Abstract
Millimolar concentrations of guanidine hydrochloride (GuHCl) are known to inhibit the replication of many plant and animal viruses having positive-sense RNA genomes. For example, GuHCl reversibly interacts with the nucleotide-binding region of poliovirus protein 2CATPase, resulting in a specific inhibition of viral negative-sense RNA synthesis. The use of GuHCl thereby allows for the spatiotemporal separation of poliovirus gene expression and RNA replication and provides a powerful tool to synchronize the initiation of negative-sense RNA synthesis during in vitro replication reactions. In the present study, we examined the effect of GuHCl on mammalian orthoreovirus (MRV), a double-stranded RNA (dsRNA) virus from the family Reoviridae. MRV growth in murine L929 cells was reversibly inhibited by 15 mM GuHCl. Furthermore, 15 mM GuHCl provided specific inhibition of viral dsRNA synthesis while sparing both positive-sense RNA synthesis and viral mRNA translation. By using GuHCl to provide temporal separation of MRV gene expression and genome replication, we obtained evidence that MRV primary transcripts support sufficient protein synthesis to assemble morphologically normal viral factories containing functional replicase complexes. In addition, the coordinated use of GuHCl and cycloheximide allowed us to demonstrate that MRV dsRNA synthesis can occur in the absence of ongoing protein synthesis, although to only a limited extent. Future studies utilizing the reversible inhibition of MRV dsRNA synthesis will focus on elucidating the target of GuHCl, as well as the components of the MRV replicase complexes.
Nonfusogenic mammalian orthoreovirus (MRV) is the prototype member of the family Reoviridae (40). Members of this family have genomes comprising 9 to 12 segments of double-stranded RNA (dsRNA), which are encased within multilayered, usually nonenveloped capsids in the virion particles (3, 40). The MRV genome totals ∼23,500 bp and is separated into 10 dsRNA segments (39, 40). In the MRV virion, the genome is densely packed within the inner of two radially concentric icosahedral capsids. The outer capsid interacts with cellular membrane components and functions to deliver the inner capsid plus genome, or viral core, to the cytoplasm of a permissive cell (21, 39, 40). The 52-MDa reovirus core particle (52) remains intact throughout the RNA replicative cycle and functions as a molecular machine (4, 5), with most or all of the core structural proteins providing the enzymatic activities necessary to synthesize capped viral mRNAs and to replicate the viral genome (4, 5, 35, 47, 51, 62). Although a great deal is known about the biochemical properties and structural interactions of individual core proteins, little is known about the functional associations of MRV core proteins with viral RNA or other viral or host proteins. As a result, almost no understanding of the molecular regulation of MRV RNA replication exists.
The five viral proteins constituting the MRV core—λ1, λ2, λ3, μ2, and σ2—serve both structural and functional roles during the replicative cycle, and all may possess enzymatic activities (60). The reovirus core lattice is made of 120 copies of λ1, arranged in 60 asymmetric dimers that give rise to a T=1 icosahedron (39). λ1 possesses canonical helicase superfamily II motifs (10) and appears to have nucleoside triphosphate phosphohydrolase (NTPase) (10, 42), RNA triphosphate phosphohydrolase (11), and RNA helicase activities (10). Although these activities are thought to be required for mRNA synthesis, capping, and dsRNA synthesis, definitive evidence for a role in each process is lacking. Sixty copies of λ2 form 12 pentameric turrets that sit at the fivefold axes of symmetry of the core particle (39). The λ2 protein possesses a guanylyltransferase activity (23, 33, 34, 50, 54), as well as 7-N and 2′-O methyltransferase activities (18, 30, 32, 53), suggesting that the λ2 turrets are the principle component of the mRNA capping machinery. Each turret encloses a central cavity that is contiguous with small pores in the λ1 lattice (39, 40, 52), and together these openings are thought to form an exit channel for viral mRNAs synthesized within the core particle. Immediately beneath each turret, probably tethered to the internal surface of the λ1 lattice, sit two stoichiometrically minor components of the core particle, proteins λ3 and μ2 (4, 39, 63). Twelve copies of λ3 and 12 to 24 copies of μ2 are found within each core. λ3 possesses RNA-dependent RNA polymerase (RdRp) motifs and adopts the cupped right-hand structure common to other viral RdRps (58). λ3 putatively possesses both a dsRNA-dependent single-stranded RNA polymerase (transcriptase) activity and a single-stranded RNA-dependent dsRNA-polymerase (replicase) activity (2, 64, 65); however, additional viral or cellular proteins may be required for each process because, to date, purified λ3 in solution demonstrates only a poly(C)-dependent poly(G)-polymerase activity (56). Data derived from temperature-sensitive mutants (24) and reassortant viruses suggest that the polymerase cofactor protein, μ2, contributes to the synthesis of mRNA and dsRNA. μ2 contains Walker A and B NTP-binding motifs (41) and has demonstrated in vitro NTPase and RTPase activities (28, 43). The positioning of the viral RdRp in association with its cofactor and the capping machinery at each fivefold axis of the core lattice suggests that mRNA capping occurs concomitantly with transcription and mRNA extrusion from the core particle and may require the enzymatic activities of λ1 and/or μ2. The core additionally contains 150 copies of the monomeric surface-nodule protein σ2 (40). Five copies of σ2 are found around the base of each of the 12 λ2 turrets, whereas the remaining 90 copies are found bound to the λ1 lattice across the two- and threefold axes of symmetry (39, 40). Because core-like particles do not assemble in the absence of σ2 (29, 59), it has been inferred that this protein serves as a clamp, holding the core particle together. Recently, however, it has been suggested that σ2 may also possess a nucleotidylyl phosphohydrolase activity (K. E. Murray, unpublished data) serving an as-yet-unknown function in the core.
It has been known since the 1960s that guanidine hydrochloride (GuHCl) (Fig. 1A) interferes with or inhibits the replication of many plant and animal viruses (19). Utilizing poliovirus (PV), the prototypical member of the family Picornaviridae, Pfister et al. demonstrated that 2 mM GuHCl reversibly inhibits the NTPase activity of PV protein 2CATPase (48, 49), while Barton and Flanegan demonstrated that GuHCl specifically inhibits the initiation of negative-sense RNA synthesis (7). PV 2CATPase, the target of GuHCl, is a 329-amino-acid protein that is a critical component of the PV replication complex. 2CATPase possesses two RNA recognition motifs in addition to three conserved helicase superfamily III NTP-binding domains (20). 2CATPase will efficiently hydrolyze both ATP and GTP and may also function as an RNA helicase (48). Although there is no MRV homolog to PV 2CATPase, μ2 and λ1 may be functional analogs. As described above, both proteins appear to hydrolyze NTPs, bind RNA (13) and are required for MRV RNA synthesis. As a result, we hypothesized that millimolar concentrations of GuHCl might reversibly inhibit MRV RNA replication by inhibiting the NTPase activities of μ2 and/or λ1. We therefore investigated the effect of various concentrations of GuHCl on MRV replication. In this study, we demonstrated that 15 mM GuHCl reversibly inhibits MRV growth in mouse L929 cells. We further demonstrate that GuHCl specifically inhibits viral dsRNA synthesis while sparing both positive-sense RNA synthesis and viral mRNA translation, allowing for the temporal separation of viral gene expression and genome replication. Utilizing 15 mM GuHCl, we demonstrate that primary transcription and translation provide sufficient protein to assemble morphologically normal viral factories containing functional replicase complexes and also that viral dsRNA synthesis has no intrinsic need for ongoing protein synthesis.
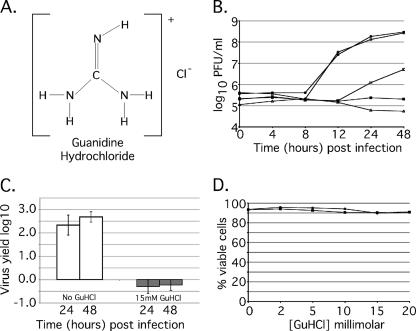
FIG. 1.
GuHCl inhibits MRV growth in mouse L929 cells. (A) Kekule structure depicting GuHCl. (B) MRV growth in the presence of increasing concentrations of GuHCl. Samples were incubated for 48 h in the presence of GuHCl at concentrations ranging from 0 to 25 mM. At 0, 4, 8, 12, 24, and 48 h p.i., samples were collected, and the virus titer was determined by plaque assay. Symbols: •, no drug added; ○, 2 mM GuHCl; □, 10 mM GuHCl; ▪, 15 mM GuHCl; ▵, 25 mM GuHCl. Approximately complete inhibition of MRV growth is achieved with 15 mM GuHCl. The experiment was repeated multiple times with similar results, and a representative plot is shown. (C) Abrogation of reovirus yield by 15 mM GuHCl. Confluent monolayers of L929 cells were infected with MRV-T1L at an MOI of 5. After virus attachment, medium with (□) or without (□) 15 mM GuHCl was added. Zero hour time points were taken immediately; the remaining samples were incubated at 37°C for either 24 h or 48 h. Virus concentration was determined by plaque assay and is presented as the virus yield. Error bars represent the standard deviation of the mean of five replicate experiments. (D) Trypan blue exclusion assays were done to evaluate the effect of GuHCl (0 to 20 mM) on cell proliferation and viability over a period of 24 h. Subconfluent monolayers of L929 cells were treated with 0, 2, 5, 10, 15, or 20 mM GuHCl and harvested either 0 h (○) or 24 h (•) after treatment. Cells were collected as described, incubated in 1% trypan blue, and loaded onto a hemacytometer, and both total and viable cells were counted. The results are presented as the percent viable cells for each concentration and time point. GuHCl exhibited no significant effect on L-cell viability. The experiment was repeated twice with similar results. A representative plot is shown.
MATERIALS AND METHODS
Cells and viruses.
