Abstract
A unique hepatitis C virus (HCV) strain JFH-1 has been shown to replicate efficiently in cell culture with production of infectious HCV. We previously developed a DNA expression system containing HCV cDNA flanked by two self-cleaving ribozymes to generate HCV particles in cell culture. In this study, we produced HCV particles of various genotypes, including 1a (H77), 1b (CG1b), and 2a (J6 and JFH-1), in the HCV-ribozyme system. The constructs also contain the secreted alkaline phosphatase gene to control for transfection efficiency and the effects of culture conditions. After transfection into the Huh7-derived cell line Huh7.5.1, continuous HCV replication and secretion were confirmed by the detection of HCV RNA and core antigen in the culture medium. HCV replication levels of strains H77, CG1b, and J6 were comparable, whereas the JFH-1 strain replicates at a substantially higher level than the other strains. To evaluate the infectivity in vitro, the culture medium of JFH-1-transfected cells was inoculated into naive Huh7.5.1 cells. HCV proteins were detected by immunofluorescence 3 days after inoculation. To evaluate the infectivity in vivo, the culture medium from HCV genotype 1b-transfected cells was inoculated into a chimpanzee and caused a typical course of HCV infection. The HCV 1b propagated in vitro and in vivo had sequences identical to those of the HCV genomic cDNA used for cell culture transfection. The development of culture systems for production of various HCV genotypes provides a valuable tool not only to study the replication and pathogenesis of HCV but also to screen for antivirals.
Hepatitis C virus (HCV) is a major public health problem and infects about 200 million people worldwide (12, 18). The majority of HCV-infected patients fail to clear the virus, and many develop chronic liver diseases, including cirrhosis and hepatocellular carcinoma. HCV does not replicate efficiently in cultured cells, and robust model systems for HCV infection have been difficult to develop. Recently, we identified a unique HCV genotype 2a strain JFH1 that can replicate and produce viral particles efficiently in cell culture and established an HCV infection model system with cell culture generated JFH-1 virus that is infectious both in vitro and in vivo (5, 7-10, 16, 22, 26).
Like other RNA viruses, HCV displays marked genetic heterogeneity and is currently classified into six major genotypes (19). Among these genotypes, genotypes 1 and 2 have worldwide distribution and are known to be associated with different clinical profiles and therapeutic responses (25). These differences in clinical features are likely to be a result of viral characteristics. Study of the molecular mechanisms underlying such differences would provide valuable information regarding the pathogenesis and therapy of hepatitis C in humans. Despite the development of the JFH1 infectious cell culture system, similar systems with other HCV strains have been difficult to establish. Recent studies have shown the production of infectious 1a strain in vitro, but multiple adaptive mutations must be introduced to confer a high level of replication (24). Therefore, a more general system that can be applied to various HCV genotypes and to antiviral testing is urgently needed. Previously, we reported a DNA expression system for efficient HCV particle production system by expressing a genomic-length HCV genotype 1b cDNA with self-cleaving ribozymes (6). This system supported HCV replication and produced and secreted HCV particles into the culture medium. In the present study, we applied this HCV-ribozyme system to various HCV genotypes. These HCV expression plasmids also contain the secreted alkaline phosphatase (SEAP) gene to control for transfection efficiency and the effects of culture conditions. Using this system, we could generate various genotypes of HCV and confirmed the infection of generated virus both in vitro (strain JFH-1) and in vivo (strain CG1b and H77). We also established permanent cell lines continuously expressing replicating HCV by transfecting the HCV-ribozyme construct with a selection marker.
MATERIALS AND METHODS
HCV expression plasmids.
Various HCV strains, H77 (genotype 1a, accession no. AF009606 [11]), CG1b (genotype 1b, accession no. AF333324 [21]), J6 (genotype 2a, accession no. AF177036 [23]), and JFH1 (genotype 2a, accession no. AB047639 [7]) were used. The HCV plasmid (pTHr) containing the HCV CG1b genomic cDNA with ribozymes was reported previously (6). As a strategy for further construction, a PmeI site was introduced in 3′ untranslated region (3′UTR) of the CG1b genome. To generate SEAP-expressing vector, the fragment encompassing the SEAP gene was amplified by PCR from pSEAP-control vector (Clontech Laboratories, Inc., Mountain View, CA). The fragment was inserted into the pEF1/Myc-His plasmid (Invitrogen, Carlsbad, CA) in place of the neomycin-resistant gene (neo) as the pEF/S plasmid. The fragment containing the CG1b-ribozyme sequence was digested with EcoRI and XbaI and cloned into the pEF/S vector as pEF/CG1b-Rz/S. This plasmid comprises the full-length CG1b genome and flanking ribozymes directed by the EF1α promoter and the SEAP gene by the simian virus 40 (SV40) promoter (Fig. 1A). The replication-deficient mutant of CG1b strain that had been reported previously (6) was also cloned into the pEF/S vector as pEF/CG1b-GND-Rz/S.
