Abstract
Recombinant human immunodeficiency virus type 1 (HIV-1) strains containing sequences from different viral genetic subtypes (intersubtype) and different lineages from within the same subtype (intrasubtype) have been observed. A consequence of recombination can be the distortion of the phylogenetic signal. Several intersubtype recombinants have been identified; however, less is known about the frequency of intrasubtype recombination. For this study, near-full-length HIV-1 subtype C genomes from 270 individuals were evaluated for the presence of intrasubtype recombination. A sliding window schema (window, 2 kb; step, 385 bp) was used to partition the aligned sequences. The Shimodaira-Hasegawa test detected significant topological incongruence in 99.6% of the comparisons of the maximum-likelihood trees generated from each sequence partition, a result that could be explained by recombination. Using RECOMBINE, we detected significant levels of recombination using five random subsets of the sequences. With a set of 23 topologically consistent sequences used as references, bootscanning followed by the interactive informative site test defined recombination breakpoints. Using two multiple-comparison correction methods, 47% of the sequences showed significant evidence of recombination in both analyses. Estimated evolutionary rates were revised from 0.51%/year (95% confidence interval [CI], 0.39 to 0.53%) with all sequences to 0.46%/year (95% CI, 0.38 to 0.48%) with the putative recombinants removed. The timing of the subtype C epidemic origin was revised from 1961 (95% CI, 1947 to 1962) with all sequences to 1958 (95% CI, 1949 to 1960) with the putative recombinants removed. Thus, intrasubtype recombinants are common within the subtype C epidemic and these impact analyses of HIV-1 evolution.
Human immunodeficiency virus type 1 (HIV-1) genetic diversity is generated during viral replication and is caused by error-prone reverse transcription, host-mediated hypermutation, and recombination. Recombination has been estimated to occur approximately 2.8 times per genome per viral replication cycle (61). Detectable recombination requires simultaneous infection with more than one virus at the cellular level. In a previous report, an average of three to four distinct proviral genomes were found in infected spleen cells (21), indicating that conditions do exist for extensive recombination between different viral genomes. Indeed, numerous intersubtype recombinants resulting in both unique forms found in single individuals and circulating forms have been observed (http://hiv-web.lanl.gov/content/hiv-db/CRFs/CRFs.html). Furthermore, recombination among sequences from the same subtype, or intrasubtype recombination, has also been observed within individual patients (36, 47, 59).
Recombination can distort the phylogenetic signal. In phylogenetic analyses, the variant sites in the recombinant portions of the genome will appear as parallel evolution (homoplasy) instead of a single lineage. This can cause longer branch lengths, overestimate the substitution rate heterogeneity, and cause false rejection of molecular clock hypotheses (26, 44). Furthermore, this effect can confound studies involving the association of viral evolutionary changes and other factors such as host genetics (3, 32). As a result of greater sequence similarity, it is more difficult to detect intrasubtype recombination than intersubtype recombination and the true amount of intrasubtype recombination is unknown. For these reasons, intrasubtype recombination is commonly ignored in phylogenetic analyses of HIV-1 sequences.
Subtype C is the most prevalent HIV-1 subtype worldwide and, thus, a critical target for vaccine design. Phylogenetic analysis of viral sequences at the host population level can allow for the estimations of ancestral sequences, rates of viral evolution, and geographic clustering, which may be important for the design of an effective vaccine. The presence of intrasubtype recombination within subtype C genomes may impact these analyses. This study addresses the prevalence of intrasubtype recombinants among near-full-length subtype C genomes at the host population level and their impact on phylogenetic analyses.
MATERIALS AND METHODS
Selection of sequences.
Sequences were selected from the Los Alamos HIV-1 sequence database (http://hiv-web.lanl.gov/content/hiv-db/mainpage.html) in August 2005 (n = 118) and from sequences obtained as part of our studies of South African subjects (n = 152) from KwaZulu-Natal Province, South Africa (42). The South African sequences we obtained were collected as part of a study approved by the University of Washington and University of KwaZulu-Natal internal review boards, and all subjects gave informed consent. The country of origin and the year of sample collection were available for most sequences from the Los Alamos database. In addition, viral load and CD4 T-cell counts were available for the 145 South African subjects (22). All subtype C sequences of ≥8 kb were included in the final data set. A total of 270 subtype C sequences were collected for analysis. All sequences were evaluated for intersubtype recombination using REGA (7) and RIP 3.0 (http://www.hiv.lanl.gov/content/hiv-db/RIP3/RIP.html). Two were found to be dissimilar intersubtype recombinants and were confirmed with bootscanning as implemented in SimPlot (27) using consensus sequences for subtypes A to H, followed by the informative site test (39). Both of these sequences were removed from the data set. Both were unique recombinants between subtypes C and A and were not similar to any known circulating recombinant form (GenBank accession numbers DQ351238 and DQ093606). Five sequences were found to be hypermutated by using the program Hypermut (41) and were also removed, leaving 263 sequences for analysis (Table 1 and http://mullinslab.microbiol.washington.edu/HIV/Rousseau2007/TableS1.html). Sequences were aligned using ClustalW, version 1.83 (55), and MacClade, version 4.08 (28), and regions of uncertain positional homology were removed (i.e., repetitious elements and the internal regions of the env variable loops). The total sequence length of the final alignment was 7,701 bp.
TABLE 1.
Geographic origin of sequences
| Country | No. of sequences |
|---|---|
| South Africa | 159 |
| Botswana | 50 |
| Tanzania | 18 |
| India | 10 |
| Brazil | 8 |
| Ethiopiaa | 6 |
| Zambia | 3 |
| Australia | 2 |
| Argentina | 1 |
| Kenya | 1 |
| Myanmar | 1 |
| Senegal | 1 |
| Somalia | 1 |
| Uruguay | 1 |
| Yemen | 1 |
| Total | 263 |
Includes five sequences from Israel that originated in Ethiopia (40).