Spinner-adapted murine L929 cells were maintained in suspension at 37°C by daily passage (5 × 105 to 1 × 106 cells/ml) in Joklik modified minimal essential medium (Irvine Scientific) supplemented with 2% fetal and 2% calf bovine serum (HyClone), 2 mM l-glutamine (Mediatech), and 100 U of penicillin and 100 μg of streptomycin/ml (Irvine Scientific). HeLa S3 cells were maintained in suspension at 37°C by daily passage (3 × 105 to 6 × 105 cells/ml) in Joklik modified minimal essential medium (MP Biomedicals) supplemented with 2% fetal and 5% calf bovine serum. HeLa cells were collected by centrifugation and resuspended in fresh medium at 3 × 105 cells/ml 20 to 24 h prior to S10 lysate preparation. Simian CV-1 cells were grown in Dulbecco modified Eagle medium (DMEM; Invitrogen) supplemented to contain 10 μg of gentamicin (Invitrogen)/ml and 10% fetal bovine serum. Third-passage L-cell lysate stocks of plaque-purified MRV type 1 Lang (T1L) clones were used to generate purified virions (22) and cores (26) as previously described. Purified virions and cores were stored at 4°C in virion buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris [pH 7.5]).
Analysis of reovirus growth.
Six-well plates were seeded with L929 cells (1.2 × 10 6 cells/well). Cells were incubated at 37°C overnight to allow nearly confluent monolayers to form (≥80% confluent). Monolayers were infected with MRV-T1L at a multiplicity of infection (MOI) of 5 PFU/cell, and virus was adsorbed for 1 h at room temperature (∼25°C). Fresh medium (2 ml/well, Joklik medium supplemented as described above) with or without 15 mM GuHCl was added to the infected monolayers, and zero time point samples were immediately harvested and frozen. The remaining samples were incubated at 37°C for the times described before harvesting and freezing. All samples were subjected to two cycles of freeze-thawing (−80°C, 37°C). Viral concentrations (in PFU/ml) were determined by plaque assay on monolayers of L929 cells. Virus yields were calculated as follows: log10 yield = log10(PFU/ml)x − log10(PFU/ml)t0, where t0 is time zero and x is the time postinfection (p.i.).
Cell viability assays.
Subconfluent monolayers of L929 cells were grown in 60-mm plates with medium containing 0, 2, 5, 10, 15, 20, 30, 40, or 50 mM GuHCl. At 0 and 24 h after treatment, the culture supernatant was removed and collected. Monolayers were washed once with phosphate-buffered saline (PBS [pH 7.5]; 137 mM NaCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 2.7 mM KCl) and treated with PBS containing 5 mM EDTA. The monolayers were incubated at 37°C for 10 min to allow cells to detach from the plate. After incubation, the cells were collected, combined with the supernatant and subjected to low-speed centrifugation (∼300 × g) to pellet the cells. After the supernatant was discarded, the cell pellet was resuspended and incubated with 0.1% trypan blue (Sigma) for 5 min at room temperature. Cells in trypan blue were loaded onto a hemacytometer, and both viable and total cells were counted for each concentration and time point (percent viable cells = viable cells/total cells × 100). Four fields per drug concentration per time point were counted, and duplicate analyses were conducted.
HeLa cell S10 extracts and translation initiation factors.
HeLa cell S10 extracts and HeLa cell translation initiation factors were prepared as previously described (9) with minor modifications. HeLa S10 extracts were prepared by collecting ∼1.5 × 109 cells (2.5 liters of culture) by centrifugation at 1,400 rpm in a Beckman Allegra 6R centrifuge with a Beckman GH-3.8 rotor (ca. 300 × g) for 5 min in four 600-ml polypropylene centrifuge bottles (Corning). The cells were washed twice with isotonic buffer (35 mM HEPES [pH 7.4], 146 mM NaCl, 5 mM dextrose), pooled, and collected in a 50-ml conical tube (Falcon). After the final wash, the cells were pelleted and resuspended in 1.5 volumes of hypotonic buffer [20 mM HEPES-KOH (pH 7.4)], 10 mM KCl, 1.5 mM Mg(CH3CO2)2, 1 mM dithiothreitol (DTT)], vortexed, and incubated on ice for 10 min. Swelled cells were transferred to a 15-ml Dounce homogenizer (Wheaton) with a tight-fitting pestle and broken with 25 to 30 firm strokes. The homogenate was transferred to a 50-ml conical tube, and a 1/10 volume of 10X S10 buffer [200 mM HEPES-KOH (pH 7.4), 1.2 M KCH3CO2, 40 mM Mg(CH3CO2)2, 52 mM DTT] was added. The mixture was centrifuged at 2,250 rpm (∼800 × g) for 10 min using a Beckman GH-3.8 rotor to pellet nuclei. The supernatant was collected, transferred to a 30-ml Corex tube, and centrifuged at 10,000 × g for 15 min at 4°C using a Sorvall SS-34 rotor in a Sorvall RC-5C centrifuge. After centrifugation, the supernatant was poured into a 50-ml conical tube, adjusted to 1 mM CaCl2, and treated with 1 U of micrococcal nuclease/ml at 20°C for 15 min. EGTA was added to 2 mM to terminate the nuclease treatment. The treated extracts were transferred to a 30-ml Corex tube and centrifuged at 10,000 × g for 15 min at 4°C by using a Sorvall SS-34 rotor. After centrifugation, the supernatant was frozen at −80°C in 150-μl aliquots.
Translation initiation factors (IFs) were prepared by harvesting HeLa cells grown in suspension and washing them with isotonic buffer as described above. Cell pellets were resuspended in 2 vol of hypotonic buffer, held on ice for 10 min, and lysed by 30 firm strokes in a 30-ml Dounce homogenizer with a tight-fitting pestle (Wheaton). Cellular debris was removed from the lysate as described above, and nuclei were removed by spinning in a Sorvall RC-5C centrifuge at 10,000 × g for 15 min at 4°C by using a Sorvall SS-34 rotor. The supernatant was decanted into a 10.4-ml Beckman type 70.1Ti polycarbonate ultracentrifuge tube and spun at 60,000 rpm (∼250,000 × g) for 60 min at 4°C in a Beckman-Coulter Optima L-100XP ultracentrifuge to pellet ribosomes. After centrifugation, the supernatant was discarded, and the ribosomal pellet was resuspended in hypotonic buffer at 250 A260 U/ml. Then, 4 M KCl was added to a final concentration of 0.5 M, and the mixture was stirred at 4°C for 15 min. The stir bar was removed, and ribosomes were repelleted by centrifugation at 60,000 rpm for 60 min at 4°C by using a type 70.1Ti rotor in a Beckman-Coulter Optima L-100XP ultracentrifuge. The supernatant was collected and dialyzed against IF dialysis buffer (5 mM Tris-HCl [pH 7.5], 75 mM KCH3CO2, 50 μM EDTA, 5% [vol/vol] glycerol, 1 mM DTT) for 2 h at 4°C. The dialyzed IFs were collected and frozen at −80°C in 50-μl aliquots.
In vitro transcription and transcription/translation reactions.
In vitro synthesis of reovirus mRNA was accomplished by programming HeLa S10/IF reactions with MRV core particles. These reactions (50 μl, final volume) contained 25 μl of HeLa S10 extract, 10 μl of HeLa cell translation IFs, 5 μl of 10× nucleotide reaction mix (10 mM ATP, 2.5 mM GTP, 2.5 mM CTP, 2.5 mM UTP, 600 mM KCH3CO2, 300 mM creatine phosphate, 4 mg of creatine kinase/ml, 155 mM HEPES-KOH [pH 7.4]), and 6.25 μl of MRV-T1L core particles (4.0 × 1010 particles). Next, 3.75 μl of 200 mM GuHCl was added to one-half of the reactions, and 3.75 μl of deionized water was added to the remaining half. We then added 3 μl of label (12.5 μCi of [α-32P]CTP [ICN] and puromycin [2.5 μg]) to each reaction. The reactions were incubated at 37°C for 90 min and terminated by adding 350 μl of 0.5% sodium dodecyl sulfate (SDS) buffer (0.5% SDS, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl); labeled RNA was extracted three times with phenol-chloroform-isoamyl alcohol (25:24:1) and twice with chloroform-isoamyl alcohol (24:1). After extraction, labeled RNA was ethanol precipitated, dried, and resuspended in 20 μl of RNase-free deionized water. Reaction products were analyzed by electrophoresis in 1.5% agarose gels containing 6 M urea and 0.025 M citrate (pH 3.0). Gels were dried, and the radiolabeled RNA was detected and quantified by phosphorimaging (Molecular Dynamics).
In vitro MRV translation was assayed by utilizing the same reaction parameters described above with the following changes. [35S]methionine (1.2 mCi/ml, Amersham) was included in HeLa S10 transcription-translation reactions in place of the [α-32P]CTP label. Reaction mixtures were incubated at 30°C for 5 h and were terminated by solubilizing [35S]methionine-labeled proteins in 1× Laemmli sample buffer (2% SDS [Sigma], 62.5 mM Tris-HCl [pH 6.8], 0.5% 2-mercaptoethanol, 0.1% bromophenol blue, 20% glycerol). Labeled MRV proteins were heated at 100°C for 5 min and separated by SDS-10% polyacrylamide gel electrophoresis (PAGE). After electrophoresis, gels were dried and [35S]methionine-labeled proteins were detected and quantified by phosphorimaging (Molecular Dynamics).
Immunoblotting.
In vitro-synthesized MRV proteins were separated by SDS-10% PAGE and transferred (100 V for 60 min at 4°C) to a 0.2-μm-pore-size, pure nitrocellulose membrane (Bio-Rad) in 1× transfer buffer (25 mM Tris, 192 mM glycine [pH 8.3]). Membranes were blocked with a 5% solution of nonfat dry milk in Tris-buffered saline (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl) containing 0.1% polyoxyethylene-sorbitan monolaurate (Tween 20). After blocking, membranes were soaked for 1 h at room temperature in a 1% solution of nonfat dry milk in TBS containing anti-T1L virion-specific rabbit polyclonal antiserum (diluted 1:5,000). The binding of primary antibodies was detected by utilizing an alkaline phosphatase-coupled goat anti-rabbit antibody (1:7,500; Bio-Rad) and the colorimetric reagents p-nitroblue tetrazolium chloride and BCIP (5-bromo-4-chloro-3-indolylphosphate) p-toluidine salt (Bio-Rad).
