Abstract
K-type major-group human rhinoviruses (HRVs) (including HRV54) share a prominent lysine residue in the HI surface loop of VP1 with all minor-group HRVs. Despite the presence of this residue, they cannot use members of the low-density lipoprotein receptor family for productive infection. Reexamining all K-type viruses for receptor usage, we noticed that HRV54 is able to replicate in RD cells that lack the major-group receptor intercellular adhesion molecule 1 (ICAM-1). By using receptor blocking assays, inhibition of sulfation, enzymatic digestion, and proteoglycan-deficient cell lines, we show here that wild-type HRV54, without any adaptation, uses heparan sulfate (HS) proteoglycan as an alternate receptor. However, infection via HS is less efficient than infection via ICAM-1. Moreover, HRV54 has an acid lability profile similar to that of the minor-group virus HRV2. In ICAM-1-deficient cells its replication is completely blocked by the H+-ATPase inhibitor bafilomycin A1, whereas in ICAM-1-expressing cells it replicates in the presence of the drug. Thus, use of a “noncatalytic” receptor requires the virus to be highly unstable at low pH.
Human rhinoviruses (HRVs), the most important pathogens in the origin of the common cold, are small, icosahedral, nonenveloped, plus-stranded RNA viruses belonging to the Picornaviridae family (27). HRVs circulate as 99 serotypes; based on comparison of the amino acid sequences of their capsid protein VP1, they were phylogenetically classified as subgenera HRV-A and HRV-B (15, 16). Independent from this classification, they are also divided into minor and major receptor groups. The minor group comprises 12 serotypes that use members of the low-density lipoprotein receptor (LDLR) family for cell entry, whereas major-group viruses attach to intercellular adhesion molecule-1 (ICAM-1) for infection (36). The LDLR family includes the LDLR, the very-low-density lipoprotein receptor (VLDLR), and LDLR-related protein 1, which are all recognized by minor-group HRVs, and many other proteins with similar architecture that probably do not function as viral receptors (20). Major-group HRVs of the two subgenera possess two respective sequence patches involved in ICAM-1 recognition (14); minor-group HRVs have only a strictly conserved lysine residue in the HI surface loop of VP1 that is necessary but not sufficient for receptor binding (36). Thus, the principles underlying receptor discrimination are still poorly understood.
There is also a species-specific discrimination in virus-receptor interaction. For example, HRV1A binds more strongly to mouse LDLR and very weakly to the human homologue, whereas HRV2 can bind equally well to both (9, 25), albeit that these serotypes belong to the minor receptor group and are of subgenus A. LDLRs presumably act only as vehicles to transport the virus across the plasma membrane within clathrin-coated vesicles; upon arrival in endosomes, the low-pH milieu destabilizes the viral capsid and the RNA is released. Minor-group viruses, at least HRV2, strictly depend on the low endosomal pH for uncoating (18, 22). ICAM-1, on the other hand, not only acts as a receptor for binding and cell entry but also facilitates uncoating (7).
As mentioned above, a single lysine residue at the capsid surface is conserved in all minor-group viruses. Interestingly, there are 9 “K-type” major-group HRVs (HRV8, -18, -24, -40, -54, -56, -58, -85, and -98; all are subgenus A) that also possess a lysine at the same position; this suggested that they might be able to use LDLR as well. However, all of these K-type viruses failed to infect HeLa cells in the presence of the ICAM-1-blocking monoclonal antibody (MAb) R6.5 (36). While screening these serotypes for their ability to infect human rhabdomyosarcoma (RD) cells that lack ICAM-1 expression (28), we were surprised to find that HRV54 is able to infect this cell line. We also noticed that increasing the 50% tissue culture infective dose (TCID50)/well (of a 96-well plate) from 103, as used in the previous study (36), to 105 resulted in infection of HeLa-H1 cells whose ICAM-1 was blocked with MAb R6.5. However, as shown previously, infection was not affected by MBP-V33333, a recombinant soluble concatemer of VLDLR repeat 3, which is a potent inhibitor of minor-group viruses (34).