Phylogenetic analysis.
Maximum-likelihood trees were inferred using PHYML (15) as follows: 100 bootstrap trees were generated from BIONJ (9) start trees; the bootstrap trees were used in turn as starting trees for a final maximum-likelihood analysis with PHYML (15). Since the results of phylogenetic analyses using heuristic searching algorithms depend on the starting trees, we used the bootstrap trees to provide a variety of starting conditions. This method was validated by using a set of 45 sequences encompassing the C2V5 region of env (549 nucleotides) (data not shown) and comparing the results of the above-described method to those of two alternate methods: (i) a maximum-likelihood tree using a single BIONJ start tree in PHYML (15) and (ii) a maximum-likelihood tree from PAUP*, version 4.0b10 (53), calculated with tree-bisection-reconnection rearrangement from both 10 random-addition orders and a neighbor-joining start. The best tree from the original method always produced a higher likelihood score than those from the two alternate methods.
Identification of reference sequences.
The viral genome length alignment was divided into a series of 16 partitions of approximately 2,000 bp, with a 1,615-bp overlap. An average of 1,057 informative sites were present in each partition (range, 855 to 1,251). The partition and overlap sizes were designed to balance the need for sufficient sequence to generate an accurate tree and to be small enough to allow topological variation based on small regions of recombination. Each partition was used to generate a maximum-likelihood phylogenetic tree as described above. Reference sequences (n = 23) were identified by comparing the trees generated from each of the partitions to each other and to the tree generated from the full-length alignment. Sequences that were in clusters with ≥70% bootstrap support in the tree from the full-length alignment and that were in the same topological position in each of the partition trees were defined as reference sequences. The most recent common ancestor of clusters consisting of three or more reference sequences was selected to represent those clusters.
Identification of recombination.
Each nonreference sequence was compared to the reference set using two passes of bootscanning as implemented in SimPlot (27). The first pass consisted of a comparison of the query sequence to the full set of reference sequences. A second-pass bootscan was performed comparing the query to a subset of references that appeared to be parental sequences based on the first pass. Putative breakpoints were identified, and the informative site test was used to evaluate the significance level supporting recombination (39). Two methods of evaluating the statistical significance of the informative site distributions were used. The first involved a chi-square value with Yates' correction. The values with a false discovery rate of ≤0.05 (2) were considered to provide significant evidence of recombination. The second method involved Fisher's exact test followed by the calculation of the q value (52), a number that indicates the proportion of false-positive results expected among the total significant results. Sequences with q values of ≤0.05 were considered to have significant evidence of recombination.
The 16 partitions of the full-length alignment were modified by removing the first and second codon position sites for nonoverlapping coding regions. In parallel, the complete sequence alignment was also divided into coding regions corresponding to p17, p24, p2/p7, protease, reverse transcriptase, RNase H, integrase, Vif, Vpr, Tat/Rev exon1, Tat/Rev exon2, Vpu, gp120, gp41, and Nef. The Shimodaira-Hasegawa test (46) was performed to compare the topologies of the phylogenetic trees from the 16 partitions of the near-full-length sequence, the 16 partitions of the third-codon-position sites, and the coding region partitions by using PAUP*.
Other statistical analysis.
The Slatkin-Maddison test (48) was performed to evaluate geographic clustering by using MacClade, version 4.08 (28). The association of recombinants with terminal branch lengths was evaluated using multivariate linear regression with year of sample as a covariate. The most recent common ancestors (MRCAs) were estimated using PAUP* (53) based on the evolutionary model estimated using PHYML (15). The estimates of the year of origin of the subtype C epidemic, the year of origin of the three sample recombinants (TV002, 00BW192113, and 98IN012), and the divergence rate were attained using previously described methods (24) which were adapted for use with sampling times known to the nearest day, month, or year. Clinical and demographic associations were evaluated in Stata 8.0 (StataCorp, College Station, TX) using linear regression and the chi-square test. The rate of recombination was estimated using RECOMBINE, version 1.4.1 (25). Because it would be computationally too intensive to analyze the complete data set with RECOMBINE, five sets of 20 sequences were randomly selected without replacement using the Random function in Microsoft Excel (version 11.2.3) and each of these sets was used for the estimation of the recombination rate.
RESULTS
Geographic clustering.
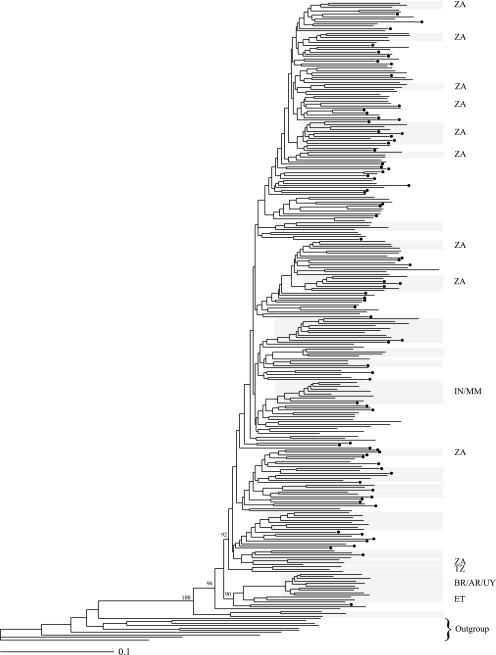
The maximum-likelihood phylogenetic tree of the near-full-length sequences revealed several clusters with ≥70% bootstrap support (Fig. 1). Several of the well-supported clusters corresponded to geographic regions and thus presumably resulted from founder events. There was significant evidence of overall geographic clustering (P < 0.0001) but no evidence of geographic clustering within South Africa (P = 0.23). Significant clusters consisting of sequences from a single geographic region included a cluster with sequences from Ethiopia, South America, India, and Botswana as well as several clusters of sequences from South Africa.
