, it is proposed that ssDNA is propelled out of the C terminus of the helicase domain owing to beta-hairpin- and ATP-dependent interactions with the sugar-phosphate backbone of ssDNA (25). The central role of the beta-hairpins and ATP hydrolysis in this process is consistent with observations made during studies of the T-ag helicase domain (27). As shown in Fig. 7B, the extrusion of ssDNA out of the central channels would result in the generation of two single-stranded loops, which could serve as templates for nascent-strand DNA synthesis (reviewed in references 13, 14, 36, and 86).
Beta-hairpins are important features of the initiators carried by members of the Papovaviridae family (1, 25, 65, 69, 71), and they are also present in the archaeal and human MCM complexes (52). Furthermore, as with the beta-hairpins in T-ag, the beta-hairpin in E1 is necessary for structural distortions of the BPV origin (18, 69). Therefore, beta-hairpin-dependent melting of origins of replication and the subsequent assembly of helicase domains on one strand of DNA may be general phenomena. Given the complexity of the initiation complex on nonviral origins of replication (8, 36), important interactions such as those between beta-hairpins and duplex DNA may be regulated by additional cellular factors. Nevertheless, it is now clear that the beta-hairpins are important not only for ATP-dependent helicase activity but also for the distortions of the origin that generate the ssDNA needed for the assembly of replication initiation complexes.
ADDENDUM IN PROOF
Omitted from Table 1 are data from B. M. Weiner and M. K. Bradley (J. Virol. 65:4973-4984, 1991) regarding the T-ag ED473/474AA double mutant situated in the Walker B motif. This mutant was defective for viral DNA replication and had only 10% of the ATPase and helicase activity of the wild-type protein.
Acknowledgments
A.K. and G.M. contributed equally to this work.
This work was supported by a grant from the National Institutes of Health to P.A.B. (9R01GM55397) and by a grant from the Canadian Institutes for Health Research to J.A., who is a senior scholar from the Fonds de la Recherche en Santé du Québec.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Abbate, E. A., J. M. Berger, and M. R. Botchan. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 18:1981-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahnert, P., and S. S. Patel. 1997. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem. 272:32267-32273. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, A. K., A. Hoss, and E. Fanning. 1988. Expression of simian virus 40 T antigen in Escherichia coli: localization of T-antigen origin DNA-binding domain to within 129 amino acids. J. Virol. 62:1999-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arunkumar, A. I., V. Klimovich, X. Jiang, R. D. Ott, L. Mizoue, E. Fanning, and W. J. Chazin. 2005. Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 12:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auborn, K., M. Guo, and C. Prives. 1989. Helicase, DNA-binding, and immunological properties of replication-defective simian virus 40 mutant T antigens. J. Virol. 63:912-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: applications to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbaro, B. A., K. R. Sreekumar, D. R. Winters, A. E. Prack, and P. A. Bullock. 2000. Phosphorylation of simian virus 40 T antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 9.Bochkareva, E., D. Martynowski, A. Seitova, and A. Bochkarev. 2006. Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. EMBO J. 25:5961-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. [DOI] [PubMed] [Google Scholar]
- 11.Borowiec, J. A., and J. Hurwitz. 1988. ATP stimulates the binding of the simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc. Natl. Acad. Sci. USA 85:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borowiec, J. A., and J. Hurwitz. 1988. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 7:3149-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brush, G. S., and T. J. Kelly. 1996. Mechanisms for replicating DNA, p. 1-43. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 15.Bullock, P. A. 1998. Viral in vitro replication systems, p. 223-243. In S. Cotterill (ed.), Eukaryotic DNA replication. A practical approach. Oxford University Press, Oxford, United Kingdom.
- 16.Bullock, P. A., W. S. Joo, K. R. Sreekumar, and C. Mello. 1997. Initiation of SV40 DNA replication in vitro: analysis of the role played by sequences flanking the core origin on initial synthesis events. Virology 227:460-473. [DOI] [PubMed] [Google Scholar]
- 17.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. [DOI] [PubMed] [Google Scholar]
- 18.Castella, S., G. Bingham, and C. M. Sanders. 2006. Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1. Nucleic Acids Res. 34:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castellino, A. M., P. Cantalupo, I. M. Marks, J. V. Vartikar, K. W. C. Peden, and J. M. Pipas. 1997. trans-Dominant and non-trans-dominant mutant simian virus 40 large T antigens show distinct responses to ATP. J. Virol. 71:7549-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins, B. S., and J. M. Pipas. 1995. T antigens encoded by replication-defective simian virus 40 mutants dl1135 and 5080. J. Biol. Chem. 270:15377-15384. [DOI] [PubMed] [Google Scholar]
- 21.Dean, F. B., M. Dodson, H. Echols, and J. Hurwitz. 1987. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc. Natl. Acad. Sci. USA 84:8981-8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA.
