Abstract
Human immunodeficiency virus type 1 (HIV-1) generates 16 alternatively spliced isoforms of env mRNA that contain the same overlapping open reading frames for Vpu and Env proteins but differ in their 5′ untranslated regions (UTR). A subset of env mRNAs carry the extra upstream Rev initiation codon in the 5′ UTR. We explored the effect of the alternative UTR on the translation of Vpu and Env proteins from authentic env mRNAs expressed from cDNA constructs. Vpu expression from the subset of env mRNA isoforms with exons containing an upstream Rev AUG codon was minimal. However, every env mRNA isoform expressed similar levels of Env protein. Mutations that removed, altered the strength of, or introduced upstream AUG codons dramatically altered Vpu expression but had little impact on the consistent expression of Env. These data show that the different isoforms of env mRNA are not redundant but instead regulate Vpu production in HIV-1-infected cells. Furthermore, while the initiation of Vpu translation conforms to the leaky ribosome-scanning model, the consistent Env synthesis infers a novel, discontinuous ribosome-scanning mechanism to translate Env.
Like most cellular pre-mRNAs, the human immunodeficiency virus type 1 (HIV-1) 9.2-kb genome-length RNA transcribed from the provirus acquires a 5′ m7GpppN cap and a 3′ poly(A) tail. Multiple splice donor (SD) and splice acceptor (SA) sites in the genome-length HIV-1 mRNA support alternative splicing to a pool of 4-kb and 2-kb mRNAs that differ in their 5′ untranslated regions (UTR) (46, 52). These mRNAs contain partly overlapping open reading frames (ORFs) and collectively encode the nine HIV-1 proteins and polyproteins. In the early phase, multiple splicing produces more than 23 different 2-kb HIV-1 mRNAs that are constitutively exported to the cytoplasm for the translation of Rev, Tat, Nef, and Vpr regulatory and accessory proteins (46, 52, 54). Later, after sufficient levels of Rev accumulate in the nucleus, Rev multimers bind to the highly structured RNA of the Rev-responsive element (RRE) that underlies the Env reading frame. This binding promotes the engagement of proteins required for the nuclear export (22, 42) and cytoplasmic accumulation of unspliced genome-length RNA and 23 different spliced 4-kb mRNAs containing the RRE (reviewed in references 15 and 26). The viral Gag and Gag-Pol structural polyproteins are translated from the 9.2-kb RNA, with a ribosome frameshift producing Gag-Pol on 5% of occasions during translational scanning (24, 36). The structural envelope (Env) glycoprotein and accessory and regulatory Vpu, Vif, and Vpr proteins and amino acids 1 through 72 of Tat (Tat72) are translated from the 4-kb spliced mRNAs (53, 54).
The high level of diversity among the spliced HIV-1 mRNAs results in part from the incorporation of the upstream noncoding exons 2 and 3 (46, 52). These exons may be included separately or together or excluded from the 5′ UTR of alternative 2-kb nef, tat, and rev and 4-kb env and tat mRNAs. Furthermore, the use of alternative SA sites can expand the diversity of spliced mRNAs (46, 52). For instance, the use of three competing alternative SA sites immediately 5′ to the Rev AUG codon, SA4b, SA4a, and SA4c, results in 12 isoforms of 2-kb rev mRNA. These alternative SA sites, together with SA5, are also used for 4-kb env mRNA, resulting in 16 env isoforms that differ in their 300- to 900-nucleotide-long 5′ UTR (see Fig. 1) (46, 53). For simplicity, the env isoforms are referred to herein as env1 to env16, as described previously (46). Selection among the various weak splice sites is controlled by interspersed exonic and intronic splice enhancer and silencer elements (2) whose activity is dictated by RNA binding proteins, including 9G8, SC35, ASF/SF2, and SRp40 (13, 49, 66).
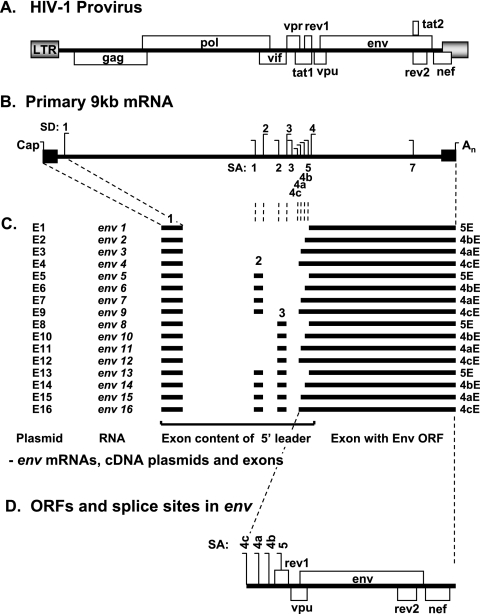
FIG. 1.
The 16 isoforms of HIV-1 env mRNA. (A) Schematic diagram of the HIV-1 provirus genome. LTR, long terminal repeat. (B) HIV-1 provirus transcribes a genome-length 9.2-kb RNA with an m7Gppp cap and a poly(A) tail (An) containing multiple SD and SA sites. (C) Sixteen different spliced forms (isoforms) of env mRNA are present in the mRNA pool following infection with HIV-1NL4.3, and these are assembled from four alternative Env-encoding exons, designated 5E, 4bE, 4aE, and 4cE, together with 5′ noncoding exon 1 and combinations of two alternatively spliced exons, exons 2 and 3 (46). E1 to E16, cDNA plasmids pDRenv1 to pDRenv16 expressing the mRNA isoforms described in this study. (D) The env mRNAs all contain ORFs for Vpu, Env, and Nef but differ in the sequence of the 5′ UTR due to the different exons spliced together to make the mRNA.
All 16 env mRNAs contain ORFs for Vpu, Env, and Nef (46, 53). The Vpu ORF lies upstream of but overlapping that of Env, and the translation of both proteins is reported to proceed by leaky scanning, in which some ribosomes bypass the weak upstream Vpu initiation codon to initiate translation at the strong Env start signal (53). HIV-1 Env is synthesized as a 160-kDa precursor (gp160) that is cleaved to a 120-kDa surface protein (gp120) and a 41-kDa transmembrane protein (gp41) that assemble into mature trimers that protrude from the virion surface (64). Interestingly, Vpu controls the level of mature Env protein available for assembling virions during viral replication (reviewed in reference 10). The Env gp160 precursor and its cellular receptor, CD4, are both translated in the endoplasmic reticulum, where they interact and potentially become entrapped (9). Vpu is also translated in the endoplasmic reticulum, where it binds and targets CD4 for degradation (63) through the ubiquitin/proteasome pathway (51). This mechanism frees the Env precursor for correct processing and expression at the cell surface, where it is incorporated into virions (reviewed in reference 10). The coordinated expression of Vpu and Env through leaky scanning translation from the same env mRNA may optimize the inclusion of mature Env into virions.
Ultimately, the purpose of the alternatively spliced HIV-1 mRNA isoforms that contain the same ORFs but differ in the 5′ UTR is largely unknown. However, changing the proportions of the alternative mRNAs by mutating redundant SA sites or by modulating the expression of splice control proteins alters the expression of HIV-1 proteins and impairs viral replication and infectivity (13, 46, 47, 49, 61, 66). Mutating specific splice sites in the HIV-1 molecular clone, NL4.3, to redistribute the mRNA isoforms dramatically alters the expression of the viral Env and regulatory Rev and Nef proteins (46). Changes in Env expression for some splice mutants were inversely related to the levels of Rev expression and instead correlated with changes in the isoforms of env mRNA (46), suggesting that individual env isoforms differing only in their 5′ UTR may express Env protein with different degrees of efficiency.
Here we tested whether the alternative env mRNAs provide a means to regulate translation. HIV-1 mRNAs are produced in the nucleus and acquire a 5′ m7GpppN cap that binds the eukaryotic initiation factor 4E (eIF4E) complex, which initiates the recruitment of 40S ribosomes that scan for a suitable AUG initiation site (reviewed in references 18 and 30). The different 5′ UTR of HIV-1 env mRNAs contain several features predicted to have an impact on ribosome-scanning efficiency. These include (i) highly structured 5′ RNA elements of various lengths (5); (ii) upstream AUG codons for Rev and/or Vpu (28, 29, 41); and (iii) binding sites for various RNA binding proteins implicated either directly in translation, like eIF2 (4), or in translation modulation, like the protein kinase regulated by RNA (PKR), transactivation-responsive RNA binding protein (TRBP), the La autoantigen, and the viral Tat protein (6, 14, 19, 38, 60). In previous work, the noncoding exon 2 of the HIV-1 5′ UTR was also reported to stimulate protein expression while noncoding exon 3 reduces the expression of a heterologous reporter, suggesting that exon 2 and/or 3 may perturb mRNA stability (31). Similar effects may occur with the env mRNA containing these noncoding exons, possibly altering Env protein expression.
The use of SA4b, SA4a, and SA4c splice acceptors during env mRNA splicing incorporates a weak translation start codon for Rev into the 5′ UTR (see Fig. 1) that may influence the progression of 40S ribosome subunits to translate the downstream Env ORF. Here we tested the effects of the different 5′ UTR of env mRNA isoforms on Vpu and Env expression. Unexpectedly, we showed that the different 5′ UTR affect Vpu synthesis, but not Env synthesis, from the env mRNA. The differences in Vpu synthesis patterns are explained by the upstream Rev translation initiation codon encountered in some 5′ UTR during ribosome scanning. However, the uniform expression of Env protein despite upstream initiation codons indicates that a discontinuous scanning mechanism akin to an internal ribosome entry site (IRES) or a ribosome shunt is used for Env translation initiation from the HIV-1 env mRNA.