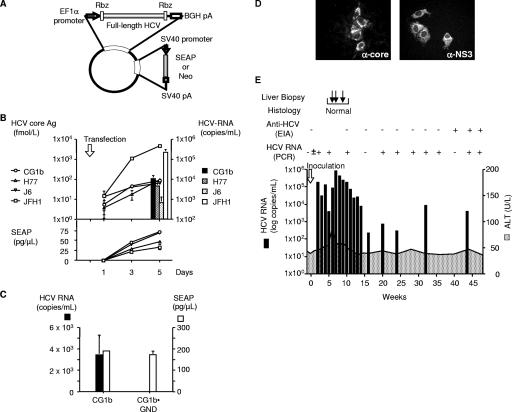
FIG. 1.
Production and infection of HCV produced in cell culture. (A) Construction of HCV-expressing plasmid with the HCV-ribozyme system. Two hammerhead ribozyme sequences were engineered at the 5′ and 3′ ends of HCV full-length cDNA. This system was expressed under the control of EF1α promoter. The SEAP reporter gene was included under the control of the SV40 promoter. (B) Production of HCV various strains after transfection of HCV-ribozyme plasmids. Various HCV-ribozyme plasmids were transfected into Huh7.5.1 cells. HCV core Ag levels and SEAP activities in the culture medium were measured at the indicated time points. HCV RNA titers in culture medium were measured at the end of the follow-up time point. (C) Comparison of HCV RNA titers of CG1b and CG1b-GND plasmid-transfected cells. HCV RNA titers and SEAP activities in the culture medium were measured 5 days after transfection. (D) Infection of naive Huh7.5.1 cells by cell culture-generated JFH-1 virus. Culture medium form JFH1-ribozyme-transfected cells were inoculated into naive Huh7.5.1 cells. The expression of HCV core and NS3 proteins was detected by immunofluorescence with appropriate antibodies. (E) Infectivity of cell culture-generated CG1b virus in a naive chimpanzee. Chimpanzee X0140 was inoculated with culture medium of CG1b plasmid-transfected cells containing 2.3 × 103 copies of HCV RNA.
For JFH1-ribozyme construction, the ribozyme sequences were introduced into the 5′ and 3′ ends of the JFH1 genomic cDNA (pJFH1) by PCR (22). The ribozyme-containing 5′UTR of JFH1 was introduced into the JFH1 genomic cDNA by cloning via the EcoRI and AgeI sites and similarly with the ribozyme-containing 3′UTR via the AscI and XbaI sites. The fragment containing the JFH1 genomic cDNA and ribozymes was inserted into the EcoRI and XbaI sites of the pEF/S vector to generate the pEF/JFH1-Rz/S. This fragment was also transferred to the pEF1/Myc-His plasmid to generate the neo-containing construct as the pEF/JFH1-Rz/N for the establishment of stable JFH1 virus-producing cells (Fig. 1A). The replication-deficient clone of JFH1 was generated by introducing a point mutation at the GDD motif of the NS5b to abolish the RNA-dependent RNA polymerase activity as the pEF/JFH1-GND-Rz/S (22).
For the H77 and J6-ribozyme construction, both full-length HCV cDNAs were cloned into the pEF/CG1b-Rz/S plasmid via the AgeI (5′UTR) and PmeI (3′UTR) sites. The ribozyme-containing 5′UTR fragment of H77 was generated by PCR and replaced the ribozyme-5′UTR sequence of the CG1b via the EcoRI and AgeI sites to generate the pEF/H77-Rz/S. The pEF/J6-Rz/S construct was similarly generated by using the ribozyme-containing 5′UTR fragment of JFH1.
Cell culture and DNA transfection.
The Huh7 derivative cell lines Huh7.5 and Huh7.5.1 were provided by C. Rice (Rockefeller University, New York, NY) and F. Chisari (Scripps Research Institute, La Jolla, CA), respectively (2, 25). Huh7 cells or these derivative cell lines were maintained at 37°C in 5% CO2 in Dulbecco modified Eagle medium containing 10% fetal bovine serum.
A 10-μg portion of HCV-ribozyme plasmids was transfected into Huh7 cells or its derivative cells in a 10-cm dish (2 × 106/dish) by using Lipofectamine and Lipofectamine Plus reagents (Invitrogen) according to the manufacturer's instructions. Alpha interferon (IFN-α) was purchased from Fitzgerald (Concord, MA).
Quantification of HCV core Ag, HCV RNA, and SEAP activity.