Heparan sulfate (HS) is ubiquitously expressed at the surfaces of mammalian cells. The natural functions of this glycosaminoglycan include cell adhesion, migration, proliferation, and differentiation; it also binds a number of signaling molecules and many other ligands (33). Recently, it has been found that many viruses can use cell surface HS proteoglycans for attachment and entry. Among the family Picornaviridae, foot-and-mouth disease virus, swine vesicular disease virus, coxsackievirus B3, Theiler's murine encephalomyelitis virus, some echoviruses, and variants of HRV89 have been shown to interact with HS for cell attachment and entry, especially in the absence of their classic receptors (4, 6, 11, 23, 35, 37). Having seen that HRV54 can infect the ICAM-1-negative RD cells but is not inhibited by the recombinant VLDLR derivative, we set out to identify the receptor used in addition to ICAM-1. We show here that it is HS proteoglycan (HSPG). In contrast to the previous report on HRV89 variants using HS as result of a tedious adaptation for growth in HEp-2 cells that express low levels of ICAM-1, HRV54 already possesses this property as the wild type (wt) without prior adaptation.
Employing the specific H+-ATPase inhibitor bafilomycin A1 (Baf), we present evidence that use of a “noncatalytic” receptor that does not facilitate uncoating, such as HS, requires the virus to be highly acid sensitive. Our data illustrate the great plasticity of these viruses with respect to receptor usage and once again demonstrate that the use of a given receptor calls for particular physicochemical properties of the virus.
MATERIALS AND METHODS
Cells, viruses, and chemicals.
HRV2 was originally obtained from the American Type Culture Collection (ATCC) and was propagated in HeLa-H1 cells. HRV54 was from the National Institute for Public Health and the Environment (RIVM), Bilthoven, The Netherlands; it was passaged twice in HeLa-OHIO cells in the Enterovirus Laboratory in Helsinki, Finland, and kindly given to us. It was further passaged six times in HeLa-H1 cells in our laboratory. All experiments with HRV54 were carried out with an isolate obtained from a single plaque whose identity was confirmed by neutralization with serotype-specific guinea pig antiserum from the ATCC. Human RD wt and RD-ICAM cells (stably transfected to express human ICAM-1) (13) were a kind gift from Darren R. Shafren, University of Newcastle, New South Wales, Australia. The wt cells do not express any ICAM-1, as verified by fluorescence-activated cell sorter analysis. These cells were grown in Dulbecco's modified Eagle medium supplemented with fetal calf serum (10% in growth medium and 2% in infection medium, which also contained 30 mM MgCl2), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. CHO-K1 cells and CHO mutants (pgsA-745 and pgsD-677) deficient in proteoglycan biosynthesis were obtained from the ATCC. These cells were grown in Ham's F-12 medium supplemented with 5% FCS, l-glutamine, and antibiotics as described above. Heparinase 1 (H2519) was purchased from Sigma-Aldrich GmbH, Vienna, Austria, and Na-chlorate was from Alfa Aesar GmbH, Germany.
Infection inhibition assays.
RD cells were grown in 96-well microtiter plates to 80% confluence. Virus (at 50 TCID50/cell) was added in the presence or absence of twofold serial dilutions of heparin or HS in the infection medium. Plates were incubated at 34°C with 5% CO2 until cells in the controls (without glycans) showed more than 90% cytopathic effect; this was generally at 36 h post infection (p.i.). Cells were fixed with paraformaldehyde (4% in phosphate-buffered saline [PBS]), washed with PBS, and stained with 0.1% crystal violet. After washing with water, the stain was eluted with methanol. Cell damage was quantified with respect to the intensity of the stain retained by living cells in a plate reader at 630 nm. Cell survival in the presence of inhibitors was calculated by setting mock-infected cells to 100% survival and cells infected without inhibitor to 0% survival (12).