Immunoblots of MRV proteins derived from infected L929 cells were prepared by collecting total proteins at the described time points. The culture medium was aspirated from infected monolayers, and the cells were washed twice with PBS and then treated with PBS containing 5 mM EDTA. The monolayers were incubated at 37°C for 10 min to allow cells to detach from the plate. After incubation, the cells were collected, counted, and subjected to low-speed centrifugation (∼300 × g). After the supernatant was discarded, the cell pellet was resuspended and lysed in 1× Laemmli sample buffer. Equal quantities of cell lysate from each time point were loaded onto 10% polyacrylamide gels. MRV proteins were separated electrophoretically and transferred (10 V for 10 h at 4°C) to a nitrocellulose membrane in 1× transfer buffer as described above. Membranes were then blocked as described above. After blocking, membranes were soaked for 6 h at room temperature in a 1% solution of nonfat dry milk in TBS containing either anti-T1L virion-specific rabbit polyclonal antiserum (diluted 1:5,000) or mouse anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; Santa Cruz Biotech) diluted 1:4,000. The binding of primary antibodies was detected photographically on Fuji medical X-ray film utilizing a horseradish peroxidase (HRP)-conjugated donkey anti-rabbit antibody (KPL Laboratories) or an HRP-conjugated goat anti-mouse antibody (Pierce) with a Western Lightning enhanced chemiluminescence kit (Perkin-Elmer Life Sciences).
Immunofluorescence microscopy.
For immunostaining, the following antibodies and dilutions were used: μNS-specific rabbit polyclonal antiserum (17) and μ2-specific rabbit polyclonal antiserum were both used at a dilution of 1:10,000 (44), goat anti-rabbit immunoglobulin G conjugated to Alexa 488 and goat anti-rabbit immunoglobulin G conjugated to Alexa 594 were obtained from Molecular Probes and used at a 1:500 dilution. L929 cells or CV-1 cells were grown overnight on glass coverslips in six-well plates (1.2 × 106 cells/well). Near-confluent monolayers were infected with MRV-T1L virions at an MOI of 5 as described above. Fresh medium (2 ml/well; Joklik medium supplemented as described above) with or without 15 mM GuHCl was added, and the MRV-infected monolayers were incubated at 37°C for 4 or 18 h as described above. When the desired time point was reached, the medium was aspirated, and the cells were fixed in 2% paraformaldehyde (for 10 min at room temperature). Unless otherwise indicated, the cells were permeabilized, blocked, incubated with antibodies and DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes), and mounted as previously described (44). Microscopy images were collected digitally using a Nikon Optiphot-2 epifluorescence upright microscope equipped with a Photometrics CoolSnapcf camera (Roper Scientific) and analyzed by using MetaVue version 5.0 software (Universal Imaging). Images were processed and presented by using Photoshop CS version 8.0 and Illustrator CS version 11.0.0 (Adobe).
Viral dsRNA extraction.
Sixty-millimeter plates were seeded with 2.5 × 106 L929 cells, followed by growth overnight at 37°C to allow confluent monolayers to form. Confluent monolayers were infected with MRV-T1L as described above. Culture medium with or without 15 mM GuHCl was added to the MRV-infected monolayers, and the cells were incubated at 37°C for various lengths of time. MRV dsRNA was extracted utilizing 1 ml of TRIzol reagent (Invitrogen). After extraction, viral RNA was ethanol precipitated, dried, and resuspended in 40 μl of RNase-free, deionized water. MRV dsRNA was analyzed by electrophoresis in 10% polyacrylamide gels. Gels were run at 14 mA for 21 h, stained with ethidium bromide (0.5 μg/ml) in 1× TAE (40 mM Tris-base, 20 mM CH3COOH, 50 mM EDTA [pH 8.0]), and visualized by UV transillumination.
GuHCl reversibility assay.
Confluent monolayers of L929 cells were grown in 60-mm plates and infected with MRV-T1L as described above. GuHCl (15 mM) was added to the culture medium, and the cells were incubated at 37°C for 20 h. GuHCl-containing medium was replaced with 5 ml of fresh medium with or without cycloheximide (20 μg/ml; Sigma). Samples were harvested (0 h postreversal) or continued to be incubated for 2, 4, 6, 8, 10, or 12 h. When the desired time point was reached, dsRNA was harvested with TRIzol reagent as described above. MRV dsRNA was analyzed by electrophoresis in 10% polyacrylamide gels run at 14 mA for 21 h. MRV dsRNA was visualized as described above.
Orthophosphate labeling of viral dsRNA synthesis.
Confluent monolayers of L929 cells were grown in 60-mm plates and infected with MRV-T1L as described above. GuHCl (15 mM) was added to the culture medium of all MRV-infected monolayers, and the cells were incubated at 37°C for 19 h. At 19 h p.i., the cells were washed three times and incubated for an additional 1 h at 37°C in serum-free, phosphate-free DMEM containing 15 mM GuHCl. At 20 h p.i., the GuHCl-containing medium was replaced with 5 ml of serum-free, phosphate-free DMEM containing 30 μl of carrier-free [32P]orthophosphate (10 μCi/μl) with or without cycloheximide (20 μg/ml). MRV-infected cells were incubated for an additional 30 min at 37°C. The cells were lysed, and labeled dsRNA was harvested with TRIzol reagent. Labeled dsRNA was analyzed by electrophoresis as described above. After ethidium bromide staining, gels were dried, and radiolabeled dsRNA was visualized by phosphorimaging on a Molecular Dynamics Typhoon.
RESULTS
MRV growth is inhibited by 15 mM GuHCl.
Given the known effect of GuHCl on PV replication, we examined its effect on MRV. We grew MRV in murine L929 cells with increasing concentrations of GuHCl added to the culture medium and found that 2 mM GuHCl, a dose that inhibits PV RNA synthesis (19), had no demonstrable effect on MRV growth (Fig. 1B). Higher concentrations of GuHCl, however, reduced MRV growth in a progressive fashion, with nearly complete inhibition achieved with 15 mM GuHCl (Fig. 1B). Infection of L929 cells with MRV at 5 PFU/cell typically yields an ∼3-log10 increase in infectious titer over a 48-h period, but when 15 mM GuHCl was added to the culture medium, this increase was consistently abrogated (Fig. 1C).
It has been previously shown that 2 mM GuHCl does not affect the viability of HeLa S3 cells (19); however, the effect of 15 mM GuHCl on L929 cells was unknown. To discern any negative effects of GuHCl on L929 cell viability, we monitored cells over a 24-h period when GuHCl was present in the culture medium at concentrations ranging from 2 to 50 mM. Cell viability was assayed by trypan blue exclusion and compared to control samples to which no GuHCl had been added. We found that the presence of GuHCl in concentrations as high as 20 mM had no adverse effects on L929 cell viability (Fig. 1D) or proliferation (data not shown). Only with GuHCl concentrations in excess of 40 mM did we see evidence of cell death at 12 to 24 h (data not shown).
MRV transcription and translation are unaffected by 15 mM GuHCl.
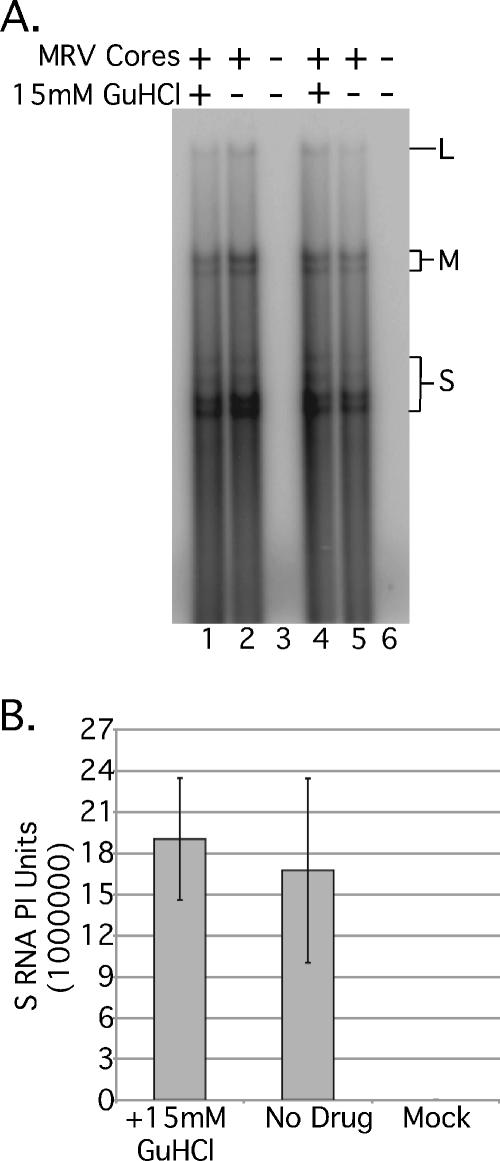
Having established that GuHCl is inhibitory to MRV growth, we next tried to identify which step in infection is blocked. We first investigated the effect of 15 mM GuHCl on viral transcription. MRV cores were generated as previously described (26), and ∼4 × 1010 particles were used to program in vitro transcription reactions. We found that cores incubated at 37°C for 90 min in the presence of [α-32P]CTP synthesized comparable quantities of labeled viral transcripts irrespective of the presence of 15 mM GuHCl (Fig. 2B), suggesting that MRV positive-sense RNA synthesis is not substantially, if at all, inhibited by 15 mM GuHCl. In each of the replicate experiments that we conducted, MRV mRNAs of consistent quality (Fig. 2A) were generated whether 15 mM GuHCl was present or not, further implying that GuHCl does not alter the short-term stability of MRV positive-sense RNAs. As expected, mock-programmed reactions produced no labeled RNA products (Fig. 2A, lanes 3 and 6).
FIG. 2.
