Abstract
Chronic hepatitis C virus (HCV) infection is frequently associated with extrahepatic manifestations, including nonmalignant and malignant B-cell lymphoproliferative disorders. It has been reported that specific changes or recurring motifs in the amino acid sequence of the HCV hypervariable region 1 (HVR1) may be associated with cryoglobulinemia. We searched for specific insertions/deletions and/or amino acid motifs within HVR1 in samples from 80 symptomatic and asymptomatic patients with and 33 patients without detectable cryoglobulins, all with chronic HCV infection. At variance with the results of a previous study which reported a high frequency of insertions at position 385 of HVR1 from cryoglobulinemic patients, we found a 6.2% prevalence of insertions in samples from patients with and a 9.1% prevalence in those without cryoglobulinemia. Moreover, statistical and bioinformatics approaches including Fisher's exact test, k-means clustering, Tree determinant-residue identification, correlation of mutations, principal component analysis, and phylogenetic analysis failed to show statistically significant differences between sequences from cryoglobulin-negative and -positive patients. Our findings suggest that cryoglobulinemia may arise by virtue of as-yet-unidentified host- rather than virus-specific factors. Specific changes in HCV envelope sequence distribution are unlikely to be directly involved in the establishment of pathological B-cell monoclonal proliferation.
Hepatitis C virus (HCV) infection is characterized by a disturbingly high propensity to persist in the host, leading to chronic liver disease, cirrhosis, and hepatocellular carcinoma (2). Besides causing liver pathologies, HCV infection is frequently associated with extrahepatic manifestations such as mixed cryoglobulinemia and non-Hodgkin's B-cell lymphoma, which are characterized by B-cell proliferation and clonal expansion (13, 33). It is currently believed that more than 50% of patients with chronic HCV infection have asymptomatic cryoglobulinemia (19, 20). HCV-related B-cell proliferative disorders are thought to be a consequence of protracted antigenic stimulation, as has been shown for several lymphomas arising from chronic microbial infections (30). An indirect demonstration of this hypothesis derives from recent observations showing remission of splenic lymphomas following eradication of HCV infection (8, 16). The mechanisms by which HCV triggers selective B-cell clonal expansion are currently unknown, but it has recently been demonstrated that engagement of CD81 on human B cells by the virus' E2 envelope protein may be a key event (25). Why this occurs in some but not all patients with chronic HCV infection is still a matter of speculation.
In an effort to identify HCV-specific motifs potentially associated with the development of cryoglobulinemia, a prototypical B-cell lymphoproliferative disorder, a recent study reported a high prevalence of a single amino acid insertion at position 385 within HCV hypervariable region 1 (HVR1), which was found exclusively in samples from patients infected with HCV genotype 1b with symptomatic cryoglobulinemia (14). However, these provocative data were not confirmed in two subsequent studies (17, 24), and in only one of them were two positions within HVR1 and three within HVR2/CD81 associated with the presence of cryoglobulinemia (17). Unfortunately, these studies suffered from small sample sizes, and in addition, with one exception (24), only patients infected with genotypes 1a and 1b were examined (14, 17). In the present study we analyzed, by using statistical and bioinformatics approaches, a large cohort of patients with HCV infection caused by genotypes 1 and 2, with and without detectable cryoglobulins, for the presence of specific motifs within HVR1. We also analyzed the entire E2 sequence in samples from a subset of patients, to verify whether properties of other regions of the protein could be correlated with the phenotype.
MATERIALS AND METHODS
Patients.
One hundred and thirty-seven patients referred to infectious disease, hematology, and rheumatology clinics of tertiary care medical centers in Pavia and Rome, Italy, by primary care physicians or other physicians attending internal medicine units because of evidence of chronic HCV infection and/or symptoms compatible with cryoglobulinemia were considered for analysis of the HCV envelope region. HVR1 sequences could be successfully amplified in samples from 113 patients, who were then enrolled in the present study. Eighty of these (median age 64 years; range 37 to 87 years) had cryoglobulins that were detectable by conventional methods: 77 had type II and 3 had type III cryoglobulinemia. Fifty-four patients showed classical symptoms of cryoglobulinemia, including purpura and/or arthralgia and/or vasculitis and/or peripheral neuropathy, while 26 were asymptomatic. Thirty-three patients without detectable cryoglobulinemia (median age 64; range 38 to 85 years) served as controls. None of these had symptoms compatible with clinically manifest cryoglobulinemia, as assessed by taking the patients' clinical histories and by physical examinations carried out by specialist physicians. Sixty-three patients with HCV infections, 46 with and 17 without cryoglobulinemia, had genotype 2a/c and 50 patients, 34 with and 16 without cryoglobulinemia, had genotype 1b. The HCV genotypes were determined by using an INNO-LiPA HCV II assay (Bayer Corp., Tarrytown, NY). Liver biopsy samples from 52 of 113 patients were available and showed histological features of mild-to-moderate chronic hepatitis, with cirrhosis in 17 (11 of these patients had HCV type 1b and 6 had type 2a/c). Only 7 of 113 patients were treated with standard interferon alpha with or without Ribavirin, and 15 patients received corticosteroid treatment prior to enrollment in our study.
Informed consent to donate a blood sample was obtained from all patients. Patients' blood samples were processed according to a rigorous protocol to maintain a constant temperature of 37°C, to avoid coprecipitation and partitioning of virions with cryoglobulins, which may interfere with HCV RNA amplification (1). Sera were immediately frozen in sterile cryovials and stored at −80°C until used, as previously described (7). To insure complete solubilization of cryoglobulins upon thawing, sera were maintained at 37°C for 30 min prior to RNA extraction with a QIAamp viral RNA mini kit (QIAGEN, Valencia, CA).
The study protocol conformed with the ethical guidelines of the 1975 Declaration of Helsinki and was specifically approved by the Institutional Review Board and Ethical Committee of Fondazione IRCCS Policlinico San Matteo, Pavia (Coordinating Center) on 28 June 2004.
Amplification of HVR1 in the E2 region of HCV genotypes 1b and 2a/c by PCR, cloning, and sequencing.