Assay for double receptor specificity.
RD-ICAM cells were seeded in 96-well plates and were infected with HRV54 at 10 TCID50/cell in the presence or absence of the ICAM-1-blocking MAb R6.5 (24) with or without heparin. Cells were incubated for 1 h with 100 μg/ml of R6.5, whereas virus was incubated with 2 mg/ml of heparin for 1 h at 34°C prior to addition to the cell monolayer. After 24 h at 34°C the cells were fixed, and cell damage was monitored as described above. For binding assays, cells grown in 12-well plates were preincubated with MAb R6.5 and challenged with 12,000 cpm of 35S-labeled HRV54 (18) in the presence or absence of heparin. After incubation for 1 h at 34°C, the cells were washed, and cell-bound radioactivity was determined by liquid scintillation counting.
Cell binding inhibition assays.
RD cells were grown in 12-well plates and washed with Hanks buffered salt solution (HBSS). 35S-labeled HRV54 at ∼12,000 cpm in HBSS was added in the presence or absence of the glycosaminoglycan, and the cells were incubated for 60 min at 34°C with gentle rocking, washed with HBSS to remove unbound virus, and detached with trypsin-EDTA. Cell-associated radioactivity was determined as described above.
Incorporation of sulfates into the proteoglycans was inhibited with chlorate as described previously (35). Briefly, RD cells were grown in 24-well plates in medium supplemented with 50 mM NaClO3 for 3 days, and binding of radiolabeled virus (in the presence or absence of 2 mg/ml of heparin, where mentioned) was assayed as described above. For digestion of proteoglycans, RD cells grown in 24-well plates to 80% confluence were washed with PBS and incubated with 4, 2, and 1 U/ml of heparinase 1 in HBSS for 2 h at 37°C. Digested material was removed by washing with ice-cold HBSS and virus binding at 4°C was determined.
To determine attachment to proteoglycan-deficient cells, CHO-K1 (wt), pgsA-745, and pgsD-677 cells were seeded in 24-well plates and grown to 90% confluence. Cells were washed with HBSS, challenged with 35S-labeled virus at 16,000 cpm, and incubated at 34°C for 1 h with gentle rocking. Cell-associated radioactivity and radioactivity in the supernatant were determined as above. The percent virus binding was calculated.
Acid sensitivity of the virus.
Virus at 107 TCID50 was incubated with 0.5 M sodium acetate buffer at pH 7.0, 6.5, 6.0, 5.6, 5.2, 4.8, and 4.4 for 30 min at room temperature and neutralized with 0.5 M Na3PO4. Infectivity was determined by end point dilution assay.
Effect of Baf on virus uncoating.
RD cells grown in 96-well plates were incubated with 200 nM of Baf (Alexis Biochemicals, Switzerland) for 1 h at 37°C. Virus at 100 TCID50/cell in infection medium containing 100 nM Baf was added to the cells, and incubation was continued for 24 h at 34°C. To investigate the role of ICAM-1 in virus uncoating, RD-ICAM cells were used in parallel. As a control, to block the ICAM-1, cells were also incubated with 100 μg/ml of R6.5 for 1 h prior to virus addition. Infection was evaluated as described before.
To test whether viral de novo synthesis occurs in the presence of Baf, RD and RD-ICAM cells were grown in 24-well plates to 80% confluence. Cells were washed with PBS and incubated with or without 200 nM Baf in 200 μl of infection medium for 1 h at 37°C. Virus at 106 TCID50 in 200 μl infection medium (about 1 TCID50/cell) was added, and incubation was continued for 1 h at 34°C. Cells were washed twice with ice-cold PBS, and 200 μl of infection medium with and without 100 nM Baf was added. Incubation was continued for a further 0, 2, 12, and 23 h. Cells were then lysed by three freeze-thaw cycles. Cell debris was pelleted, and the virus titer in the supernatant was determined.