MATERIALS AND METHODS
Plasmid construction.
The HIV-1 molecular clone pNL4.3 was provided by Malcolm Martin (National Institutes of Health, Bethesda, MD) (1). The derivative clone pDRNL4.3, with a deletion of 1.5-kb of cellular DNA flanking the proviral sequence, was constructed by joining the 5′ and 3′ half clones provided by Ron Desrosiers (New England Primate Research Center, Southborough, MA) (20). Klaus Strebel (National Institutes of Health, Bethesda, MD) generously provided the constructs pNLA1U− (containing a Vpu ATG-to-GTG mutation) and pNLΔU35 (containing an 8-bp insertion at Vpu amino acid 32, inducing the premature termination of Vpu at amino acid 35) (57).
pEGFP-NI that expresses green fluorescent protein (GFP) was obtained from Clontech (BD Biosciences). The Rev reporter plasmid pDM128 expresses chloramphenicol acetyltransferase (CAT) in response to Rev activity in the cell (23). pHIVlacZ containing the Escherichia coli β-galactosidase gene cloned downstream of the HIV-1 long terminal repeat promoter was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health, from Joseph Maio (35). The human β-actin plasmid was provided by Johnson Mak (Burnet Institute, Prahran, Australia).
Cloning and mutagenesis.
To generate a library of NL4.3 cDNA clones representing 1.8-kb spliced mRNAs, HeLa cells were transfected with 20 μg of pNL4.3, poly(A)+ RNA was harvested, and the viral cDNA was reverse transcribed as described previously (46). cDNA representing spliced, HIV-1 1.8-kb mRNA was PCR amplified using primers Odp.001 and Odp.032 (sequences are given in Table 1) with the Expand high-fidelity PCR system per the instructions of the manufacturer (Roche). The DNA was purified (Progen PCR Spinclean kit), digested with BssHII plus BamHI, and cloned into the analogous region of pNL4.3 to produce a library of 1.8-kb cDNA clones.
TABLE 1.
Primer sequences
| Primer | 5′→3′ sequencea |
|---|---|
| Odp.001 | CTCTAGCAGTGGCGCCCGAAC |
| Odp.002 | CTCTGGTAACTAGAGATCCCTCAG |
| Odp.031 | AGTAGAGCAAAATGGAATGCCAC |
| Odp.032 | CCGCAGATCGTCCCAGATAAG |
| Odp.067 | GCTCATCAGAACAGTCAGACTCA |
| Odp.080 | GCTGCCTTGTAAGTCATTGGTCTTAA |
| Odp.081 | TCAGCTCGTCTCATTCTTTCCCTTAC |
| Odp.084 | TCATTGCCACTGTCTTCTGCTCT |
| Odp.087 | TACCCCATAATAGACTGTGACCCAC |
| Odp.092 | CCAGACTGTGAGTTGCAACAG |
| Odp.151 | GGGACCCGACAGGCCCGAAGGAA |
| Odp.263 | GCGGCGACTGGCATCTCCTACGGCAGGAAGAAGCGGAGACAG |
| Odp.264 | CTGTCTCCGCTTCTTCCTGCCGTAGGAGATGCCAGTCGCCGC |
| Odp.267 | GACCCGACAGGCCTGAAGGAATAGAAG |
| Odp.268 | CTTCTATTCCTTCAGGCCTGTCGGGTC |
| Odp.281 | GCTGTAAGGGAAAGATGAAGACGAGCTGAGCCAGC |
| Odp.282 | GCTGGCTCAGCTCGTCTTCATCTTTCCCTTACAGC |
| Odp.330 | GCGAGGGGCGGCGACTGGCAGCCACCATGGCAGGAAGAAGCGGAGAC |
| Odp.331 | GTCTCCGCTTCTTCCTGCCATGGTGGCTGCCAGTCGCCGCCCCTCGC |
| Odp.513 | GACAGCAGAGATCCAGTGGCACCATGGCCAGCTAAGCTCCTCTGGAAAG |
| Odp.514 | CTTTCCAGAGGAGCTTAGCTGGCCATGGTGCCACTGGATCTCTGCTGTC |
| Odp.515 | GACAGCAGAGATCCAGTGGCACCAGGGCCAGCTAAGCTCCTCTGGAAAG |
| Odp.516 | CTTTCCAGAGGAGCTTAGCTGGCCCTGGTGCCACTGGATCTCTGCTGTC |
| 5′ human β-actin gene fragment | CCCAAGCTTGCGGCCGCCCATGTGCAAGGCCGGCTTCG |
| 3′ human β-actin gene fragment | GCGGGCCCGAAGGTGTGGTGCCAGATC |
Mutations are shown in bold.
cDNA clones representing specific HIV-1 1.8-kb spliced mRNAs were identified from the library by using restriction enzyme analysis, sequencing (ABI Prism kit; Perkin Elmer), and colony lifts (colony/plaque screen hybridization transfer membranes; DU PONT) probed with [γ-32P]dATP-labeled oligonucleotides spanning different HIV-1 exon-exon junctions. This allowed the following 1.8-kb cDNA clones to be isolated: pNLnef2 (exons 1, 5, and 7 [1/5/7]), pNLnef3 (1/2/5/7), pNLnef4 (1/3/5/7), pNLnef5 (1/2/3/5/7), pNLrev1 (1/4b/7), pNLrev2 (1/4a/7), pNLrev3 (1/4c/7), pNLrev4 (1/2/4b/7), pNLrev5 (1/2/4a/7), pNLrev7 (1/3/4b/7), pNLrev8 (1/3/4a/7), pNLrev10 (1/2/3/4b/7), and pNLtat1 (1/4/7).
HIV-1 env cDNA clones were then generated using a three-fragment ligation strategy. The BssHII-SacI 5′ UTR from the appropriate 1.8-kb cDNA was cloned with the SacI-BamHI and BssHII-BamHI fragments from pDRNL4.3. The following env cDNA constructs were thus generated, and their exon contents (shown in parentheses) were verified by restriction enzyme analysis and sequencing: pDRenv1 (1/5E), pDRenv2 (1/4bE), pDRenv3 (1/4aE), pDRenv4 (1/4cE), pDRenv5 (1/2/5E), pDRenv6 (1/2/4bE), pDRenv7 (1/2/4aE), pDRenv8 (1/3/5E), pDRenv10 (1/3/4bE), pDRenv11 (1/3/4aE), pDRenv13 (1/2/3/5E), and pDRenv14 (1/2/3/4bE).
Mutations that prematurely terminated Rev translation at an arginine residue at amino acid 38 (R38), removed the Rev AUG codon (Rev−) and/or the Vpu AUG codon (Vpu−), increased the Kozak strength of the Rev AUG codon, added a strong Kozak AUG codon to exon 2, or mutated this AUG codon to AGG were all synthesized by PCR mutagenesis by using the Expand high-fidelity PCR system (Roche) and the following protocol with the template and primer sets outlined in Table 2. Briefly, 5′ and 3′ mutagenic fragments were PCR amplified using the plasmid template plus wild-type forward and mutagenic reverse primers or mutagenic forward and wild-type reverse primers, respectively. Both fragments were purified, pooled together, and PCR amplified with wild-type forward and wild-type reverse primers, producing a larger fragment containing the mutation used in subsequent cloning. The pNL-M2A2 construct that introduces a translation stop codon corresponding to Vpu amino acid 2 was the result of a random PCR mutation arising during the preparation of an SA4b proviral splice site mutant (46). All oligonucleotides used throughout this study were synthesized by Invitrogen and are outlined in Table 1.
TABLE 2.
Primers and initial plasmid template for PCR mutagenesis
| Mutation | Initial PCR template | Primera
|
|||
|---|---|---|---|---|---|
| WT F | WT R | MUT F | MUT R | ||
| R38 | pNL4.3 | Odp.031 | Odp.081 | Odp.267 | Odp.268 |
| N20 | pDRenv2 | Odp.151 | Odp.080 | Odp.281 | Odp.282 |
| Rev− | pDRenv2 | Odp.002 | Odp.092 | Odp.263 | Odp.264 |
| Strengthening of Rev AUG codon | pDRenv2 | Odp.002 | Odp.092 | Odp.330 | Odp.331 |
| Addition of Kozak AUG codon | pDRenv5 or pDRenv6 | Odp.001 | Odp.092 | Odp.513 | Odp.514 |
| Change from Kozak AUG to AGG | pDRenv5 or pDRenv6 | Odp.001 | Odp.092 | Odp.515 | Odp.516 |
WT, wild-type; F, forward; R, reverse; MUT, mutagenic.