In vitro MRV mRNA synthesis is not affected by 15 mM GuHCl. (A) In vitro transcription reactions were programmed with ca. 4 × 1010 MRV-T1L core particles (lanes 1, 2, 4, and 5) or were mock treated (no cores, lanes 3 and 6). Reactions were incubated with 15 mM GuHCl (lanes 1 and 4) or without GuHCl (lanes 2, 3, 5, and 6) at 37°C for 90 min. Transcription reactions were stopped by the addition of 0.5% SDS buffer, and [32P]CTP-labeled RNA was phenol-chloroform extracted and ethanol precipitated. Labeled MRV mRNA was separated by electrophoresis in 1.5% agarose containing 6 M urea and 0.025 M citrate (pH 3.0) and detected by phosphorimaging. L, MRV large mRNAs; M, MRV medium mRNAs; S, MRV small mRNAs (B) Full-length MRV S mRNA synthesis was quantified by phosphorimaging and plotted as the mean of duplicate samples ± the standard deviation. Similar results were obtained with the M and L mRNA bands (not shown).
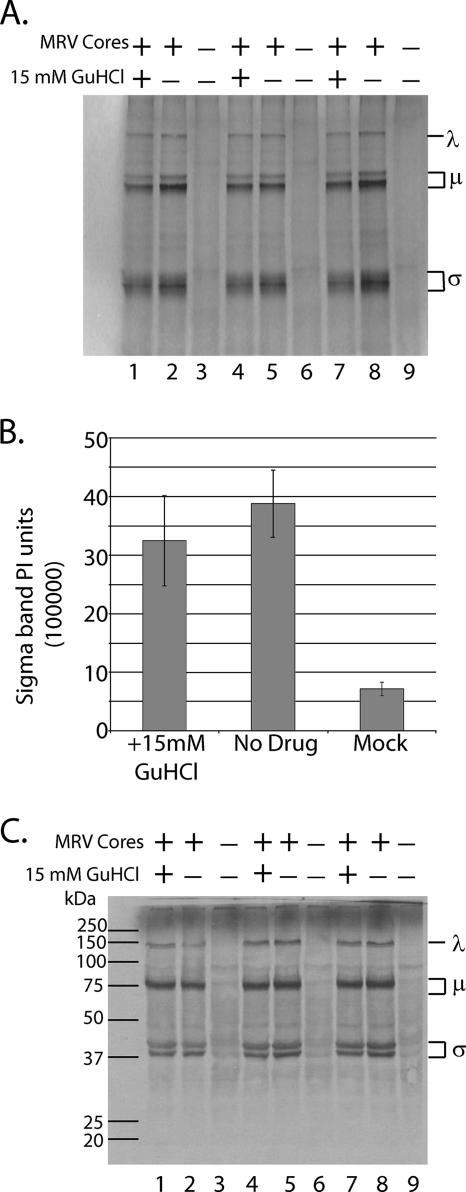
Because millimolar concentrations of GuHCl are not known to inhibit viral mRNA translation (9, 48), we utilized an in vitro transcription-translation system to demonstrate that MRV positive-sense RNAs synthesized in the presence of 15 mM GuHCl are intact and functional. Reactions were established as described above except that products were labeled with [35S]methionine during a 5-h incubation at 30°C. Labeled proteins were solubilized in 1× Laemmli sample buffer and separated by electrophoresis in 10% polyacrylamide gels. Labeled proteins were visualized (Fig. 3A) and quantified (Fig. 3B) by phosphorimaging. As expected, no significant inhibition of MRV protein synthesis was observed (Fig. 3A, compare lanes 1, 4, and 7 to lanes 2, 5, and 8). To confirm that the labeled proteins were MRV in origin, we electroblotted proteins from a second gel (run in parallel, using the same reaction products shown in Fig. 3A) to a nitrocellulose membrane and probed with a polyclonal rabbit anti-MRV virion serum. Bound MRV proteins were detected colorimetrically using a goat anti-rabbit immunoglobulin G antibody conjugated to alkaline phosphatase (Fig. 3C). Virus-specific proteins were seen only in reactions programmed with MRV cores; mock-programmed reactions consistently failed to produce any labeled viral protein products (Fig. 3A and C).
FIG. 3.
In vitro MRV transcription and translation are not affected by 15 mM GuHCl. (A) In vitro transcription-translation reactions were programmed with ca. 4 × 1010 MRV-T1L core particles (lanes 1, 2, 4, 5, 7, and 8) or mock treated (no cores; lanes 3, 6, and 9). Reactions were incubated with 15 mM GuHCl (lanes 1, 4, and 7) or without GuHCl (lanes 2, 3, 5, 6, 8, and 9) at 30°C for 5 h. Transcription-translation reactions were terminated by solubilizing [35S]methionine-labeled proteins in 1× Laemmli sample buffer. Labeled MRV proteins were separated by SDS-PAGE. Gels were dried, and labeled proteins were visualized by phosphorimaging (Molecular Dynamics) λ, MRV proteins λ1, λ2, and λ3; μ, MRV proteins μ1, μ2, and μNS/μNSC; σ, MRV proteins σ1, σ2, σNS, and σ3. (B) MRV σ protein bands were quantified by phosphorimaging and plotted as the mean of triplicate samples ± the standard deviation. Similar results were obtained with the λ and μ protein bands (data not shown). (C) Parallel in vitro transcription-translation reactions lacking [35S]methionine were prepared as for those described above. After a 5-h incubation at 30°C, unlabeled MRV proteins were electroblotted onto a nitrocellulose membrane and probed with a rabbit polyclonal anti-MRV virion serum. MRV proteins on the blot were detected colorimetrically using a goat anti-rabbit antibody conjugated to alkaline phosphatase. Molecular masses are indicated in kilodaltons on the left; the protein size classes are indicated on the right.
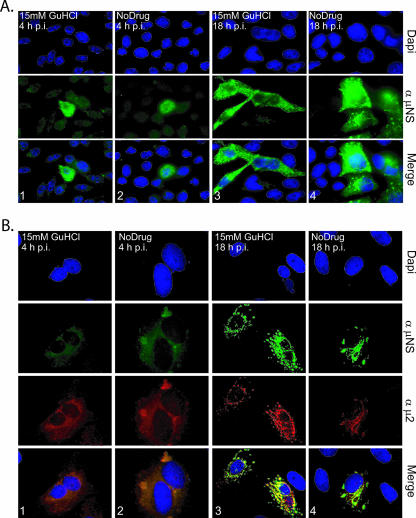
To extend the evidence that 15 mM GuHCl does not inhibit MRV transcription or translation, we evaluated expression of the MRV nonstructural protein μNS in infected L929 cells both at early (4 h) and later (18 h) times after infection. The MRV μNS protein is known to be present in large phase-dense inclusions in the cytoplasm of infected cells (14, 17). These inclusions are termed viral factories and are the putative locations of viral RNA synthesis and nascent core morphogenesis. Immunostaining of μNS at 4 h p.i. revealed diffuse cytoplasmic staining in the majority of infected cells, accompanied by many small punctate inclusions (Fig. 4A and B, columns 1 and 2) as have been previously described (17). We found that the presence of 15 mM GuHCl did not alter the distribution or synthesis of μNS, nor did it affect the number of cells infected by MRV at 4 h p.i. (Fig. 4). The μNS protein putatively recruits numerous viral and cellular proteins to the factories to facilitate MRV RNA replication and core morphogenesis (15, 16, 37) and, as a result, the factories grow in both size and number over the course of an infection. At 18 h p.i., we found no discernible differences in the number of cells infected or in the size, number, or distribution of the MRV factories in infected cells as a consequence of 15 mM GuHCl having been added to the culture medium (Fig. 4A and B, columns 3 and 4). Thus, 15 mM GuHCl did not alter the continued development of the MRV factories.
FIG. 4.
GuHCl at 15 mM does not inhibit MRV entry, mRNA synthesis, or viral translation in mouse L929 cells. (A). L929 cells were infected at with MRV-T1L (MOI = 5). Medium with (columns 1 and 3) or without (columns 2 and 4) 15 mM GuHCl was added, and MRV-infected cells were incubated at 37°C for 4 h (columns 1 and 2) or 18 h (columns 3 and 4). At each time point, the cells were fixed, permeabilized, blocked, incubated with antibodies and DAPI, and mounted as described in the text. α-μNS, μNS-specific rabbit polyclonal antiserum; Merge, overlay of DAPI- and α-μNS-stained images. (B) MRV factory morphology is not affected by 15 mM GuHCl. CV-I cells were infected with MRV-T1L (MOI = 5). After virus adsorption, medium with (columns 1 and 3) or without (columns 2 and 4) 15 mM GuHCl was added, and MRV-infected cells were incubated at 37°C for 4 h (columns 1 and 2) or 18 h (columns 3 and 4). At each time point cells were fixed, permeabilized, blocked, incubated with antibodies and DAPI, and mounted as described in the text. α-μNS, μNS-specific rabbit polyclonal antiserum; α-μ2, μ2-specific rabbit polyclonal antiserum; Merge, overlay of DAPI-, α-μNS-, and α-μ2-stained images.
The factories of MRV strain T1L, which we used in all of these experiments, adopt a distinctive morphology at later times after infection (44). The nonstructural protein μNS, through an association with the RdRp cofactor protein μ2, associates with microtubules, giving rise to elongated or filamentous factories (44). Using simian CV-1 cells for enhanced immunofluorescence imaging of subcellular structures (44), we found that the presence of 15 mM GuHCl did not alter the morphology of MRV-T1L factories at 18 h p.i. (Fig. 4B, columns 3 and 4), suggesting that GuHCl did not affect the synthesis of viral protein μ2 or its capacity to interact with microtubules and μNS. In addition to the more direct implications, the results in this section suggest that 15 mM GuHCl has little or no influence on cell entry by MRV (no reduction in the number of cells infected) or on the expression of the MRV proteome (no effect on μNS or μ2 synthesis).
MRV dsRNA synthesis is inhibited by 15 mM GuHCl.