FIG. 1.
Maximum-likelihood tree of the complete set of subtype C sequences. Terminal clusters with more than two sequences and ≥70% bootstrap support are highlighted. Additional bootstrap values are given at the corresponding node. Clusters with sequences from a single geographic region are labeled to the right of the cluster, with the letters corresponding to the nations represented according to the ISO3166 code list (http://www.iso.ch/iso/en/prods-services/iso3166ma/02iso-3166-code-lists/list-en1.html). Dots indicate putative recombinant sequences. The outgroup is indicated with a bracket. ZA, South Africa; IN, India; MM, Myanmar; TZ, Tanzania; BR, Brazil; AR, Argentina; UY, Uruguay; ET, Ethiopia.
Recombination in the data set.
The percent difference in log likelihoods of the tree topologies given the sequence data for each of the 16 sliding window partitions were compared to each other and to that of the tree generated from the full-length alignment. A total of 272 comparisons were performed (Table 2). Ninety-one percent of the comparisons showed a significant difference in likelihood, suggesting that these regions evolved along different topologies. The tree topologies of each genetic region were similarly compared, and 99.6% of the comparisons were significant (data not shown). The presence of recombination in the data set is one explanation for the observed high levels of divergence. Because convergent evolution due to parallel replacement of amino acids may also lead to incongruence in phylogenetic tree topologies, the same analysis was repeated with only the third position of each codon included (data not shown), as it is less likely to be under selective pressure. Similarly, 90% of comparisons were found to be significant, suggesting that recombination was likely to have taken place among the sequences in this data set and that recombination takes place throughout the genome.
TABLE 2.
Results of Shimodaira-Hasegawa test of log-likelihood scores of phylogenetic trees from 16 HIV-1 sequence alignment partitionsa
| Topology | % Difference in log-likelihood scores between indicated topology and partition:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | Full | |
| 1 | Bestb | 0.7209 | 1.1930 | 1.9674 | 2.1753 | 2.2974 | 2.3512 | 2.6015 | 2.7271 | 2.7048 | 2.6335 | 2.7620 | 2.5882 | 2.6108 | 2.5713 | 2.5361 | 1.3891 |
| 2 | 0.5185 | Best | 0.2557 | 0.9859 | 1.1623 | 1.2799 | 1.7866 | 2.1588 | 2.6553 | 2.6878 | 2.5347 | 2.5648 | 2.3750 | 2.3361 | 2.1135 | 2.0083 | 1.1230 |
| 3 | 1.0678 | 0.5327 | Best | 0.6109 | 0.8745 | 0.9897 | 1.5571 | 2.4391 | 2.9920 | 2.9947 | 2.7849 | 2.9575 | 2.7233 | 2.7536 | 2.5049 | 2.4218 | 1.3669 |
| 4 | 1.0561 | 0.7319 | 0.2932 | Best | 0.2855 | 0.4307 | 0.8002 | 1.6009 | 2.5731 | 2.7184 | 2.5613 | 2.7427 | 2.6035 | 2.6045 | 2.4672 | 2.4265 | 1.2074 |
| 5 | 1.5933 | 1.0578 | 0.5012 | 0.2280 | Best | 0.0630 | 0.7739 | 1.6692 | 2.5656 | 2.7227 | 2.6942 | 2.9002 | 2.6727 | 2.7123 | 2.6138 | 2.5240 | 1.3184 |
| 6 | 2.0691 | 1.5322 | 0.8054 | 0.5802 | 0.2133 | Best | 0.5049 | 1.4456 | 2.3298 | 2.5709 | 2.4989 | 2.7465 | 2.5415 | 2.7267 | 2.5447 | 2.5870 | 1.3661 |
| 7 | 2.0241 | 1.9965 | 1.6533 | 1.3221 | 0.9574 | 0.6126 | Best | 0.4764 | 1.3834 | 1.6688 | 1.8100 | 2.3133 | 2.2885 | 2.4053 | 2.2186 | 2.1311 | 1.1313 |
| 8 | 2.0890 | 2.2356 | 2.2325 | 1.9515 | 1.5627 | 1.2564 | 0.4859 | Best | 0.7663 | 1.0350 | 1.2561 | 1.8467 | 2.1894 | 2.4730 | 2.2735 | 2.2648 | 1.1607 |
| 9 | 2.3840 | 2.7783 | 2.9452 | 3.0665 | 2.6967 | 2.3794 | 1.3562 | 0.5137 | Best | 0.2814 | 0.6077 | 1.2922 | 1.6419 | 2.4848 | 2.4826 | 2.3586 | 1.2749 |
| 10 | 2.3853 | 2.7809 | 2.7679 | 3.0004 | 2.5331 | 2.2114 | 1.1801 | 0.5814 | 0.1744 | Best | 0.3394 | 0.9424 | 1.2240 | 1.9605 | 2.0840 | 2.0594 | 1.0873 |
| 11 | 2.0515 | 2.3241 | 2.4578 | 2.7522 | 2.5574 | 2.3529 | 1.6246 | 1.0726 | 0.6188 | 0.3250 | Best | 0.4112 | 0.5662 | 1.1464 | 1.3853 | 1.6641 | 0.8758 |
| 12 | 1.9734 | 2.3448 | 2.4497 | 2.7742 | 2.7011 | 2.5509 | 2.0917 | 1.6131 | 1.1423 | 0.7546 | 0.2168 | Best | 0.0246 | 0.5643 | 0.7589 | 1.1675 | 0.7813 |
| 13 | 2.1229 | 2.4303 | 2.5165 | 2.8453 | 2.7626 | 2.6427 | 2.4491 | 2.3765 | 1.9560 | 1.5132 | 0.9095 | 0.5765 | Best | 0.5025 | 0.7633 | 1.1289 | 1.0132 |
| 14 | 2.3206 | 2.7398 | 2.8867 | 3.2141 | 3.1136 | 2.9320 | 2.7095 | 2.7783 | 2.7982 | 2.3112 | 1.4976 | 1.0486 | 0.4448 | Best | 0.3187 | 0.7565 | 1.1685 |
| 15 | 2.0514 | 2.3911 | 2.4271 | 2.8182 | 2.6858 | 2.4941 | 2.2069 | 2.2631 | 2.4310 | 2.1594 | 1.4966 | 1.2172 | 0.6932 | 0.3354 | Best | 0.3885 | 0.9242 |
| 16 | 2.2006 | 2.5360 | 2.5980 | 2.9949 | 2.8646 | 2.6873 | 2.3829 | 2.5254 | 2.6265 | 2.4253 | 2.0910 | 1.7298 | 1.2809 | 0.8847 | 0.3880 | Best | 1.0002 |
| Full | 0.7028 | 0.8927 | 0.8995 | 1.1256 | 1.0281 | 0.9316 | 0.5759 | 0.5808 | 0.6830 | 0.6131 | 0.4665 | 0.5974 | 0.5677 | 0.6368 | 0.4246 | 0.3091 | Best |
Full, full-length alignment. Different fonts indicate significance: boldface, P < 0.001; underline, P < 0.01; italic, P < 0.05; roman, not significant.