E2-HVRI sequences were amplified by nested reverse transcription-PCR using AmpliTaq DNA polymerase (Applied Biosystems) as previously reported (5). Two sets of primers were used for separate amplification of the genotype 1b and 2a/c sequences, yielding final DNA fragments of 176 bp, spanning nucleotides (nt) 1428 to 1603, numbered according to the sequence of reference strain H77 (GenBank accession no. AF009606) in agreement with the recommendations of an international panel of experts (29). The genotype-1b HVR1 primer set was Outer AS (nt 1612 to 1633) 5′-TCATTGCAGTTCAGGGCAGTCC-3′, Outer S (nt 1395 to 1413) 5′-CACTGGGGAGTCCTGGCGG-3′, Inner AS (nt 1584 to 1603) 5′-TGCCAGCTGCCGTTGGTGTT-3′, and Inner S (nt 1428 to 1447) 5′-TCCATGGTGGGGAACTGGGC-3′. The genotype 2a/c primer set was Outer AS (nt 1614 to 1634) 5′-GTCATTGCAATTCAGGGCAGT-3′, Outer S (nt 1395 to 1414) 5′-CACTGGGGCGTGATGTTTGG-3′, Inner AS (nt 1585 to 1603) 5′-TGCCAACTGCCATTGGTGT-3′, and Inner S (nt 1428 to 1446) 5′-TCCATGCAGGGAGCGTGGG-3′.
The amplification products were analyzed by gel electrophoresis, purified by using a QIAquick PCR purification kit (QIAGEN, Valencia, CA), and cloned using a TA cloning kit (Invitrogen, Leek, The Netherlands). Sequencing was performed on PCR products from at least 10 to 12 clones per patient, for a total number of 1,207 clones, by using the Big Dye terminator cycle sequencing kit on an ABI 310 genetic analyzer (Applied Biosystems, Foster City, CA) according to the manufacturers' instructions. The deduced amino acid sequences of 58 residues from all clones were aligned with published HVR1 sequences from HCV genotypes 1b and 2a/c and examined for mutations, insertions, or deletions and for specific amino acid motifs. From the 1,207 clones, 449 unique, nonrepetitive HVR1 sequences were obtained.
Denaturing polyacrylamide gel electrophoresis of E2-HVR1 nested-PCR products.
The HVR1-amplification products of 176 bp obtained after the second round of nested PCR were immediately analyzed using a rapid screening method that would solve differences of at least 3 nt, i.e., insertions or deletions of 1 or more amino acids (aa). For optimal separation of the PCR amplicons, a 5% denaturing polyacrylamide gel electrophoresis (PAGE) solution was set up as follows: 3.75 ml acrylamide/bisacrylamide (19:1 vol/vol) 40% (Sigma-Aldrich, St. Louis, MO), 15 g urea (Fluka; Sigma-Aldrich), 3 ml of 10-fold-concentrated Tris-borate-EDTA buffer (Invitrogen), 150 μl 10% ammonium persulfate (Sigma, Sigma-Aldrich), 20 μl N,N,N′,N«-tetramethylethylenediamine (Sigma, Sigma-Aldrich) in a final solution of 30 ml. The gel solution was poured in a layer of 0.4 mm between two glass plates measuring 25 × 20 cm. A small amount of each PCR product was loaded and run near a specific molecular marker 176 bp in length (representing wild-type HVR1) and markers of 176 bp plus multiples of 3 nt. The electrophoresis was run for approximately 90 min at 950 V, and the cDNA bands were visualized by silver staining.
PCR titration.
Since previous findings suggested that a significant number of patients with cryoglobulinemia showed a 1-aa insertion at position 385 within HVR1, two fragments of different lengths, 176 bp and 179 bp, were ligated into a vector using the TA cloning kit (Invitrogen, Leek, The Netherlands). To assess the sensitivity of our PCR assay for detecting insertion/deletion variants of HVR1, gel titration experiments were performed using variable starting ratios of a wild-type (176 bp) and an insertion variant (179 bp) cloned sequence. The mixture was amplified by PCR and loaded on a denaturing PAGE gel under the same conditions mentioned above. The intensities of the two bands of 176 bp and 179 bp obtained from the PCR were subsequently analyzed. These experiments showed that it was possible to detect a 179-bp variant within a pool of 30 wild-type sequences. The ratio of staining intensities of wild-type and variant HVR1 from each patient was used to assess the number of HVR1 clones for a model to appropriately represent each pool of sequences.
Amplification by PCR, cloning, and sequencing of complete E2 of HCV.
The complete envelope 2 region (E2) region of HCV was also analyzed for 58 of the patients: 29 patients with genotype 1b (21 with and 8 without detectable cryoglobulins) and 29 patients with genotype 2a/c (21 with and 8 without detectable cryoglobulins). Two primer sets spanning the entire E2 region were used to separately amplify genotype 1b and 2a/c sequences, yielding final PCR products of 1,170 nt for genotype 1b and 1,192 nt for genotype 2a/c (nt 1428 to 2579; numbered according to reference sequence H77, GenBank accession no. AF009606). The genotype 1b E2 primer set was Outer AS (nt 2637 to 2659) 5′-CAGAAGAACACAAGGAAGGAGAG-3′,Outer S (nt 1395 to 1413) 5′-CACTGGGGAGTCCTGGCGG-3′, Inner AS (nt 2574 to 2597) 5′-CACCAGGTTCTCTAAGGCGGCCTC-3′, and Inner S (nt 1428 to 1447) 5′-TCCATGGTGGGGAACTGGGC-3′. The genotype 2a/c primer set was Outer AS (nt 2661 to 2682) 5′-GACCTTTAATACACCAAGCGGC-3′ and 5′-GACCCTTGATGTACCAAGCA(T)GC-3′, Outer S (nt 1395 to 1414) 5′-CACTGGGGCGTGATGTTTGG-3′, Inner AS (nt 2586 to 2607) 5′-CATGCAAGAT(C)GACCAG(A)CTTCTC-3′, and Inner S (nt 1428 to 1446) 5′-TCCATGCAGGGAGCGTGGG-3′.