To assess HCV replication, the HCV core antigen (Ag) in culture supernatant was quantified by a highly sensitive enzyme immunoassay (Ortho HCV antigen ELISA kit; Ortho Clinical Diagnostics, Tokyo, Japan) (1). To determine the amount of HCV RNA in culture supernatant, RNA in the culture medium was extracted from 100 to 250 μl of culture medium by TRIzol LS reagent (Invitrogen) and treated with DNase (TURBO DNase; Ambion, Austin, TX) at 37°C for 1 h. Extracted RNA was purified by using an RNeasy minikit, which includes another step of RNase-free DNase digestion (QIAGEN, Valencia, CA). Copy numbers of HCV RNA were determined by real-time quantitative reverse transcription-PCR as described previously (20). The detection limit was estimated as 300 copies/ml. When necessary, the culture medium was concentrated by ultrafiltration concentrator (Vivaspin 20, molecular weight cutoff of 100,000; Vivascience, Hannover, Germany). SEAP activity in the culture medium was detected by using a Great EscAPe SEAP detection kit (Clontech Laboratories, Inc.).
Titration of HCV infectivity.
Huh7.5.1 cells were seeded at 104 cells/well in 96-well flat bottom plate 24 h before inoculation. A total of 100 μl of serially 10-fold diluted culture media or gradient fractions were inoculated to the cells in the 96-well plate. After 4 h of incubation, the inocula were replaced with fresh media, and the cells were incubated for 72 h. Intracellular expression of HCV core protein were assayed by indirect immunofluorescence with α-core C1 antibody (provided by H. Greenberg, Stanford Medical School, Palo Alto, CA) and Alexa Flour 488-conjugated goat anti-mouse immunoglobulin G (Invitrogen). Clusters of core protein-positive cells were counted as a single infection focus, and infectivity titers were represented as focus-forming units (FFU). Infection of the generated virus in culture medium was also confirmed by the use of α-NS3 antibody (provided from G. Luo, University of Kentucky College of Medicine, Lexington).
Density gradient analysis.
A total of 50 ml of culture medium harvested 5 days after transfection and passed through a 0.45-μm-pore-size filter was precipitated with one-fourth volume of 40% (wt/vol) polyethylene glycol 8000 in phosphate-buffered saline by overnight incubation at 4°C. Virus precipitates were collected by centrifugation, resuspended with HEPES-NaCl buffer (10 mM HEPES [pH 7.55], 0.85% NaCl, 0.02% bovine serum albumin), and purified by 20% (wt/vol) iodixanol (Optiprep; Axis-Shield, Oslo, Norway) cushion by centrifugation for 6 h at 40,000 rpm at 4°C in an SW41 Ti rotor. Purified virus was layered on top of 10 to 40% iodixanol gradient and centrifuged for 16 h at the same condition for iodixanol cushion. Fractions were collected from the bottom of gradient. The HCV RNA titer of each fraction was measured after DNase treatment and RNA extraction as described above. The HCV core Ag level and HCV infectivity titer of each fraction were assayed. Negative stain electron microscope was performed on each fraction.
Infection study in chimpanzee.
The chimpanzee experiment, approved by the Institutional Animal Care and Use Committee and the NIH Interagency Animal Models Committee, was conducted in the Southwest Foundation for Biomedical Research, an American Association of Laboratory Animal Care-accredited animal facility. A naive chimpanzee (X0140) was first inoculated with culture medium from mock-transfected cells that were exposed to the pEF/CG1b-Rz/S plasmid without transfection reagent. This inoculation serves as a control for the potential infectivity of any residual plasmid DNA in the culture medium. After inoculation of this control medium, the chimpanzee was observed for 8 weeks. The chimpanzee was then inoculated with culture medium from pEF/CG1b-Rz/S plasmid-transfected cells. After inoculation, the chimpanzee was monitored weekly with blood samples for HCV RNA (Roche Amplicor Monitor II with a lower limit of detection of 200 copies/ml), anti-HCV (Bayer Anti-HCV EIA II), and alanine aminotransferase (ALT). Liver biopsy was performed for histology after the demonstration of infection. To evaluate the in vivo infectivity of cell culture generated H77 virus, the culture medium from pEF/H77-Rz/S plasmid-transfected cells was also inoculated into another chimpanzee (X0199) who had previously recovered from HCV CG1b infection.
RT-PCR and sequencing.
The cDNA of the CG1b virus in chimpanzee was synthesized from RNA extracted from serum at 4 weeks after inoculation using reverse primer at the 3′UTR or 3′ X region. The cDNA was subsequently amplified with DNA polymerase (TaKaRa LA Taq; Takara Mirus Bio, Madison, WI). Four separate PCR primer sets were used to amplify the fragments of nucleotides [nt] 152 to 2777, nt 2743 to 5098, nt 4923 to 7670, and nt 7611 to 9390 covering the entire open reading frame and part of 5′UTR and 3′UTR of the CG1b strain. The sequence of each amplified fragment was determined.
Statistical analysis.
Data from repeated experiments were averaged and are expressed as means ± the standard deviations. Statistical analysis was performed by using the Student t test, Welch's t test, or one-factor analysis of variance. P values of <0.05 were considered statistically significant.
