Abstract
Marek's disease virus (MDV) is an alphaherpesvirus that induces a highly malignant T-lymphoma in chickens. The viral genome encodes two identical copies of a viral telomerase RNA subunit (vTR) that exhibits 88% sequence identity to its chicken ortholog chTR. The minimal telomerase ribonucleoprotein complex consists of a protein subunit with reverse transcriptase activity (TERT) and an RNA subunit (TR). The active complex compensates for the progressive telomere shortening that occurs during mitosis and is involved in the cell immortalization process. We show here that the upregulation of telomerase activity is associated with an increase in vTR gene expression in chickens infected with the highly oncogenic MDV strain RB-1B. A comparative functional analysis of the viral and chicken TR promoters, based on luciferase reporter assays, revealed that the vTR promoter was up to threefold more efficient than the chTR promoter in avian cells. We demonstrated, by directed mutagenesis of the vTR promoter region, that the stronger transcriptional activity of the vTR promoter resulted largely from an E-box located two nucleotides downstream from the transcriptional start site of the vTR gene. Furthermore, transactivation assays and chromatin immunoprecipitation assays demonstrated the involvement of the c-Myc oncoprotein in the transcriptional regulation of vTR. Finally, an Ets binding site was specifically implicated in the transcriptional regulation of vTR in the MDV-transformed lymphoblastoid cell line MSB-1.
Marek's disease virus (MDV), also referred to as gallid herpesvirus type 2 (GaHV-2), is an oncogenic alphaherpesvirus responsible for a highly malignant T-lymphoma in chickens that occurs within 2 or 3 weeks of infection. Upon primary lytic replication in B cells, MDV establishes latency in activated CD4+ T cells, which are also the primary target for oncogenic transformation (3, 27). The viral genome consists of a linear 180-kb double-stranded DNA molecule comprising a unique long (UL) and a unique short (US) region, each flanked by inverted terminal repeats (TRL and TRS) and internal repeats (IRL and IRS), respectively (5).
The detailed molecular mechanisms by which MDV transforms T cells are still unknown. The rapid onset of MDV-induced lymphoma formation, however, suggests that one or more virus-encoded oncogenes are involved in the transformation process. The MDV-encoded 339-amino-acid protein Meq, a basic leucine zipper protein (bZIP), is the only member of the Jun/Fos oncoprotein family present in herpesviruses. This protein has previously been reported to be involved in MDV-induced tumorigenesis (2, 21). Through homodimerization or heterodimerization with c-Jun, JunB, or Fos, Meq can bind to promoters containing AP-1 or to so-called Meq-responsive element (MERE) sites, thereby regulating the transcription of other viral and cellular genes (20). Another viral gene encoding the viral RNA subunit of telomerase (vTR) has been reported to have a major impact on the malignancy of MDV-induced lymphoma (30). We previously identified the vTR gene in the repeat regions flanking the UL (TRL and IRL) region. Thus, as for the gene encoding Meq, the vTR gene is present as two copies in the MDV genome (10). The deletion of both copies of the vTR gene from an infectious clone of the highly oncogenic MDV strain RB-1B reduced the tumor incidence by 60% in infected chickens compared with that of parental virus, with no effect on lytic MDV replication or the establishment of latency (30). This study also showed that the constitutive expression of vTR in the chicken fibroblast line DF-1 resulted in a phenotype consistent with transformation similar to that of DF-1 cells transformed by the MDV oncoprotein Meq.
The most straightforward interpretation of the tumor-promoting properties of vTR is that this subunit enhances telomerase activity, leading to the stabilization of telomeres, the nucleoprotein structures constituting the ends of eukaryotic chromosomes. Indeed, telomerase is a ribonucleoprotein composed of TERT, the telomerase reverse transcriptase, and TR, the telomerase RNA subunit encoding the template sequence reverse transcribed by the telomerase enzyme to telomeres. Telomerase upregulation in human cancer plays a key role in cell immortalization, and abnormally high levels of telomerase activity are detectable in over 85% of human cancers (17, 1). A role for vTR in the T-cell immortalization process would be due to the capacity of this subunit to enhance telomerase activity during MDV infection. Indeed, a comparative functional study has shown that vTR yields higher levels of telomerase activity than its chicken ortholog, chTR, when combined with recombinant chicken TERT protein in vitro (10). This greater efficiency of vTR over chTR seems to be due to mutations affecting the pseudoknot core domain, which presumably stabilizes the pseudoknot P2 helix (11).
Human TR (hTR) seems to be constitutively expressed in a broad range of human cells, but hTR levels are generally higher in cancer cells, and there is evidence that the upregulation of hTR is an early event during multistage tumorigenesis (35). TERT expression in humans is otherwise correlated with telomerase activity. Several transcription factors regulating human TERT (hTERT) transcription have been identified. These factors include c-Myc and Sp1, which have been described as the principal activators of hTERT transcription, with responsive elements located within the proximal core hTERT promoter (19, 33). Human cancers caused by infections with oncogenic viruses, e.g., human papilloma virus 16 (HPV16), Kaposi's sarcoma-associated herpesvirus (KSHV), and Epstein-Barr virus (EBV), have been associated with the upregulation of telomerase activity through a virus-mediated upregulation of hTERT expression (18, 15). Thus, all these viruses increase cellular telomerase activity, but MDV is currently the only one known to encode a functional telomerase subunit in its genome.
We provide the first demonstration that telomerase activity is strongly upregulated in the peripheral blood mononuclear cells (PBMC) of chickens infected with the highly oncogenic MDV strain RB-1B. The observed increase in telomerase activity was positively correlated with high levels of vTR expression. We also identified a TATA-like box as the main element controlling the basal transcriptional activity of promoters vTR and chTR. Finally, we carried out a functional analysis of the vTR promoter-specific cis elements. We found that vTR expression was specifically regulated by an E-box located two nucleotides downstream from the transcriptional initiation site in two different transformed avian cell lines. Chromatin immunoprecipitation (ChIP) assays demonstrated that the vTR promoter region encompassing this E-box binds the oncoprotein c-Myc. We also found that the Ets binding site (EBS) was specifically involved in regulating vTR transcription in the MDV-transformed lymphoblastoid cell line MSB-1.
MATERIALS AND METHODS
Cell lines.
We used two chicken cell lines, the LMH cell line derived from hepatocellular carcinoma and the MDV-transformed MSB-1 chicken T cells. The LMH cell line was cultured in gelatin-coated dishes, as previously described (10). The MSB-1 cell line was cultured in RPMI 1640 medium (Cambrex Bio Science, Paris, France) supplemented with 10% fetal bovine serum and maintained at 41°C in an atmosphere containing 5% CO2.
Experimental in vivo assay and PBMC isolation.
We used White Leghorn specific pathogen-free B13B13 chickens highly susceptible to Marek's disease virus strain. These animals were bred, raised, and housed in isolated accommodations. Chickens were injected intramuscularly at the age of 8 weeks with 1,000 PFU of the highly oncogenic MDV strain RB-1B, using a chicken embryo fibroblast suspension as previously described (8). PBMC were collected as previously described (9) at various times after inoculation to measurement telomerase activity and gene expression.
All the surviving chickens were killed 45 days after inoculation, and necropsies were performed. Mortality and macroscopic tumors were recorded. All experimental procedures were conducted in compliance with approved protocols for the use of animals in research.