The R38 PCR mutation was cloned into the analogous regions of pDRenv1 to pDRenv4 by using MfeI and BamHI to generate the pDRenvR38 cDNA constructs. The Rev− mutation was cloned into the analogous region of pDRenv2 by using BssHII and MfeI to generate pDRenv2Rev− cDNA. The same strategy was used to clone the mutation increasing the Kozak strength of the Rev AUG codon into pDRenv2 and pDRenv2R38, producing pDRenv2RevK and pDRenv2R38RevK, respectively. The pDRenv1R38 to pDRenv3R38 constructs containing the Vpu− mutation and the mutations terminating Vpu translation at amino acid 2 and amino acid 35 were cloned by ligating three fragments: the BssHII-SacI 5′ UTR from pDRenv1, pDRenv2, or pDRenv3; the SacI-MfeI fragment of pNLA1U−, pNLM2A2, or pNLΔU35; and the BssHII-MfeI plasmid backbone from pDRenv1R38. The same cloning strategy was used to generate pDRenv2Rev−Vpu− (except that the 5′ UTR was derived from pDRenv2Rev−, the SacI-MfeI fragment from pNLA1U−, and the plasmid backbone from pDRenv1). The exon 2 mutations adding a strong Kozak AUG codon or mutating the codon to AGG were cloned into pDRenv5 and pDRenv6 by using BssHII and MfeI to generate pDRenv5KAUG and pDRenv6KAUG and pDRenv5KAGG and pDRenv6KAGG. Finally, the pDRenv1ΔS clone, containing a deletion of the splice sites SD4, SD5, SA6, SA7, SA7a, and SA7b, was generated by a multistep assembly of fragments mutated using PCR templates and primers listed in Tables 1 and 2. All clones were verified by restriction enzyme analysis and sequencing. A derivative clone of pEGFP-N1, pCMVempty, carried a deletion of the GFP gene between the AgeI and NotI sites; overhangs were end-filled with Klenow DNA polymerase before blunt-end ligation. The pCMVTat2X construct was generated by inserting the SalI-BamHI fragment from pNLtat1 (1/4/7) into pCMVempty.
Cells and transfections.
HeLa cells (CCL-2; American Type Culture Collection) were propagated in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal calf serum, 100 U/ml penicillin, and 100 U/ml streptomycin. Plasmids for transfection were generated with QIAGEN Mega prep kits. HeLa cells were transfected using calcium phosphate coprecipitation (62). Cells (1 × 106) were typically transfected in 25-cm2 flasks with 7.5 μg of env cDNA, 5 μg of pCMVTat2X, 2.5 μg of pEGFP-N1, and either 8 or 4 μg of pNLrev1 for wild-type or R38 mutant env cDNA, respectively. To quantify Rev, 5 μg of the pDM128 reporter was also included in the DNA mix. pHIVlacZ and pCMVempty were used to normalize for differences in DNA amounts and promoter activities among samples.
Antibodies.
The HIV-1-positive patient serum used to detect Env (gp160) was kindly provided by Johnson Mak (Burnet Institute, Prahran, Australia). The Vpu antibody raised in rabbits by using E. coli-derived Vpu amino acids 31 to 81 (37) was generously donated by Klaus Strebel (National Institutes of Health, Bethesda, MD). The rabbit anti-human, donkey anti-rabbit, and rabbit anti-sheep sera conjugated to horseradish peroxidase (HRP) were obtained from Dako (Denmark), Amersham Pharmacia Biotech (England), and Zymed (United States), respectively.
Immunoblots.
Forty hours after transfection, 106 cells were washed twice in phosphate-buffered saline (PBS) and lysed on ice for 2 h in a solution containing 150 μl of 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.5% (vol/vol) Triton X-100 supplemented with 1 μg/ml aprotinin and 2 mM phenylmethylsulfonyl fluoride. Cell debris and nuclei were removed by centrifugation at 12,000 × g for 2 min. GFP in the cell extracts was quantified to measure transfection efficiency by using opaque 96-well plates (Corning) and a FLA-2000 fluorescent-image analyzer (Fujifilm, Japan). Extracts with equivalent levels of GFP were resolved on 10% (Env) or 12.5% (Vpu) sodium dodecyl sulfate-polyacrylamide gels. Proteins were transferred onto polyvinylidene difluoride membranes (DU PONT) by using a semidry electroblotting apparatus (Integrated Separation Systems, Enprotech). Membranes were blocked in PBS containing either 5% (wt/vol) skim milk powder (for Env detection) or 3% (wt/vol) bovine serum albumin (for Vpu detection). To detect Env, blots were probed with HIV-1-positive patient sera (1:5,000) followed by HRP-conjugated anti-human sera (1:4,000). Vpu sera (1:500) and HRP-conjugated anti-rabbit sera (1:10,000) were used to detect Vpu. Probed bands were visualized using enhanced chemiluminescence plus autoradiography and the LAS-1000 luminescent-image analyzer (Fujifilm, Japan). The densities of protein bands were quantified using accompanying MacBAS version 3.3 software.
CAT assays.
Rev activity was indirectly measured by transfecting cells with the pDM128 Rev reporter plasmid and analyzing cell extracts for CAT activity 40 h posttransfection. Cells (106) were washed in PBS, resuspended in 120 μl of 0.25 M Tris-HCl (pH 7.8)-0.5% (vol/vol) Nonidet P-40, and lysed by three consecutive rounds of rapid freeze-thawing with a 10-s vortexing step after each thaw. Cell debris was removed by centrifugation at 15,300 × g for 10 min at 4°C, and extracts were heat inactivated for 10 min at 65°C and centrifuged as before. Final lysates were normalized for transfection efficiency by quantifying GFP, and the samples were diluted with 0.25 M Tris (pH 7.8)-0.5% (vol/vol) Nonidet P-40 to contain equivalent levels of GFP. Normalized extract (12.5 μl) was mixed with 12.5 μl of 0.5 M Tris-HCl (pH 7.8), 5 μl of acetyl coenzyme A (3.5 mg/ml; Roche), and 2.5 μl of d-threo-dichloroacetyl-[1,2-14C]chloramphenicol (52.8 mCi/mmol; NEN Life Science Products). Samples were incubated for 2 to 4 h at 37°C, and the extracts were typically diluted 1:10 for analysis to keep the assay samples within the linear range (below 30% conversion). The chloramphenicol forms were extracted with ethyl acetate, resolved using thin-layer chromatography in 5% (vol/vol) methanol and 95% (vol/vol) chloroform, and visualized using the FLA-2000 fluorescent-image analyzer (Fujifilm, Japan). The acetylated and unacetylated forms were quantified using MacBAS version 3.3 software.
RPA.
A T7 promoter was added to DNA templates required for RNA probe synthesis by using the Lig'nScribe RNA polymerase promoter addition kit (Ambion). The env probe template was PCR amplified from pDRenv1 by using primers Odp.067 and Odp.087 (Table 1). The β-actin probe template was PCR amplified by using a human β-actin plasmid and PCR primers (Table 1) provided by Johnson Mak (Burnet Institute, Prahran, Australia). RNA probes were synthesized using the MAXIscriptTM in vitro transcription kit (Ambion) with the supplied T7 polymerase and [α-32P]UTP (NEN Life Science Products). The RNase protection assay (RPA) was performed using the RPA II TM kit per the instructions of the manufacturer (Ambion). Total RNA was isolated from transfected cells by using TRIzol (Invitogen). In the RPA, the probe and total RNA were both hybridized at 42°C for 14 h, followed by digestion with either RNase A-RNase T1 or RNase T1 at a 1:50 dilution. Samples were resolved on a prewarmed, 8 M urea-6% polyacrylamide gel (Shelton Scientific-IBI). The gel was dried on 3MM chromatography paper (Whatman International Ltd., England), and bands were visualized using autoradiography and the FLA-2000 fluorescent-image analyzer (Fujifilm, Japan).
RNA stability analysis.
Twenty-four hours after transfection, medium containing 5 μg/ml actinomycin D was added to cells. Total RNA was harvested 0, 4, 8, and 12 h later using TRIzol (Invitrogen). The amount of env mRNA present in 5 μg of total RNA was analyzed via the RNA protection assay and quantitated using the FLA-2000 (Fujifilm, Japan) and MacBAS version 3.3 software.
RESULTS
Alternative isoforms of env mRNA express similar levels of Env but different levels of Vpu.
To determine the effects on protein synthesis of the different 5′ UTR of the 16 env mRNA isoforms, we generated a panel of cDNA constructs that each expressed one isoform of env mRNA. These subgenomic cDNA constructs expressed native env isoforms with the 5′ UTR exons already spliced together. Therefore, these env mRNA isoforms had the same structures as those arising naturally after alternative splicing of the 9.2-kb genomic RNA (Fig. 1). These env mRNAs all contained exon 1 and included either, neither, or both exon 2 and/or exon 3 and interchange exon 5E, 4bE, 4aE, or 4cE (Fig. 1C) from the HIV-1 NL4.3 molecular clone. These mRNAs all carried the same Vpu, overlapping Env, and downstream Nef ORFs (Fig. 1D) but differed in the 5′ UTR upstream of the region corresponding to Vpu depending on their exon contents (Fig. 1C).
To express the individual env mRNAs from these cDNA plasmids in HeLa cells, cells were cotransfected with an HIV-1 Tat expression plasmid to transactivate the viral long terminal repeat promoter together with the pEGFP-N1 transfection efficiency reporter expressing GFP. Protein expression from env mRNA that contains the RRE is known to be acutely reliant on the level of Rev protein for nucleocytoplasmic export (22, 42). Because the subset of env mRNA transcripts containing exon 4bE, 4aE, or 4cE, such as those produced by the pDRenv2, pDRenv3, and pDRenv4 cDNA plasmids, can actively splice to SA7, leading to Rev expression (31a; Fig. S1 in the supplemental material), it was important to develop a system to equalize the levels of Rev activity among the samples. Two approaches were used to equalize the levels of Rev activity in experiments with the env isoforms, allowing us to focus on the effects of the 5′ UTR on protein synthesis.