RESULTS
Production of various HCV genotypes in cell culture.
Various HCV-ribozyme expression plasmids (pEF/H77-Rz/S, pEF/CG1b-Rz/S, pEF/J6-Rz/S, and pEF/JFH1-Rz/S) were transfected into the Huh7.5.1 cells. Culture media were harvested from days 1, 3, and 5 after transfection and assayed for HCV core Ag level and SEAP activity. HCV RNA titers were measured in the culture medium on day 5. The HCV core Ag levels of strains H77, CG1b, and J6 on day 5 were 97.1 ± 28.7, 89.8 × 10.6, and 67.3 ± 5.1 fmol/liter, respectively. JFH-1 produced a much higher HCV core Ag level (4686.6 ± 287.1 fmol/liter on day 5) than the other strains (P < 0.05). Likewise, the HCV RNA titer in culture supernatant of strain JFH1 transfected cell was 2.10 × 105 ± 7.55 × 104 copies/ml on day 5, significantly higher than for strains H77, CG1b, and J6 (4.28 × 103 ± 3.00 × 103, 9.94 × 103 ± 4.64 × 103, and 6.29 × 102 ± 5.76 × 102 copies/ml, respectively; P < 0.05) (Fig. 1B). The SEAP activities in the culture supernatants were comparable among all strains, indicating similar transfection efficiencies. These data indicate that the JFH-1 strain replicates and produces viral particles more efficiently than the other strains in the HCV-ribozyme system. This observation is not unexpected because of the higher replication potential of JFH-1 in the HCV subgenomic replicon system (9). To confirm CG1b virus replication in this system, the CG1b-ribozyme expression plasmid (pEF/CG1b-Rz/S) and its replication-deficient mutant expression plasmid (pEF/CG1b-GND-Rz/S) were transfected into the Huh7.5.1 cells. At 5 days after transfection, the HCV RNA titer in the culture supernatant of the CG1b-transfected cells was 3.42 × 103 ± 1.84 × 103 copies/ml. The HCV RNA titer in the culture supernatant of CG1b-GND-transfected cells was undetectable, although the SEAP activities in both culture supernatants were comparable (Fig. 1C). To test the infectivity of the cell culture-produced HCV, culture supernatants were inoculated into naive Huh7.5.1 cells. By immunofluorescence microscopy, HCV core and NS3 proteins were detected in JFH-1 supernatant-infected cells. The HCV core protein showed spotty perinuclear and cytoplasmic distribution, and NS3 protein showed cytoplasmic distribution (Fig. 1D). However, no HCV-positive cells were detected in naive cells incubated with culture medium from H77-, CG1b-, and J6-transfected cells.
Infection of cell culture-generated CG1b virus in a chimpanzee.
JFH-1 virus generated in a cell culture was shown previously to be infectious in a chimpanzee (22). To assess the in vivo infection of cell culture-produced CG1b virus, a naive chimpanzee was inoculated with culture medium first from mock-transfected cells (control medium) and then from pEF/CG1b-Rz/S plasmid-transfected cells (2.3 × 103 copies of HCV RNA). After inoculation of the control medium, the chimpanzee showed no signs of infection for 8 weeks. However, 2 weeks after the inoculation of culture medium from CG1b plasmid-transfected cells, HCV RNA became positive in the chimpanzee serum and persisted for up to 48 weeks after inoculation (Fig. 1E). The highest virus titer was 7.5 × 105 copies/ml, and there was a transient mild elevation in ALT. Anti-HCV was detected 40 weeks after inoculation. To demonstrate that the virus generated in cell culture did not acquire adaptive mutations in cultured cells, the entire HCV open reading frame from serum at 4 weeks after inoculation was sequenced. The sequence was completely identical to that of the CG1b strain used for transfection. Culture medium of pEF/H77-Rz/S-transfected cells (1.4 × 104 copies of HCV RNA) was also inoculated into a chimpanzee who had previously recovered from HCV CG1b infection. The chimpanzee developed viremia but quickly resolved the infection (data not shown). This attenuated infection was typically observed in rechallenge experiments of chimpanzees that had recovered from a previous HCV infection (15).
Suppression of JFH-1 replication by IFN-α.