Best indicates the comparison of trees generated from the same data.
The rate of recombination was estimated for five sets of 20 randomly selected sequences. The 95% confidence interval (CI) of each estimate did not overlap zero, providing further evidence for the presence of recombination among this set of sequences. The per-site ratio of the recombination rate to the mutation rate ranged from 0.0023 (95% CI, 0.0010 to 0.0044) to 0.0067 (95% CI, 0.0039 to 0.010). Using these recombination rates, an average divergence value of 0.3170 in our tree, and a previously reported mutation rate of HIV (2.5 × 10−5) (29), we calculated an expected 5.6 to 16.4 recombination events per genome.
Identification of recombinants.
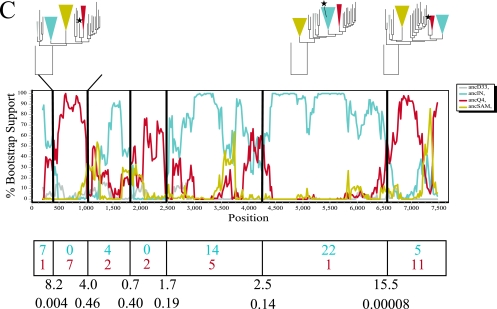
A total of 23 reference sequences (including ancestral states) that maintained consistent topological positioning in the 16 partition trees were identified. Nonreference sequences (n = 167) were compared to the reference set using two passes of bootscanning, followed by the informative site test (Fig. 2). The number of informative sites matching each parental sequence was used in a contingency table, and either a Yates-corrected chi-square test or a Fisher exact test was performed to determine whether significant evidence of recombination existed. At a false discovery rate of ≤0.05, the Yates-corrected chi-square values showed that 77 (47%) of the sequences had significant evidence of recombination while the Fisher exact test showed the same 77 sequences plus an additional 40 (25%), for a total of 72%, with a q value of ≤0.05. Among the 77 putative recombinants defined from both analyses, 16% had putative parental sequences that were ancestors from the South American cluster, Indian cluster, or Ethiopian cluster. From the estimated time of origin of the parental fragments (regions flanking the most significant breakpoints) (Fig. 2), the recombination events were estimated to have taken place after 1982 for TV002, after 1980 for 00BW192113, and after 1974 for 98IN012.
FIG. 2.
Phylogenetic, bootscan, and informative site analyses of three sample recombinant sequences: (A) TV002, (B) 00BW192113, and (C) 98IN012. The maximum-likelihood phylogenetic trees, constrained on phylogenetically defined clusters based on the ancestral reference sequences used in bootscanning (the ancestor to the Indian cluster [ancIN], the ancestor to the South American cluster [ancSAM], and ancestors to other arbitrarily named clusters [ancD33, ancG3, ancQ4, ancT13, and ancT17]), are shown above regions flanking significant breakpoints. The colors of the branches and constrained groups match the colors of the representative sequences in the bootscan analysis. The putative recombinant sequence is labeled with a star. The bootscan analysis was performed using a window size of 400 bp and a step size of 20 bp. The graph represents the bootstrap support for clustering with each of the putative parental sequences (ancIN, ancSAM, ancD33, ancG3, ancQ4, ancT13, ancT17, and SK213). Putative recombination breakpoints are shown as black vertical lines. The number of informative sites of the query sequence matching each putative parental sequence for each region is shown below the graph in the corresponding color. The Yates-corrected chi-square value and the corresponding P value are shown below each breakpoint. The times of origin of the most recent ancestors to the recombinant sequences were as follows: (A) 1982 (95% CI, 1963 to 1988) for the portion of TV002 most related to ancT13 and 1973 (95% CI, 1956 to 1977) for the portion most related to SK213, (B) 1970 (95% CI, 1954 to 1976) for the central portion of 00BW192113 most related to ancT17 and 1980 (95% CI, 1968 to 1983) for the 3′ portion most related to ancG3, and (C) 1974 (95% CI, 1943 to 1979) for the central portion of 98IN012 most related to ancIN and 1970 (95% CI, 1960 to 1978) for the 3′ portion most related to ancQ4.
Impact of the putative recombinants.