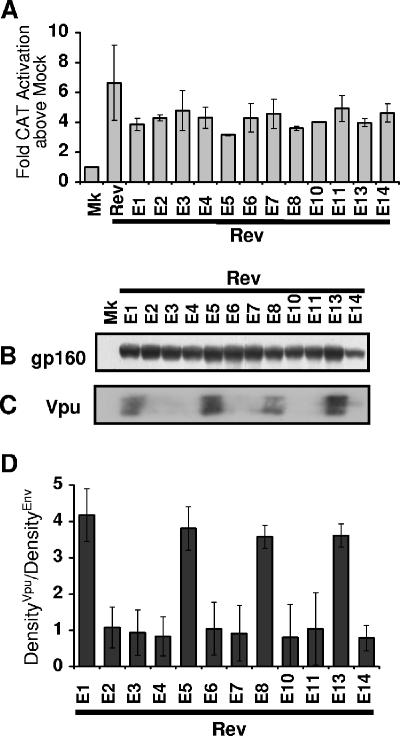
Firstly, cells were cotransfected with individual env isoform cDNA plasmids and high levels of the Rev expression plasmid pNLrev1 to produce similar levels of saturating Rev among the samples (Fig. 2A). When similar levels of Rev activity among the samples with the native env isoforms were induced using this method (Fig. 2A), the different isoforms expressed similar levels of gp160 Env protein but dramatically different levels of Vpu (Fig. 2B and C). A densitometry analysis of the protein synthesized from each Env cDNA plasmid revealed that constructs producing env mRNA isoforms containing exon 5E, such as pDRenv1, pDRenv5, pDRenv8, and pDRenv13, consistently expressed approximately fourfold more Vpu than Env relative to their exon 4bE-, 4aE-, and 4cE-containing counterparts (Fig. 2D). This protein expression profile was conserved irrespective of whether noncoding exon 2 and/or exon 3 was included in the env mRNA. The differences in levels of Vpu expression measured by immunoblotting were significant because titrating lysate from cells transfected with pDRenv2 cDNA revealed that this quantification method detected these protein levels in the linear range, supporting its use here for Env and Vpu quantification (Fig. S2 in the supplemental material). Hence, this analysis revealed that the env isoforms expressed similar levels of Env but different levels of Vpu, coincident with the presence in the mRNAs of exon 5E, which has no upstream Rev AUG codon, or that of exon 4bE, 4aE, or 4cE, which has an upstream AUG codon.
FIG. 2.
Similar levels of Env but different levels of Vpu coexpressed by the env isoforms with saturating Rev in trans. (A) Rev activity 40 h after triplicate transfections of HeLa cells with pLTRrev1 (8 μg) along with the env cDNA construct, pCMVTat2x, pEGFP, and pDM128 was determined by CAT assays. (B and C) Samples normalized for transfection efficiency by using GFP were immunoblotted for gp160 Env (B) and Vpu (C), with representative blots shown. (D) The density of the Vpu band was divided by the density of the Env band for each env isoform, and the average ratio from three independent experiments was graphed. Mk, mock (no env or pLTRrev1) transfection control; Rev, pLTRrev1 (no env) transfection control; E1 to E14, plasmids expressing isoforms env1 to env14.
To overcome any discrepancy in levels of endogenous Rev expression among cDNA constructs (Fig. S1 in the supplemental material), we adopted a second method of equalizing levels of Rev activity obtained with the Env constructs by introducing a premature translation termination codon in the Rev ORF in the env mRNAs to terminate Rev translation at the arginine residue at amino acid position 38 (Fig. 3A). This mutation led to the expression of a nonfunctional truncated Rev protein after splicing (Fig. 3B). The cotransfection of cells with these R38 mutant env cDNA plasmids and equal amounts of the Rev expression plasmid equalized the levels of Rev activity between the different env isoforms (Fig. 3B). Neither Vpu nor Env protein was expressed from the env R38 mutant mRNAs unless wild-type Rev was supplied in trans (Fig. 3B). When Rev was supplied in trans, the env1 to env4 R38 mutant cDNA plasmids expressed similar levels of Env protein but the env1 R38 mutant cDNA plasmid that expressed an exon 5E-containing env1 mRNA isoform produced two- to fourfold more Vpu than env2, env3, and env4 R38 mutant cDNA plasmids that expressed 4bE-, 4aE-, and 4cE-containing env2, env3, and env4 isoforms, respectively (Fig. 3B and C). Titrating the env1 and env2 R38 cDNA mutants with increasing amounts of Rev further revealed that at each Rev concentration tested, both env1 and env2 mRNA isoforms expressed similar amounts of Env but that the env1 R38 mutant isoform that used exon 5E always synthesized more Vpu than the env2 R38 mutant isoform that used exon 4bE (Fig. 3D). Using the two methods for equalizing levels of Rev activity, we consistently demonstrated that the env isoforms expressed similar levels of Env protein but different levels of Vpu, depending upon the structure of the 5′ UTR resulting from the use of exon 5E, 4bE, 4aE, or 4cE. Furthermore, equivalent levels of Env were also expressed from the env1 to env4 mRNA isoforms following the transfection of the Env cDNA clones into Jurkat T cells, which express CD4 viral receptor (33) (Fig. S3 in the supplemental material). Nef expression did not have an impact on the amounts of Env and Vpu expressed from these Env cDNA clones (Fig. S4 in the supplemental material). While Vpu translation conformed to the leaky ribosome-scanning model, Env expression was not influenced by upstream AUG codons.
FIG. 3.
Rev R38 mutant env isoforms express different levels of Vpu but similar levels of Env irrespective of the Rev titer. (A) The R38 mutation introduces a premature translation termination codon corresponding to amino acid 38 in the second Rev exon of the env cDNA constructs. (B) The activity of Rev R38 env isoform mutants was measured by CAT assays 40 h after the cotransfection of HeLa cells with pCMVTat2X, pEGFP, and pDM128 with or without pLTRrev1. (C) Densitometry analysis of Env and Vpu expression from the env cDNA constructs. (D) Immunoblot assay of gp160 Env and Vpu synthesized in HeLa cells cotransfected with env cDNA, pCMVTat2X, pEGFP, pDM128, and increasing amounts of pLTRrev1. wt, wild-type sequence; R, R38 mutant sequence; Mk, mock (no env or pLTRrev1) transfection control; NL, HIVNL4.3 proviral plasmid control; E1R to E4R, env1 to env4 R38 mutant plasmids.
Consistent Env expression from the env isoforms is not due to cryptic splicing or increased RNA stability.
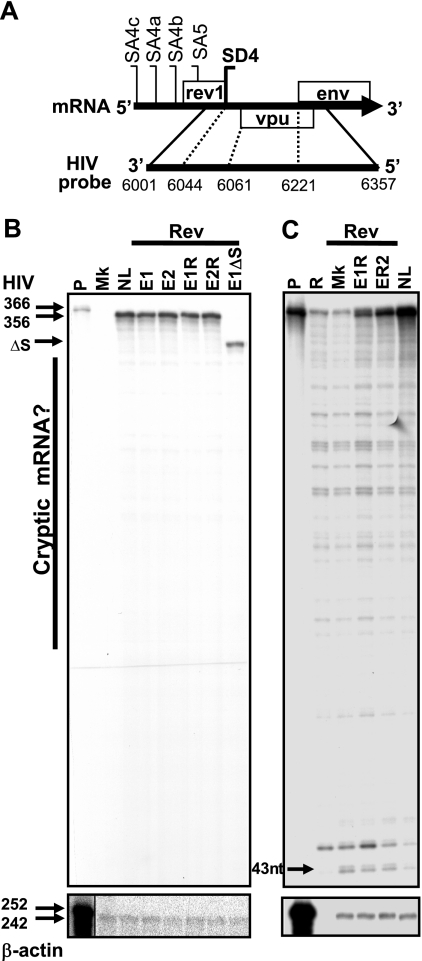
The leaky scanning model of translation predicts that ribosomes engaging the m7Gppp cap of mRNA at a constant rate proportionally initiate protein synthesis at the weak upstream Vpu AUG codon (53). While the inclusion of the weak upstream Rev AUG codon in the 5′ UTR of some isoforms of env mRNA altered Vpu synthesis, consistent with a leaky ribosome-scanning model, constant levels of Env synthesis from the alternative isoforms of env mRNA were not easily explained. We anticipated that 5′ UTR-induced changes in Vpu synthesis would alter the number of ribosomes available for Env translation and also cause changes in Env production. But the level of Env expression by the different env isoforms remained consistent (Fig. 2 and 3). It was possible that the env isoforms transcribed from our cDNA constructs underwent cryptic splicing to an mRNA monocistronic for the Env ORF, leading to similar levels of Env expression by the different isoforms despite the changes in Vpu production. To explore if such cryptic splicing occurred, RNA harvested from cells transfected with Env cDNA constructs expressing different env isoforms was analyzed by RPA (Fig. 4). For the RPA, a probe binding the NL4.3 env mRNA after SA5 and along the first 136 nucleotides of the Env ORF was employed (Fig. 4A). This probe protects a 356-nucleotide fragment corresponding to the native env isoforms. Importantly, this probe can also detect any cryptic mRNA species that remove the Vpu ORF and are thus monocistronic for the Env ORF. Treating the hybridized RNA-probe complexes with either a mix of RNaseA-RNase T1 (Fig. 4B) or RNase T1 alone (Fig. 4C) allowed the 356-nucleotide fragments of native env isoforms to be seen. However, no extra cryptic mRNA species indicative of an mRNA monocistronic for the Env ORF could be observed above the background radiolytic decay of the probe. A 313-nucleotide protected band from a mutant env construct which cannot actively splice was also included as a control to demonstrate that this method will detect aberrant env mRNA species (Fig. 4B). A 43-nucleotide band was also seen, representing a protected fragment from the SD4 splicing event in the 1.8-kb mRNA of pCMVTat2X (Fig. 4C). Therefore, the Env cDNA constructs did not appear to produce a cryptic mRNA monocistronic for the Env ORF that could explain the consistent Env production by the various env mRNA isoforms despite changes in Vpu levels.