The PCR products were cloned as above and approximately 8 clones for each of the 58 patients were isolated and sequenced, for a total of 449 sequences which yielded 269 unique E2 amino acid sequences (about 4 to 5 different clones for each patient). Of these, 114 sequences were extracted and added to the previous 449 unique HVR1 sequences obtained with the HVR1 primer sets, giving a total of 563 different HVR1 sequences; 548 of these were used for bioinformatics analysis as described below.
Statistical and bioinformatics analysis.
We analyzed both the complete data set including all available nonredundant protein sequences and a reduced subset including 111 representative HVR1 sequences, one for each patient studied (data from 2 patients with cryoglobulinemia infected with genotype 1b were lost during electronic transfer of the database). The latter was constructed using, for each patient, the sequence with the highest sequence identity to each other sequence from the same patient.
Fisher's exact test was employed to examine whether there was any position within HVR1 that showed a composition that differed statistically between the sets of positive and negative data, i.e., to discriminate between patients with and without cryoglobulinemia; between the sets of data for genotypes 1b and 2a/c; and between patients with and without cryoglobulinemia within each genotype. The Fisher exact test, applied to two independent samples, was used to provide a measure for the probability that the data belonged to the same distribution. The k-means clustering method, with k varying from 1 to 5, was employed to detect clusters of sequences that discriminated significantly between patients with and without cryoglobulinemia. To determine whether the sequences could be divided into families by searching for positions that had a specific distribution in some members of our data set, the tree determinant-residue identification (Treedet) method (11), which can detect such cases on the basis of a statistical analysis of multiple-sequence alignments, was used. Correlations of mutations were also sought in the alignment of positive and negative samples, in order to see whether second-order effects could be responsible for the phenotype, i.e., whether there was any pair of positions that would vary in a correlated fashion in each of the two data sets (sequences from cryoglobulinemic versus noncryoglobulinemic patients), with the aim of comparing them (15). Principal component analysis (PCA) (9) was also applied to the frequency table obtained from our data. This mathematical procedure transforms a number of (possibly) correlated variables into a (smaller) number of uncorrelated variables called principal components. The first principal component accounts for as much of the variance in the data as possible, and each succeeding component accounts for as much of the remaining variance as possible. With this approach, it is possible to examine how many and which of the input variables are noncorrelated and can therefore be used to best separate the data. Each data set was first considered as a single string obtained by concatenating the frequency of occurrence for all 20 amino acids in the 27 positions. The first PCA components should indicate which independent variables, i.e., position and amino acid, can better separate the positive and negative samples. A PCA analysis was also performed on the frequency tables for each of the 27 positions in the two data sets. A sequence logo representation of the positions that appear more distant in the PCA analysis (see Results) was obtained by using Weblogo version 2.8.2 (http://weblogo.berkeley.edu/) (10, 27) for the 350 sequences from patients with and the 198 sequences from patients without detectable cryoglobulinemia, using default parameters and a bitmap resolution enhanced to 600 dpi. Phylogenetic trees were constructed using the neighbor-joining method (26) implemented in the Phylip package, version 3.66 (12). One hundred trials of bootstrap analysis were performed.
Nucleotide sequence accession numbers.
All 563 nonrepetitive HVR1 nucleotide sequences have been submitted to GenBank and were assigned accession nos. EF198910 to EF199472.
RESULTS
Rapid analysis by PAGE of nucleotide insertions/deletions within E2-HVR1 nested-PCR products.
Insertions or deletions of 3 nt within nested amplicons of E2-HVR1 of both genotype 1b and 2a/c in each of the 113 patients ' samples tested could be reliably and efficiently identified by denaturing PAGE. As expected, we found bands of 176 bp (wild-type HVR1) and insertions/deletions of 3 nt or multiples thereof. Insertions/deletions not multiples of 3 nt were not found, as this would result in a sequence frameshift in the protein. Of 113 patients' samples analyzed by denaturing PAGE, only 7 samples, 2 from patients with genotype 1b and 5 from patients with genotype 2a/c, showed a band pattern corresponding to insertions/deletions. Of the two patients infected with genotype 1b, one (patient 17) showed one 185-bp extra band (insertion of 9 nt) and one (patient 171) showed a 173-bp extra band (deletion of 3 nt). The samples from the five patients with genotype 2a/c showed different nucleotide insertions: that from patient 28 had a 188-bp extra band (insertion of 12 nt); that from patient 169 showed a 191-bp band exclusively (insertion of 15 nt); that from patient 183 had a 185-bp extra band (insertion of 9 nt); that from patient 191 had two extra bands, 185 bp (insertion of 9 nt) and 182 bp (6 nt); and that from patient 193 showed only a single 185-bp band (insertion of 9 nt).
These results were confirmed by cloning and sequencing (see below). PAGE was also tested as a possible approach for the rapid identification of the previously reported insertion at position 385, and Fig. 1 shows representative findings from three patients with HVR1-length polymorphisms and 10 with wild-type HVR1 sequences. Sequencing of 10 to 12 clones from each patient also revealed a correspondence between the intensity of the extra bands and the relative number of clones carrying the insertion, although the sequencing data were obviously more accurate.
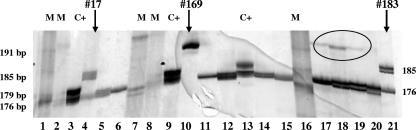
FIG. 1.
E2-HVR1 amplified PCR products analyzed by denaturing PAGE. Lanes: 1, 2, 7, 8, and 16, molecular markers; 3, 9, and 13, 176-bp and 179-bp positive controls; 4, result for patient 17 (genotype 1b; cryoglobulin negative) showing two bands corresponding to mutated (insertion of 9 nt; 185 bp) and wild-type (176 bp) HVR1 amplicons; 5 and 6, results for patients 16 and 22 (wild-type genotype 1b); 10, result for patient 169 (genotype 2a/c; cryoglobulin positive) showing a single band of mutated HVR1 (insertion of 15 nt; 191 bp); 11 and 12, results for patients 6 and 8 (wild-type genotype 2a/c); 14, 15, and 17 to 20, results for patients with wild-type E2-HVR1 (176 bp); and 21, result for patient 183 (genotype 2a/c; cryoglobulin negative) showing two bands, one of which has an insertion of 9 nt (185 bp). M, molecular marker; C+, positive control (176 and 179 bp). Arrows indicate products from patients with mutated HVR1 fragments; the circle indicates fragments including one of the outer primers. Molecular sizes are indicated on the left.