The 77 putative recombinants identified using both methods of multiple comparison were removed from the alignment, and the maximum-likelihood phylogenetic tree was regenerated using PHYML (15). Evidence of geographic clustering remained significant (P < 0.0001), and clustering within South Africa remained nonsignificant (P = 0.08). The MRCA estimated from the tree generated from the complete set of sequences differed slightly from the MRCA generated from the set of sequences with the putative recombinants removed (0.23% nucleotide distance and 0.43% amino acid difference). The consensus sequence of the complete data set was less impacted by the removal of the recombinant sequences (0.05% nucleotide distance and 0.09 to 0.17% amino acid difference).
To estimate the time of origin of the subtype C epidemic, we used only the third position site from each codon, as this position does not change the encoded amino acid as often and is therefore under less selective pressure than the first and second positions. The rate of HIV-1 divergence was then estimated with and without the 77 putative recombinants. The estimated rates were slightly shifted without the recombinants: 0.51%/year (95% CI, 0.39 to 0.53%) with all sequences versus 0.46%/year (95% CI, 0.38 to 0.48%) with putative recombinants removed. The estimates of the year of origin of the subtype C epidemic were slightly different between the two groups: 1961 (95% CI, 1947 to 1962) with all sequences versus 1958 (95% CI, 1949 to 1960) with the recombinants removed. These results were more dramatic when we removed the additional 25% of the sequences that had been identified as recombinants in the Fisher exact test with a q value of >0.05 (divergence rate, 0.37%/year [95% CI, 0.27 to 0.41%]; origin, 1954 [95% CI, 1932 to 1956]).
The 77 putative recombinant genomes that had been identified using both methods of correction for multiple comparisons were significantly associated with longer branch lengths after we adjusted for the year of sampling (P = 0.001) but were not significantly associated with any particular country, year, or geographic region (P > 0.05). Nor were recombinants significantly associated with viral load or CD4 T-cell counts (P > 0.05).
DISCUSSION
This study demonstrates that intrasubtype recombination is frequently detected among near-full-length HIV-1 subtype C genomes. For observable recombination to take place in lentiviral infections, an individual must be infected with two different viral strains (dual infection) and these strains must then infect the same cell, be copackaged, and form an infectious new virion. Dual infection with different subtypes or the same subtype has been reported to be common (1, 5, 12-14, 17-19, 49, 54, 60), and newly generated recombinants have been observed within some (8, 10, 31, 35, 36) but not all (4, 6, 23) dually infected patients. Furthermore, recombination has been observed in multiply infected cells (21) and the observation of circulating recombinant forms demonstrates that recombinant HIV-1 genomes are capable of being transmitted (http://hiv-web.lanl.gov/content/hiv-db/CRFs/CRFs.html). Thus, the results we describe from our study are consistent with the biology and epidemiology of HIV-1.
Although we can conclude that recombination is extensive, the rate of recombination does not need to be high for a high number of recombinants to be present. Using the lowest recombination rate per mutation rate estimated in our study (0.0023), we calculated at least 5.6 recombination events per genome. This suggests that even with a low recombination rate, most genomes are predicted to be recombinants. Furthermore, our estimated recombination rate was much lower than the mutation rate, despite the fact that we know that recombination occurs 2 to 3 times per replication cycle per genome, a frequency 10 times greater than for the generation of point mutations (20, 29, 61). We expect that we are not detecting all recombinant genomes, as some of them may have parental sequences that are genetically too similar to distinguish using the methods employed in this report. Because recombination between two very similar viral strains is difficult to detect, many HIV-1 sequences will never be identified as recombinants. The limiting factors in the identification of recombinants are thus the occurrence of dual infection and the degree of divergence of the dually infecting strains.
We demonstrated that recombination had a modest impact on the inferred MRCA and a greater influence on the estimated rate of HIV-1 evolution. Because recombinant regions of a genome can be incorrectly scored as parallel evolution, it is not surprising that our estimate of the rate of HIV-1 evolution with the recombinants removed was lower than that with all sequences. Because of this, it was also not surprising that the estimated date of origin of the subtype C epidemic with the recombinants removed was earlier than that with all sequences. From a previous analysis of envelope sequences, the MRCA of the HIV-1 epidemic in humans is thought to have taken place near 1930 (95% CI, 1913 to 1944) (24). Another report based on an analysis of envelope and gag sequences showed the estimated origin of the subtype C epidemic to have taken place in 1962 (95% CI, 1956 to 1968 based on the full envelope alignment and genetic distance) (56). Using the most conservative definition of recombinant (resulting in 47% recombinants), our estimate of the origin of the subtype C epidemic (1958 [95% CI, 1949 to 1960]) places the date of origin of the subtype C epidemic several years after the origin of the HIV-1 epidemic and within the 95% CI of the previous estimate of the origin of the subtype C epidemic. Removing the more broadly defined recombinant sequences (77%), we found that the estimate of the origin was even earlier (1954 [95% CI, 1932 to 1956]) and barely within the CI of the previous estimate based on gag sequences. Both of our estimates of the divergence rate for subtype C without the putative recombinants (0.46%/year [95% CI, 0.38 to 0.48%] and 0.37%/year [95% CI, 0.27 to 0.41%]) were higher than that previously reported for all subtypes (0.24%/year [95% CI, 0.18 to 0.28%]) (24). This may be due to the use of different evolutionary models, third-site positions (versus all codon positions), and entire genome sequences (versus gp160). We conclude that the presence of intrasubtype recombination has implications for future phylogenetic analyses using these or other sequences, because taking recombination into account may increase estimate accuracy.
Using additional data and analyses compared to previous reports, we were also able to further identify evidence of geographic clustering within the subtype C epidemic (11, 16, 30, 33, 34, 37, 40, 43, 45, 51, 57, 58, 62). Vaccine candidates designed on regional HIV-1 subtype C strains may be able to provide greater sequence coverage than those designed on all subtype C sequences detected to date. The impact of recombination was negligible in the context of overwhelming evidence of geographic clustering.