FIG. 4.
Consistent Env expression among env isoforms is not due to cryptic splicing to monocistronic env mRNA. (A) Location of the probe in env mRNA used for RPA to explore RNA species generated from our env cDNA constructs. Numbers at the bottom indicate nucleotide positions. (B and C) Five micrograms of total RNA harvested from HeLa cells transfected with either NL4.3 or env cDNA plasmids plus pLTRrev1 was hybridized with the probe and digested with either an RNase A-RNase T1 mix (B) or RNase T1 alone (C) during the RPA. Protected fragments detected by autoradiography are shown. A β-actin-specific probe was used as a loading control. Molecular sizes in nucleotides are shown on the left. P, undigested probe; R, probe plus RNase; Mk, mock (no env or pLTRrev1); NL, HIVNL4.3 proviral plasmid control; E1 and E2, plasmids expressing isoforms env1 and env2; E1R and E2R, plasmids expressing env1 and env2 R38 mutant isoforms; E1ΔS, env1Δ splice site control; ΔS, deletion of SD4, SD5, SA7, SA7a, and SA7b; nt, nucleotides.
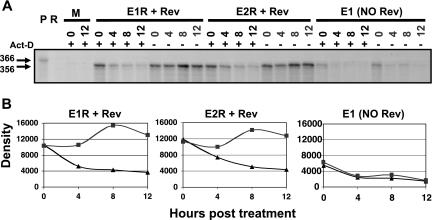
To test whether the consistent Env expression was instead due to an increased stability of the exon 5E-containing isoforms relative to that of the exon 4bE-, 4aE-, and 4cE-containing isoforms, we compared the degrees of stability of the env1 (1/5E) and env2 (1/4bE) mRNAs with or without actinomycin D treatment when normalized for Rev activity. Adding actinomycin D prevents transcription by RNA polymerase II so that the fate of the existing pool of mRNA over time can be examined. The amount of env mRNA present in cells expressing the env1 or env2 isoform was measured 0, 4, 8, and 12 h after treatment by using RPA (Fig. 5A) and densitometry for quantification (Fig. 5B). The similar accumulation and degradation profiles of both env1 and env2 isoforms normalized for Rev activity clearly showed that env1 mRNA containing exon 5E expressed by the env1 R38 mutant plasmid was no more stable than the env2 mRNA isoform containing exon 4bE expressed by the env2 R38 mutant plasmid. The similar profiles of these isoforms in the presence of Rev also contrasted with that of the no-Rev control, which displayed limited accumulation and rapid degradation of the mRNA with and without actinomycin D treatment. These data clearly show that the similar levels of Env synthesis from the env isoforms cannot be attributed to any increased stability of env isoforms containing exon 5E. If anything, the greater slope of the decay curve over the first 4 h for the env1 mRNA produced from the env1 R38 mutant plasmid than for the env2 mRNA produced from the env2 R38 mutant plasmid would indicate that the exon 5E-containing env1 mRNA may have a shorter half-life.
FIG. 5.
Exon 5E isoforms do not have greater stability than exon 4bE isoforms. (A) HeLa cells were transfected with env cDNA plasmids with or without pLTRrev1, and 24 h later, cells were either treated (+) or untreated (−) with actinomycin D (Act-D) for a further 0, 4, 8, and 12 h prior to RNA harvesting. Five micrograms of RNA per sample was analyzed for native env mRNA via RPA using RNase A-RNase T1 digestion and autoradiography. Molecular sizes in nucleotides are shown on the left. (B) Densitometry analysis of protected bands (untreated cells, ▪; treated cells, ▴). P, undigested probe; R, probe plus RNase; M, mock (no env or pLTRrev1); Rev, pLTRrev1; E1R and E2R, plasmids expressing env1 and env2 R38 mutant isoforms; E1, pDRenv1. Density equals photostimulated luminescence minus background.
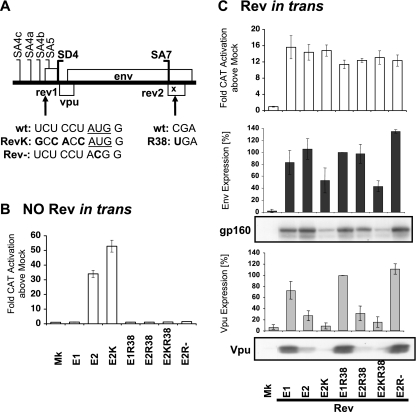
The effects of the Rev AUG codon upon Vpu expression are consistent with leaky ribosome scanning.
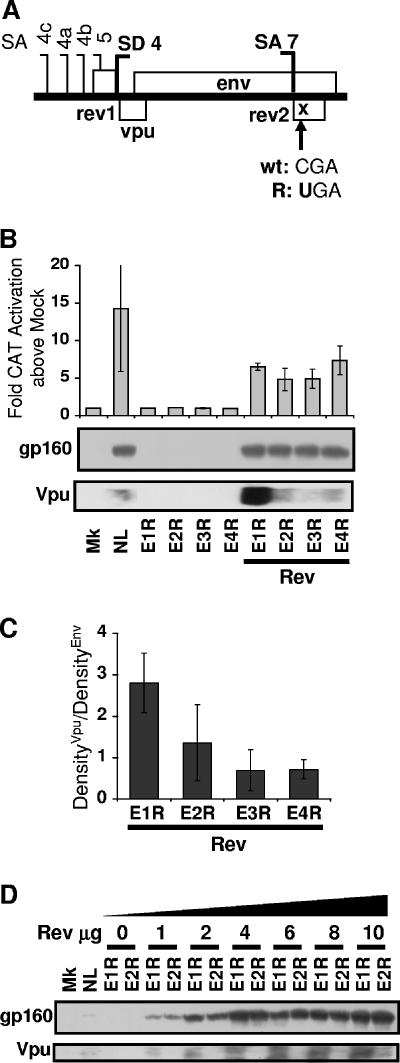
The env isoforms containing exons 4bE, 4aE, and 4cE from pDRenv2, pDRenv3, and pDRenv4 cDNA constructs, respectively, that expressed diminished levels of Vpu also incorporated the weak, upstream Rev AUG codon in the 5′ UTR. This Rev AUG codon was excluded from the exon 5E env isoforms produced by pDRenv1 cDNA due to the position of SA5. As Vpu is reported to be translated by ribosome scanning (53), we reasoned that including this weak, upstream Rev AUG codon in the 5′ UTR of env mRNA may impede scanning 40S ribosomes from reaching the Vpu AUG codon and diminish Vpu expression. To explore the role of the upstream Rev AUG codon in downstream Vpu expression, we constructed the pDRenv2RevK cDNA mutant, which carried a strengthened Rev AUG codon, and the pDRenv2Rev− mutant, which had the Rev AUG codon removed from the env mRNA (Fig. 6A). Strengthening the Kozak consensus of the Rev AUG codon increased Rev activation in the pDM128 Rev-dependent CAT reporter assay by almost twofold above that from the wild-type env2 isoform (Fig. 6B), suggesting that Rev expression was increased by improving the AUG start context. In contrast, deleting the Rev AUG codon extinguished the production of functional Rev, similar to the result for env R38 mutant constructs containing the Rev premature termination codon corresponding to amino acid 38 (Fig. 6B). Combining the enhanced Rev Kozak AUG consensus and the Rev R38 premature termination codon mutations in the same construct also blocked Rev function (Fig. 6B), allowing levels of Rev activity to be equalized in subsequent experiments (Fig. 6C). The Rev AUG Kozak translation initiation mutation that increased Rev expression from env2 cDNA decreased Vpu expression from this cDNA (Fig. 6C). Conversely, removing the Rev AUG codon increased Vpu synthesis from env2 to match the high levels of Vpu from env1 (Fig. 6C). These data indicate that Vpu synthesis from the native mRNA is affected by the upstream Rev AUG codon in a leaky ribosome-scanning manner because (i) increasing initiation at the Rev AUG codon decreased Vpu output, indicating that fewer ribosomes escape the Rev AUG codon to translate downstream Vpu, and (ii) removing the Rev AUG codon increased Vpu output, as scanning ribosomes were no longer lost to the upstream Rev AUG codon. Furthermore, this result indicates that the upstream Rev AUG codon in the 5′ UTR of the exon 4bE, 4aE, and 4cE env isoforms produced from pDRenv2, pDRenv3, and pDRenv4 cDNA constructs was responsible for their diminished Vpu synthesis relative to that of the exon 5E isoform from pDRenv1 cDNA clones that lacked the upstream Rev AUG codon.
FIG. 6.
Increasing the strength of or removing the Rev AUG modulates Vpu expression. (A) Mutations in the env2 cDNA constructs that either strengthened the Rev AUG codon (underlined; RevK) or removed it (Rev−). (B and C) HeLa cells were transfected with the env cDNA constructs, pCMVTat2X, pEGFP, and pDM128 without (B) or with (C) pLTRrev1. Rev activity was measured by CAT assay. Env and Vpu were detected by immunoblotting followed by densitometry analysis, and the percentage of expression relative to the expression of the env1 R38 mutant (E1R38; set at 100%) is graphed. wt, wild type; Mk, mock (no env or pLTRrev1); E1 and E2, pDRenv1 and pDRenv2; E2K, env2 cDNA construct expressing RevK mutation; E2R38, env2 cDNA construct expressing R38 mutation; E2KR38, env2 cDNA construct expressing RevK and R38 mutations; E2R−, env2 cDNA construct expressing Rev− mutation.