HVR1 sequences in samples from patients with and without detectable cryoglobulins.
The E2-HVR1 sequences from 34 patients infected with genotype 1b and 46 patients with genotype 2a/c with detectable cryoglobulinemia were cloned, and 10 to 12 clones from each patient were sequenced and translated into the deduced amino acid sequence. The 58-aa E2-HVR1 sequences (residues 363 to 420) encompassing HVR1 (residues 384 to 410) derived from all clones were aligned and analyzed for amino acid insertions or deletions and types of substitution. For the sake of clarity, the results are summarized in Table 1. Of the samples from the two patients infected with genotype 1b showing amino acid insertions, one (from patient 171) revealed an uncommon amino acid pattern which is compatible with two hypotheses: a highly conserved threonine shifted to position 384 and a proline at position 385, or a deletion at position 384 such that the threonine would be shifted to position 385, resulting in one amino acid insertion (proline) at position 386. Three of the samples from the 46 patients infected with genotype 2a/c showed insertions within HVR1 (Table 1).
TABLE 1.
Amino acid insertions in HVR1 in patients with and without cryoglobulinemia
| Group and patient (HCV genotype) | Insertion | Position | No. of clones carrying changes/total no. of clones |
|---|---|---|---|
| Cryoglobulinemic | |||
| 117 (1b) | TR | 387 | 1/20 |
| 171 (1b) | (See text) | (See text) | (See text) |
| 28 (2a/c) | GLSL | 404 | 9/11 |
| GLTL | 404 | 1/11 | |
| 169 (2a/c) | ASSSM | 384 | 8/12 |
| SSPTA | 385 | 3/12 | |
| SSPMA | 385 | 1/12 | |
| 193 (2a/c) | GAG | 385 | 9/10 |
| GTV | 385 | 1/10 | |
| Noncryoglobylinemic | |||
| 17 (1b) | GPG | 385 | 4/12 |
| ELG | 385 | 2/12 | |
| 191 (2a/c)a | ARY | 385 | 6/11 |
| TRQ | 385 | 1/11 | |
| ARQ | 385 | 3/11 | |
| TRR | 385 | 1/11 | |
| 183 (2a/c) | RTV | 384 | 2/10 |
| RKT | 384 | 1/10 | |
| RTA | 384 |
For four more sequences, alignments are ambiguous. An insertion of 3 aa can, however, be localized in the position comprising aa 384 to aa 387.
Thirty-three HCV-positive patients without detectable cryoglobulinemia served as negative controls for this analysis. Insertions were identified in sequences from three patients from this group: one with genotype 1b and two with genotype 2a/c (Table 1).
In summary, 1,656 clones from 113 patients were sequenced; 1,207 sequences were amplified using HVR1-specific primers, whereas 449 were obtained after amplification with primers of the entire E2-region sequence. From these, 548 nonrepetitive HVR1 sequences from 111 of the 113 patients enrolled (15 sequences from 2 patients with cryoglobulinemia infected with genotype 1b were lost during electronic transfer of the database) were extracted and subjected to further analysis. This extensive analysis of a large number of HVR1 sequences revealed the occurrence of insertions in sequences from 6.2% of cryoglobulinemic patients and from 9.1% of noncryoglobulinemic ones. The occurrence was higher in sequences from patients with genotype 2a/c (6.5% of those with and 11.8% of those without cryoglobulins) than in sequences from patients infected with genotype 1b (5.9% with versus 6.2% without cryoglobulins, respectively). There was no correlation between the presence of mutations within HVR1 and viral load, serum alanine aminotransferase and gammaglobulin, patient age, sex, and the presence or absence of symptoms of cryoglobulinemia.
Analysis of the hydropathicity profile (18) of HVR1 revealed a hydrophobicity plot common to all sequences of both genotype 1b and 2a/c, with a variable N-terminal region and a more conserved C-terminal region, in agreement with previously published findings showing conformational conservation of HVR1 (22 and data not shown).
Complete E2-region sequences.
Complete sequences of the E2 region (aa 384 to 746) were derived from 58 patients. A total of 449 clones were sequenced, 269 of which showed amino acid changes in the E2 region (mean of 4.5 clones per patient). The sequences were aligned and compared to the published HCV genotype 1b (J, BK, and H, the latter having a recognized higher affinity for CD81) and 2a/c (BEBE1/2c and JCH-2/2a) strains. Amino acid changes within E2 were compared for the two groups of patients with and without detectable cryoglobulins. A single amino acid insertion (proline) was found at position 576 for a single patient (patient 162) with cryoglobulinemia. As expected, conserved and nonconserved amino acid substitutions were found in the E2 region, predominantly within HVR1 and HVR2 and between these two regions, but no clustering of changes was observed for cryoglobulinemic patients. In particular, specific sequence changes within the CD81-binding site were not observed for cryoglobulinemic subjects. Conserved regions previously described by others were confirmed (the WHY motif, the 502-to-520-aa region, and all cysteine residues).
Analysis of HVR1 region by bioinformatics tools.
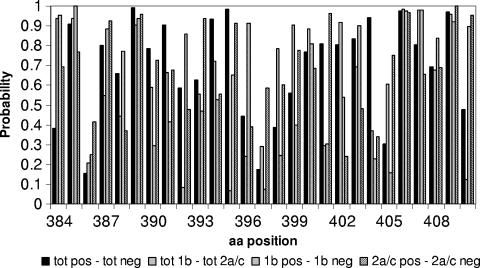
The aim of this analysis was to evaluate whether a correlation exists between a cryoglobulinemic phenotype in patients with chronic HCV infection and changes within the HVR1. The large set of sequences from 111 of the 113 patients enrolled in this study and the subset containing only one representative sequence per patient (see Materials and Methods) were investigated using several methods. It is important to emphasize that our data set contained a substantial number of sequences, so that our results cannot simply be ascribed to sampling limitations. As a first step, we performed Fisher's exact test to determine whether there was any position within HVR1 that showed a composition that differed statistically between the sets of positive (i.e., sequences from cryoglobulinemic patients) and negative (i.e., sequences from noncryoglobulinemic patients) data. For the sake of comparison, the same analysis was performed to discriminate between patients infected with genotypes 1b and 2a/c, and, within each genotype, between patients with and without cryoglobulinemia. The F-test, when applied to two independent samples, provides an estimate of the probability that they belong to the same distribution. All the positions have probability values well above the significance threshold (5% divided by the 27 comparisons performed) (Fig. 2); thus, the compared samples cannot be assigned to different distributions.