Statistical analysis was carried out with the nonparametric Kruskal-Wallis test, as implemented in Simstat version 2.04 (Provalis Research). We compared the telomerase activity and the relative gene expression levels of PBMC from infected and from control chickens. We considered P values of ≤0.045 to be statistically significant.
TRAP assay.
We quantified the telomerase activity of 600 ng of protein extracted from PBMC, using the semiquantitative fluorescence-based telomeric repeat amplification protocol (TRAP) assay, as previously described (10). Briefly, the PCR steps were carried out using the 6-carboxytetramethylrhodamine-labeled forward primers TS and CX-ext as the reverse primers. An internal amplification standard was also added to the PCR mixture. PCR products were analyzed by capillary electrophoresis (ABI Prism 310; PerkinElmer Life Sciences). The telomerase activity level of each protein extract was estimated by adding the integrated value of each telomerase elongation product normalized to the integrated value of the internal amplification standard.
Reverse transcription-PCR analysis.
Levels of chTERT, chTR, and vTR expression were quantified by fluorescence-based reverse transcription (RT)-PCR, as previously described (10). The primers used for reverse transcription were an oligodT(15) primer for chTERT and the DS4 primer (5′-GCCCGCTGAAAGTCAGCGAGTA-3′) for chTR/vTR. The resulting cDNA was amplified by PCR using specific tetrachlorofluorescein phosphoramidite-labeled forward primers and specific reverse primers (Table 1). An internal control (IC) was included in the PCR for each sample (sizes of the IC amplification products are shown in Table 1). The PCR protocol consisted of 35 cycles of 45 s at 94°C, 45 s at 55°C, and 45 s at 72°C. Amplification products were analyzed with an automated ABI Prism 310 fragment analyzer (Perkin-Elmer Corp., MA). Gene expression was calculated as the ratio of the integrated value for the gene product normalized to the integrated IC value.
TABLE 1.
Primers used for RT-PCR and size of PCR products
| Name | Type | Sequence | PCR product size (bp)
|
|
|---|---|---|---|---|
| RNA | IC | |||
| chTERT | Forward | 5′TET-CATTGTCAAACTGTCCAACCAC3′ | 121 | 175 |
| Reverse | 5′CACCGTCTTCAGCAGTTCCAT3′ | |||
| chTR | Forward | 5′TET-CGTGGCGGGTGGAAGGCTCCGC3′ | 110 | 160 |
| Reverse | 5′GCCCGCTGAAAGTCAGCGAGTA3′ | |||
| vTR | Forward | 5′TET-CGTGGCGGGTGGAAGGCTCCGC3′ | 105 | 160 |
| Reverse | 5′GCCCGCTGAAAGTCAGCGAGTA3′ | |||
Plasmid construction.
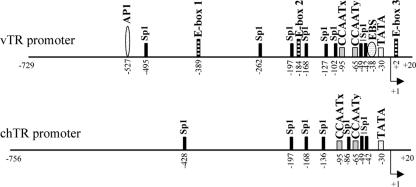
The vTR promoter region extending from −729 to +20 nucleotides from the transcription start site (+1) (as shown in Fig. 2) was amplified by PCR from the genomic DNA of MDV strain RB-1B, using primers 802 and 729 (Table 2). The corresponding region of the chTR promoter (−756 to +20 nucleotides from the transcription start site, +1) (Fig. 2) was obtained by PCR with genomic DNA extracted from PA-12 chickens, using the primers M165 and M166 (Table 2). The amplified promoters were inserted in a sense orientation into the XhoI/HindIII sites of the firefly luciferase reporter vector pGL3-Basic (Promega Corp., Madison, WI) to generate pchTR and pvTR. A truncated vTR promoter was obtained by the amplification of pvTR by PCR, using primers 782 and 729 to generate pvS-TR (−104 to +20 nucleotides from the transcription start site, +1). Various site-specific mutations (poly-T-replacing sequence) were introduced into the chTR and vTR promoters, using a PCR-based protocol, as previously described (11). We used two approaches to generate mutations according to the targeted cis element location within the promoter sequence. Internal mutations were introduced into the TR promoter sequences by overlap extension. This method was used to generate constructs pvTRΔAP1, pvTRΔE1, pvTRΔE2, pvTRΔCAATx, pvTRΔCAATy, pchTRΔCAATx, and pchTRΔCAATy. The primers and templates used in this approach are shown in Table 2 and 3. Distal mutations were introduced into the promoter directly via the primer sequence used for amplification. This method was used to generate the pvTRΔE3 construct (Table 2 and 4). The same method was used to generate the pvTRΔTATA, pvS-TRΔSp1, pvTRΔEBS, and pchTRΔTATA constructs, using two successive PCR amplifications (Table 2 and 4). The pvTRΔE2ΔE3 and pvTRΔEBSΔE3 constructs were generated by amplification by PCR from pvTRΔEBS and pvTRΔE2 (Table 2 and 4). All these modified promoters were inserted into the pGL3-Basic vector, as described above.
FIG. 2.
Comparison of the promoter regions of the viral and the chicken TR genes. Potential transcription factor binding sites are indicated in the promoter sequences. The transcriptional start site (+1) is indicated on each sequence by an arrow. Sites present in viral TR promoter sequences but absent from the chicken TR promoter include AP-1, E-boxes, and the EBS. Four Sp1 motifs are found in identical locations in the vTR and chTR promoter sequences. The vTR promoter contained four Sp1 sites in addition to these four sites, whereas the chTR promoter contained three additional Sp1 sites.
TABLE 2.