- 23.Dixon, R. A. F., and D. Nathans. 1985. Purification of simian virus 40 large T antigen by immunoaffinity chromatography. J. Virol. 53:1001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egelman, E. H., X. Yu, R. Wild, M. M. Hingorani, and S. S. Patel. 1995. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl. Acad. Sci. USA 92:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enemark, E. J., and L. Joshua-Tor. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270-275. [DOI] [PubMed] [Google Scholar]
- 26.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 27.Gai, D., R. Zhao, D. Li, C. V. Finkielstein, and X. S. Chen. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119:47-60. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goetz, G. S., F. B. Dean, J. Hurwitz, and S. W. Matson. 1988. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263:383-392. [PubMed] [Google Scholar]
- 30.Gomis-Ruth, F. X., G. Moncallan, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 31.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 32.Hacker, K. J., and K. A. Johnson. 1997. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry 36:14080-14087. [DOI] [PubMed] [Google Scholar]
- 33.Höss, A., I. F. Moarefi, E. Fanning, and A. K. Arthur. 1990. The finger domain of simian virus 40 large T antigen controls DNA-binding specificity. J. Virol. 64:6291-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang, X., V. Klimovich, A. I. Arunkumar, E. B. Hysinger, Y. Wang, R. D. Ott, G. D. Guler, B. Weiner, W. J. Chazin, and E. Fanning. 2006. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 25:5516-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao, J., and D. T. Simmons. 2003. Nonspecific double-stranded DNA binding activity of simian virus 40 large T antigen is involved in melting and unwinding of the origin. J. Virol. 77:12720-12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson, A., and M. O'Donnell. 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74:283-315. [DOI] [PubMed] [Google Scholar]
- 37.Joo, W. S., H. Y. Kim, J. D. Purviance, K. R. Sreekumar, and P. A. Bullock. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaplan, D. L., M. J. Davey, and M. O'Donnell. 2003. Mcm4,6,7 uses a ‘pump in ring’ mechanism to unwind DNA by steric exclusion and actively translocates along a duplex. J. Biol. Chem. 278:49171-49182. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan, D. L., and M. O'Donnell. 2004. Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol. Cell 15:453-465. [DOI] [PubMed] [Google Scholar]
- 40.Kim, H.-Y., B. Y. Ahn, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, H. Y., B. A. Barbaro, W. S. Joo, A. Prack, K. R. Sreekumar, and P. A. Bullock. 1999. Sequence requirements for the assembly of simian virus 40 T antigen and T-antigen origin binding domain on the viral core origin of replication. J. Virol. 73:7543-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyl, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumor antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 43.Li, J., and T. Kelly. 1984. Simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. USA 81:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, H. J. L., R. H. Upson, and D. T. Simmons. 1992. Nonspecific DNA binding activity of simian virus 40 large T antigen: evidence for the cooperation of two regions for full activity. J. Virol. 66:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, J.-Y., and D. T. Simmons. 1991. Stable T-p53 complexes are not required for replication of simian virus 40 in culture or for enhanced phosphorylation of T antigen and p53. J. Virol. 65:2066-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loeber, G., R. Parsons, and P. Tegtmeyer. 1989. The zinc finger region of simian virus 40 large T antigen. J. Virol. 63:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loeber, G., J. E. Stenger, S. Ray, R. E. Parsons, M. E. Anderson, and P. Tegtmeyer. 1991. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J. Virol. 65:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loeber, G., M. J. Tevethia, J. F. Schwedes, and P. Tegtmeyer. 1989. Temperature-sensitive mutants identify crucial structural regions of simian virus 40 large T antigen. J. Virol. 63:4426-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat. Struct. Biol. 3:1034-1039. [DOI] [PubMed] [Google Scholar]