RESULTS
HRV54 infection is inhibited by soluble glycosaminoglycans.
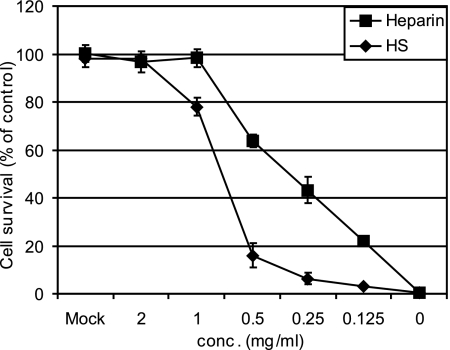
Recently, several representatives of the Picornaviridae family have been shown to use HSPG as an alternate receptor. Moreover, the rhinovirus serotype HRV89 was adapted to grow in cells devoid of ICAM-1, and these variants also use HSPG as a receptor (35). After having noticed the ability of HRV54 to infect ICAM-1-negative RD cells and the lack of inhibition by the VLDLR derivative MBP-V33333 (data not shown), we asked whether this serotype might also use HSPG for infection. We thus tested glycosaminoglycans for their ability to inhibit infection. Cells were challenged with HRV54 in the presence of serial twofold dilutions of heparin and HS, and cell survival was evaluated. As shown in Fig. 1, both glycosaminoglycans reduced the cytopathic effect of the virus. The more strongly sulfated heparin showed more inhibition than HS. The former, at 1 mg/ml, was sufficient to completely protect the cells. For HS, complete inhibition was observed at 2 mg/ml, with almost no inhibition below 0.5 mg/ml.
FIG. 1.
Heparin and HS inhibit cytopathic effect upon infection of RD cells with HRV54 in a concentration-dependent manner. RD cells grown in 96-well plates were infected with HRV54 (50 TCID50/cell) in the presence of the glycans at the concentrations indicated. Plates were incubated at 34°C until cells in the control (no glycan added) showed >90% damage. Cells were fixed and stained with crystal violet, and the dye was eluted with methanol and quantified in a plate reader. Survival of mock-infected cells was set to 100%, and that in the absence of the glycans was set to 0%. Values are means from three parallel experiments ± standard deviations.
To exclude a role of heparin other than competing with cell surface glycosaminoglycans, we carried out a cell protection experiment in three different ways (4): (i) virus was incubated with heparin for 30 min at 34°C, and the mixture was then transferred onto the cells; (ii) cells were incubated with virus at 4°C for 30 min, unbound virus was removed by washing, and heparin was added; and (iii) cells were incubated with heparin for 30 min at 37°C and washed, and virus was added. As expected, we observed maximum inhibition of infection when the virus was incubated with heparin prior to challenging the cells. We also observed an inhibition of up to 40% when heparin was added after virus attachment at 4°C but only at the highest concentration (1 mg/ml). Possibly, already attached virus could be eluted from the cell surface by the heparin. Finally, incubation of the cells with heparin did not modify infection to any significant extent (data not shown). These results clearly show that the soluble derivatives of the cell surface proteoglycan physically interact with the virus and thus block its binding sites for the cell surface receptors.
Heparin and HS suppress HRV54 binding to RD cells.