Primers used to perform mutated TR promoters
| Primer no. | type | Primer sequencesa |
|---|---|---|
| 802 | Forward | 5′CTCGAGCCCTAACCCTAACCCCCCAAATTTTCACC3′ |
| 729 | Reverse | 5′AAGCTTGCCTTCCACCCGCCACGTGTG3′ |
| M165 | Forward | 5′CTCGAGCAAGAGATCGGCGTTGCTTTC3′ |
| M166 | Reverse | 5′AAGCTTGCCTTCCACCCGCCACGCGTG3′ |
| 782 | Forward | 5′CTCGAGTCCCCGCCGCCAATAGCTAC3′ |
| 730 | Reverse | 5′AAGCTTGCCTTCCACCCGCTTTTTTTGCCGGGGGAACCCCGCGTGGGGCTCTTG3′ |
| 718 | Reverse | 5′GGGGAACCCCGCGTGGGGCTCTTGTAGCTTTTTTCGCCTACGCCCACCGCGC3′ |
| 719 | Reverse | 5′AAGCTTGCCTTCCACCCGCCACGTGTGCCGGGGGAACCCCGCGTGGGGCTCTTGTA3′ |
| 700 | Reverse | 5′GAACCCCGCGTGGGGCTTTTTTTGCTTCCTCCGCCTAC3′ |
| 701 | Reverse | 5′AAGCTTGCCTTCCACCCGCCACGTGTGCCGGGGGAACCCCGCGTGGGGC3′ |
| M270 | Forward | 5′CTCGAGTCTTTTTTGCCAATAGCTACGCGGCAGC3′ |
| M271 | Reverse | 5′CCCCGCGTGGGGCTCTTGTAGCTTCCTCTTTTTTTTTTCACCGCGCGCCTATTG3′ |
| M171 | Reverse | 5′GAACCCCGCGTGGGGCTTTTTTTGCCTCCTCCGCCTAC3′ |
| M178 | Reverse | 5′AAGCTTGCCTTCCACCCGCCACGCGTGCCGGGGGAACCCCGCGTGGGGC3′ |
| 713 | Forward | 5′AAACTTGGATTATGCAAGTG3′ |
| 712 | Reverse | 5′CACTTGCATAATCCAAGTTTTTTTTACATCACAGGTGGTATGTG3′ |
| 715 | Forward | 5′GTACACCTGCCTGCACTACT3′ |
| 714 | Reverse | 5′AGTAGTGCAGGCAGGTGTACTTTTTCCTGTCGGCCGCGAGAGG3′ |
| 717 | Forward | 5′GCATGGGGCGTGGCGGGAGA3′ |
| 716 | Reverse | 5′TCTCCCGCCACGCCCCATGCTTTTTCCCCGCCCCTTCCTGTGG3′ |
| 703 | Forward | 5′AGCTACGCGGCAGCGTACAGCCCGG3′ |
| 702 | Reverse | 5′ACGCTGCCGCGTAGCTTTTTTCGGCGGGGAGGAGAGCG3′ |
| 705 | Forward | 5′AGGCGCGCGGTGGGCGTAGG3′ |
| 704 | Reverse | 5′TACGCCCACCGCGCGCCTTTTTTCCGGGCTGTACGCTGCC3′ |
| M168 | Forward | 5′AGCGGGGCGGCAGCGTGCAGCCCGG3′ |
| M167 | Reverse | 5′ACGCTGCCGCCCCGCTTTTTTCCGCGGGGAGGAGAGCGGG3′ |
| M170 | Forward | 5′GGGCGCCCGGTGGGCGTAGG3′ |
| M169 | Reverse | 5′TACGCCCACCGGGCGCCCTTTTTCCGGGCTGCACGCTGCC3′ |
| 804 | Forward | 5′TGGCGGGAGATGAATGACCG3′ |
| M180 | Forward | 5′GCATCGGACCCCGCGGGCCCACAGGAAGGGG3′ |
Point mutations with respect to the wild-type sequences of the vTR and chTR promoters are in bold and restriction sites are underlined.
TABLE 3.
Production of mutant constructs: internal mutations
| Constructsa | PCRb
|
||
|---|---|---|---|
| 1st amplification
|
2nd amplification
|
||
| Template | Primers | Primers | |
| pvTRΔAP1 | pvTR | 802/712 | 802/729 |
| pvTR | 713/729 | ||
| pvTRΔE1 | pvTR | 802/714 | 802/729 |
| pvTR | 715/729 | ||
| pvTRΔE2 | pvTR | 802/716 | 802/729 |
| pvTR | 717/729 | ||
| pvTRΔCAATx | pvTR | 802/702 | 802/729 |
| pvTR | 703/729 | ||
| pvTRΔCAATy | pvTR | 802/704 | 802/729 |
| pvTR | 705/729 | ||
| pchTRΔCAATx | pchTR | M165/M167 | M165/M166 |
| pchTR | M168/M166 | ||
| pchTRΔCAATy | pchTR | M165/M169 | M165/M166 |
| pchTR | M170/M166 | ||
The Δ symbol means that the following site specified is mutated. E1 and E2 correspond to E-box 1 and E-box 2, respectively.
Primer sequences are described in Table 2. Primers which comprise desired mutations are in bold.
TABLE 4.
Production of mutant constructs: distal mutations
| Constructsa | PCRb
|
||
|---|---|---|---|
| 1st amplification
|
2nd amplification
|
||
| Template | Primers | Primersb | |
| pvTRΔE3 | pvTR | 802/730 | |
| pvTRΔTATA | pvTR | 802/700 | 802/701 |
| pvS-TRΔSp1 | pvTR | M270/M271 | M270/719 |
| pvTRΔEBS | pvTR | 802/718 | 802/719 |
| pchTRΔTATA | pchTR | M165/M171 | M165/M178 |
| pvTRΔE2ΔE3 | pvTRΔE2 | 802/730 | |
| pvTRΔEBSΔE3 | pvTRΔEBS | 802/730 | |
The Δ symbol means that the following site specified is mutated. E2 and E3 correspond to E-box 2 and E-box 3, respectively.
Primer sequences are described in Table 2. Primers which comprise desired mutations are in bold.
The avian c-Myc expression vector (pCDNAMyc) consisted of the avian c-Myc coding sequence obtained by PCR amplification from PA-12 chicken genomic DNA inserted into the pCDNA3 expression vector downstream from the cytomegalovirus (CMV) promoter.
All of the intermediate and final constructs were checked by sequencing with appropriate primers.
Luciferase assay.
For the luciferase assay, Renilla luciferase plasmids (pRL) were used for cotransfection to standardize transfection efficiency. PCDNAMLuc, which carries the firefly luciferase gene downstream from the CMV promoter, and the pGL3-Basic vector were also used for transfection as positive and negative controls, respectively.
LMH cells were seeded on 96-well-plates (5 × 104 cells/well). They were cultured overnight and cotransfected with 300 ng of luciferase reporter plasmids and 3.75 ng of pRL plasmids, using Lipofectamine 2000 transfection reagent (Invitrogen-Life Technologies, Cergy Pontoise, France). For c-Myc overexpression assays, cells were cotransfected with 150 ng of c-Myc expression vector, the reporter plasmids, and pRL.
MSB-1 cells were electroporated using an Equibio “EasyjecT Plus” electroporator (single pulse, 400 V, 1500 μF) and aluminum electrodes (4-mm cuvette;Eurogentec). For all assays, 8 × 106 cells were electroporated in the presence of 40 μg of luciferase reporter plasmids and 500 ng of pRL diluted in RPMI 1640 medium. After electroporation, cells were plated in 6-well plates containing 2.5 ml of RPMI 1640 medium supplemented with 10% fetal bovine serum. The plates were then incubated at 41°C.
Luciferase assays were performed 24 h after transfection, using a Dual-Luciferase reporter assay system (Promega, Madison, WI). Firefly luciferase activities were normalized with respect to Renilla luciferase activity. Each transfection reaction was carried out at least three times. Mean relative luciferase activity is presented. The significance of differences between promoter constructs was assessed with the nonparametric Kruskal-Wallis test implemented in Simstat version 2.04 (Provalis Research). We compared the luciferase activity for each mutated promoter with that for the corresponding wild-type promoter. We considered P values of ≤0.045 to be statistically significant.
Western blot analysis.
The whole LMH cell extracts used for Western blotting to assess c-Myc levels were prepared by osmotic shock followed by sonication. Proteins (40 μg) were subjected to electrophoresis in 10% polyacrylamide gels containing SDS and electrotransferred onto nitrocellulose membranes (Macherey-Nagel, Düren, Germany). The c-Myc antibody (Ab) (sc-42X; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used as the primary antibody. Proteins were visualized using a BCIP/NBT substrate kit (Zymed Laboratories, CA).
ChIP assays.