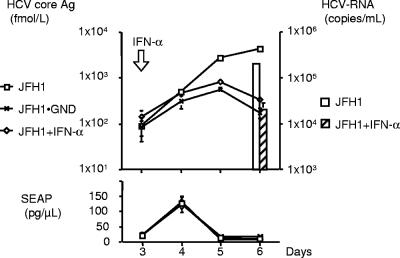
To test the sensitivity of HCV production to antiviral in this system, JFH1-ribozyme plasmid-transfected cells were treated with IFN-α (100 IU/ml) 3 days after transfection, and HCV core Ag and HCV RNA were measured in the culture supernatants (Fig. 2). JFH1-GND mutant that has an inactivating mutation in the NS5b RdRp site was used as a replication deficient control. IFN-α significantly suppressed the HCV core Ag level in the supernatant (4050.3 ± 310.8 to 331.3 ± 14.1 fmol/liter, P < 0.01) to a level that is similar to that of JFH1-GND-transfected cells (171.5 ± 42.0 fmol/liter). The HCV RNA titer of the supernatant was also suppressed by IFN-α (1.92 × 105 ± 1.69 × 104 to 1.93 × 104 ± 8.25 × 103 copies/ml, P < 0.0001). The HCV RNA titer of JFH1-GND mutant transfected was under the detection limit. The SEAP levels were not significantly affected by IFN-α.
FIG. 2.
Inhibition of HCV replication and production by IFN-α. IFN-α (100 IU/ml) was administered to the JFH1-ribozyme-transfected cells 3 days after transfection. The HCV core Ag level, HCV RNA titer, and SEAP activity in the culture supernatants were monitored at the indicated time points. The JFH1-GND mutant was used as a replication-deficient control.
Density gradient analysis.
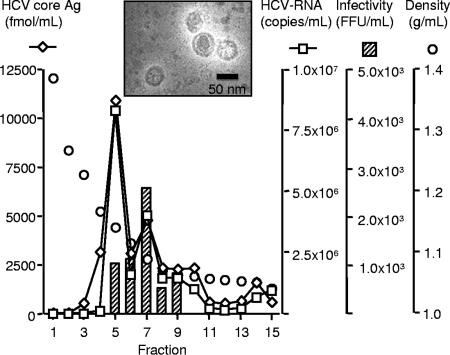
To confirm HCV particle production of this system, the culture medium of JFH1 expression plasmid-transfected cells was concentrated, purified, and subjected to iodixanol density gradient centrifugation. Distributions of HCV core Ag and HCV RNA showed similar profiles and peaked in the fraction with a density of 1.14 g/ml (Fig. 3). The infectivity titer of each fraction to naive Huh7.5.1 cells was also evaluated. The peak infectivity titer located in the fraction at a density of 1.09 g/ml. By negative-stain electron microscopy, the HCV particle was observed as a spherical structure measuring about 50 nm in diameter (Fig. 3, inset). The presence of the particles was detected primarily in the peak fraction of the infectivity. We have previously reported the production and secretion of HCV particles in CG1b-ribozyme plasmid-transfected cells (6). The density gradient analysis and electron microscopic morphology of the particles are similar to those of the JFH1 particles described here.
FIG. 3.
Iodixanol density gradient analysis of the cell culture-generated JFH-1 HCV. Huh7.5.1 cells were transfected with JFH-1-ribozyme plasmid, and culture medium was collected and analyzed by iodixinol density gradient as described in Materials and Methods. Fractions were collected from the bottom of the gradient, and the HCV core Ag, HCV RNA, and infectivity titers were determined. The HCV particles visualized by negative-stain electron microscopy in the peak fraction of infectivity titer is shown in the inset.
Long-term culture of JFH1-expressing cells.
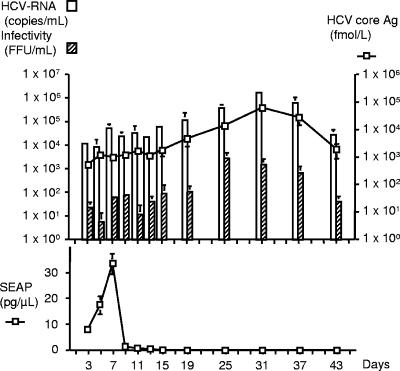
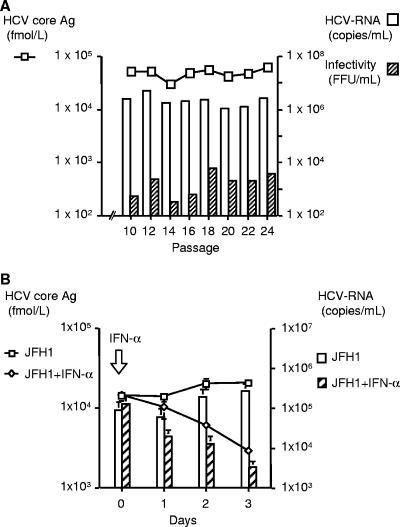
To evaluate the continuous production of HCV particles, JFH1-transfected Huh 7.5 cells were cultured and passaged for more than 6 weeks. The production of HCV core Ag and secretion of HCV RNA were maintained during the observed period (Fig. 4). The highest titer of HCV core Ag and level of HCV RNA were observed on day 31 after transfection in this experiment: 6.35 × 104 fmol/liter and 1.48 × 106 copies/ml, respectively. The SEAP production disappeared about 2 weeks after transfection, supporting the continuous replication and production of the JFH-1 virus. The infectivity titers of culture medium were also determined. The peak infectivity titer of 2.50 × 103 FFU/ml was observed at 25 days after transfection, but in general the HCV core Ag, HCV RNA, and infectivity titers correlated reasonably well in this experiment. However, the transfected cells gradually lost the ability to support HCV replication and production beyond this time period.