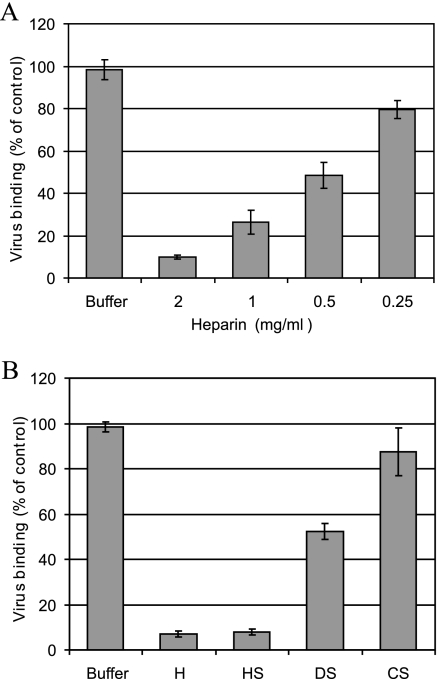
To obtain additional evidence for HRV54 using HS as a receptor, we carried out binding inhibition assays using 35S-labeled virus. RD cells were incubated with radiolabeled virus in the presence or absence of the glycosaminoglycans at 34°C for 60 min. Unbound virus was removed, and cell-associated radioactivity was determined. Heparin showed a concentration-dependent inhibition of binding (Fig. 2A). We also tested HS, chondroitin sulfate (CS), and dermatan sulfate (DS) for their ability to compete with the natural receptor. Only heparin and HS reduced HRV54 binding to the background level, while DS showed some weak inhibition and the effect of CS was only marginal (Fig. 2B). As these proteoglycans differ in monosaccharide moiety and degree of sulfation, these results indicate specificity for particular sulfation sites and/or oligosaccharide structures. It is likely that the interaction preferentially relies on the high concentration of sulfate groups present in HS. At very high, physiologically irrelevant concentrations, DS and CS possibly also inhibit virus binding.
FIG. 2.
Heparin and HS inhibit HRV54 binding to RD cells. Cells were grown in 12-well plates, washed, and challenged with 35S-labeled HRV54 at ∼16,000 cpm in 0.4 ml HBSS with gentle rocking for 60 min at 34°C in the presence of glycans as indicated. Inhibition by heparin (H) was assessed at concentrations of between 0.25 and 2 mg/ml (A), and inhibition by the respective glycans was assessed at 2 mg/ml (B). Cells were washed to remove unbound virus, and cell-associated radioactivity was measured by liquid scintillation counting. Virus binding in the absence of the glycans (buffer) was set to 100%. Values are means from three independent experiments ± standard deviations.
Sulfation is required for virus binding.
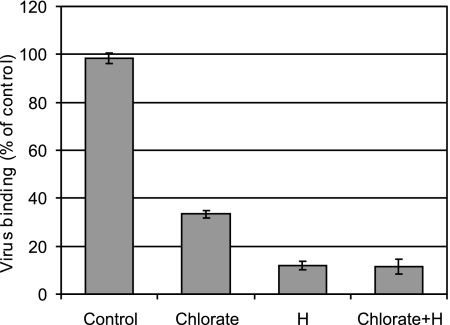
Further evidence for the involvement of sulfated proteoglycans was obtained by the suppression of sulfate incorporation into proteoglycans. RD cells were grown for 3 days in medium with and without NaClO3, washed, and incubated with radiolabeled virus. Unbound virus was removed, and cell-associated radioactivity was quantified. As shown in Fig. 3, HRV54 binding to cells grown in the presence of chlorate was reduced by 65% compared to that to control cells. However, binding further diminished to about 10% in the presence of heparin. This is probably due to incomplete suppression of sulfate incorporation by the chlorate.
FIG. 3.
RD cells grown in medium containing 50 mM NaClO3 exhibit reduced HRV54 binding. Cells were grown in the presence and absence (control) of 50 mM NaClO3 for 3 days, washed, and incubated with virus at ∼16,000 cpm for 60 min at 34°C. Unbound virus was washed away, and cell-associated radioactivity was quantified. Binding is shown as a percentage of the control value (cells grown in the absence of chlorate and without addition of heparin [H]). Values are means from three independent experiments ± standard deviations.
Heparinase 1 treatment of RD cells reduces HRV54 attachment.