ChIP assays were carried out as previously described (24). Briefly, chromatin from 107 MSB-1 cells was cross-linked, washed, resuspended in lysis buffer, and sonicated 12 times for 6 s each, with an 18-W pulse (Vibra Cell 75455; Bioblock Scientific). A fraction of total chromatin was taken as the total input DNA control. The protein/DNA complexes were immunoprecipitated by incubation at 4°C overnight with a mixture of anti-c-Myc Abs (5 μg and 5 μg; sc-42X and sc-764X; Santa Cruz Biotechnology, Santa Cruz, CA) or with mouse immunoglobulin G1κ (IgG1κ) Abs (2 μg; Sigma-Aldrich, Lyon, France) as a negative control. Immunoprecipitated complexes were collected with protein A/protein G beads (1:1). Immunoprecipitation products were washed with low-salt washing buffer, high-salt washing buffer, lithium chloride washing buffer, and finally with TE (Tris-EDTA) buffer. Immunocomplexes were extracted, and the cross-linking was reversed by incubation at 65°C overnight. Chromatin fragments purified with a QIAquick PCR purification kit (QIAGEN, Courtaboeuf, France) were eluted in 50 μl of TE buffer. PCR was initiated by incubation at 94°C for 3 min, followed by 22 cycles of 94°C for 45 s, 58°C for 45 s, 72°C for 45 s, and 72°C for 10 min. PCRs targeting the vTR and chTR promoters were carried out in 1.1× ReddyMix PCR Master Mix (Abgene, Courtaboeuf, France) with primer pairs 804/729 and M180/M166, respectively (Table 2).
RESULTS
Telomerase activity and vTR transcription increase during MDV infection in vivo.
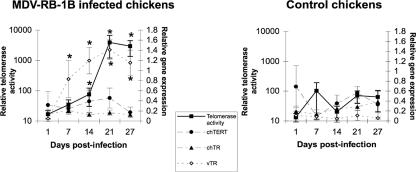
We examined the effects of MDV infection in vivo on telomerase activity in PBMC, using a semiquantitative TRAP assay. PBMC were isolated from 12 chickens infected with the highly oncogenic MDV strain RB-1B on days 1, 7, 14, 21, and 27 postinfection. PBMC were also isolated from eight noninfected chickens as a control. In the MDV strain RB-1B-infected group, three chickens died during the experiment, from day 27 onward. Necropsies performed on the 20 chickens revealed a 100% incidence of tumors in infected animals and no tumor development in noninfected chickens. Similarly, high levels of telomerase activity were detected only in PBMC from MDV-infected chickens (Fig. 1). Telomerase activity in infected chickens increased strongly from day 14 onward, reaching a maximum on day 21 (Fig. 1).
FIG. 1.
Telomerase activity and relative expression of telomerase subunit genes during MDV strain RB-1B (MDV-RB-1B) infection. Eight-week-old chickens were inoculated intramuscularly with 1,000 PFU of highly oncogenic MDV strain RB-1B. Telomerase activity and gene expression were analyzed on days 1, 7, 14, 21, and 27 postinfection, using PBMC from infected and control chickens. Means and standard deviations are given. Values for infected chickens significantly different from those for control chickens are indicated by an asterisk.
We investigated the possible association between MDV infection and the dysregulation of the chicken telomerase counterpart expression by measuring the levels of transcription of chTERT and chTR by using semiquantitative RT-PCR. We found no significant differences in the expression of chTERT and that of chTR between infected and control chickens (Fig. 1), suggesting that the increase in telomerase activity in infected chickens was not due to changes in the level of transcription of the two chicken genes. The vTR gene was strongly expressed from day 7 onward in infected chickens, with expression peaking on day 21. Then, tumor formation in infected chickens seems to be associated with an increase in telomerase activity in PBMC, and vTR seems to be the only telomerase subunit displaying high levels of expression in vivo during MDV-induced lymphomagenesis.
Basal transcriptional activities of viral and chicken TR promoters are essentially driven by the TATA-like box element.
As vTR was the only telomerase subunit displaying high levels of expression during MDV tumorigenesis in chickens, we compared the transcriptional regulation of vTR with that of chTR. We identified several transcription factor binding sites common to the chicken and the viral TR promoter sequences and other cis elements specific to the viral promoter sequence (Fig. 2; (10). Most of the common cis regulatory elements were elements generally thought to control basal transcription, such as the TATA-like box (at −30), two CCAAT boxes (at −65 and −95), and several GC boxes that serve as consensus binding sites for Sp1 (at −42, −49, −168, and −197).
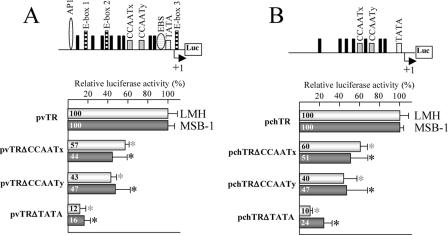
We analyzed the role of the TATA-like box and the two CCAAT boxes (CCAATx and CCAATy) in the two promoters, using site-directed mutagenesis to generate a set of mutations within the vTR and the chTR promoters. The TATA-like box (−30 to −27) was replaced by a thymine hexamer in the pvTRΔTATA and the pchTRΔTATA constructs. Similarly, the CCAATx box (−95 to −91) and the CCAATy box (−65 to −61) were replaced by thymine pentamers in the pvTRΔCCAATx, pchTRΔCCAATx, pvTRΔCCAATy, and pchTRΔCCAATy constructs. We then used a luciferase assay to assess the ability of each promoter construct to drive the firefly luciferase reporter gene expression by transiently transfecting LMH and MSB-1 avian cells with the mutated constructs (Fig. 3A and B).
FIG. 3.
Luciferase promoter assay for vTR (A) and chTR (B) in LMH and MSB-1 cells. Wild-type and mutant vTR/chTR promoter fragments were inserted upstream from the firefly luciferase reporter gene in the pGL3-Basic vector to generate pvTR and pchTR. Schematic diagrams of pvTR and pchTR are shown at the top. Black rectangles indicate Sp1 sites. An arrow indicates the transcription start sites of the vTR and chTR genes. Firefly luciferase activity was normalized with respect to the Renilla luciferase activity obtained from the cotransfected pRL plasmid. The luciferase activities of the pvTR and pchTR wild-type plasmids were normalized to 100%. The means and standard deviations of at least three independent experiments are shown. Values for the mutated promoter significantly different from those for the corresponding wild-type promoter are indicated by an asterisk.
Mutations of the TATA-like box caused the largest decrease in viral (Fig. 3A) and cellular (Fig. 3B) promoter activities in both cell types. The promoter activity of the pvTRΔTATA and the pchTRΔTATA constructs was 12% and 10%, respectively, of the activity levels observed for pvTR and pchTR in LMH cells and 16% and 24%, respectively, of the levels observed for pvTR and pchTR in MSB-1 cells. Mutations of the CCAATx and CCAATy boxes of the vTR promoter reduced promoter activity levels to 57% and 43%, respectively, of that in LMH cells and to 44% and 47%, respectively, of that in MSB-1 cells. An identical pattern was observed for the chTR promoter (Fig. 3B). The promoter activity level of the pchTRΔCCAATx and pchTRΔCCAATy constructs was 40% and 60%, respectively, lower than that of the wild-type chTR promoter in LMH cells and 49% and 53%, respectively, lower than that of the wild-type chTR promoter in MSB-1 cells. These results suggest that the CCAATx box and the CCAATy box are involved in vTR and chTR promoter regulation and that the TATA-like box element is critical for the basal transcription activity associated with the chicken and viral TR promoters.
E-box 3 is essential for vTR promoter activity in LMH and MSB-1 cells.