FIG. 4.
Long-term culture of JFH1-ribozyme-transfected cells. Huh7.5 cells were transfected with JFH1-ribozyme plasmid as described previously. The transfected cells were passaged 3 to 4 days, and culture medium was collected at various time points. HCV core Ag, HCV RNA, and infectivity titers and SEAP activities in the culture medium were monitored for up to 43 days after transfection.
Establishment of cell lines stably producing JFH-1 virus.
To establish stable JFH-1 virus-producing cells, pEF/JFH1-Rz/N plasmid was transfected into Huh7 and Huh7.5 cells. After 3 weeks of culture with G418 at a concentration of 1.0 mg/ml, visible colonies were identified in both transfected cell lines. A total of 23 Huh7 colonies and 26 Huh7.5 colonies were selected and screened by HCV core Ag production and indirect immunofluorescence with α-core and α-NS3 antibodies. Four Huh7-derived clones and three Huh7.5-derived clones were identified to produce HCV proteins (Table 1) . Three of four Huh7 clones and one of three Huh7.5 clones were found to produce high levels of HCV core Ag and detectable HCV RNA and infectivity titers in culture medium. Among these clones, one Huh 7.5-derived clone (clone 7.5-20) with the highest HCV production was monitored for an extended period of time under drug selection. This clone showed continuous and stable production of HCV core Ag (2.96 × 104 to 6.20 × 104) and HCV RNA (1.03 × 106 to 4.73 × 106) in the medium for up to 24 passages (Fig. 5A). The infectivity titers of culture medium were also determined in this period and ranged from 3.25 × 102 to 6.00 × 103 FFU/ml. To assess the sensitivity of HCV production to IFN-α, 7.5-20 cells were treated with IFN-α (100 IU/ml), and HCV core Ag and HCV RNA were measured in the culture supernatants (Fig. 5B). IFN-α significantly suppressed the HCV core Ag (2.04 × 104 ± 1.85 × 103 to 2.90 × 103 ± 1.51 × 102 fmol/liter, P < 0.005) and HCV RNA titer (2.52 × 105 ± 1.25 × 105 to 3.13 × 103 ± 1.26 × 103 copies/ml, P < 0.05) in the supernatant.
TABLE 1.
Established clones of stable JFH1 virus-producing cells
| Clone | Origin | HCV core Ag (fmol/liter) | HCV RNA (copies/ml) | Infectivity (FFU/ml) |
|---|---|---|---|---|
| 7-09 | Huh7 | 112 | <3.00 × 102a | <1.00 × 101b |
| 7-10 | Huh7 | 5,053 | 1.78 × 105 | 1.50 × 102 |
| 7-19 | Huh7 | 4,438 | 9.72 × 104 | 3.00 × 101 |
| 7-20 | Huh7 | 1,025 | 4.32 × 104 | 2.00 × 101 |
| 7.5-01 | Huh7.5 | 354 | <3.00 × 102 | <1.00 × 101 |
| 7.5-15 | Huh7.5 | 158 | <3.00 × 102 | <1.00 × 101 |
| 7.5-20 | Huh7.5 | 51,397 | 2.32 × 106 | 5.30 × 102 |
Under the detection limit of quantitative reverse transcription-PCR.
Under the detection limit of indirect immunofluorescence.
FIG. 5.
Production of HCV in the stable JFH1-virus producing cell line. (A) Huh7.5 cells were transfected with the pEF/JFH1-Rz/N plasmid and exposed to 1 mg of G418/ml. Clones were isolated as described in the text. The 7.5-20 clone was passaged every 3 to 4 days and monitored continuously for an extended period of time. The HCV core Ag, HCV RNA, and infectivity titers in the culture medium at various time points were analyzed and are shown. (B) Suppression of HCV replication and production in the stable JFH1 virus-producing cell line (clone 7.5-20) by IFN-α. The production of HCV core Ag and HCV RNA in the culture medium was assessed at various time points after the administration of IFN-α (100 IU/ml).