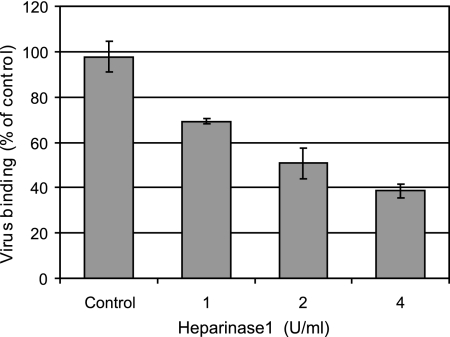
In order to obtain direct evidence for HRV54 binding to HSPG, HS was enzymatically removed. RD cells grown in 24-well plates were incubated with increasing concentrations of heparinase 1 for 2 h at 37°C and washed to remove digested material, and radiolabeled virus was added. After incubation for 60 min at 4°C, unbound virus was removed, and cell-associated radioactivity was determined. As seen in Fig. 4, virus binding was significantly decreased with increasing concentration of the enzyme used to digest cell surface-exposed HS.
FIG. 4.
Heparinase 1 treatment of RD cells decreases HRV54 binding in a concentration-dependent manner. Cells grown in 24-well plates were washed with HBSS and incubated with the indicated concentrations of heparinase 1 for 2 h at 37°C. Digested material was removed with ice-cold buffer, cells were challenged with 16,000 cpm of HRV54, and incubation was continued for 1 h at 4°C. Unbound virus was washed away, cells were trypsinized, and cell-associated radioactivity was determined. The value for virus bound to nontreated cells (control) was set to 100%. Values are means from three independent experiments ± standard deviations.
CHO mutant cells deficient in proteoglycan synthesis fail to bind HRV54.
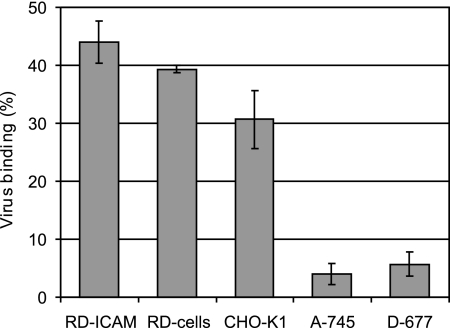
To further confirm the involvement of cell surface HSPG in virus binding, we made use of CHO mutant cells deficient in proteoglycan synthesis. These cells have been extensively used for the investigation of virus-HSPG interactions (17). CHO pgsA-745 cells are deficient in xylosyltransferase synthesis and are unable to produce any glycosaminoglycans, and pgsD-677 cells are doubly deficient in N-acetylglucosaminyltransferase and glucuronyltransferase and fail to synthesize HS but produce threefold-higher levels of CS than the wt. These cells, along with wt CHO-K1, were challenged with 35S-labeled HRV54, and bound virus was determined as described above. A significant difference in binding to the different cell lines was evident (Fig. 5). RD-ICAM cells possessing both receptors, ICAM-1 and HS, strongly bound HRV54, followed by the cells devoid of ICAM-1. wt CHO-K1 cells also bound HRV54 significantly (about 77% compared to RD cells), whereas the mutant cell lines showed only background binding. These results again confirm that HRV54 binds to cell surface HSPG.
FIG. 5.
CHO mutant cells deficient in proteoglycan synthesis show only background binding of HRV54. Cells were seeded in 24-well plates, grown to 90% confluence, and washed with HBSS. 35S-labeled HRV54 at 12,000 cpm was, added and the plates were incubated at 34°C for 60 min. Unbound virus was washed away. Cells were trypsinized, and cell-associated radioactivity and radioactivity in the supernatant were quantified. The percentage of bound virus with respect to total input virus is shown. Values are means from three independent experiments ± standard deviations.
HRV54 has double receptor specificity.