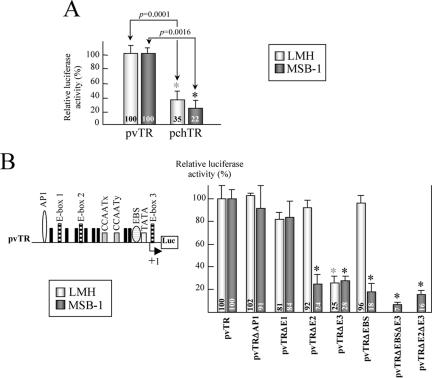
We compared the transcriptional activities of the chicken and viral TR promoter regions in LMH and MSB-1 cells, with a luciferase assay (Fig. 4A). The vTR promoter was 2.8-fold and 4.5-fold more active than the chTR promoter in LMH and MSB-1 cells, respectively. These results suggest that some cis elements present only on the viral promoter sequence may be responsible for the promoter's higher transcriptional efficiency. Several cis elements, one AP-1 site (at −527), three E-boxes (at −389, −184 and +2), and one EBS (at −38), were found only in the vTR promoter. As outlined above, AP-1 sites are potential binding sites for Meq, E-boxes are involved in the transcriptional regulation of hTERT (19), and Ets binding sites are involved in the transcriptional regulation of cell proliferation (28). These cis elements are therefore potential candidates for a major role in regulating the malignancy-promoting expression of vTR. We therefore generated a set of mutations within the vTR promoter sequence, using site-directed mutagenesis as described above (each targeted site was replaced by a thymine polymer), to enable us to identify the responsive elements involved in increasing activity. We constructed five mutated promoters from pvTR as follows: pvTRΔAP1, pvTRΔE1, pvTRΔE2, pvTRΔE3, and pvTRΔEBS, with mutations in the AP-1 site (−527 to −521), in E-box 1 (−389 to −383), E-box 2 (−184 to −178), and E-box 3 (+2 to + 7), and in the EBS (−38 to −32), respectively. These mutated plasmids were used to transfect LMH and MSB-1 cells, and a luciferase assay was performed (Fig. 4B). Mutations of the AP-1 site and E-box 1 had no effect on promoter activity, whereas the mutation of E-box 3 decreased transcriptional activity by 75% and 72% with respect to the activity of pvTR in LMH and MSB-1 cells, respectively. E-box 3 is, thus, a critical cis-acting element in the transcriptional activation of vTR in both cell lines.
FIG. 4.
Characterization of responsive elements involved in vTR transcription. (A) Comparison of vTR and chTR promoter activities in LMH and MSB-1 cells. The luciferase activity of the pvTR plasmid is normalized to 100%. The means and standard deviations from at least three independent experiments are shown. All the values obtained for pchTR are significantly different from those obtained for pvTR (indicated by an asterisk), and the P values for nonparametric Kruskal-Wallis tests are shown. (B) Identification of the DNA elements responsible for vTR transcriptional regulation in LMH and MSB-1 cells. Schematic diagram of pvTR is shown. Five mutations were generated within the vTR promoter sequence by site-directed mutagenesis. The promoter activity of each construct was measured by the luciferase assay, normalized with respect to Renilla luciferase activity, and expressed as a value relative to pvTR (wild-type) activity. The means and standard deviations of at least three independent experiments are shown. Values for mutated promoters significantly different from those for the vTR wild-type promoter are indicated by an asterisk.
E-box 2 and the EBS are functional elements of the vTR promoter in the MSB-1 cell line.
Studies of the involvement of the various cis elements in vTR promoter regulation revealed that E-box 2 and the EBS were functional regulatory elements specific to MSB-1 cells. Indeed, the mutations of E-box 2 and the EBS reduced transcriptional activity levels by 76% and 82%, respectively, only in MSB-1 cells (Fig. 4B). We therefore generated two double-mutant reporter plasmids, pvTRΔEBSΔE3 and pvTRΔE2ΔE3, the first with mutations within the EBS and E-box 3 and the second with mutations within E-box 2 and E-box 3, respectively (both sites were replaced by a thymine polymer), to determine whether the EBS and E-box 2 act in synergy with E-box 3 in MSB-1 cells. These plasmids were used in luciferase assays with the MSB-1 cell line (Fig. 4B). The pvTRΔEBSΔE3 and pvTRΔE2ΔE3 constructs reduced transcriptional activity levels by 93% and 84%, respectively, whereas mutations of the EBS, E-box 2, or E-box 3 alone led to 82%, 76%, and 72% decreases in activity levels, respectively. These results suggest that the EBS and E-box 2 cooperate with E-box 3 to regulate the promoter activity of vTR in the MSB-1 cell line.
Sp1 binding sites are involved in the transcriptional regulation of vTR.
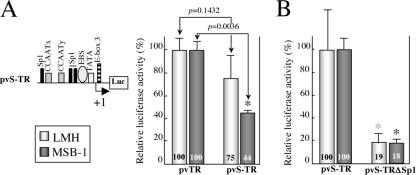
Our results show that the four cis elements of the vTR promoter functional in the two cell types tested (the CCAATx and CCAATy boxes, the TATA-like box, and E-box 3) are located close to the transcriptional initiation site (between −95 and +7). Moreover, the EBS, which is specifically functional in MSB-1 cells, is also located in this promoter region. Three of the eight Sp1 sites identified in the vTR promoter sequence are also located in this region. We investigated whether these Sp1 sites were involved in the transcriptional regulation of vTR by studying this proximal promoter region using a short vTR promoter comprising the −104 to +20 region of the promoter sequence (pvS-TR). The transcriptional efficiency of pvS-TR was then compared with that of an equivalent promoter region in which the three Sp1 sites had been replaced by a thymine polymer (pvS-TRΔSp1). The transcriptional activity of the truncated pvS-TR construct was not significantly different from that of the pvTR construct in LMH cells (P = 0.1432) (Fig. 5A). The 56% lower level of transcriptional activity observed for the truncated promoter, pvS-TR, than for the entire promoter, pvTR, in MSB-1 cells seems to be due to the absence of E-box 2 from this construct. In both types of cells, mutation of the three Sp1 sites led to levels of transcriptional activities at least 81% lower than those for pvS-TR (Fig. 5B). This result suggests that the three Sp1 sites, located at −42, −49, and −102, are involved in the transcriptional regulation of vTR.
FIG. 5.
Functional study of Sp1 sites located close to the transcriptional start site in LMH and MSB-1 cells. (A) Comparison of pvTR and pvS-TR activities in LMH and MSB-1 cells. Schematic diagram of pvS-TR is shown. The promoter activity of the pvS-TR construct was assessed with the luciferase assay, normalized with respect to Renilla luciferase activity, and expressed relative to the activity of pvTR (the entire vTR promoter). The means and standard deviations of at least three independent experiments are shown. Values for pvS-TR significantly different from those for pvTR (indicated by an asterisk) and P values for nonparametric Kruskal-Wallis tests are shown. (B) Involvement of Sp1 sites in vTR transcriptional regulation. The promoter activity of pvS-TRΔSp1 was assessed with the luciferase assay, normalized with respect to Renilla luciferase activity, and expressed relative to the activity of pvS-TR (wild type). The means and standard deviations of at least three independent experiments are shown. Values for mutated promoters significantly different from those for the short vTR wild-type promoter are indicated by an asterisk.
Avian c-Myc binds and activates the vTR promoter.