The primary limitation to recombination analysis is the difficulty in identifying a null distribution for comparison, as all present-day viruses are recombinants at some level. The Fisher exact test used in this study was a conservative test for evaluating the distribution of informative sites surrounding a putative breakpoint. A conservative false discovery rate (q ≤ 5%) was then applied to the resulting P values as a mechanism to take multiple comparisons into account. However, the chi-square values adjusted with false discovery rate correction resulted in a lower estimate of the number of recombinants (47% versus 72%). In either case, a large proportion of the sequences were identified as intrasubtype recombinants. An alternate method suggested by Maynard Smith (29a) was to compare the results of the informative site analysis to those of a distribution of identical tests performed on a set of nonrecombinant genomes. Nonrecombinant genomes were simulated by randomizing the nucleotide columns in the sequence alignment. The identification of putative recombinants was then repeated many times with the randomized data set to establish a null distribution for comparison. An automated version of this method is available that allows a single breakpoint and equally spaced flanking regions (29a, 38). However, our larger-scale analysis involved the selection of up to three breakpoints that were spaced to maximize the chi-square value. Establishing this type of null distribution with several breakpoints is computationally more complex and not currently possible in an automated manner using the number of sequences analyzed here. An additional inherent limitation to intrasubtype recombination analysis is that recombinant sequences whose parental sequences were very similar or not present in our data set would not have been detectable. Despite these limitations, our analysis using the best available methods and conservative statistical correction clearly demonstrates that intrasubtype recombinants are common among HIV-1 subtype C genomes.
In this study, the space between two informative sites flanking a putative breakpoint often spanned several hundred to several thousand base pairs; thus, uncertainty existed in most breakpoint estimates. Additional methods or more intensely sampled sequences would be needed to better define these breakpoints in order to compare them across genomes and identify hot/cold spots of recombination. Such analyses may provide additional information about the mechanism(s) of recombination.
Our study demonstrated that intrasubtype recombination can be detected and is common among circulating HIV-1 subtype C strains throughout the genome. This work underscores the importance of recombination as a significant contributor to HIV-1 diversity as well as the value of taking intrasubtype recombination into account in phylogenetic analyses of HIV-1. Accurate phylogenetic inferences may be important for future vaccine designs based on most central or ancestral sequences.
Acknowledgments
We thank all of the South African individuals who contributed samples for our study and the HIV-1 Pathogenesis Program staff at the Doris Duke Medical Research Institute in Durban, South Africa. We also thank Feng Gao, Vijay K. Chaudhary, and Shigeru Kusagawa for contributing unpublished sequences and David Lockhart for statistical consultation.
This work was funded by NIH contract N01-A1-11514 to B.D.W., the University of Washington Center for AIDS Research New Investigator Award NIH R37 AI047734 to C.M.R., and the University of Washington Center for AIDS Research Genomics Core Facility P01 AI27757.
Footnotes
Published ahead of print on 21 February 2007.
REFERENCES
- 1.Becker-Pergola, G., J. L. Mellquist, L. Guay, F. Mmiro, C. Ndugwa, P. Kataaha, J. B. Jackson, and S. H. Eshleman. 2000. Identification of diverse HIV type 1 subtypes and dual HIV type 1 infection in pregnant Ugandan women. AIDS Res. Hum. Retrovir. 16:1099-1104. [DOI] [PubMed] [Google Scholar]
- 2.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 3.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science, in press. [DOI] [PubMed]
- 4.Brenner, B., J. P. Routy, Y. Quan, D. Moisi, M. Oliveira, D. Turner, and M. A. Wainberg. 2004. Persistence of multidrug-resistant HIV-1 in primary infection leading to superinfection. AIDS 18:1653-1660. [DOI] [PubMed] [Google Scholar]
- 5.Chohan, B., L. Lavreys, S. M. Rainwater, and J. Overbaugh. 2005. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J. Virol. 79:10701-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curlin, M. E., G. S. Gottlieb, S. E. Hawes, P. S. Sow, I. Ndoye, C. W. Critchlow, N. B. Kiviat, and J. I. Mullins. 2004. No evidence for recombination between HIV type 1 and HIV type 2 within the envelope region in dually seropositive individuals from Senegal. AIDS Res. Hum. Retrovir. 20:958-963. [DOI] [PubMed] [Google Scholar]
- 7.de Oliveira, T., K. Deforche, S. Cassol, M. Salminen, D. Paraskevis, C. Seebregts, J. Snoeck, E. J. van Rensburg, A. M. Wensing, D. A. van de Vijver, C. A. Boucher, R. Camacho, and A. M. Vandamme. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797-3800. [DOI] [PubMed] [Google Scholar]
- 8.Fang, G., B. Weiser, C. Kuiken, S. M. Philpott, S. Rowland-Jones, F. Plummer, J. Kimani, B. Shi, R. Kaul, J. Bwayo, O. Anzala, and H. Burger. 2004. Recombination following superinfection by HIV-1. AIDS 18:153-159. [DOI] [PubMed] [Google Scholar]