FIG. 2.
Graph reporting the probability (y axis) that the observed distribution frequency at each position (x axis) of HVR1 is statistically indistinguishable between the two compared distributions, derived from Fisher's exact test applied to the sets of nonredundant protein sequences.
Next, we asked whether we could detect clusters to which negative and positive samples could be assigned. However, k-means clustering methods (see Materials and Methods), with k varying from 1 to 5, could not detect any significant separation of the two sets. We then tried to determine whether sequences could be divided into families by searching for positions that had a specific distribution in some members of our data set. Using the Treedet method, no significant position could be highlighted in our sample. In addition, analysis for correlated mutations in the complete E2-sequence alignment of positive and negative samples failed to detect any correlated positions in both alignments.
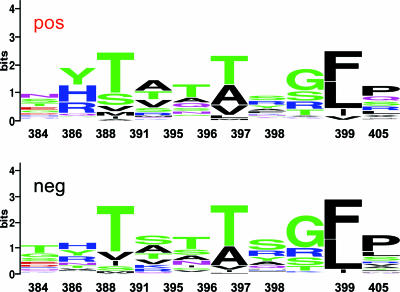
Next, we performed several PCAs (see Materials and Methods). In our setting, the first component was sufficient to explain most of the variance of the data and none of the variables could be considered significantly more discriminatory than the others. We report here the most meaningful ones, with the caveat that they are not statistically more significant than the others: A in position 493, N in position 384, Y in position 386, G in position 398, K in position 410, P in position 405, S in position 391, T in position 384, and T in position 396. We also repeated the PCA on the frequency tables for each of the 27 positions in the two data sets (sequences derived from samples from cryoglobulinemia-positive and -negative patients). In this case, several components (at least five) need to be used to explain 80% of the variance of the data. The positions that appear more distant between the two sets of data in the PCA (i.e., the positions where the amino acid composition differs more) are 384, 386, 389, 392, 396, 397, 398, 399, and 405. We concentrated on these positions and manually analyzed the frequency matrix. As can be seen from Table 2 and Fig. 3, the differences between negative and positive sequences in these positions were not statistically significant, and specifically, there was no case where the composition was completely nonoverlapping between positive and negative samples. Therefore, even if this effect were significant, it would not have sufficient predictive power to identify patients with cryoglobulinemia.
TABLE 2.
Frequency of occurrence of the 20 amino acids in the subset of positions highlighted by PCA of the 27 positions in the two data setsa
| Amino acid | % of amino acid in indicated position for positive and negative dataset
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 384
|
386
|
388
|
391
|
395
|
396
|
397
|
398
|
399
|
405
|
|||||||||||
| Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | |
| A | 3 | 2 | 0 | 2 | 3 | 2 | 27 | 21 | 14 | 14 | 31 | 27 | 10 | 11 | 3 | 0 | 0 | 0 | 4 | 6 |
| C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| D | 8 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| E | 12 | 10 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 4 | 0 | 0 | 44 | 54 | 3 | 4 |
| G | 10 | 15 | 1 | 1 | 0 | 0 | 1 | 0 | 5 | 6 | 0 | 0 | 5 | 2 | 43 | 56 | 0 | 0 | 0 | 0 |
| H | 6 | 2 | 32 | 24 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| I | 0 | 0 | 0 | 2 | 4 | 8 | 1 | 0 | 1 | 7 | 1 | 4 | 0 | 0 | 0 | 1 | 10 | 5 | 1 | 1 |
| K | 1 | 4 | 1 | 0 | 0 | 0 | 3 | 9 | 1 | 0 | 0 | 0 | 6 | 3 | 2 | 3 | 0 | 0 | 0 | 0 |
| L | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 5 | 2 | 3 | 3 | 1 | 1 | 38 | 40 | 10 | 12 |
| M | 0 | 0 | 0 | 4 | 6 | 4 | 4 | 1 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 3 | 3 |
| N | 18 | 5 | 0 | 10 | 0 | 0 | 1 | 1 | 9 | 13 | 0 | 0 | 5 | 2 | 1 | 1 | 0 | 0 | 1 | 0 |
| P | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 30 | 43 |
| Q | 5 | 5 | 5 | 2 | 0 | 0 | 4 | 5 | 13 | 2 | 0 | 0 | 4 | 9 | 2 | 0 | 0 | 0 | 14 | 3 |
| R | 4 | 6 | 20 | 19 | 0 | 0 | 8 | 9 | 1 | 0 | 0 | 0 | 17 | 17 | 15 | 16 | 0 | 0 | 13 | 7 |
| S | 17 | 13 | 1 | 2 | 18 | 7 | 11 | 25 | 9 | 15 | 0 | 1 | 21 | 33 | 19 | 14 | 0 | 0 | 14 | 12 |
| T | 13 | 29 | 2 | 9 | 60 | 68 | 21 | 16 | 37 | 35 | 45 | 60 | 1 | 0 | 12 | 9 | 0 | 0 | 2 | 5 |
| V | 1 | 1 | 2 | 6 | 9 | 13 | 19 | 14 | 2 | 6 | 15 | 6 | 1 | 0 | 1 | 1 | 6 | 1 | 5 | 6 |
| W | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Y | 2 | 1 | 33 | 20 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 |
The data sets comprise the sequences derived from samples from cryoglobulinemia-positive (Pos), and -negative (Neg) patients. The values in bold refer to amino acids in specific positions identified as meaningful, albeit with low weight, by the PCA performed on the entire set of data.
FIG. 3.
WebLogo (7, 25) of the positions highlighted by the PCA. Top, positive (cryoglobulinemic) sequences; bottom, negative (noncryoglobulinemic) sequences. The taller the letter, the higher the frequency of the amino acid in that position.