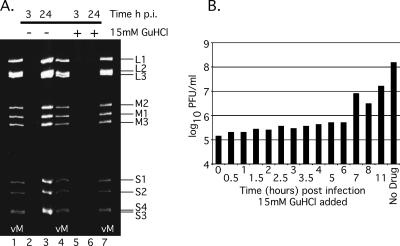
Given our evidence that the inhibition of MRV replication by GuHCl is not the consequence of a defect in entry or viral protein synthesis, we suspected that, as with PV, negative-sense RNA synthesis (i.e., dsRNA synthesis in the case of MRV) was being inhibited (7). To examine this possibility, we evaluated the synthesis of MRV dsRNA when 15 mM GuHCl was added to the culture medium of infected cells. Using TRIzol reagent, we isolated total RNA from MRV-infected cells at 3 and 24 h after infection. As expected, MRV dsRNA was not detected in infected cells at 3 h p.i. irrespective of the presence of 15 mM GuHCl (Fig. 5A, lanes 2 and 5). At 24 h p.i., however, untreated cells (no GuHCl in the culture medium) yielded large quantities of MRV dsRNA (Fig. 5A, lane 3), whereas GuHCl-treated cells repeatedly failed to yield any detectable MRV dsRNA (Fig. 5A, lane 6).
FIG. 5.
MRV dsRNA synthesis is inhibited by 15 mM GuHCl. (A) L929 cells were infected with MRV-T1L (MOI = 5). After virus adsorption, medium with (lanes 5 and 6) or without (lanes 2 and 3) 15 mM GuHCl was added, and MRV-infected cells were incubated at 37°C for 3 h (lanes 2 and 5) or 24 h (lanes 3 and 6). At each time point, cells were lysed, and the total RNA was extracted with TRIzol reagent. dsRNA was separated by electrophoresis in 10% polyacrylamide gels, stained with ethidium bromide, and visualized by UV transillumination. vM, virion marker (purified MRV-T1L genomic RNA). Individual MRV genome segments are identified on the right. (B) GuHCl bypass temporally correlates with dsRNA synthesis. L929 cells were infected with MRV-T1L (MOI = 5). After virus adsorption, cells were incubated at 37°C, and 15 mM GuHCl was added at 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 5, 6, 7, 8, or 11 h p.i. Incubation at 37°C was continued to 48 h p.i., when the infected cells were lysed, and virus yield was determined by plaque assay. The plot shown is representative of duplicate experiments.
To extend the evidence that MRV dsRNA synthesis is targeted by 15 mM GuHCl, we conducted GuHCl bypass experiments. In these experiments, we attempted to determine the time after infection at which MRV could bypass the GuHCl-imposed block to replication and to correlate this temporally with the synthesis of MRV dsRNA. To accomplish this, we delayed the addition of 15 mM GuHCl to the culture medium of infected cells by 0.5 h increments to 4 h p.i., and by 1 h increments from 4 to 8 h p.i.; in addition, we allowed one infection to proceed for 11 h prior to treatment with 15 mM GuHCl (Fig. 5B). We found that delaying the addition of 15 mM GuHCl by 7 h resulted in a significant increase in viral yield (Fig. 5B). Delaying the addition of GuHCl by 11 h reliably produced viral yields reduced by less than 1 log10 PFU/ml (Fig. 5B). Previous investigations of the MRV replicative cycle have reported that dsRNA synthesis initiates at ∼6 h p.i. and peaks at 10 to 11 h p.i (31, 62). The results of our bypass experiments are thus temporally consistent with the conclusion that GuHCl inhibits MRV dsRNA synthesis.
Primary MRV transcripts continuously express viral proteins during the GuHCl blockade of dsRNA synthesis.
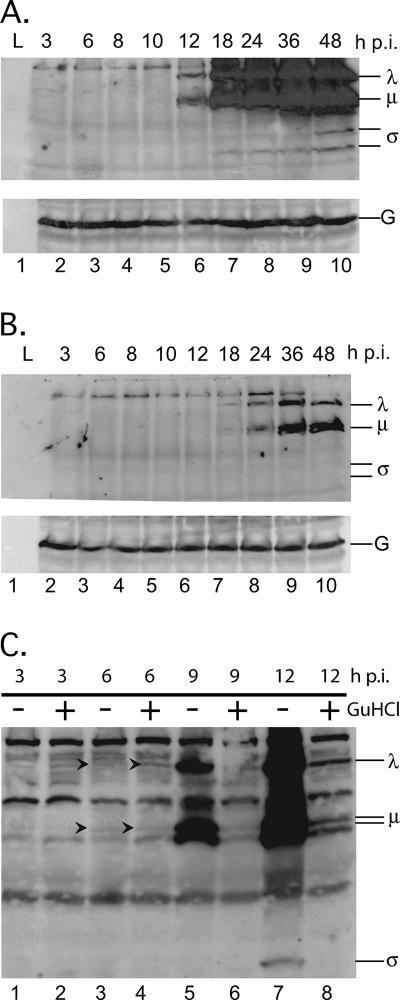
Previous reports have suggested that 80% or more of MRV protein molecules are expressed from secondary transcripts, i.e., transcripts synthesized by nascently formed core particles (24, 61). The data presented above, however, infer that 15 mM GuHCl inhibits MRV secondary transcription by blocking dsRNA synthesis and nascent core morphogenesis (Fig. 5A), even though it does not prevent the assembly of morphologically normal viral factories (Fig. 4). We therefore sought to determine whether continued translation of primary transcripts, in cells treated with 15 mM GuHCl, yields viral protein concentrations similar to those produced in untreated cells or whether submaximal concentrations of viral proteins are produced and are sufficient for formation of the normal-looking viral factories. To address these questions, we infected L929 cells with T1L virions (MOI = 5) and incubated in the presence or absence of 15 mM GuHCl. Cell lysates were collected at intervals between 3 and 48 h p.i., and total proteins were separated by electrophoresis in 10% polyacrylamide gels, transferred to nitrocellulose membranes, and probed with an anti-T1L polyclonal antiserum. In untreated cells, newly synthesized viral proteins were readily observable by 12 h p.i. and accumulated to maximal concentrations by 36 to 48 h p.i. (Fig. 6A). When 15 mM GuHCl was present, however, MRV proteins were not readily observable until 18 to 24 h p.i., with significantly smaller quantities having accumulated by 48 h (Fig. 6B). When we examined early time points after infection more carefully, by overdeveloping the blots, we observed (consistent with results from in vitro translation assays [Fig. 3]) that comparable quantities of MRV proteins were synthesized by 6 h p.i. irrespective of the presence of 15 mM GuHCl (Fig. 6C, arrowheads, lanes 3 and 4). By 9 h p.i., when dsRNA synthesis and secondary transcription had likely begun in the absence of GuHCl (see Fig. 5), less protein accumulation was seen in the presence of GuHCl, as would be expected from the lack of secondary transcripts. In sum, these data provide further support to the conclusion that GuHCl does not directly influence translation of the MRV proteins. These and preceding data also suggest that translation of primary transcripts over an extended period of time produces enough viral protein to assemble and maintain morphologically normal viral factories but that, as previously reported, secondary transcripts produce the majority of MRV proteins in infected cells in the absence of GuHCl.
FIG. 6.
Primary MRV transcripts continuously express proteins for extended time periods. L929 cells were infected with MRV-T1L (MOI = 5). After virus adsorption, infected cells were fed medium without GuHCl (A) or with 15 mM GuHCl (B). Cells were then incubated at 37°C for 3, 6, 8, 10, 12, 18, 24, 36, or 48 h. At each time point, total proteins were harvested, separated by SDS-PAGE, and electroblotted onto nitrocellulose. MRV proteins on the blot were probed with a rabbit polyclonal anti-MRV virion serum and were detected by exposure to film after treatment with a mouse anti-rabbit antibody conjugated to HRP. Protein size classes are indicated on the right. G, GAPDH loading control. (C) To evaluate protein synthesis at early time points after infection, L929 cells were infected with MRV-T1L (MOI = 5). After virus adsorption, infected cells were fed medium with or without 15 mM GuHCl as indicated and incubated at 37°C for 3, 6, 9, or 12 h. At each time point, total proteins were harvested and processed as described above. Blots A and B were simultaneously exposed to film for 10 s, whereas blot C was exposed to film for 10 min.
Inhibition of MRV dsRNA synthesis by 15 mM GuHCl is fully reversible.
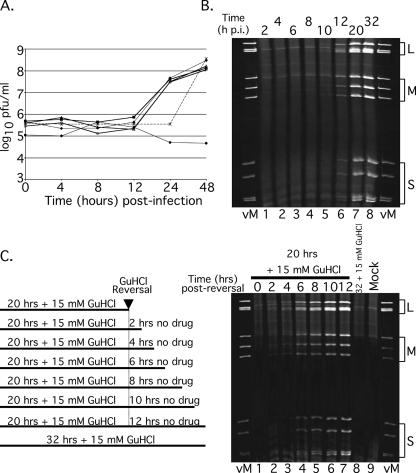
To be most useful as a tool for studying aspects of MRV gene expression and RNA replication, the GuHCl-induced inhibition of MRV growth should be fully reversible. To evaluate the reversibility of the GuHCl-dependent inhibition of MRV dsRNA synthesis, we treated MRV-infected L929 cells with 15 mM GuHCl for various times (4, 8, 12, or 24 h) beginning at the time of infection. At each time point, the GuHCl-treated culture medium was replaced by untreated medium, and the infections were allowed to proceed until 48 h p.i. When we plotted the growth curves for each treatment regime, we discovered that the presence of 15 mM GuHCl during the first 4, 8, or 12 h of infection resulted in no detectable defect in MRV growth over a 48-h period (Fig. 7A). The presence of 15 mM GuHCl in MRV-infected cell culture for 24 h abolished the burst in virus growth normally seen between 12 and 24 h p.i. but resulted in no reduction in the MRV yield at 48 h p.i. (Fig. 7A), suggesting that the GuHCl block to MRV dsRNA synthesis is fully reversible. In subsequent experiments, we found that the GuHCl-dependent inhibition of MRV dsRNA synthesis is fully reversible after inhibitory periods as long as 32 h (data not shown). As in experiments described earlier, the continuous presence of 15 mM GuHCl in the culture medium completely abolished any increase in virus titer over a 48-h period (Fig. 7A).