- 9.Gascuel, O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 10.Gerhardt, M., D. Mloka, S. Tovanabutra, E. Sanders-Buell, O. Hoffmann, L. Maboko, D. Mmbando, D. L. Birx, F. E. McCutchan, and M. Hoelscher. 2005. In-depth, longitudinal analysis of viral quasispecies from an individual triply infected with late-stage human immunodeficiency virus type 1, using a multiple PCR primer approach. J. Virol. 79:8249-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, M., T. De Oliveira, K. Bishop, H. M. Coovadia, L. Madurai, S. Engelbrecht, E. Janse van Rensburg, A. Mosam, A. Smith, and S. Cassol. 2003. Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: implications for vaccine and antiretroviral control strategies. J. Virol. 77:2587-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlieb, G. S., D. C. Nickle, M. A. Jensen, K. G. Wong, J. Grobler, F. Li, S. L. Liu, C. Rademeyer, G. H. Learn, S. S. Karim, C. Williamson, L. Corey, J. B. Margolick, and J. I. Mullins. 2004. Dual HIV-1 infection associated with rapid disease progression. Lancet 363:619-622. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb, G. S., P. S. Sow, S. E. Hawes, I. Ndoye, A. M. Coll-Seck, M. E. Curlin, C. W. Critchlow, N. B. Kiviat, and J. I. Mullins. 2003. Molecular epidemiology of dual HIV-1/HIV-2 seropositive adults from Senegal, West Africa. AIDS Res. Hum. Retrovir. 19:575-584. [DOI] [PubMed] [Google Scholar]
- 14.Grobler, J., C. M. Gray, C. Rademeyer, C. Seoighe, G. Ramjee, S. Abdool Karim, L. Morris, and C. Williamson. 2004. Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J. Infect. Dis. 190:1355-1359. [DOI] [PubMed] [Google Scholar]
- 15.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 16.Harris, M. E., S. Maayan, B. Kim, M. Zeira, G. Ferrari, D. L. Birx, and F. E. McCutchan. 2003. A cluster of HIV type 1 subtype C sequences from Ethiopia, observed in full genome analysis, is not sustained in subgenomic regions. AIDS Res. Hum. Retrovir. 19:1125-1133. [DOI] [PubMed] [Google Scholar]
- 17.Herbinger, K. H., M. Gerhardt, S. Piyasirisilp, D. Mloka, M. A. Arroyo, O. Hoffmann, L. Maboko, D. L. Birx, D. Mmbando, F. E. McCutchan, and M. Hoelscher. 2006. Frequency of HIV type 1 dual infection and HIV diversity: analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res. Hum. Retrovir. 22:599-606. [DOI] [PubMed] [Google Scholar]
- 18.Hu, D. J., S. Subbarao, S. Vanichseni, P. A. Mock, A. Ramos, L. Nguyen, T. Chaowanachan, F. Griensven, K. Choopanya, T. D. Mastro, and J. W. Tappero. 2005. Frequency of HIV-1 dual subtype infections, including intersubtype superinfections, among injection drug users in Bangkok, Thailand. AIDS 19:303-308. [PubMed] [Google Scholar]
- 19.Iversen, A. K., G. H. Learn, L. Fugger, J. Gerstoft, J. I. Mullins, and P. Skinhoj. 1999. Presence of multiple HIV subtypes and a high frequency of subtype chimeric viruses in heterosexually infected women. J. Acquir. Immune Defic. Syndr. 22:325-332. [DOI] [PubMed] [Google Scholar]
- 20.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung, A., R. Maier, J.-P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 22.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 23.Koelsch, K. K., D. M. Smith, S. J. Little, C. C. Ignacio, T. R. Macaranas, A. J. Brown, C. J. Petropoulos, D. D. Richman, and J. K. Wong. 2003. Clade B HIV-1 superinfection with wild-type virus after primary infection with drug-resistant clade B virus. AIDS 17:F11-F16. [DOI] [PubMed] [Google Scholar]
- 24.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 25.Kuhner, M. K., J. Yamato, and J. Felsenstein. 2000. Maximum likelihood estimation of recombination rates from population data. Genetics 156:1393-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Y., D. C. Nickle, D. Shriner, M. A. Jensen, G. H. Learn, Jr., J. E. Mittler, and J. I. Mullins. 2004. Molecular clock-like evolution of human immunodeficiency virus type 1. Virology 329:101-108. [DOI] [PubMed] [Google Scholar]
- 27.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison, W. P., and D. R. Maddison. 2001. MacClade: analysis of phylogeny and character evolution, version 4.08. Sinauer Associates, Inc., Sunderland, MA.
- 29.Mansky, L. M. 1996. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res. Hum. Retrovir. 12:307-314. [DOI] [PubMed] [Google Scholar]
- 29a.Maynard Smith, J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 30.McCormack, G. P., J. R. Glynn, A. C. Crampin, F. Sibande, D. Mulawa, L. Bliss, P. Broadbent, K. Abarca, J. M. Ponnighaus, P. E. Fine, and J. P. Clewley. 2002. Early evolution of the human immunodeficiency virus type 1 subtype C epidemic in rural Malawi. J. Virol. 76:12890-12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCutchan, F. E., M. Hoelscher, S. Tovanabutra, S. Piyasirisilp, E. Sanders-Buell, G. Ramos, L. Jagodzinski, V. Polonis, L. Maboko, D. Mmbando, O. Hoffmann, G. Riedner, F. von Sonnenburg, M. Robb, and D. L. Birx. 2005. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J. Virol. 79:11693-11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 33.Novitsky, V., U. R. Smith, P. Gilbert, M. F. McLane, P. Chigwedere, C. Williamson, T. Ndung'u, I. Klein, S. Y. Chang, T. Peter, I. Thior, B. T. Foley, S. Gaolekwe, N. Rybak, S. Gaseitsiwe, F. Vannberg, R. Marlink, T. H. Lee, and M. Essex. 2002. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J. Virol. 76:5435-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung'u, M. Rahman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pernas, M., C. Casado, R. Fuentes, M. J. Perez-Elias, and C. Lopez-Galindez. 2006. A dual superinfection and recombination within HIV-1 subtype B 12 years after primoinfection. J. Acquir. Immune Defic. Syndr. 42:12-18. [DOI] [PubMed] [Google Scholar]