Given that the native weak Rev AUG codon had no impact upon Env expression from env isoforms (Fig. 2), it was surprising that Env expression was also affected by the Rev AUG codon mutations in a pattern consistent with effects on leaky scanning (Fig. 6C). The pDRenv2Rev− cDNA construct that removed the Rev AUG codon in the env2 isoform led to a 1.3-fold increase in Env expression, and the pDRenv2R38RevK cDNA construct that increased the strength of the Rev AUG codon led to a twofold reduction in Env expression (Fig. 6C). However, env1, env5, env8, and env13 cDNA constructs that produced env mRNA isoforms containing exon 5E, which lacks the Rev AUG codon, expressed levels of Env protein similar to those expressed by the other isoforms with the Rev AUG codon (Fig. 2), not fitting the leaky scanning mechanism model. These data suggest that the translation mechanism for the Env ORF is more complex than the standard leaky ribosome-scanning mechanism.
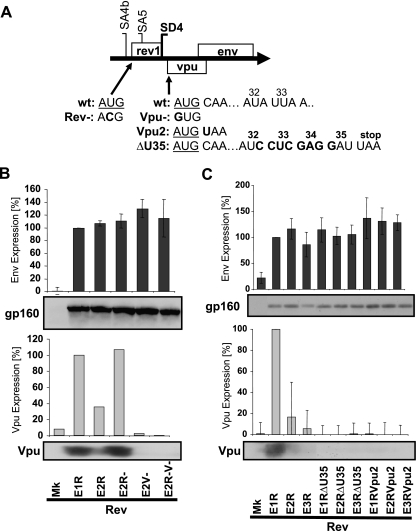
Upstream AUG codons and the length of the Vpu ORF do not alter Env expression.
To further explore potential elements in the 5′ UTR, we reasoned that removing both the upstream Rev and Vpu AUG codons should allow more scanning ribosomes to reach the Env AUG codon and thus induce greater synthesis of Env than if either AUG codon was removed in isolation. env2 R38 cDNA mutants which had the Vpu and/or Rev AUG codon removed were generated for this purpose (Fig. 7A). Removing the Rev or Vpu AUG codon in isolation caused minor increases in Env protein synthesis (Fig. 7B), but no greater increase in Env synthesis from a construct with the double AUG codon mutation was observed (Fig. 7B). Again, Env synthesis from these env2 mRNAs was clearly not consistent with leaky ribosome-scanning translation.
FIG. 7.
Removing upstream AUG codons and changing Vpu ORF length does not alter consistent Env expression. (A) Mutations in the env cDNA constructs that removed the upstream Rev and/or Vpu AUG codon (underlined; Rev− and/or Vpu−) or shortened the Vpu ORF at the position corresponding to amino acid 2 (Vpu2) or 35 (ΔU35). Numbers indicate amino acid positions corresponding to the codons. (B and C) HeLa cells were transfected with the env cDNA constructs, pCMVTat2X, pLTRrev1, and pEGFP. Env and Vpu expression was analyzed by immunoblotting and then densitometry. The percentage of expression relative to that of the env1 R38 mutant (E1R; set at 100%) is graphed for the env isoforms lacking upstream AUG codons (B) or containing altered Vpu ORF lengths (C). wt, wild type; Mk, mock (no env); E1R to E3R, pDRenv1 to pDRenv3 cDNA constructs containing the Rev R38 premature termination codon mutation; E2−, env2 cDNA construct expressing the Rev− mutation; E2V−, env2 cDNA construct expressing the Vpu− mutation; E2R−V−, env2 cDNA construct expressing the Rev− and Vpu− mutations; E1RΔU35 to E3RΔU35, env1 to env3 R38 mutant cDNA constructs with the Vpu ORF shortened at the position corresponding to amino acid 35; E1RVpu2 to E3RVpu2, env1 to env3 R38 mutant cDNA constructs with the Vpu ORF shortened at the position corresponding to amino acid 2.
To explore the importance for Env synthesis of the overlap of the Vpu ORF with the 5′ portion of the Env ORF, the Vpu ORF was prematurely terminated to produce Vpu mutants of two different lengths, one ending at amino acid 2 and the other terminating at amino acid 35 following an 8-nucleotide insertion at Vpu amino acid 32 (57) (Fig. 7A). As anticipated, these premature termination mutations in the Vpu ORF eliminated Vpu expression (Fig. 7C). However, minimal impact on the consistent expression of Env was observed. The constructs with the mutation at amino acid 35 expressed wild-type levels of Env protein (Fig. 7C), while Env synthesis by the Vpu mutants with the termination mutation at amino acid 2 increased merely 1.3-fold (Fig. 7C). Therefore, Env expression was not perturbed by changes to the length of the Vpu ORF, ruling out this parameter as an influence on consistent Env expression.
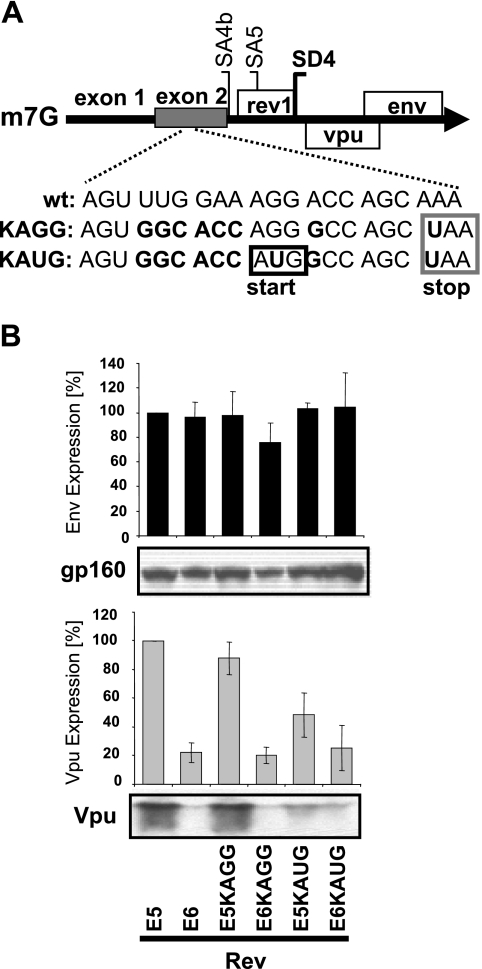
Env translation uses discontinuous ribosome scanning.
To delineate whether the translation mechanism for Env protein involved a continuous or discontinuous ribosome-scanning mechanism, we developed Env cDNA constructs producing mRNAs containing a strong Kozak AUG codon (Fig. 8A) in the upstream noncoding exon 2 that was shown not to affect Env expression from the env5 and env6 isoforms from pDRenv5 and pDRenv6 cDNA constructs (Fig. 2B). If HIV-1 Env protein is translated via a continuous ribosome-scanning mechanism, then adding a strong, upstream Kozak AUG codon should usurp the scanning 40S ribosomes and block the downstream translation of Env (and Vpu). Conversely, if Env uses a discontinuous scanning mechanism for translation initiation, this upstream AUG codon could be bypassed and no decline in downstream Env translation would occur. Mutants of cDNA plasmids pDRenv5 and pDRenv6 with the addition of an upstream, strong Kozak AUG codon and a translation termination signal in exon 2 of the env5 (1/2/5E) and env6 (1/2/4bE) mRNAs (Fig. 8A) showed reduced Vpu expression but no effect on Env production, supporting a discontinuous initiation mechanism for Env (Fig. 8B). Importantly, the control constructs lacking the exon 2 AUG start signal (Fig. 8A) did not show effects on Vpu or Env expression relative to the wild-type env5 and env6 isoforms (Fig. 8B). This finding indicates that changes to protein translation in the constructs with the strong Kozak AUG codon were specifically the result of the Kozak AUG motif and not of other nucleotide changes added to exon 2. Collectively, our data indicate that the Env ORF is translated mostly via a discontinuous ribosome-scanning mechanism, as introducing an upstream strong Kozak codon (Fig. 8) did not diminish the synthesis of Env.
FIG. 8.
Adding an upstream, strong Kozak AUG codon in exon 2 of the env isoforms does not diminish Env expression. (A) Mutations in exon 2 of the env cDNA constructs that introduced either a strong Kozak AUG codon (KAUG) or the same mutations minus the AUG codon (KAGG). A UAA translation termination motif was included at codon 4 to avoid aberrant protein synthesis. (B) HeLa cells transfected with the env cDNA constructs, pCMVTat2X, pLTRrev1, and pEGFP were analyzed for Env and Vpu by immunoblotting and densitometry. The percentage of expression relative to that of the env5 construct (E5; set at 100%) is graphed. wt, wild type; E6, pDRenv6; E5KAGG and E6KAGG, env5 and env6 constructs without the Kozak AUG codon; E5KAUG and E6KAUG, env5 and env6 constructs with the Kozak AUG codon.