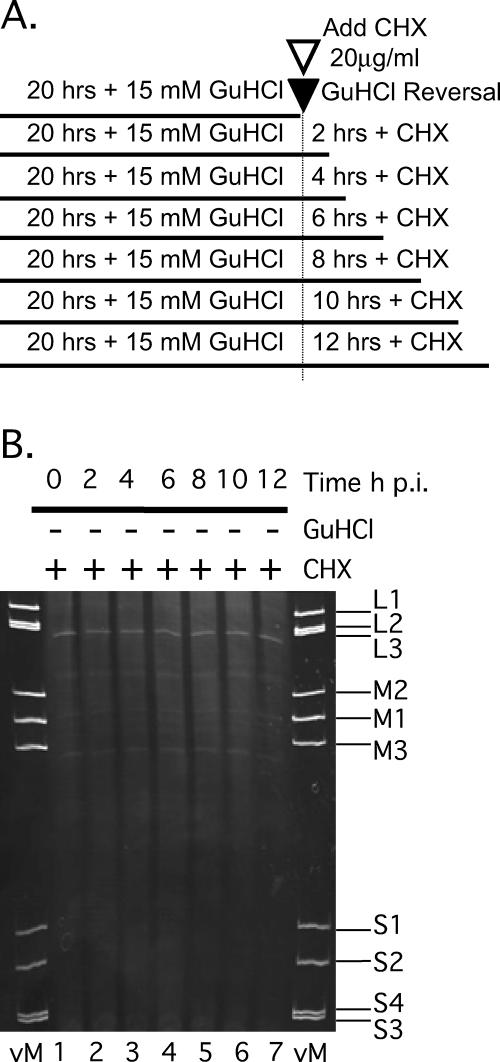
FIG. 7.
The inhibitory effect of 15 mM GuHCl on MRV replication is reversible. (A) L929 cells were infected with MRV-T1L (MOI = 5). After virus adsorption, infected cells were fed medium without GuHCl (×) or with 15 mM GuHCl and were then incubated at 37°C for a total of 48 h. Medium containing 15 mM GuHCl was removed and replaced with fresh medium lacking GuHCl (▪) at 4 (⧫), 8 (○), 12 (▵), or 24 (*) h p.i. Control cells were infected with MRV-T1L as described and incubated with 15 mM GuHCl for 48 h (×). At 48 h p.i., all cells were lysed, and viral yields were determined by plaque assay. (B) Time course of MRV dsRNA synthesis in the absence of GuHCl. Monolayers of L929 cells infected with MRV-T1L (MOI = 5) were incubated at 37°C for 2, 4, 6, 8, 10, 12, 20, or 32 h. At each time point after infection, total RNA was extracted with TRIzol reagent. Viral dsRNA was separated by electrophoresis in 10% polyacrylamide gels, stained with ethidium bromide, and visualized by UV transillumination. vM, virion marker (purified T1L mORV genomic dsRNA); L, MRV large genome segments; M, MRV medium genome segments; and S, MRV small genome segments. (C) The left panel depicts the time course of GuHCl reversal reaction. Infected cells were incubated at 37°C with 15 mM GuHCl for 20 or 32 h. At 20 h p.i., GuHCl-containing medium was aspirated and replaced with GuHCl-free medium. The cells were either lysed (0 h postreversal) or allowed to continue incubating at 37°C for an additional 2, 4, 6, 8, 10, or 12 h. At each designated time point after GuHCl reversal, the cells were lysed and total RNA was extracted with TRIzol reagent. For the right panel, MRV dsRNA was separated by electrophoresis in 10% polyacrylamide gels, stained with ethidium bromide, and visualized by UV transillumination. Gel labels are as described for panel B. Mock, mock-infected cells; 32+15 mM GuHCl, no reversal of GuHCl inhibition.
To gain a better understanding of MRV growth after GuHCl reversal, we evaluated the kinetics of MRV dsRNA synthesis in untreated cells compared to cells after GuHCl reversal. In untreated cells, we found that MRV dsRNA is detectable by 8 h p.i. (Fig. 7B, lane 4), a finding consistent with previous reports, as well as the results described above (Fig. 5B). By 20 h p.i., robust quantities of MRV dsRNA had accumulated in untreated cells and were observed to continue accumulating for as long as 32 h p.i., the latest time point we investigated (Fig. 7B, lanes 7 and 8). To evaluate the kinetics of dsRNA synthesis after GuHCl reversal, we infected L929 cells with MRV and treated the culture medium with 15 mM GuHCl for 20 h. At 20 h p.i., the GuHCl-containing medium was removed and replaced with GuHCl-free medium. Total RNA samples were collected at 2-h intervals from the time of reversal continuing through 12 h (32 h p.i.) (Fig. 7C, left panel). Analysis of postreversal samples revealed that dsRNA synthesis initiates very rapidly after the removal of GuHCl from the culture medium. By 2 h after reversal, dsRNA production was comparable to that seen by 10 h p.i. in the absence of GuHCl (compare Fig. 7B, lane 5, to Fig. 7C, right panel, lane 2). Our interpretation of these data is that “pre- or postinitiation replicase complexes” are formed following the normal progression of the MRV replicative cycle through cell entry, viral transcription, mRNA translation into viral proteins, factory formation, and assembly of the replicase complexes; however, the initiation and/or elongation of negative-sense RNAs into MRV progeny dsRNAs is prohibited by an as-yet-unknown GuHCl-sensitive mechanism. Removing the GuHCl-induced block then allows for the rapid synthesis and accumulation of MRV dsRNA.
MRV dsRNA synthesis proceeds in the absence of ongoing protein synthesis but is limited in scope.
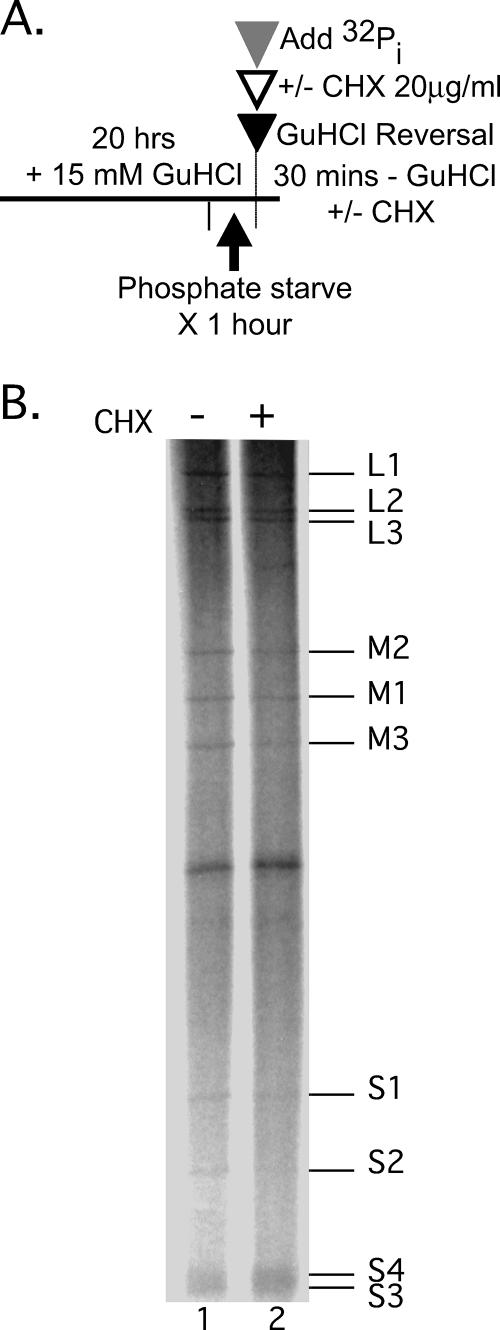
Previous reports (27, 31, 61, 62) have concluded that ongoing protein synthesis is required for MRV dsRNA synthesis. To readdress this question, we took advantage of the capacity for inhibition followed by reversal of inhibition of dsRNA synthesis by GuHCl. Specifically, we treated MRV-infected L929 cells with 15 mM GuHCl for 20 h. At 20 h p.i., we replaced the GuHCl-containing culture medium with medium containing carrier-free [32P]orthophosphate plus or minus 20 μg of cycloheximide/ml to inhibit protein synthesis (Fig. 8A). Infected cells were incubated at 37°C for 30 min, and total RNA was collected as previously described. We found that all 10 MRV positive-sense RNAs were copied into dsRNA within 30 min of GuHCl reversal irrespective of the presence of cycloheximide. Furthermore, we observed that approximately equal amounts of each dsRNA gene segment were synthesized in both cases (Fig. 8B). Thus, in this setting, at least one initial round of MRV dsRNA synthesis is not dependent on ongoing protein synthesis. These findings are consistent with the hypothesis that GuHCl-sensitive preinitiation replicase complexes are being assembled on MRV positive-sense RNA templates in the presence of GuHCl and can give rise to dsRNA in the absence of further protein synthesis once the GuHCl block is removed.
FIG. 8.
MRV dsRNA synthesis can occur in the absence of ongoing protein synthesis. (A) Experimental design. (B) Infected cells were incubated at 37°C with 15 mM GuHCl for 19 h. At 19 h p.i., GuHCl-containing medium was aspirated and replaced with serum-free, phosphate-free medium containing 15 mM GuHCl. The cells were incubated for an additional 1 h at 37°C. At 20 h p.i., the medium was aspirated and replaced with serum-free, phosphate-free medium containing 30 μl of [32P]orthophosphate (300 μCi/reaction) ± 20 μg of cycloheximide/ml. The cells were then incubated for an additional 30 min at 37°C. The cells were lysed, and the total RNA was extracted with TRIzol reagent. Labeled MRV dsRNA was separated by electrophoresis in 10% polyacrylamide gels and visualized by phosphorimaging. The positions of individual MRV dsRNA genome segments are indicated at right.