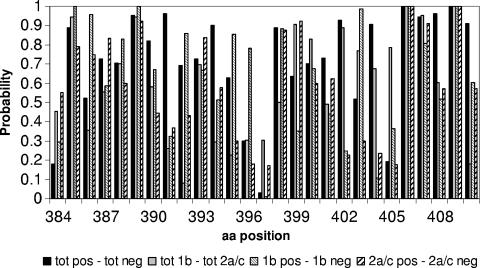
The same analyses, performed on the subset of 111 representative HVR1 sequences, gave analogous results; the results of Fisher's exact test are shown in Fig. 4).
FIG. 4.
Graph reporting the probability (y axis) that the observed distribution frequency at each position (x axis) of HVR1 is statistically indistinguishable between the two compared distributions, derived from Fisher's exact test applied to the sets of protein sequences representative of the 111 patients.
Neighbor-joining trees have been constructed both for the whole set of nonredundant sequences and for the subset of 111 representative sequences. One hundred bootstrap trials were performed for each data set. Most of the sequences isolated from the same patient clustered together in the majority of the bootstrap trials. The phylogenetic analysis did not reveal any pattern that could be associated with the cryoglobulinemic phenotype. No clustering of the nonredundant sequences from patients with the same phenotype (or genotype) was observed in the majority of trials. The only exceptions were represented by six sequences from four patients (8, 9, 102, and 142; all cryoglobulinemic and infected with genotype 2ac—2 sequences of the 10 isolated from patient 8 gave two separated clusters with 2 of the 3 sequences isolated from patient 102, and 1 of the 4 sequences from patient 9 clusters with one of the 4 sequences from patient 142). Similarly, when the representative sequences were considered, no significant clustering of sequences from patients sharing the same phenotype could be observed, but one group of three sequences (from patients 9, 142, and 172) and three groups of two sequences (from patients 61 and 91, 26 and 156, and 112 and 193) all occurred in cryoglobulinemic patients infected with genotype 2a/c (data not shown).
DISCUSSION
We report here an extensive analysis of a large HCV HVR1-sequence database obtained from carefully characterized patients with cryoglobulinemia. Unfortunately, only limited HCV sequence data are available from accurately characterized patients. Previous studies reported on small series of 14 to 21 patients (14, 17, 24), and the viral genotype could only be clearly assigned in two papers (14, 24). In this study, we recruited a large number of patients with HCV genotypes 1b and 2a/c, the two most prevalent genotypes in Italy (4). Our data clearly indicate that the single amino acid insertion at position 385 of HVR1, which was reported to occur at significantly greater frequency in patients with cryoglobulinemia (14), was never detected in our series. Longer insertions of 2 to 5 aa were found at position 384 or 385 in samples from approximately 6% of patients but were similarly represented in those with and without evidence of cryoglobulinemia. Moreover, no difference was found between samples from patients with symptomatic and asymptomatic cryoglobulinemia.
In consideration of these negative findings, we set out to analyze the possible existence and significance of HVR1-sequence-specific motifs that would discriminate between patients with and without cryoglobulinemia. Using several bioinformatics approaches, no statistically significant differences were found, indicating that HVR1 sequencing is not useful for the identification of a cryoglobulinemic phenotype. Our findings are in contrast with those of Hofmann and coworkers (17), who reported that positions 386, 387, and 396 within HVR1 could be predictive of cryoglobulinemia in a rather small number of patients with detectable cryoprecipitate. However, the authors used a correlation coefficient analysis, which is less sensitive than Fisher's exact test, and a classifier that is not described in detail, which may have influenced their conclusions. Our data do not support these results and lead to the conclusion that the observed differences likely reflect biases in their data set. Our data are instead comparable with those of Rigolet and associates (24), who failed to detect molecular features associated with B-cell clonal expansion and cryoglobulinemia. In this study, phylogenetic analysis did not reveal clustering associated with lymphoproliferative disorders, nor were the N-terminal insertions or residues at positions 4 and 13 discriminative for such conditions. Moreover, the high frequency of the insertion at position 385 of the HVR1 deduced amino acid sequence could not be ascribed to genotype 1, since in our study we examined more than twice as many patients infected with the same genotype, from whom a large number of cloned sequences were derived, and found only a low prevalence of insertions in that position. The frequency of changes in sequences from genotype 2-infected patients was higher than that found in sequences from genotype 1, particularly in clones derived from patients without cryoglobulinemia. These findings most likely reflect a stronger positive selection of HVR1 variants in patients infected by genotype 2 than in those infected by genotype 1, as reported previously by our group (6, 21). Evidence supporting the contention that HVR1 may be under positive selection is currently derivative rather than direct, although we are not aware of alternative evidence supporting random variation or genetic drift as being responsible for the emergence of HCV variants. Supportive evidence for HVR1 being under selective pressure in the context of a relatively stable HCV replication rate comes from the following: (i) rapid and continuous build-up of quasispecies within individual patients; (ii) higher variation in HVR1 than in other HCV regions; (iii) HVR1 evolutionary rates that differ among patients; and (iv) a higher dN/dS (nonsynonymous mutation/synonymous mutation) ratio within HVR1 than in other regions. Studies on the pathogenetic mechanisms responsible for variant selection in this setting emphasized the importance of B-cell responses (21, 31) in the active selection process, in agreement with a study which showed lower nucleotide and amino acid variation in HCV isolates from patients with genetic immunoglobulin defects than in isolates from control HCV patients (5), but failed to provide a plausible biological explanation for this finding beyond the well-recognized associations of genotype 2 with mild liver disease (23, 28) and better responses to antiviral treatment (3).
According to the results of our analysis, and because of the large number of sequences examined, it is highly unlikely that HVR1-sequence-specific motifs are associated with an initial lymphoproliferative disorder such as that associated with cryoglobulinemia. Recently, significant compartmentalization of HVR1 variants was observed in the peripheral blood mononuclear cells and cryoprecipates from a small number of patients with cryoglobulinemia (32). Interestingly, a large insertion of 5 aa encoded by codons 385 to 389, akin to that from 1 of our patients, was detected for 1 of the 10 patients with cryoglobulinemia that were studied. Also, phylogenetic analysis of the HVR1 quasispecies revealed a significantly greater distance between variants for genotype 2-infected patients than for those infected with genotype 1, irrespective of the presence or absence of cryoglobulins, in agreement with our previously published data (6). Although our study was not specifically designed to examine differences between HVR1 quasispecies compositions in the supernatants and cryoprecipitates, an issue which was carefully addressed in other studies such as that mentioned above (32), the methodological approach we followed with this large series of patients allowed for the amplification of variants representative of the entire serum virus population.