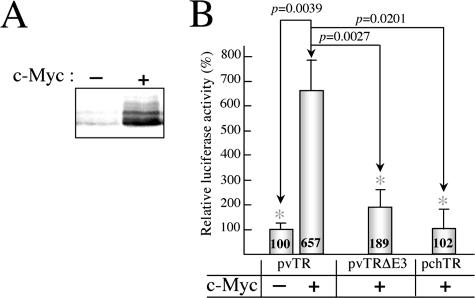
The transcription factor c-Myc can bind canonical E-box elements. The presence of functional c-Myc binding elements in the vTR promoter is of particular interest, given the role of c-Myc in cell proliferation and transformation. We studied the effects of c-Myc overexpression in LMH cells, using the avian c-Myc expression plasmid pCDNAMyc for cotransfection together with the pvTR, the pvTRΔE3, or the pchTR plasmid carrying the wild-type vTR promoter sequence, the corresponding E-box 3 mutant, and the E-box-free chTR promoter sequence, respectively. Western blotting analysis confirmed that there was an effective ectopic expression of c-Myc protein in c-Myc-cotransfected cells (Fig. 6A). The overexpression of c-Myc protein increased wild-type vTR promoter activity by a factor of 6.5 in LMH cells (Fig. 6B). Reporter plasmids carrying the E-box 3-mutated vTR promoter and pchTR responded similarly to c-Myc overexpression, with a response that was significantly different from that obtained for the wild-type vTR promoter (P = 0.0027 and P = 0.0201, respectively). These results indicate that c-Myc transactivates vTR and that E-box 3 is involved in this activity.
FIG. 6.
Effect of c-Myc overexpression on vTR promoter activity in LMH cells. (A) Effective overexpression of c-Myc in LMH cells was checked by Western blotting. (B) Effect of c-Myc overexpression on the transcriptional efficiency of the vTR promoter. LMH cells were cotransfected with the c-Myc expression vector (pCDNA3Myc) and reporter plasmids (pvTR, pvTRΔE3 or pchTR) using Lipofectamine 2000. Each transfection was carried out at least in triplicate and standard deviation bars are shown. The promoter activity of each construct was compared with that of the wild-type vTR promoter, which was defined as 100. Assays with a significantly different value for cotransfection with pvTR and pCDNA3Myc are indicated by an asterisk, and the corresponding P values from nonparametric Kruskal-Wallis tests are shown.
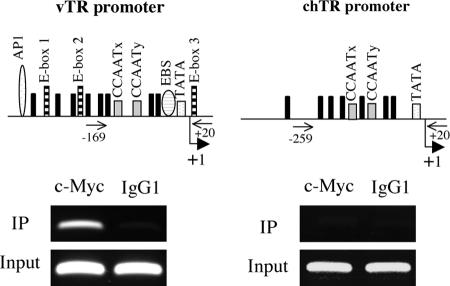
As pchTR and pvTRΔE3 displayed minimal responses to c-Myc overexpression, we assessed the specific and direct association of c-Myc with the vTR promoter by carrying out ChIP experiments with MSB-1 cells. We analyzed immunoprecipitated DNA by PCR, using primer pairs for the vTR and chTR promoters. We found that c-Myc was recruited to the vTR promoter in MSB-1 cells, whereas this factor did not bind to the chTR promoter, which contains no E-box elements (Fig. 7). These results suggest that the endogenously expressed c-Myc protein mediates the transcriptional regulation of vTR by interacting with the region encompassing the functional cis elements.
FIG. 7.
Direct association of c-Myc with the vTR promoter. Schematic diagrams of the vTR promoter (left) and the chTR promoter (right) are shown at the top. The pairs of primers used for the PCR amplification are indicated on the schematic diagrams of the two promoters. Electrophoretic analysis of the PCR amplification products, using immunoprecipitated DNA (IP) or total input DNA (Input), is shown below. Mouse IgG1 antibodies (right column) were used in ChIP assays as a negative control. The column on the left shows the PCR amplification products from DNA immunoprecipitated with c-Myc antibodies. vTR promoter-specific primers amplified a 189-bp product, whereas chTR promoter-specific primers amplified a 279-bp product.
DISCUSSION
Several studies have reported an association between TR upregulation and malignancy (6, 26), and one recent study reported that TR is required for the tumor-promoting effects of TERT overexpression (4). TR is constitutively expressed in somatic cells, but there is evidence that the upregulation of telomerase activity in immortalized and cancer cells involves the transcriptional regulation of TR (14, 22). Consistent with these data, we report here a correlation between the high level of vTR expression in PBMC during MDV infection and the upregulation of telomerase activity in these cells, which is itself associated with lymphoma development in chickens (Fig. 1). Previous functional analyses have shown that vTR can reconstitute telomerase activity by interacting with TERT more efficiently than with chTR (10, 11). The high level of vTR expression observed during MDV infection in lymphocytes, the target population for transformation, may therefore lead to an increase in telomerase activity, promoting cell immortalization. These findings are consistent with a recent study reporting that vTR promotes malignant T-cell lymphomagenesis during MDV infection (30). Indeed, pathogenesis studies in chickens have clearly shown that the incidence of lymphoma was drastically reduced in birds inoculated with an infectious clone of the highly oncogenic MDV strain RB-1B lacking both copies of the vTR gene compared with that of the parental virus. Moreover, lymphomas induced by vTR double-deletion viruses were significantly less disseminated and generally smaller in size than those induced by the parental virus. Thus, vTR is required for efficient MDV-induced lymphomagenesis, and the tumor-promoting effects of vTR seem to involve its capacity to increase telomerase activity.
We found that vTR expression was significant on day 7 postinfection and that it increased during the latent period and lymphoma development (Fig. 1). Consistent with the results of a previous study (30), an analysis of endogenous vTR expression in cells revealed that vTR was expressed in MDV-infected primary chicken embryo cells and in MDV-transformed lymphoblastoid cell lines (MSB-1, MDCC-PA9, and MDCC-PA5) (data not shown). The vTR gene therefore seems to be both a lytic and a latent gene, as it is expressed during both of these phases of MDV infection. However, semiquantitative RT-PCR analysis revealed that vTR levels were up to 17-fold higher in T cells sustaining latent infection (MSB-1, MDCC-PA9, and MDCC-PA5) than in permissive cells undergoing viral replication (MDV-infected chicken embryo cells) (data not shown). These results are of particular interest given the functionality of vTR. Indeed, Trapp et al. (30) have shown that the tumor-promoting effects of vTR expression cannot be attributed to a functional role in MDV replication or in the establishment or reactivation from latency. Instead, these effects seem to involve events downstream from the primary establishment of infection. Thus, vTR seems to display its oncogenic properties only during latent infection in lymphocytes, which sustain high levels of vTR expression.