Because data in this report indicate that translation from MRV primary transcripts is not sensitive to GuHCl inhibition and continues for at least 48 h p.i. (see Fig. 4 and 6B and C), we questioned the need for ongoing protein synthesis during an extended period of MRV dsRNA synthesis following the reversal of inhibition by GuHCl. We wanted to determine whether a large enough pool of protein components was synthesized and/or assembled during the GuHCl-induced block to support multiple rounds of dsRNA synthesis or if complete inhibition of protein synthesis would limit the scope and duration of dsRNA synthesis. To this end, we treated MRV-infected L929 cells with 15 mM GuHCl for 20 h and added 20 μg of cycloheximide/ml to the culture medium at the time of GuHCl reversal. Total RNA was collected every 2 h from the time of reversal and evaluated (Fig. 9A) by electrophoresis in 10% polyacrylamide gels. When protein synthesis was inhibited by cycloheximide we did not observe the normal accumulation of MRV dsRNA in infected cells (Fig. 9B), suggesting that multiple rounds of MRV replication can occur after GuHCl reversal but require ongoing protein synthesis for all but the initial round(s). These data suggest that a limited pool of replicase components and possibly only preformed replicase complexes are giving rise to the dsRNA products seen in Fig. 8B.
FIG. 9.
MRV dsRNA synthesis in the absence of ongoing protein synthesis is limited in scope. (A) Experimental design. (B) Infected cells were incubated at 37°C with 15 mM GuHCl for 20 h. At 20 h p.i., GuHCl-containing medium was aspirated and replaced with medium containing 20 μg of cycloheximide/ml. The cells were either lysed immediately (0 h postreversal) or allowed to continue incubating at 37°C for an additional 2, 4, 6, 8, 10, or 12 h before being lysed. Total RNA was extracted from each sample with TRIzol reagent. MRV dsRNA was separated by electrophoresis in 10% polyacrylamide gels, stained with ethidium bromide, and visualized by UV transillumination. vM, virion marker. The positions of the individual MRV dsRNA genome segments are indicated on the right.
DISCUSSION
In vitro translation-replication assays for members of the family Picornaviridae such as PV have proven invaluable in the investigation of viral RNA synthesis. These assays have been especially useful because they allow for the spatiotemporal dissection of the complete RNA replicative cycle (6, 8). In vitro translation-replication assays utilizing HeLa S10 (6, 8, 38) or Krebs 2 (57) cytoplasmic extracts have accomplished this by permitting reversible inhibition of negative-sense RNA synthesis while sparing the preceding steps of viral gene expression (7). Negative-sense RNA synthesis has been reversibly inhibited in these assays by using millimolar concentrations of GuHCl (7, 19, 48) or micromolar concentrations of dipyridamole (25). Similarly to PV, successful completion of the MRV RNA replicative cycle appears likely to require the formation of at least two distinct, multicomponent ribonucleoprotein (RNP) complexes: the messenger RNP (mRNP) complex that is required for translation of the viral proteome and the replicase RNP complex that functions to replicate the viral dsRNA genome (62). Putatively, the replicase RNP complex is also involved in recognizing and sorting the 10 different genome segments to ensure that one of each is incorporated per particle (2). Because targeted manipulation of the reovirus genome remains difficult, the specific RNA-protein interactions required to form these two different RNP complexes remain largely unknown. Toward this end, we have turned our efforts toward developing an in vitro translation-replication assay for MRV in line with those previously developed and used to such advantage for PV.
In the present study, we investigated the effect of GuHCl on MRV growth, primarily in murine L929 cells. The results presented here provide strong evidence that 15 mM GuHCl specifically inhibits MRV replication by reversibly blocking the initiation and/or elongation of MRV dsRNA synthesis. To our knowledge, this is the first report demonstrating the inhibition of RNA synthesis by GuHCl for any dsRNA virus. The specific inhibition of MRV dsRNA synthesis by 15 mM GuHCl suggests that MRV positive-sense and negative-sense RNA (i.e., dsRNA) synthesis are in some way(s) distinct. Because λ3 is the only MRV protein with a confirmed RdRp activity (56, 58, 60), we suspect that the specific inhibition of dsRNA synthesis may stem from an interaction of GuHCl with one or more accessory proteins required for processive dsRNA synthesis. Investigations of other genera of Reoviridae have shown that the viral polymerases are regulated by temporally specific associations with other proteins (12, 36). Rotavirus dsRNA synthesis, for example, requires VP1 (the rotavirus RdRp) to interact with VP2 (the core lattice protein) in a specific stoichiometric VP1/VP2 ratio of ∼1:10 prior to VP1 activation (45, 46). Similarly, the orbivirus RdRp requires the association of an unknown specificity factor to mediate the exclusive synthesis of viral dsRNA (12). In light of these data and the seemingly redundant NTPase activities of MRV proteins μ2 and λ1 (10, 28), one possibility is that λ3 may be differentially regulated by these two proteins during positive- and negative-sense RNA synthesis. If true, this would suggest that GuHCl inhibition of dsRNA synthesis may be mediated by a specific interaction with one or the other of these proteins. In an analogy to GuHCl inhibition of PV 2CATPase, we suspect that GuHCl may disrupt the NTPase activity of these proteins through an interaction with the nucleotide-binding motifs.
There are other possibilities for the mechanism by which GuHCl specifically inhibits MRV dsRNA synthesis. One of these invokes structural constraints within reovirus particles. MRV positive-sense RNA synthesis occurs within a closed core particle, whereas the exact nature of the replicase RNP complex is unknown. It has been suggested that, at the time of initiation of dsRNA synthesis, MRV RNA is susceptible to treatment by RNase but that shortly thereafter it becomes resistant to RNase treatments (1). These findings have led to the hypothesis that dsRNA synthesis and core morphogenesis are linked. If true, assembly of the core particle may obfuscate the target of GuHCl, thus rendering positive-sense RNA synthesis resistant without regard to any required cofactors for each process. There is also the possibility that the inhibition of MRV dsRNA synthesis by GuHCl may arise through the manipulation of a viral protein other than λ3, μ2, or λ1, possibly σ2 or one of the MRV nonstructural proteins or through manipulation of a host protein(s) involved in the replicase RNP complex.
Previous investigations have suggested that positive-sense RNA synthesis by MRV occurs in two phases, denoted as primary and secondary transcription (61). Primary transcripts are synthesized by infecting, parent particles, whereas secondary transcripts are synthesized by nascently formed, progeny particles. Evidence gathered independently through the study of numerous temperature-sensitive mutants has suggested that 80 to 95% of MRV positive-sense RNAs over the course of an infection are synthesized as secondary transcripts (24, 61). Moreover, it has been inferred from these and other data that the majority of viral protein is expressed from the secondary transcripts (61). Because treatment of MRV-infected L929 cells with 15 mM GuHCl prevents dsRNA synthesis and thus the morphogenesis of progeny core particles, only primary transcripts can be synthesized in GuHCl-treated cells. We show in this report, nonetheless, that during GuHCl blockade of dsRNA synthesis, reasonably large amounts of viral proteins are produced, implying that these proteins are being translated from primary transcripts. In fact, so much protein is synthesized from these transcripts that morphologically normal viral factories can be discerned by light microscopy. Our favored interpretation of these observations posits that the mRNP complexes assembled with primary transcripts are stable and long-lived. As such, these complexes continue to translate primary transcripts for the duration of the GuHCl blockade, giving rise to significantly larger amounts of viral protein than may have been previously expected. Related possibilities are that GuHCl may in some way promote the mRNP complexes assembled with primary transcripts to be longer-lived or may promote a larger number of primary transcripts to be assembled into mRNP complexes than normally occurs.
In the presence of cycloheximide, only limited dsRNA synthesis occurs upon the release of the GuHCl blockade. This strongly suggests that in the presence of 15 mM GuHCl only a limited number of primary transcripts are incorporated into pre- or postinitiation replicase RNP complexes. While several possibilities may explain this observation, our preferred explanation is that primary transcription is being turned off after a limited number of primary transcripts are synthesized, probably as a result of newly translated μ1-σ3 complexes being reassembled onto the primary transcriptase (core) particles. Due to the initial absence of viral proteins in a newly infected cell, all of the primary transcripts first synthesized will be incorporated into mRNP complexes. Once viral protein synthesis has produced the correct concentrations of necessary proteins, primary transcripts can then be incorporated into replicase RNP complexes, probably within the setting of a nascent viral factory (55). We suspect, however, that shutoff of the primary transcription occurs with kinetics that approximate replicase RNP formation and therefore that the shutoff of viral transcription prevents the assembly of large quantities of replicase RNP complexes, as well as the assembly of additional mRNP complexes. This interpretation further suggests that the replicase RNP complexes, like the mRNP complexes, are stable and long-lived, with the former being held static until the release of the GuHCl blockade and the latter continuing to produce large amounts of proteins as suggested above.
Once the GuHCl blockade is released, it is presumed that progeny core morphogenesis also proceeds. Progeny core morphogenesis generates secondary transcriptase particles with subsequent synthesis of secondary transcripts. Secondary transcripts can assemble into new replicase RNP complexes, which in turn can give rise to new dsRNA. Note, however, that one or more of the steps between the initial burst of dsRNA synthesis (that is, dsRNA synthesis from the primary transcripts in replicase RNP complexes) and the subsequent round(s) of dsRNA synthesis from secondary transcripts requires new protein synthesis since dsRNA synthesis after the initial burst is blocked by cycloheximide. This suggests that a limited pool of functional protein components is available to give rise to new replicase RNP complexes once secondary transcripts become available. Our favored explanation for this is that during the GuHCl-induced blockade of dsRNA synthesis one or more of the viral proteins that continue to be translated from the primary transcripts in mRNP complexes become lost to functional replicase assembly by some manner of trapping into a nonproductive complex. However, we have seen little if any increase in the number of non-genome-containing particles in infections performed in the presence, versus the absence, of GuHCl (K. E. Murray, unpublished data), and thus the manner of trapping of the essential protein(s) remains unknown. In any case, based on the findings to date, we propose that it is the assembly of replicase RNP complexes, not dsRNA synthesis per se, that requires ongoing protein synthesis, which furthermore suggests that replicase assembly and translation may be directly coupled in some manner.
We have presented here our favored explanations for a number of new and interesting observations in this report, as well as several other reasonable alternatives. An important point is that several of these explanations have testable features, which we plan to pursue in future work.
Acknowledgments
We thank David J. Barton, University of Colorado Health Sciences Center, for the kind gift of spinner adapted HeLa S3 cells (CCL 2.2). We are grateful to Michelle Arnold for instruction and assistance with immunofluorescence microscopy. We thank Elaine Freimont for excellent lab management and technical assistance with tissue culture. We also thank Sean P. J. Whelan for generously allowing us the use of equipment and reagents and for valuable discussions.