- 50.Manos, M. M., and Y. Gluzman. 1984. Simian virus 40 large T-antigen point mutants that are defective in viral DNA replication but competent in oncogenic transformation. Mol. Cell. Biol. 4:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McEntee, K., G. M. Weinstock, and I. R. Lehman. 1980. RecA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc. Natl. Acad. Sci. USA 77:857-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGeoch, A. T., M. A. Trakselis, R. A. Laskey, and S. D. Bell. 2005. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 12:756-762. [DOI] [PubMed] [Google Scholar]
- 53.McVey, D., S. Ray, Y. Gluzman, L. Berger, A. G. Wildeman, D. R. Marshak, and P. Tegtmeyer. 1993. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J. Virol. 67:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meinke, G., P. A. Bullock, and A. Bohm. 2006. The crystal structure of the T-antigen origin binding domain. J. Virol. 80:4304-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meinke, G., P. J. Phelan, S. Moine, E. Bochkareva, A. Bochkarev, P. A. Bullock, and A. Bohm. 2007. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PloS Biol. 5:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyata, T., K. Yamada, H. Iwasaki, H. Shinagawa, K. Morikawa, and K. Mayanagi. 2000. Two different oligomeric states of the RuvB branch migration motor protein as revealed by electron microscopy. J. Struct. Biol. 131:83-89. [DOI] [PubMed] [Google Scholar]
- 57.Moarefi, I. F., D. Small, I. Gilbert, M. Hopfner, S. K. Randall, C. Schneider, A. A. R. Russo, U. Ramsperger, A. K. Arthur, H. Stahl, T. J. Kelly, and E. Fanning. 1993. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J. Virol. 67:4992-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohr, I. J., M. P. Fairman, B. Stillman, and Y. Gluzman. 1989. Large T-antigen mutants define multiple steps in the initiation of simian virus 40 DNA replication. J. Virol. 63:4181-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O' Reilly, D. R., and L. K. Miller. 1988. Expression and complex formation of simian virus 40 large T antigen and mouse p53 in insect cells. J. Virol. 62:3109-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ott, R. D., Y. Wang, and E. Fanning. 2002. Mutational analysis of simian virus 40 T-antigen primosome activities in viral DNA replication. J. Virol. 76:5121-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peden, K. W. C., A. Srinivasan, J. M. Farber, and J. M. Pipas. 1989. Mutants with changes within or near a hydrophobic region of simian virus 40 large tumor antigen are defective for binding cellular protein p53. Virology 168:13-21. [DOI] [PubMed] [Google Scholar]
- 62.Peden, K. W. C., A. Srinivasan, J. V. Vartikar, and J. M. Pipas. 1998. Effects of mutations within the SV40 large T antigen ATPase/p53 binding domain on viral replication and transformation. Virus Genes 16:153-165. [DOI] [PubMed] [Google Scholar]
- 63.Ray, S., M. E. Anderson, G. Loeber, D. McVey, and P. Tegtmeyer. 1992. Functional characterization of temperature-sensitive mutants of simian virus 40 large T antigen. J. Virol. 66:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reese, D. K., G. Meinke, A. Kumar, S. Moine, K. Chen, J. L. Sudmeier, W. Bachovchin, A. Bohm, and P. A. Bullock. 2006. Analyses of the interaction between the origin binding domain from simian virus 40 T antigen and single-stranded DNA provide insights into DNA unwinding and initiation of DNA replication. J. Virol. 80:12248-12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reese, D. K., K. R. Sreekumar, and P. A. Bullock. 2004. Interactions required for binding of simian virus 40 T antigen to the viral origin and molecular modeling of initial assembly events. J. Virol. 78:2921-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynisdottir, I., and C. Prives. 1992. Two conditional tsA mutant simian virus 40 T antigens display marked differences in thermal inactivation. J. Virol. 66:6517-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 68.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schuck, S., and A. Stenlund. 2005. Assembly of a double hexamer helicase. Mol. Cell 20:377-389. [DOI] [PubMed] [Google Scholar]
- 70.SenGupta, D. J., and J. A. Borowiec. 1992. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science 256:1656-1661. [DOI] [PubMed] [Google Scholar]
- 71.Shen, J., D. Gai, A. Patrick, W. B. Greenleaf, and X. S. Chen. 2005. The roles of the residues on the channel β-hairpin and loop structures of simian virus 40 hexameric helicase. Proc. Natl. Acad. Sci. USA 102:11248-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simanis, V., and D. P. Lane. 1985. An immunoaffinity purification procedure for SV40 large T antigen. Virology 144:88-100. [DOI] [PubMed] [Google Scholar]