In this study, we identified several cis elements specifically involved in vTR transcriptional regulation. Comparative analysis of the vTR and chTR promoters showed that the transcriptional efficiency of the viral promoter is up to fourfold higher than that of the chTR promoter. Our analysis of the functional significance of the TATA-like box and the two CCAAT boxes, using luciferase assays in two avian cell lines, showed that basal transcriptional regulation was similar for the two promoters. Indeed, the two CCAAT boxes had equivalent functions, and the TATA-like box was found to be essential for the basal transcriptional activity of both promoters (Fig. 3). Thus, as previously described for the hTR promoter (36, 37), the vTR and chTR promoter sequences have cis regulatory elements in common, and these elements are involved in maintaining basal TR expression levels. However, consistent with our comparative analysis of the activities of the vTR and the chTR promoters, quantification of the endogenous levels of vTR and chTR expression in MSB-1 cells by semiquantitative RT-PCR demonstrated that vTR expression was 14-fold stronger than chTR expression in these MDV-transformed T cells (data not shown). The functionality of E-box 3, located 2 bp downstream from the transcription start site, in both cell types studied suggests that this cis element is the principal element responsible for the higher levels of activity observed for the vTR promoter. A similar canonical E-box element, located 22 bp downstream from the transcription initiation site, is involved in hTERT transcriptional regulation (16, 29). Moreover, as described for the hTERT promoter (19), Sp1 binding sites located near E-box 3 are involved in vTR transcriptional activation. Thus, the vTR and hTERT promoter sequences include similar cis regulatory elements, and these elements are the main elements controlling vTR and hTERT expression in cells. Our findings indicate that vTR transcription is regulated by responsive elements involved in the regulation of basal transcription, as is the case for the chTR and hTR promoters, and that the E-box 3 cis element is required for high levels of transcription, as previously reported for hTERT. The mutagenesis assays carried out with LMH and MSB-1 cell lines identified no negative cis elements in the vTR promoter sequence. Thus, the vTR promoter seems to be the homolog of the original avian cellular chTR promoter but with certain responsive elements related to oncogenesis, particularly the E-box located in the transcribed region of vTR, introduced into the viral genome during the course of evolution, resulting in high levels of vTR expression during infection.
E-boxes serve as binding sites for proteins of the bHLH-Zip transcription factor superfamily, including the Myc/Mad/Max family of transcription factors (http://www.myccancergene.org/) (7). bHLH-Zip proteins and their E-box recognition sites act as crucial positive regulators of diverse biological processes, including cell proliferation and death (12). Previous studies have shown that c-Myc can induce telomerase activity through the transcriptional activation of hTERT (13, 32). Our overexpression assays, in which the reporter plasmid carrying the wild-type vTR promoter sequence was used for cotransfection with pCDNA3Myc, showed that c-Myc activates transcription of the vTR gene. Moreover, our ChIP assays showed that endogenously expressed c-Myc binds to the vTR promoter sequence in an MDV-transformed cell line. Thus, the transactivation of vTR expression induced by the interaction of c-Myc with the E-boxes located within the vTR promoter sequence is involved in the high levels of vTR expression observed during MDV-induced lymphomagenesis. The c-Myc oncoprotein is also involved in the increase in telomerase activity due to HPV16 infection. Indeed, the E6 oncoprotein encoded by HPV16 has been shown to interact directly with c-Myc, and c-Myc/E6 complexes have been shown to activate hTERT expression, increasing telomerase activity in infected tumor cells (31). The c-Myc oncoprotein therefore seems to be a common element of the pathways leading to higher levels of telomerase activity in MDV- and HPV16-transformed tumor cells.
In this study, we have shown that E-box 2 and the EBS, located 184 bp and 38 bp upstream, respectively, from the transcription initiation site of vTR are specifically functional in MSB-1 cells but not in LMH cells (Fig. 4B). This finding is of particular interest because the MSB-1 cell line is a T-lymphoblastoid line transformed by MDV, whereas the epithelioid LMH cell line represents a cell population which is not susceptible to transformation during MDV infection. This result therefore provides evidence that vTR is specifically regulated in transformed T cells. A previous study describing the crystal structures of c-Myc-Max heterodimers selectively bound to duplex oligonucleotides containing E-boxes revealed that two c-Myc-Max heterodimers tightly associate to form a bivalent heterotetramer, providing a substantial platform for the assembly of an additional protein factor (25). As the EBS and E-box 2 act together with E-box 3 to regulate vTR expression (Fig. 4B), an interaction between the EBS-bound factor and a Myc-Max heterotetramer bound to E-boxes 2 and 3 may be involved in the specific transcriptional regulation of vTR observed for MSB-1 cells. Moreover, as the functional EBS is located between the E-boxes, the formation of a specific chromatin structure should involve the specific location of those cis elements on the promoter sequence. Thereby, the assembly of a large nuclear complex on vTR promoter, particularly in MDV-transformed lymphoblastoid cells, may account for the restricted, cell type-dependent activity of the vTR promoter. Tess software (www.cbil.upenn.edu/cgi-bin/tess/tess) analysis of the vTR promoter sequence revealed that the EBS contains specific binding domains for three Ets proteins, PU.1, Ets-1, and PEA3. ChIP assays using a rabbit polyclonal antibody against the chicken PU.1 protein (Santa Cruz Biotechnology, CA) (23) failed to precipitate the vTR promoter sequence (data not shown). A previous study showed that PEA3 is primarily expressed in epithelial cells and not in hematopoietic cells (34) and that high levels of Ets-1 expression are restricted to lymphoid tissues in adults. Ets-1 may therefore be the factor responsible for driving vTR transcriptional regulation through the EBS located within its promoter sequence. Future studies should characterize the interaction between bHLH-Zip transcription factors and Ets proteins leading to the specific coregulation of vTR in MDV-transformed lymphoblastoid cells.
Finally, our study suggests that the viral oncoprotein Meq has no direct effect on vTR transcriptional regulation. Meq has a structure similar to that of the Jun/Fos family oncogenes and is a transactivator that can heterodimerize with c-Jun and bind AP1 sites (20). In our luciferase assays, mutations in the AP-1 site located in the vTR promoter sequence had no effect on the transcriptional activity of the vTR promoter in Meq-expressing MSB-1 cells (Fig. 4B). This result is consistent with a previous study in which a ChIP-based approach scanning the entire MDV genome for Meq binding sites failed to identify the vTR promoter sequence as a target binding site for Meq (20). Thus, although vTR and Meq may cooperate during lymphoma formation and dissemination, the oncogenic properties of Meq required for MDV-induced lymphomagenesis do not seem to involve the direct regulation of vTR transcription by Meq. Our results therefore identify EBS factors and the oncoprotein c-Myc as the main elements involved in the high levels of vTR expression specific to the MDV-transformed T-lymphoblastoid cell line. Indeed, the binding of a factor to the EBS in the vTR promoter specifically in lymphoblastoid cells may induce Myc-Max heterotetramer complex formation, leading to high levels of vTR expression. Thus, high levels of vTR expression in these cells, due to cooperation between factors bound to the TATA-like box, E-box, and the EBS, may be an essential prerequisite for the full expression of the oncogenic properties of vTR. Consistent with this idea, the constitutive expression of vTR in the nontransformed chicken cell line DF-1 resulted in a phenotype consistent with transformation (30). Thus, the tumor-promoting effects of vTR must involve its specific overexpression in lymphoblastoid cells during the latent period and the transcriptional regulation of vTR seems to be essential for the expression of its oncogenic properties.
Acknowledgments
We thank Gérard Benoit (UMR 5161, ENS Lyon, France) for helpful advice concerning ChIP assays. We also thank Sascha Trapp (TLVI, INRA de Tours, France) for critical reading of the manuscript.
This work was supported by grants from the Ligue Nationale Contre Le Cancer-Comité du Cher and from the Région Centre.
Footnotes
Published ahead of print on 21 February 2007.