This study was supported by grants from the Life Sciences Research Foundation and the Schering Plough Corp. (to K.E.M.) and by Public Health Service grant R01 AI47904 from the National Institutes of Health (to M.L.N.).
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Acs, G., H. Klett, M. Schonberg, J. Christman, D. H. Levin, and S. C. Silverstein. 1971. Mechanism of reovirus double-stranded ribonucleic acid synthesis in vivo and in vitro. J. Virol. 8:684-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760-776. [DOI] [PubMed] [Google Scholar]
- 3.Attoui, H., F. Mohd Jaafar, M. Belhouchet, P. Biagini, J.-F. Cantaloube, P. de Micco, and X. de Lamballerie. 2005. Expansion of family Reoviridae to include nine-segmented dsRNA viruses: isolation and characterization of a new virus designated Aedes pseudoscutellaris reovirus assigned to a proposed genus (Dinovernavirus). Virology 343:212-223. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, D. H. 2000. Virus structures: those magnificent molecular machines. Curr. Biol. 10:558-561. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, D. H., and L. Mindich. 2004. Viral molecular machines: replication systems within the inner cores of dsRNA viruses. Virus Res. 101:1. [Google Scholar]
- 6.Barton, D., E. Black, and J. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D., and J. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton, D. J., and J. B. Flanegan. 1993. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 67:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., B. Joan Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3DPol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 10.Bisaillon, M., J. Bergeron, and G. Lemay. 1997. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus λ1 protein. J. Biol. Chem. 272:18298-18303. [DOI] [PubMed] [Google Scholar]
- 11.Bisaillon, M., and G. Lemay. 1997. Characterization of the reovirus λ1 protein RNA 5′-triphosphatase activity. J. Biol. Chem. 272:29954-29957. [DOI] [PubMed] [Google Scholar]
- 12.Boyce, M., J. Wehrfritz, R. Noad, and P. Roy. 2004. Purified recombinant bluetongue virus VP1 exhibits RNA replicase activity. J. Virol. 78:3994-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brentano, L., D. L. Noah, E. G. Brown, and B. Sherry. 1998. The reovirus protein μ2, encoded by the M1 gene, is an RNA-binding protein. J. Virol. 72:8354-8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broering, T. J., M. M. Arnold, C. L. Miller, J. A. Hurt, P. L. Joyce, and M. L. Nibert. 2005. Carboxyl-proximal regions of reovirus nonstructural protein μNS necessary and sufficient for forming factory-like inclusions. J. Virol. 79:6194-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broering, T. J., J. Kim, C. L. Miller, C. D. S. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. L. Parker. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broering, T. J., J. S. L. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bujnicki, J., and L. Rychlewski. 2001. Reassignment of specificities of two cap methyltransferase domains in the reovirus λ2 protein. Genome Biol. 2:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caliguiri, L. A., and I. Tamm. 1973. Guanidine and 2-(α-hydroxylbenzyl)-benzimidazole) (HBB): selective inhibitors of picornavirus multiplication, p. 257-293. In W. Carter (ed.), Selective inhibitors of viral function. CRC Press, Inc., Cleveland, OH.
- 20.Carrasco, L., R. Guinea, A. Irurzun, and A. Barco. 2002. Effects of viral replication on cellular membrane metabolism and function, p. 337-354. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 21.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleveland, D. R., H. Zarbl, and S. Millward. 1986. Reovirus guanylyltransferase is L2 gene product λ2. J. Virol. 60:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coombs, K. M. 1998. Temperature-sensitive mutants of reovirus. Curr. Top. Microbiol. Immunol. 233/I:69-107. [DOI] [PubMed] [Google Scholar]
- 25.Fata-Hartley, C. L., and A. C. Palmenberg. 2005. Dipyridamole reversibly inhibits mengovirus RNA replication. J. Virol. 79:11062-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. σ1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomatos, P. J. 1967. RNA synthesis in reovirus-infected L929 mouse fibroblasts. Proc. Natl. Acad. Sci. USA 58:1798-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J., J. S. L. Parker, K. E. Murray, and M. L. Nibert. 2004. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. J. Biol. Chem. 279:4394-4403. [DOI] [PubMed] [Google Scholar]
- 29.Kim, J., X. Zhang, V. E. Centonze, V. D. Bowman, S. Noble, T. S. Baker, and M. L. Nibert. 2002. The hydrophilic amino-terminal arm of reovirus core shell protein λ1 is dispensable for particle assembly. J. Virol. 76:12211-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koonin, E. V. 1993. Computer-assisted identification of a putative methyltransferase domain in ns5 protein of flaviviruses and λ2 protein of reoviruses. J. Gen. Virol. 74:733-740. [DOI] [PubMed] [Google Scholar]
- 31.Kudo, H., and A. F. Graham. 1966. Selective inhibition of reovirus induced RNA in L cells. Biochem. Biophys. Res. Commun. 24:150-155. [DOI] [PubMed] [Google Scholar]
- 32.Luongo, C. L., C. M. Contreras, D. L. Farsetta, and M. L. Nibert. 1998. Binding site for S-adenosyl-l-methionine in a central region of mammalian reovirus λ2 protein. Evidence for activities in mRNA cap methylation. J. Biol. Chem. 273:23773-23780. [DOI] [PubMed] [Google Scholar]
- 33.Luongo, C. L., K. M. Reinisch, S. C. Harrison, and M. L. Nibert. 2000. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein λ2. J. Biol. Chem. 275:2804-2810. [DOI] [PubMed] [Google Scholar]
- 34.Mao, Z., and W. K. Joklik. 1991. Isolation and enzymatic characterization of protein λ2, the reovirus guanylyltransferase. Virology 185:377-386. [DOI] [PubMed] [Google Scholar]
- 35.Mertens, P. 2004. The dsRNA viruses. Virus Res. 101:3-13. [DOI] [PubMed] [Google Scholar]
- 36.Mertens, P. P. C., and J. Diprose. 2004. The bluetongue virus core: a nano-scale transcription machine. Virus Res. 101:29-43. [DOI] [PubMed] [Google Scholar]
- 37.Miller, C. L., T. J. Broering, J. S. L. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino terminus. J. Virol. 77:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 39.Nibert, M. L. 1998. Structure of mammalian orthoreovirus particles. Curr. Top. Microbiol. Immunol. 233/I:1-30. [DOI] [PubMed] [Google Scholar]
- 40.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 41.Nibert, M. L., and J. Kim. 2004. Conserved sequence motifs for nucleoside triphosphate binding unique to turreted Reoviridae members and coltiviruses. J. Virol. 78:5528-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noble, S., and M. L. Nibert. 1997. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J. Virol. 71:2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noble, S., and M. L. Nibert. 1997. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 71:7728-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker, J. S. L., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patton, J. 1996. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J. Virol. 70:7940-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patton, J., M. Jones, A. Kalbach, Y. He, and J. Xiaobo. 1997. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 71:9618-9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patton, J. T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277:217-225. [DOI] [PubMed] [Google Scholar]
- 48.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 49.Pincus, S. E., and E. Wimmer. 1986. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J. Virol. 60:793-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu, T., and C. L. Luongo. 2003. Identification of two histidines necessary for reovirus mRNA guanylyltransferase activity. Virology 316:313-324. [DOI] [PubMed] [Google Scholar]
- 51.Reinisch, K. M. 2002. The dsRNA Viridae and their catalytic capsids. Nat. Struct. Biol. 9:714-716. [DOI] [PubMed] [Google Scholar]
- 52.Reinsch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nat. Struct. Biol. 404:960-967. [DOI] [PubMed] [Google Scholar]
- 53.Seliger, L. S., K Zheng, and A. J. Shatkin. 1987. Complete nucleotide sequence of reovirus L2 gene and deduced amino acid sequence of viral mRNA guanylyltransferase. J. Biol. Chem. 262:16289-16293. [PubMed] [Google Scholar]
- 54.Shatkin, A. J., Y. Furuichi, A. J. LaFiandra, and M. Yamakawa. 1983. Initiation of mRNA synthesis and 5′ terminal modification of reovirus transcripts, p. 43-54. In R. Compans and D. Bishop (ed.), Double-stranded RNA viruses. Elsevier, New York, NY.
- 55.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Starnes, M. C., and W. K. Joklik. 1993. Reovirus protein λ3 is a poly(C)-dependent poly(G) polymerase. Virology 193:356-366. [DOI] [PubMed] [Google Scholar]
- 57.Svitkin, Y. V., and N. Sonenberg. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 77:6551-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao, Y., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage: structural studies of reovirus polymerase λ3. Cell 111:733-745. [DOI] [PubMed] [Google Scholar]
- 59.Xu, P., S. E. Miller, and W. K. Joklik. 1993. Generation of reovirus core-like particles in cells infected with hybrid vaccinia viruses that express genome segments L1, L2, L3, and S2. Virology 197:726-731. [DOI] [PubMed] [Google Scholar]
- 60.Yue, Z., and A. J. Shatkin. 1998. Enzymatic and control functions of reovirus structural proteins. Curr. Top. Microbiol. Immunol. 233/I:31-56. [DOI] [PubMed] [Google Scholar]
- 61.Zarbl, H., D. Skup, and S. Millward. 1980. Reovirus progeny subviral particles synthesize uncapped mRNA. J. Virol. 34:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zarbl, H. J., and S. Millwood. 1983. The reovirus multiplication cycle, p. 107-196. In W. K. Joklik (ed.), The Reoviridae. Plenum Press, Inc., New York, NY.
- 63.Zhang, X., S. B. Walker, P. R. Chipman, M. L. Nibert, and T. S. Baker. 2003. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 10:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zweerink, H. J. 1974. Multiple forms of SS → DS RNA polymerase activity in reovirus-infected cells. Nature 247:313-315. [DOI] [PubMed] [Google Scholar]
- 65.Zweerink, H. J., Y. Ito, and T. Matsuhisa. 1972. Synthesis of reovirus double-stranded RNA within virionlike particles. Virology 50:349-358. [DOI] [PubMed] [Google Scholar]