Analysis of the entire E2 sequence for a proportion of patients also failed to establish a link between HCV envelope motifs and cryoglobulinemia, suggesting that, at least in serum, no clustering of specific mutations, including potentially important regions such as the CD81-binding site, exists in this setting.
Why, then, do over two-thirds of patients with chronic HCV infection develop B-cell lymphoproliferative disorders which, in a proportion of cases, may be clinically relevant? It is conceivable that the initially nonneoplastic monoclonal expansion characteristic of this condition may be a consequence of protracted antigenic stimulation, as frequently observed in several chronic viral infections (30). This phenomenon is thought to occur via the engagement of a widely distributed tetraspanin, CD81, with the HCV E2 protein, which would reduce the B-cell activation threshold (25), providing a plausible explanation for the antigen-dependent autoantibody production and B-cell clonal expansion which may occur as extrahepatic manifestations of chronic HCV infection.
In summary, extensive analysis of the HCV E2 envelope protein and, in particular, of HVR1, which is known to be the target of immune selection contributing to the characteristic HCV quasispecies distribution, failed to reveal sequence-specific motifs which were allegedly associated with cryoglobulinemia. Since B-cell activation appears to be a general feature of HCV infection (25 and our own unpublished data), B-cell lymphoproliferative disorders and the related inappropriate generation of cryoglobulins may arise following as-yet-unidentified host rather than virus-specific factors. Sequence variation in the HVR1 region of HCV most likely results from Darwinian selection regulated by the host immune response. HVR1 sequence changes are therefore secondary to the establishment of a specific immune selection. Skewing in HVR1-sequence distribution in cryoglobulinemia may reflect skewed immune responses, which probably follow rather than precede the establishment of pathological B-cell monoclonal proliferation.
Acknowledgments
This work was supported by a grant from the Italian Ministry of Universities and Research MiUR (Progetti di Ricerca di Interesse Nazionale-PRIN), by Research Funds of the Italian Ministry of Health (Ricerca Corrente, Fondazione IRCCS Policlinico San Matteo), by an unrestricted research grant from Schering-Plough, Italy, and by a generous donation from CANTEL, S.p.A, Fino Mornasco, Italy. G.B. was supported by a contract from the Ministry of Health and A.C. by a contract from Schering-Plough. C.B. was supported by a postdoctoral grant from Istituto Pasteur-Fondazione Cenci Bolognetti. R.O. is a recipient of a postdoctoral grant from Centro Linceo Interdisciplinare “Beniamino Segre,” Accademia dei Lincei. A.T. and R.O. thank “BICG—Bioinformatics tools for identifying, understanding, and attacking targets in cancer,” AIRC 2004/IFOM, for financial support.
We thank Stefania Varchetta, Area Infettivologica, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy, for help with submission of cDNA sequences to the NCBI database; Massimo Cugno, Department of Internal Medicine, University of Milan, for providing control sera; and Lara Firmo for editorial assistance.
Footnotes
Published ahead of print on 21 February 2007.
REFERENCES
- 1.Agnello, V., R. T. Chung, and L. M. Kaplan. 1992. A role for hepatitis C virus infection in type II cryoglobulinemia. N. Engl. J. Med. 327:1490-1493. [DOI] [PubMed] [Google Scholar]
- 2.Alberti, A., L. Chemello, and L. Benvegnù. 1999. Natural history of hepatitis C. J. Hepatol. 31(suppl. 1):17-24. [DOI] [PubMed] [Google Scholar]
- 3.Alberti, A. 2005. Towards more individualised management of hepatitis C virus patients with initially or persistently normal alanine aminotransferase levels. J. Hepatol. 42:266-274. [DOI] [PubMed] [Google Scholar]
- 4.Ansaldi, F., B. Bruzzone, S. Salmaso, M. C. Rota, P. Durando, R. Gasparini, and G. Icardi. 2005. Different seroprevalence and molecular epidemiology patterns of hepatitis C virus infection in Italy. J. Med. Virol. 76:327-332. [DOI] [PubMed] [Google Scholar]
- 5.Booth, J. C., U. Kumar, D. Webster, J. Monjardino, and H. C. Thomas. 1998. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology 26:223-227. [DOI] [PubMed] [Google Scholar]
- 6.Brambilla, S., G. Bellati, M. Asti, A. Lisa, M. E. Candusso, M. D'Amico, G. Grassi, M. Giacca, A. Franchini, S. Bruno, G. Ideo, M. U. Mondelli, and E. M. Silini. 1998. Dynamics of hypervariable region 1 variation in hepatitis C virus infection and correlation with clinical and virological features of liver disease. Hepatology 27:1678-1686. [DOI] [PubMed] [Google Scholar]
- 7.Casato, M., V. Agnello, L. P. Pucillo, G. B. Knight, M. Leoni, S. Del Vecchio, C. Mazzilli, G. Antonelli, and L. Bonomo. 1997. Predictors of long-term response to high-dose interferon therapy in type II cryoglobulinemia associated with hepatitis C virus infection. Blood 90:3865-3873. [PubMed] [Google Scholar]
- 8.Casato, M., C. Mecucci, V. Agnello, M. Fiorilli, G. B. Knight, C. Matteucci, L. Gao, and K. Jay. 2002. Regression of lymphoproliferative disorder after treatment for hepatitis C virus infection in a patient with partial trisomy 3, Bcl-2 overexpression, and type II cryoglobulinemia. Blood 99:2259-2261. [DOI] [PubMed] [Google Scholar]
- 9.Cox, T. 2005. An introduction to multivariate data analysis. Hodder Arnold, London, United Kingdom.
- 10.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Sol Mesa, A., F. Pazos, and A. Valencia. 2003. Automatic methods for predicting functionally important residues. J. Mol. Biol. 326:1289-1302. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1995. PHYLIP (phylogenetic inference package): version 3.57. Department of Genetics, University of Washington, Seattle.