REFERENCES
- 1.Blasco, M. A., and W. C. Hahn. 2003. Evolving views of telomerase and cancer. Trends Cell Biol. 13:289-294. [DOI] [PubMed] [Google Scholar]
- 2.Brown, A. C., S. J. Baigent, L. P. Smith, J. P. Chattoo, L. J. Petherbridge, P. Hawes, M. J. Allday, and V. Nair. 2006. Interaction of MEQ protein and C-terminal-binding protein is critical for induction of lymphomas by Marek's disease virus. Proc. Natl. Acad. Sci. USA 103:1687-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calnek, B. W. 1986. Marek's disease-a model for herpesvirus oncology. Crit. Rev. Microbiol. 12:293-320. [DOI] [PubMed] [Google Scholar]
- 4.Cayuela, M. L., J. M. Flores, and M. A. Blasco. 2005. The telomerase RNA component Terc is required for the tumour-promoting effects of Tert overexpression. EMBO Rep. 6:268-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cebrian, J., C. Kaschka-Dierich, N. Berthelot, and P. Sheldrick. 1982. Inverted repeat nucleotide sequences in the genomes of Marek disease virus and the herpesvirus of the turkey. Proc. Natl. Acad. Sci. USA 79:555-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, L. Y., S. C. Lin, C. S. Chang, Y. K. Wong, Y. C. Hu, and K. W. Chang. 1999. Telomerase activity and in situ telomerase RNA expression in oral carcinogenesis. J. Oral Pathol. Med. 28:389-396. [DOI] [PubMed] [Google Scholar]
- 7.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 8.Djeraba, A., N. Bernardet, G. Dambrine, and P. Quere. 2000. Nitric oxide inhibits Marek′s disease virus replication but is not the single decisive factor in interferon-γ-mediated viral inhibition. Virology 277:58-65. [DOI] [PubMed] [Google Scholar]
- 9.Djeraba-AitLounis, A., D. Soubieux, W. Klapper, and D. Rasschaert. 2004. Induction of telomerase activity in avian lymphoblastoid cell line transformed by Marek's disease virus, MDCC-MSB1. Vet. Pathol. 41:405-407. [DOI] [PubMed] [Google Scholar]
- 10.Fragnet, L., M. A. Blasco, W. Klapper, and D. Rasschaert. 2003. The RNA subunit of telomerase is encoded by Marek's disease virus. J. Virol. 77:5985-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fragnet, L., E. Kut, and D. Rasschaert. 2005. Comparative functional study of the viral telomerase RNA based on natural mutations. J. Biol. Chem. 280:23502-23515. [DOI] [PubMed] [Google Scholar]
- 12.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg, R. A., R. C. O'Hagan, H. Deng, Q. Xiao, S. R. Hann, R. R. Adams, S. Lichtsteiner, L. Chin, G. B. Morin, and R. A. DePinho. 1999. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 18:1219-1226. [DOI] [PubMed] [Google Scholar]
- 14.Guilleret, I., P. Yan, L. Guillou, R. Braunschweig, J. M. Coindre, and J. Benhattar. 2002. The human telomerase RNA gene (hTERC) is regulated during carcinogenesis but is not dependent on DNA methylation. Carcinogenesis. 23:2025-2030. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 16.Horikawa, I., P. L. Cable, S. J. Mazur, E. Appella, C. A. Afshari, and J. C. Barrett. 2002. Downstream E-box-mediated regulation of the human telomerase reverse transcriptase (hTERT) gene transcription: evidence for an endogenous mechanism of transcriptional repression. Mol. Biol. Cell 13:2585-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 18.Kyo, S., T. Kanaya, M. Takakura, M. Tanaka, and M. Inoue. 1999. Human telomerase reverse transcriptase as a critical determinant of telomerase activity in normal and malignant endometrial tissues. Int J. Cancer 80:60-63. [DOI] [PubMed] [Google Scholar]
- 19.Kyo, S., M. Takakura, T. Taira, T. Kanaya, H. Itoh, M. Yutsudo, H. Ariga, and M. Inoue. 2000. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 28:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy, A. M., Y. Izumiya, P. Brunovskis, L. Xia, M. S. Parcells, S. M. Reddy, L. Lee, H.-W. Chen, and H.-J. Kung. 2003. Characterization of the chromosomal binding sites and dimerization partners of the viral oncoprotein Meq in Marek's disease virus-transformed T cells. J. Virol. 77:12841-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupiani, B., L. F. Lee, X. Cui, I. Gimeno, A. Anderson, R. W. Morgan, R. F. Silva, R. L. Witter, H. J. Kung, and S. M. Reddy. 2004. Marek's disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 101:11815-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maitra, A., K. Yashima, A. Rathi, C. F. Timmons, B. B. Rogers, J. W. Shay, and A. F. Gazdar. 1999. The RNA component of telomerase as a marker of biologic potential and clinical outcome in childhood neuroblastic tumors. Cancer 85:741-749. [PubMed] [Google Scholar]
- 23.Matsudo, H., A. Otsuka, Y. Ozawa, and M. Ono. 2003. Disruption of the PU.1 gene in chicken B lymphoma DT40 cells and its effect on reported target gene expression. Gene 322:169-174. [DOI] [PubMed] [Google Scholar]
- 24.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 25.Nair, S. K., and S. K. Burley. 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112:193-205. [DOI] [PubMed] [Google Scholar]
- 26.Rathi, A., K. Hur, A. F. Gazdar, J. S. Bae, J. J. Jang, and D. Y. Kim. 1999. Telomerase RNA expression during progression of gastric cancer. Hum. Pathol. 30:1302-1308. [DOI] [PubMed] [Google Scholar]
- 27.Schat, K. A., C.-L. Chen, B. W. Calnek, and D. Char. 1991. Transformation of T-lymphocyte subsets by Marek's disease herpesvirus. J. Virol. 65:1408-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seth, A., and D. K. Watson. 2005. ETS transcription factors and their emerging roles in human cancer. Eur. J. Cancer 41:2462-2478. [DOI] [PubMed] [Google Scholar]
- 29.Takakura, M., S. Kyo, T. Kanaya, H. Hirano, J. Takeda, M. Yutsudo, and M. Inoue. 1999. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 59:551-557. [PubMed] [Google Scholar]
- 30.Trapp, S., M. S. Parcells, J. P. Kamil, D. Schumacher, B. K. Tischer, P. M. Kumar, V. K. Nair, and N. Osterrieder. 2006. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 203:1307-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veldman, T., X. Liu, H. Yuan, and R. Schlegel. 2003. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc. Natl. Acad. Sci. USA 100:8211-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, J., L. Y. Xie, S. Allan, D. Beach, and G. J. Hannon. 1998. Myc activates telomerase. Genes Dev. 12:1769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, K. J., C. Grandori, M. Amacker, N. Simon-Vermot, A. Polack, J. Lingner, and R. Dalla-Favera. 1999. Direct activation of TERT transcription by c-MYC. Nat Genet. 21:220-224. [DOI] [PubMed] [Google Scholar]
- 34.Xin, J. H., A. Cowie, P. Lachance, and J. A. Hassell. 1992. Molecular cloning and characterization of PEA3, a new member of the Ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev. 6:481-496. [DOI] [PubMed] [Google Scholar]
- 35.Yi, X., V. M. Tesmer, I. Savre-Train, J. W. Shay, and W. E. Wright. 1999. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 19:3989-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, J., A. Bilsland, S. F. Hoare, and W. N. Keith. 2003. Involvement of NF-Y and Sp1 binding sequences in basal transcription of the human telomerase RNA gene. FEBS Lett. 536:111-119. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, J. Q., R. M. Glasspool, S. F. Hoare, A. Bilsland, I. Szatmari, and W. N. Keith. 2000. Activation of telomerase rna gene promoter activity by NF-Y, Sp1, and the retinoblastoma protein and repression by Sp3. Neoplasia 2:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]