DISCUSSION
The discovery of the JFH-1 strain enabled us to develop a robust system for HCV replication and infection in culture cells (13, 22, 26). However, the JFH-1 strain is unique among the HCV strains and not necessarily representative of HCV biology (7). Studies in chimpanzees suggest that the virus is not particularly infectious in vivo, causing an attenuated and transient infection, which is atypical for the general behavior of HCV (22). It is indeed interesting that this virus was originally isolated from a patient with fulminant hepatitis C (7). It is likely that the fulminant hepatitis is associated more with the clinical setting of the patient than with the virus. Further studies are necessary to resolve the question of whether a viral factor(s) plays a role in the severity of acute HCV infection. Currently, no other natural HCV strain has yet been shown to replicate efficiently and demonstrate robust infectivity in cell culture without adaptive mutations. Although the establishment of a genotype 1a (H77 strain) infectious system in cell culture is important, introduction of several adaptive mutations is clearly required (24). These adaptive mutations have been shown to confer unusual biological properties to the viral strain in vivo (3). Chimeric viruses containing the structural region of other genotypes and the JFH-1 nonstructural genes have been generated and showed in vitro infectivity (13, 14, 17), but the biological relevance of these chimeric viruses is difficult to assess.
In the present study, we established an HCV particle production system with various HCV genotypes by exploiting a recently established DNA transfection cell culture system (6). By engineering two hammerhead ribozyme sequences at the both 5′ and 3′ ends of the HCV genomic cDNA, DNA expression plasmids of multiple HCV strains could be constructed. These plasmids also contain the SEAP gene under the control of the SV40 promoter. By measuring the SEAP activity in the culture medium, the transfection efficiency and the effect of the culture conditions, such as antiviral treatment, could be monitored. Using this system, we demonstrated that the HCV particle production of H77 strain (genotype 1a), CG1b strain (genotype 1b), and J6 strain (genotype 2a) were comparable. Viral particles resembling HCV virions were visualized by electron microscopy in CG1b- and JFH1-transfected cells (6) (Fig. 3). JFH-1 strain showed substantially higher virus production; the HCV core Ag was 50 times and HCV RNA production was 1 to 2 logs higher than those of the other strains. JFH-1 virus released into the culture medium was infectious in naive Huh7 cells. Consistent with previous reports, the infectivity titer (in FFU/ml) was about 3 logs lower than the HCV RNA titer (4, 13, 22, 26). The infectivity of the culture medium of other strain-transfected cells was also assessed. However, evidence of in vitro infection of these strains by immunofluorescence was not detected. The absence of infection is likely a result of the sensitivity limit of the detection method and a much lower replication efficiency of the other strains compared to the JFH-1 strain. The JFH-1 replicates about 1 to 2 logs more than the other strains, and the infectivity titer of the JFH-1 was about 102 FFU/ml in this experiment. Therefore, it is not surprising that we could not detect infection by the other strains. Perhaps by optimizing the culture and infection conditions, using more sensitive methods to detect infection, and concentrating the virus produced in the culture medium, we could detect infection by other genotypes.
To demonstrate the infectivity of strains other than JFH-1, the cell culture-generated CG1b virus was inoculated into a naive chimpanzee and caused a typical course of infection as the infectious CG1b RNA or serum (21). Similarly, H77 virus generated in cell culture was also infectious in vivo. The sequence of CG1b virus replicating in the chimpanzee was completely identical to that of the CG1b strain used for transfection. CG1b virus seems to be capable of replicating more efficiently than the JFH-1 virus in chimpanzees without adaptive mutations, although it has a lower replication efficiency in vitro. This observation is consistent with the contrasting effects of the described adaptive mutations on in vitro replication and in vivo productive infection (3). This discrepancy may be explained by the possibility that lower replication efficiency in vitro may be essential for productive infection and persistence in vivo.
In the iodixanol density gradient analysis, we demonstrated the colocalization of HCV RNA and core Ag proteins. Their peaks were in the identical fraction at a density of about 1.14 g/ml, which is consistent with our previous reports (6, 22). However, the peak of infectivity titer was at a less dense fraction (a density of 1.09 g/ml). This discrepancy has also been reported previously (4, 13). We could detect HCV particles in the peak infectivity fraction; about 50 nm of spherical structures resembling putative HCV were observed by electron microscopy. Thus, infection-competent HCV particles probably exist mainly in the peak fraction of the infectivity titer but not in the peak fraction of the HCV RNA and HCV core Ag. This observation explains the difference between HCV infectivity and the RNA titer of cell culture-generated HCV (an ∼1,000-fold difference). The forms of the viral RNA and protein in the peak fraction are unknown. It is possible that they represent defective viral particles and/or nucleocapsids. Further studies are needed to clarify this point. It is also interesting that the ratio of HCV RNA titer to HCV core Ag level is higher in these gradient fractions than in the unfractionated culture medium. This could be explained by the presence of nonparticulate core protein or empty nucleocapsid in the medium.