- 36.Philpott, S., H. Burger, C. Tsoukas, B. Foley, K. Anastos, C. Kitchen, and B. Weiser. 2005. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J. Virol. 79:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollakis, G., A. Abebe, A. Kliphuis, T. F. De Wit, B. Fisseha, B. Tegbaru, G. Tesfaye, H. Negassa, Y. Mengistu, A. L. Fontanet, M. Cornelissen, and J. Goudsmit. 2003. Recombination of HIV type 1C (C′/C«) in Ethiopia: possible link of EthHIV-1C′ to subtype C sequences from the high-prevalence epidemics in India and Southern Africa. AIDS Res. Hum. Retrovir. 19:999-1008. [DOI] [PubMed] [Google Scholar]
- 38.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249-259. [DOI] [PubMed] [Google Scholar]
- 40.Rodenburg, C. M., Y. Li, S. A. Trask, Y. Chen, J. Decker, D. L. Robertson, M. L. Kalish, G. M. Shaw, S. Allen, B. H. Hahn, and F. Gao. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retrovir. 17:161-168. [DOI] [PubMed] [Google Scholar]
- 41.Rose, P. P., and B. T. Korber. 2000. Detecting hypermutations in viral sequences with an emphasis on G → A hypermutation. Bioinformatics 16:400-401. [DOI] [PubMed] [Google Scholar]
- 42.Rousseau, C., B. A. Birditt, A. R. McKay, J. N. Stoddard, T. C. Lee, S. McLaughlin, S. W. Moore, N. Shindo, G. H. Learn, B. T. Korber, C. Brander, P. J. Goulder, P. Kiepiela, B. D. Walker, and J. I. Mullins. 2006. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J. Virol. Methods 136:118-125. [DOI] [PubMed] [Google Scholar]
- 43.Sanabani, S., W. K. Neto, D. J. de Sa Filho, R. S. Diaz, P. Munerato, L. M. Janini, and E. C. Sabino. 2006. Full-length genome analysis of human immunodeficiency virus type 1 subtype C in Brazil. AIDS Res. Hum. Retrovir. 22:171-176. [DOI] [PubMed] [Google Scholar]
- 44.Schierup, M. H., and J. Hein. 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics 156:879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shankarappa, R., R. Chatterjee, G. H. Learn, D. Neogi, M. Ding, P. Roy, A. Ghosh, L. Kingsley, L. Harrison, J. I. Mullins, and P. Gupta. 2001. Human immunodeficiency virus type 1 Env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. J. Virol. 75:10479-10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 47.Shriner, D., A. G. Rodrigo, D. C. Nickle, and J. I. Mullins. 2004. Pervasive genomic recombination of HIV-1 in vivo. Genetics 167:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slatkin, M., and W. P. Maddison. 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123:603-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, D. M., J. K. Wong, G. K. Hightower, C. C. Ignacio, K. K. Koelsch, E. S. Daar, D. D. Richman, and S. J. Little. 2004. Incidence of HIV superinfection following primary infection. JAMA 292:1177-1178. [DOI] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Soares, M. A., T. De Oliveira, R. M. Brindeiro, R. S. Diaz, E. C. Sabino, L. Brigido, I. L. Pires, M. G. Morgado, M. C. Dantas, D. Barreira, P. R. Teixeira, S. Cassol, and A. Tanuri. 2003. A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS 17:11-21. [DOI] [PubMed] [Google Scholar]
- 52.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swofford, D. L. 1999. PAUP* 4.0: phylogenetic analysis using parsimony (* and other methods), version 4.0b10. Sinauer Associates, Inc., Sunderland, MA.
- 54.Taylor, J. E., and B. T. Korber. 2005. HIV-1 intra-subtype superinfection rates: estimates using a structured coalescent with recombination. Infect. Genet. Evol. 5:85-95. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Travers, S. A., J. P. Clewley, J. R. Glynn, P. E. Fine, A. C. Crampin, F. Sibande, D. Mulawa, J. O. McInerney, and G. P. McCormack. 2004. Timing and reconstruction of the most recent common ancestor of the subtype C clade of human immunodeficiency virus type 1. J. Virol. 78:10501-10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Harmelen, J., C. Williamson, B. Kim, L. Morris, J. Carr, S. S. Karim, and F. McCutchan. 2001. Characterization of full-length HIV type 1 subtype C sequences from South Africa. AIDS Res. Hum. Retrovir. 17:1527-1531. [DOI] [PubMed] [Google Scholar]
- 58.Van Harmelen, J. H., E. Van der Ryst, A. S. Loubser, D. York, S. Madurai, S. Lyons, R. Wood, and C. Williamson. 1999. A predominantly HIV type 1 subtype C-restricted epidemic in South African urban populations. AIDS Res. Hum. Retrovir. 15:395-398. [DOI] [PubMed] [Google Scholar]
- 59.van Rij, R. P., M. Worobey, J. A. Visser, and H. Schuitemaker. 2003. Evolution of R5 and X4 human immunodeficiency virus type 1 gag sequences in vivo: evidence for recombination. Virology 314:451-459. [DOI] [PubMed] [Google Scholar]
- 60.Yerly, S., S. Jost, M. Monnat, A. Telenti, M. Cavassini, J. P. Chave, L. Kaiser, P. Burgisser, and L. Perrin. 2004. HIV-1 co/super-infection in intravenous drug users. AIDS 18:1413-1421. [DOI] [PubMed] [Google Scholar]
- 61.Zhuang, J., A. E. Jetzt, G. Sun, H. Yu, G. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.zur Megede, J., S. Engelbrecht, T. de Oliveira, S. Cassol, T. J. Scriba, E. J. van Rensburg, and S. W. Barnett. 2002. Novel evolutionary analyses of full-length HIV type 1 subtype C molecular clones from Cape Town, South Africa. AIDS Res. Hum. Retrovir. 18:1327-1332. [DOI] [PubMed] [Google Scholar]