DISCUSSION
We examined the significance of the different 5′ UTR for Env and Vpu synthesis from the multiple env isoforms produced by RNA splicing during HIV-1 replication. We found that the different 5′ UTR that arise from alternative splicing regulate Vpu synthesis while curiously maintaining a uniform level of Env production. Our analysis used cDNA plasmids expressing a single env mRNA isoform with the unique 5′ UTR exon combination already spliced together to reproduce native env mRNA isoforms made during viral replication. This method is likely to avoid changes to 5′ UTR mRNA elements and protein binding sites potentially important for translation control (4). This issue may have complicated the interpretation from earlier cDNA constructs and reporters (53). We found that only pDRenv1, pDRenv5, pDRenv8, and pDRenv13 cDNA constructs that produced env mRNA isoforms containing exon 5E, and not exons 4bE, 4aE, and 4cE, efficiently expressed Vpu.
This differential expression of Vpu by the env isoforms indicates the purpose of the alternately spliced isoforms of env mRNA. While factors influencing the preferential selection among the various weak splice sites by interspersed exonic and intronic splice enhancer and silencer elements (2), such as the levels of RNA binding proteins including 9G8, SC35, ASF/SF2, and SRp40 (13, 49, 66), have been identified, the effects of this regulation on HIV-1 replication and pathogenesis have been unclear. Our results show that alternative splice control mechanisms may modulate Vpu production during HIV-1 infection. Vpu plays two important roles in the natural replication cycle of HIV-1, reviewed in references 3 and 10. Firstly, Vpu promotes CD4 degradation in infected cells, allowing Env to efficiently accumulate at the cell surface for incorporation into new virions, thereby enhancing virus infectivity (10, 51, 63). Secondly, Vpu enhances virion release (27, 56), overcoming a restriction on virion release in human producer cells (58). Hence, changing the ratio of the env mRNA isoforms during viral infection may allow the virus to control Vpu-directed CD4 degradation, the accumulation of Env at the cell surface, and/or particle release, adjusting viral replication to suit the cellular environment. The alternative env isoforms are not redundant and appear, at the very least, to regulate Vpu production to perhaps aid HIV-1 replication in specific cellular environments.
Vpu expression by the native env isoforms clearly depended on the translation initiation signals in the 5′ UTR. The env mRNA isoforms containing exons 4bE, 4aE, and 4cE, such as those expressed by the pDRenv2, pDRenv3, and pDRenv4 cDNA constructs, respectively, contained the weak upstream AUG codon for Rev that would allow a proportion of scanning 40S ribosomes to initiate at the Rev AUG codon, thus depleting the number of ribosomes available to initiate at the downstream Vpu codon. The mutation of the Rev AUG codon confirmed its involvement in controlling Vpu expression from the env isoforms in a leaky ribosome-scanning manner (Fig. 6 and 7). Previous studies that described efficient Vpu translation from in vitro-transcribed env RNA isoforms containing exon 4bE and 4aE used the rabbit reticulocyte lysate translation system that ignores known control elements such as the 5′ m7Gppp cap structure and the 3′ poly(A) tail (17, 53). Rabbit reticulocyte lysate translation also ignores powerful upstream ORFs such as the Tat AUG codon in 4-kb tat mRNA, where the upstream Tat ORF otherwise blocks Env translation in vivo (53).
The most perplexing finding of this study was that even though the Rev AUG codon in the 5′ UTR controlled Vpu synthesis, the levels of Env synthesis by the env isoforms remained uniform. Therefore, contrary to that of Vpu, this consistent expression of Env protein does not lend itself to the leaky scanning translation mechanism previously reported by others (53). The native env isoforms expressed similar levels of Env irrespective of the 5′ UTR and Rev AUG codon. Furthermore, removing the upstream Rev and/or Vpu AUG codon or adding a strong Kozak consensus AUG codon in the 5′ UTR had little impact on the uniform synthesis of Env protein (Fig. 7 and 8). Explanations involving cryptic splicing to a monocistronic Env mRNA or highly stable RNA were also ruled out experimentally (Fig. 4 and 5). Instead, we propose that the consistent expression of Env from these isoforms arises from a discontinuous translation initiation mechanism that can bypass the effects of upstream AUG codons on downstream ORFs, enabling consistent Env production irrespective of the upstream Rev, Vpu, or strong AUG codon inserted into exon 2.
Examples of translation initiation by discontinuous initiation methods include the use of an IRES and ribosome shunting. Both mechanisms allow ribosomes to bypass upstream AUG codons and ORFs, targeting translation initiation to the correct, downstream start codon. For initiation by an IRES, ribosome components are recruited directly to the initiation codon mRNA without the 40S subunits engaging the 5′ cap structure, hence the description of this process as cap independent translation. In contrast, ribosome shunting is cap dependent, but the scanning 40S ribosome subunit can jump over or bypass large regions of RNA between shunt “donor” and “acceptor” motifs and does not require continuous scanning to locate downstream initiation codons (reviewed in references 18 and 40). Common to both of these translation initiation mechanisms are long 5′ UTR rich in secondary structures, a feature of HIV-1 5′ UTR (5, 7).
The translation of Env via the discontinuous IRES or ribosome shunt initiation would fit with the timing of Env translation late in HIV-1 replication, when viral Gag and Gag-Pol structural polyproteins are also synthesized. The Pol polyprotein contains the viral protease that was shown to cleave eIF4G and inhibit cap-dependent translation (59). Furthermore, IRES elements for the translation of both HIV-1 and simian immunodeficiency virus Gag have been defined, as well as elements for the translation of Env protein from other retroviruses (8, 11, 12, 16, 34, 39). Therefore, HIV-1 Env may use a discontinuous translation mechanism at a time late in replication if cap-dependent translation is diminished by the cleavage of eIF4G by the viral protease (59). The HIV Vpr accessory protein is also translated from mRNA containing RRE late in HIV-1 replication (53, 54). Vpr arrests the cell cycle in the G2/M phase (25, 48), which coincides with limiting cellular levels of the eIF4E, a critical 5′ cap binding translation factor that restricts cellular translation during cell division (50). Therefore, HIV-1 Env may also use an IRES or ribosome shunt translation mechanism to maximize Env production during G2/M cell cycle arrest. In a cellular environment in which viral protease and/or Vpr G2/M arrest represses the cap-dependent translation of Vpu and CD4, efficient Env synthesis may potentially overwhelm entrapment by CD4, ensuring efficient Env incorporation into virions.
Env translation may use cap-dependent initiation, but thereafter short ORFs in the 5′ UTR of the env isoforms may direct discontinuous ribosome shunting, similar to that in the cauliflower mosaic virus (43-45). Short ORFs are present in the env isoforms just upstream of but overlapping the Vpu ORF. Mutation studies of the highly conserved short ORF (32) suggest that it is a promising candidate for a donor site for potential Env ribosome shunt translation (31a).
The introduction of a strong upstream AUG codon can usurp all the scanning 40S ribosome subunits and block the translation of downstream ORFs (21). For example, in HIV-1 tat mRNAs, the high level of Kozak strength of the Tat ORF blocks downstream Rev and Nef translation in 1.8-kb mRNA (55). We found that adding the strong Kozak AUG codon to upstream exon 2 in env5 and env6 mRNAs produced from pDRenv5 and pDRenv6 cDNA plasmids did not block consistent Env production, despite significant declines in the levels of Vpu, which is translated by leaky scanning (Fig. 8). This result supports a discontinuous ribosome-scanning mechanism for Env translation to avoid the effect of the strong, upstream, artificial AUG codon in exon 2. In contrast, when the Kozak strength of the Rev AUG codon in the env2 isoform was increased nearly twofold (Fig. 6B), Env synthesis from the same isoform declined twofold (Fig. 6C), as would be expected if Env translation conformed to leaky ribosome-scanning initiation. But this mutation may also perturb the RNA structure involved in a discontinuous IRES or shunt initiation mechanism, for which RNA structure is often critical, and thus cause Env production to decline. The deletion of the Rev or Vpu AUG codon in isolation also led to minor, 1.3-fold increases in Env production (Fig. 6 and 7), consistent with leaky-scanning initiation, although the significance of such minor changes is questionable. Perhaps instead, these data indicate that HIV-1 Env is translated via dual mechanisms of discontinuous scanning (like the use of an IRES or shunt) and continuous leaky scanning after the translation of Vpu, analogous to the translation of adenovirus late mRNAs. With adenovirus late mRNAs, proportions of 40S ribosome subunits either shunt or continuously scan to translate downstream ORFs (65). A similar fate for ribosomes on HIV-1 env mRNAs would allow Env synthesis by a discontinuous IRES or shunt mechanism and Vpu (and, to a lesser extent, Env) synthesis by continuous leaky scanning. Therefore, the decline in Env levels corresponding to the Kozak Rev AUG codon mutation (Fig. 6C) may reflect a decline in Env production from leaky scanning while Env production by the discontinuous IRES or shunt mechanism proceeds.
HIV-1 env mRNAs are a compelling example in which separate translation mechanisms seem likely to translate the viral Vpu and Env genes. Perhaps lessons from the adenovirus late mRNAs, where 40S subunits scan continuously or shunt (65), will provide a useful model for understanding Vpu and Env production from HIV-1 env mRNAs. We have identified a mode of HIV-1 translational control in which two different translation initiation mechanisms enable the virus to regulate Vpu expression while maintaining a constant supply of Env protein. Delineating the translation initiation mechanism for Env may reveal key features that may benefit future DNA vaccines that seek efficient expression of diverse HIV-1 Env proteins in vivo without synthetically optimizing the codons in the mRNA.
Supplementary Material
Acknowledgments
We are grateful to K. Strebel and S. Bour for Vpu sera, plasmids, and critical discussions and to H. Schaal and C. M. Stoltzfus for review of the manuscript.