This system has the advantage in that it is based on DNA expression plasmids, it is much easier to manipulate, and it contains a reporter gene to monitor various culture conditions. This approach can be extended to various HCV genotypes and strains as well as to the generation of stable cell lines expressing replicating HCV. In the transient-transfection system, robust HCV replication could be observed for up to 6 weeks posttransfection (Fig. 4). It is interesting that the transfected cells eventually lost their ability to support HCV replication. It is possible that some major alterations in the biology of the infected cultured cells, such as the activation of endogenous antiviral mechanisms and/or genetic or epigenetic changes, eventually occurred to shut off the viral replication. However, using a selectable marker such as the neo gene, we could develop stable JFH-1 virus-producing cells. We have generated one clone that supports continuous and stable high-level viral replication and production with multiple passages for an extended period of time. A recent study reported the application of a similar DNA expression system in generating a JFH-1-producing cell line (4). The system used the HDV ribozyme sequence at the 3′ end without any ribozyme sequence at the 5′ end of the JFH-1 genomic cDNA. It is not clear whether the 5′ end of the resulting HCV RNA contains the correct sequence. Furthermore, we also tested the HDV ribozyme sequence in our DNA expression system and found it to be not as efficient in generating the cleaved product as the hammerhead ribozyme sequence we have designed (unpublished data).
This system is also applicable to antiviral testing. In both transient-transfection and stable cell lines, HCV production was substantially suppressed by IFN-α (Fig. 2 and 5B). Both HCV core Ag and HCV RNA titers were suppressed to the baseline level of the JFH1-GND-transfected cells. The absence of a significant change in SEAP activity in IFN-α-treated cells indicates that the effect is not of a general toxicity but specific to HCV replication. Because this system represents the complete replication cycle of HCV, it could prove very useful for high-throughput antiviral screening.
In summary, we have established an HCV production system with various genotypes by using DNA expression plasmids with HCV genomic cDNA flanked by self-cleaving ribozymes. HCV generated in this system showed infection both in vitro and in vivo. This system provides a valuable tool not only to study the replication and pathogenesis of HCV but also to screen for antivirals against multiple HCV strains.
Acknowledgments
We thank Z. Hong Zhou and his coworkers at the University of Texas, Houston, for performing the electron microscopy, and Charles Rice, Francis Chisari, Harry Greenberg, Robert Purcell, Jens Bukh, and Guangxiang George Luo for providing various valuable reagents.
This study was supported in part by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. T. W. is supported by grants from the Japan Society for the Promotion of Science; the Ministry of Health, Labor, and Welfare of Japan; and the Research on Health Sciences Focusing on Drug Innovation program of the Japan Health Sciences Foundation.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Aoyagi, K., C. Ohue, K. Iida, T. Kimura, E. Tanaka, K. Kiyosawa, and S. Yagi. 1999. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J. Clin. Microbiol. 37:1802-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Date, T., T. Kato, M. Miyamoto, Z. Zhao, K. Yasui, M. Mizokami, and T. Wakita. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371-22376. [DOI] [PubMed] [Google Scholar]
- 6.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. USA 102:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64:334-339. [DOI] [PubMed] [Google Scholar]
- 8.Kato, T., M. Miyamoto, A. Furusaka, T. Date, K. Yasui, J. Kato, S. Matsushima, T. Komatsu, and T. Wakita. 2003. Processing of hepatitis C virus core protein is regulated by its C-terminal sequence. J. Med. Virol. 69:357-366. [DOI] [PubMed] [Google Scholar]
- 9.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 10.Kato, T., T. Date, M. Miyamoto, Z. Zhao, M. Mizokami, and T. Wakita. 2005. Nonhepatic cell lines HeLa and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 79:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 12.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 13.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 14.Lindenbach, B. D., P. Meuleman, A. Ploss, T. Vanwolleghem, A. J. Syder, J. A. McKeating, R. E. Lanford, S. M. Feinstone, M. E. Major, G. Leroux-Roels, and C. M. Rice. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA 103:3805-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Major, M. E., K. Mihalik, M. Puig, B. Rehermann, M. Nascimbeni, C. M. Rice, and S. M. Feinstone. 2002. Previously infected and recovered chimpanzees exhibit rapid responses that control hepatitis C virus replication upon rechallenge. J. Virol. 76:6586-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto, M., T. Kato, T. Date, M. Mizokami, and T. Wakita. 2006. Comparison between subgenomic replicons of hepatitis C virus genotypes 2a (JFH-1) and 1b (Con1 NK5.1). Intervirology 49:37-43. [DOI] [PubMed] [Google Scholar]
- 17.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeff, L. B., and J. H. Hoofnagle. 2003. Appendix: the National Institutes of Health Consensus Development Conference Management of Hepatitis C 2002. Clin. Liver Dis. 7:261-287. [DOI] [PubMed] [Google Scholar]
- 19.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, T. Shin-I, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 21.Thomson, M., M. Nascimbeni, S. Gonzales, K. K. Murthy, B. Rehermann, and T. J. Liang. 2001. Emergence of a distinct pattern of viral mutations in chimpanzees infected with a homogeneous inoculum of hepatitis C virus. Gastroenterology 121:1226-1233. [DOI] [PubMed] [Google Scholar]
- 22.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 24.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zein, N. N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]