- 13.Ferri, C., F. Caracciolo, and L. Zignego. 1994. Hepatitis C virus infection in patients with non-Hodgkin's lymphoma. Br. J. Haematol. 88:392-394. [DOI] [PubMed] [Google Scholar]
- 14.Gerotto, M., F. Dal Pero, S. Loffreda, F. B. Bianchi, A. Alberti, and M. Lenzi. 2001. A 385 insertion in the hypervariable region 1 of hepatitis C virus E2 envelope protein is found in some patients with mixed cryoglobulinemia type 2. Blood 98:2657-2663. [DOI] [PubMed] [Google Scholar]
- 15.Gobel, U., C. Sander, R. Schneider, and A. Valencia. 1994. Correlated mutations and residue contacts in proteins. Proteins 18:309-317. [DOI] [PubMed] [Google Scholar]
- 16.Hermine, O., F. Lefrere, J. P. Bronowicki, X. Mariette, K. Jondeau, V. Eclache-Saudreau, B. Delmas, F. Valensi, P. Cacoub, C. Bréchot, B. Varet, and X. Troussard. 2002. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N. Engl. J. Med. 347:89-94. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann, W. P., E. Herrmann, B. Kronenberg, C. Merkwirth, C. Welsch, T. Lengauer, S. Zeuzem, and C. Sarrazin. 2004. Association of HCV-related mixed cryoglobulinemia with specific mutational pattern of the HCV E2 protein and CD81 expression on peripheral B lymphocytes. Blood 104:1228-1229. [DOI] [PubMed] [Google Scholar]
- 18.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 19.Lunel, F., L. Musset, P. Cacoub, L. Frangeul, P. Cresta, M. Perrin, P. Grippon, C. Hoang, D. Valla, and J. C. Piette. 1994. Cryoglobulinemia in chronic liver diseases: role of hepatitis C virus and liver damage. Gastroenterology 106:1291-1300. [DOI] [PubMed] [Google Scholar]
- 20.Mondelli, M. U., I. Zorzoli, A. Cerino, A. Cividini, M. Bissolati, L. Segagni, V. Perfetti, E. Anesi, P. Garini, and G. Merlini. 1998. Clonality and specificity of cryoglobulins associated with HCV: pathophysiological implications. J. Hepatol. 29:879-886. [DOI] [PubMed] [Google Scholar]
- 21.Mondelli, M. U., A. Cerino, A. Lisa, S. Brambilla, L. Segagni, A. Cividini, M. Bissolati, G. Missale, G. Bellati, A. Meola, B. Bruni Ercole, A. Nicosia, G. Galfrè, and E. Silini. 1999. Antibody responses to hepatitis C virus hypervariable region 1: evidence for cross-reactivity and immune-mediated sequence variation. Hepatology 30:537-545. [DOI] [PubMed] [Google Scholar]
- 22.Penin, F., C. Combet, C. Germanidis, P. O. Frainais, G. Deleage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puoti, C., R. Castellacci, F. Montagnese, S. Zaltron, G. Stornaiuolo, N. Bergami, L. Bellis, D. F. Precone, P. Corvisieri, M. Puoti, E. Minola, and G. B. Gaeta. 2002. Histological and virological features and follow-up of hepatitis C virus carriers with normal aminotransferase levels: the Italian prospective study of the asymptomatic C carriers (ISACC). J. Hepatol. 37:117-123. [DOI] [PubMed] [Google Scholar]
- 24.Rigolet, A., P. Cacoub, A. Schnuriger, L. Vallat, A. Cahour, P. Ghillani, F. Davi, Y. Benhamou, J. C. Piette, and V. Thibault. 2005. Genetic heterogeneity of the hypervariable region I of hepatitis C virus and lymphoproliferative disorders. Leukemia 19:1070-1076. [DOI] [PubMed] [Google Scholar]
- 25.Rosa, D., G. Saletti, E. De Gregorio, F. Zorat, C. Comar, U. D'Oro, S. Nuti, M. Houghton, V. Barnaba, G. Pozzato, and S. Abrignani. 2005. Activation of naïve B lymphocytes via CD81, a pathogenetic mechanism for hepatitis C virus-associated B lymphocyte disorders. Proc. Natl. Acad. Sci. USA 102:18544-18549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silini, E., F. Bono, A. Cividini, A. Cerino, S. Bruno, S. Rossi, G. Belloni, B. Brugnetti, E. Civardi, L. Salvaneschi, and M. U. Mondelli. 1995. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology 21:285-290. [PubMed] [Google Scholar]
- 29.Simmonds, P., J. Bukh, C. Combet, G. Deléage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspé, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J.-M. Pawlotsky, F. Penin, E. Sablon, T. Shin-I, L. J. Stuyver, H.-J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 30.Suarez, F., O. Lortholary, O. Hermine, and M. Lecuit. 2006. Infection-associated lymphomas derived from marginal zone B cells: a model of antigen-driven lymphoproliferation. Blood 107:3034-3044. [DOI] [PubMed] [Google Scholar]
- 31.Yoshioka, K., T. Aiyama, A. Okomura, M. Takayanagi, K. Iwata, T. Ishikawa, Y. Nagai, and S. Kakumu. 1997. Humoral immune response to the hypervariable region of hepatitis C virus differs between genotype 1b and 2a. J. Infect. Dis. 175:505-510. [DOI] [PubMed] [Google Scholar]
- 32.Zehender, G., C. De Maddalena, F. Bernini, E. Ebranati, G. Monti, P. Pioltelli, and M. Galli. 2005. Compartmentalization of hepatitis C virus quasispecies in blood mononuclear cells of patients with mixed cryoglobulinemic syndrome. J. Virol. 79:9145-9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuckerman, E., T. Zuckerman, A. M. Levine, D. Douer, K. Gutekunst, M. Mizokami, D. G. Quian, M. Velankar, B. N. Nathwani, and T. L. Fong. 1997. Hepatitis C virus infection in patients with B cell non-Hodgkin's lymphoma. Ann. Intern. Med. 127:423-438. [DOI] [PubMed] [Google Scholar]