A CARG Postgraduate Research Scholarship to J. L. Anderson and grants 111700, 299907, and 400302 to D. F. J. Purcell from the National Health and Medical Research Council of Australia supported this research.
Footnotes
Published ahead of print on 28 February 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt, B. A., D. Hesslein, L. J. Chang, and C. M. Stoltzfus. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 14:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. L., and T. J. Hope. 2004. HIV accessory proteins and surviving the host cell. Curr. HIV/AIDS Rep. 1:47-53. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Asouli, Y., Y. Banai, H. Hauser, and R. Kaempfer. 2000. Recognition of 5′-terminal TAR structure in human immunodeficiency virus-1 mRNA by eukaryotic translation initiation factor 2. Nucleic Acids Res. 28:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout, B. 2000. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol. 48:29-73. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout, B., and J. L. B. Van Wamel. 2000. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA 6:282-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berlioz, C., and J.-L. Darlix. 1995. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 69:2214-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bour, S., F. Boulerice, and M. A. Wainberg. 1991. Inhibition of gp160 and CD4 maturation in U937 cells after both defective and productive infections by human immunodeficiency virus type 1. J. Virol. 65:6387-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bour, S., and K. Strebel. 2000. HIV accessory proteins: multifunctional components of a complex system. Adv. Pharmacol. 48:75-119. [DOI] [PubMed] [Google Scholar]
- 11.Brasey, A., M. Lopez-Lastra, T. Ohlmann, N. Beerens, B. Berkhout, J. L. Darlix, and N. Sonenberg. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 77:3939-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck, C. B., X. Shen, M. A. Egan, T. C. Pierson, C. M. Walker, and R. F. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75:181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caputi, M., M. Freund, S. Kammler, C. Asang, and H. Schaal. 2004. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J. Virol. 78:6517-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, Y.-N., D. J. Kenan, J. D. Keene, A. Gatignol, and K.-T. Jeang. 1994. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 68:7008-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419-424. [DOI] [PubMed] [Google Scholar]
- 16.Deffaud, C., and J.-L. Darlix. 2000. Characterization of an internal ribosome entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 74:846-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furtado, M. R., R. Balachandran, P. Gupta, and S. M. Wolinsky. 1991. Analysis of alternatively spliced human immunodeficiency virus type-1 mRNA species, one of which encodes a novel TAT-ENV protein. Virology 185:258-270. [DOI] [PubMed] [Google Scholar]
- 18.Gale, M., S.-L. Tan, and M. G. Katze. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatignol, A., A. Buckler-White, B. Berkhout, and K. T. Jeang. 1991. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 251:1597-1600. [DOI] [PubMed] [Google Scholar]
- 20.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in nonessential genes. AIDS Res. Hum. Retrovir. 10:343-350. [DOI] [PubMed] [Google Scholar]
- 21.Gray, N. K., and M. Wickens. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399-458. [DOI] [PubMed] [Google Scholar]
- 22.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365:186-191. [DOI] [PubMed] [Google Scholar]
- 23.Hope, T. J., X. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 25.Jowett, J. B., V. Planelles, B. Poon, N. P. Shah, M. L. Chen, and I. S. Y. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 69:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kjems, J., and P. Askjaer. 2000. Rev protein and its cellular partners. Adv. Pharmacol. 48:251-298. [DOI] [PubMed] [Google Scholar]
- 27.Klimkait, T., K. Strebel, M. D. Hoggan, M. A. Martin, and J. M. Orenstein. 1990. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J. Virol. 64:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak, M. 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196:947-950. [DOI] [PubMed] [Google Scholar]
- 29.Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283-292. [DOI] [PubMed] [Google Scholar]
- 30.Kozak, M. 1992. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8:197-225. [DOI] [PubMed] [Google Scholar]
- 31.Krummheuer, J., C. Lenz, S. Kammler, A. Scheid, and H. Schaal. 2001. Influence of the small leader exons 2 and 3 on human immunodeficiency virus type 1 gene expression. Virology 286:276-289. [DOI] [PubMed] [Google Scholar]
- 31a.Krummheuer, J., A. T. Johnson, I. Hauber, S. Kammler, J. L. Anderson, J. Hauber, D. F. Purcell, and H. Schaal. 27 February 2007, posting date. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology. doi: 10.1016/j.virol.2007.01.022. [DOI] [PubMed]
- 32.Kuiken, C. L., B. Foley, E. O. Freed, B. Hahn, B. Korber, P. A. Marx, F. McCutchan, J. W. Mellors, and S. M. Wolinsky. 2002. HIV Sequence Compendium 2002. Los Alamos National Laboratory, Los Alamos, NM.
- 33.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Lastra, M., C. Gabus, and J.-L. Darlix. 1997. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum. Gene Ther. 8:1855-1865. [DOI] [PubMed] [Google Scholar]
- 35.Maio, J. J., and F. L. Brown. 1988. Regulation of expression driven by human immunodeficiency virus type 1 and human T-cell leukemia virus type 1 long terminal repeats in pluripotential human embryonic cells. J. Virol. 62:1398-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mak, J., M. Jiang, M. A. Wainberg, M. L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maldarelli, F., M.-Y. Chen, R. L. Willey, and K. Strebel. 1993. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type 1 integral membrane protein. J. Virol. 67:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMillan, N. A. J., R. F. Chun, D. P. Siderovski, J. Galabru, W. M. Toone, C. E. Samuel, T. W. Mak, A. G. Hovanessian, K. T. Jeang, and B. R. G. Williams. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213:413-424. [DOI] [PubMed] [Google Scholar]
- 39.Ohlmann, T., M. Lopez-Lastra, and J.-L. Darlix. 2000. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 275:11899-11906. [DOI] [PubMed] [Google Scholar]
- 40.Pe'ery, T., and M. B. Mathews. 2000. Viral translational strategies and host defense mechanisms, p. 371-424. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. T. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98:7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 43.Pooggin, M. M., J. Futterer, K. G. Skryabin, and T. Hohn. 2001. Ribosome shunt is essential for infectivity of cauliflower mosaic virus. Proc. Natl. Acad. Sci. USA 98:886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pooggin, M. M., J. Futterer, K. G. Skryabin, and T. Hohn. 1999. A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J. Gen. Virol. 80:2217-2228. [DOI] [PubMed] [Google Scholar]
- 45.Pooggin, M. M., T. Hohn, and J. Fütterer. 2000. Role of a short upstream open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J. Biol. Chem. 275:17288-17296. [DOI] [PubMed] [Google Scholar]
- 46.Purcell, D. F. J., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riggs, N. L., S. J. Little, D. D. Richman, and J. C. Guatelli. 1994. Biological importance and cooperativity of HIV-1 regulatory gene splice acceptors. Virology 202:264-271. [DOI] [PubMed] [Google Scholar]
- 48.Rogel, M. E., L. I. Wu, and M. Emerman. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 69:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ropers, D., L. Ayadi, R. Gattoni, S. Jacquenet, L. Damier, C. Branlant, and J. Stevenin. 2004. Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J. Biol. Chem. 279:29963-29973. [DOI] [PubMed] [Google Scholar]
- 50.Sachs, A. B. 2000. Cell cycle-dependent translation initiation: IRES elements prevail. Cell 101:243-245. [DOI] [PubMed] [Google Scholar]
- 51.Schubert, U., L. C. Antón, I. Bacík, J. H. Cox, S. Bour, M. BenniOrlowski, K. Strebel, and J. W. Yewdell. 1998. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz, S., B. K. Felber, D. M. Benko, E.-M. Fenyö, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz, S., B. K. Felber, E. M. Fenyo, and G. N. Pavlakis. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 64:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1991. Expression of human immunodeficiency virus type 1 vif and vpr mRNAs is rev-dependent and regulated by splicing. Virology 183:677-686. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1992. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol. Cell. Biol. 12:207-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strebel, K., T. Klimkait, F. Maldarelli, and M. A. Martin. 1989. Molecular and biochemical analyses of human immunodeficiency virus type 1 Vpu protein. J. Virol. 63:3784-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strebel, K., T. Klimkait, and M. Martin. 1988. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 241:1221-1223. [DOI] [PubMed] [Google Scholar]
- 58.Varthakavi, V., R. M. Smith, S. P. Bour, K. Strebel, and P. Spearman. 2003. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA 100:15154-15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ventoso, I., R. Blanco, C. Perales, and L. Carrasco. 2001. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl. Acad. Sci. USA 98:12966-12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waysbort, A., S. Bonnal, S. Audigier, J.-P. Estève, and A.-C. Prats. 2001. Pyrimidine tract binding protein and La autoantigen interact differently with the 5′ untranslated regions of lentiviruses and oncoretrovirus mRNAs. FEBS Lett. 490:54-58. [DOI] [PubMed] [Google Scholar]
- 61.Wentz, M. P., B. E. Moore, M. W. Cloyd, S. M. Berget, and L. A. Donehower. 1997. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type 1 induces dominant aberrant splicing and arrests virus production. J. Virol. 71:8542-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wigler, M., A. Pellicer, S. Silverstein, and R. Axel. 1978. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as a donor. Cell 14:725-731. [DOI] [PubMed] [Google Scholar]
- 63.Willey, R., F. Maldarelli, M. A. Martin, and K. Strebel. 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 66:7193-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 65.Yueh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557-1567. [DOI] [PubMed] [Google Scholar]
- 66.Zahler, A. M., C. K. Damgaard, J. Kjems, and M. Caputi. 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279:10077-10084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.