- 73.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 74.Simmons, D. T., G. Loeber, and P. Tegtmeyer. 1990. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J. Virol. 64:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sreekumar, K. R., B. A. Barbaro, A. Prack, and P. A. Bullock. 2000. Methods for studying interactions between simian virus 40 T-antigen and the viral origin of replication, p. 49-67. In L. Raptis (ed.), Methods in molecular biology: SV40 protocols. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- 76.Sreekumar, K. R., A. E. Prack, D. R. Winters, B. A. Barbaro, and P. A. Bullock. 2000. The simian virus 40 core origin contains two separate sequence modules that support T-antigen double-hexamer assembly. J. Virol. 74:8589-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stahl, H., P. Droge, and R. Knippers. 1986. DNA helicase activity of SV40 large tumor antigen. EMBO J. 5:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stenlund, A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. 4:777-785. [DOI] [PubMed] [Google Scholar]
- 79.Stillman, B., R. D. Gerard, R. A. Guggenheimer, and Y. Gluzman. 1985. T antigen and template requirement for SV40 DNA replication in vitro. EMBO J. 4:2933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stillman, B. W., and Y. Gluzman. 1985. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol. Cell. Biol. 5:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tack, L. C., C. A. Cartwright, J. H. Wright, W. Eckhart, K. W. C. Peden, A. Srinivasan, and J. M. Pipas. 1989. Properties of a simian virus 40 mutant T antigen substituted in the hydrophobic region: defective ATPase and oligomerization activities and altered phosphorylation accompany an inability to complex with cellular p53. J. Virol. 63:3362-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Titolo, S., E. Welchner, P. W. White, and J. Archambault. 2003. Characterization of the DNA-binding properties of the origin-binding domain of SV40 large T antigen by fluorescence anisotropy. J. Virol. 77:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valle, M., X. S. Chen, L. E. Donate, E. Fanning, and J. M. Carazo. 2006. Structural basis for the cooperative assembly of large T antigen on the origin of replication. J. Mol. Biol. 357:1295-1305. [DOI] [PubMed] [Google Scholar]
- 84.Valle, M., C. Gruss, L. Halmer, J. M. Carazo, and L. E. Donate. 2000. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 20:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.VanLoock, M. S., A. Alexandrov, X. Yu, N. R. Cozzarelli, and E. H. Egelman. 2002. SV40 large T antigen hexamer structure: domain organization and DNA-induced conformational changes. Curr. Biol. 12:472-476. [DOI] [PubMed] [Google Scholar]
- 86.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 87.Walmacq, C., A. R. Rahmouni, and M. Boudvillain. 2006. Testing the steric exclusion model for hexameric helicases: substrate features that alter RNA-DNA unwinding by the transcription terminator factor Rho. Biochemistry 45:5885-5895. [DOI] [PubMed] [Google Scholar]
- 88.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wiekowski, M., M. W. Schwarz, and H. Stahl. 1988. Simian virus 40 large T antigen DNA helicase. J. Biol. Chem. 263:436-442. [PubMed] [Google Scholar]
- 90.Wobbe, C. R., F. Dean, L. Weissbach, and J. Hurwitz. 1985. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc. Natl. Acad. Sci. USA 82:5710-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu, C., R. Roy, and D. T. Simmons. 2001. Role of single-stranded DNA binding activity of T antigen in simian virus 40 DNA replication. J. Virol. 75:2839-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu, H., N. Strater, W. Schroder, C. Bottcher, K. Ludwig, and W. Saenger. 2003. Structure of DNA helicase RepA in complex with sulfate at 1.95 A resolution implicates structural changes to an ‘open’ form. Acta Crystallogr. D 59:815-822. [DOI] [PubMed] [Google Scholar]
- 93.Yoon-Robarts, M., A. G. Blouin, S. Bleker, J. A. Kleinschmidt, A. K. Aggarwal, C. R. Escalante, and R. M. Linden. 2004. Residues within the B′ motif are critical for DNA binding by the superfamily 3 helicase Rep40 of the adeno-associated virus type 2. J. Biol. Chem. 279:50472-50481. [DOI] [PubMed] [Google Scholar]