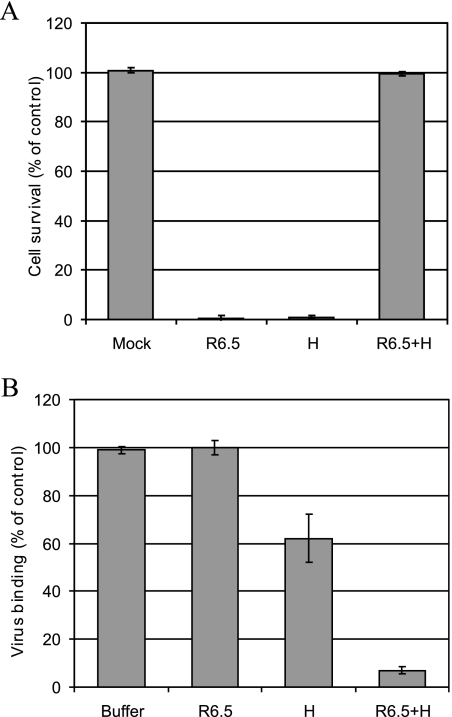
Human ICAM-1 is the receptor for major-group viruses. This receptor not only is responsible for cell attachment but also facilitates virus uncoating. Since HS is not expected to possess any catalytic activity, we asked whether HRV54 can use both receptors independently. RD-ICAM cells were preincubated with R6.5. This is an ICAM-1-specific MAb that blocks the virus binding site as exemplified with HRV14 (29). On the other hand, virus was incubated with heparin. Infection was then monitored in the presence of MAb R6.5 alone or in combination with heparin. After 24 h, cell survival was quantified (Fig. 6A). As expected, neither MAb R6.5 nor heparin alone prevented cell damage, while in combination almost 100% of the cells remained alive. These results clearly show that HRV54 can use either of the receptors independently for productive infection. The same experimental setup was used to monitor virus binding. RD-ICAM cells were preincubated with MAb R6.5 and challenged with radiolabeled HRV54 in the presence or absence of heparin as described above. Surprisingly, MAb R6.5 failed to inhibit virus binding, whereas heparin suppressed attachment by 40%. This indirectly suggests that the remaining 60% binding is due to ICAM-1. The lack of binding inhibition by MAb R6.5 indicates that the cellular HS provides many more attachment sites than ICAM-1; with the former receptor blocked, the limited ICAM-1 binding sites become quickly saturated, and only 60% of input virus can be accommodated. Virus binding was reduced to background values when both inhibitors were used together (Fig. 6B). This excludes the existence of a third receptor.
FIG. 6.
HRV54 can use ICAM-1 and HS independently for binding and productive infection. (A) Cells grown in 96-well plates were incubated with MAb R6.5 for 1 h at 37°C to block ICAM-1. To block heparin binding sites, virus was incubated for 30 min at 34°C with heparin (H). Cells were challenged, and after 24 h at 34°C, cell survival was determined as described for Fig. 1. Note that only the combination of MAb R6.5 and heparin completely protects RD-ICAM cells from viral damage. (B) Cells grown in 12-well plates were treated as described above and challenged with about 15,000 cpm of radiolabeled HRV54 with or without heparin for 60 min at 34°C. Binding in plain buffer (in the absence of R6.5 and heparin) was set to 100%. Values are means from three independent experiments ± standard deviations.
HRV54 is highly acid labile.
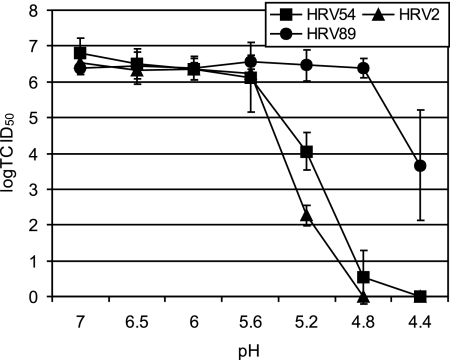
A prime characteristic of HRVs is their acid lability, which has been widely used to differentiate them from enteroviruses (30). HRVs usually become inactivated at a pH of <3 (10). However, some serotypes (e.g., HRV2) undergo structural changes associated with uncoating already at a pH of ≤5.6, and most probably all minor-group viruses are uncoated at a pH of around 5.5, which prevails in late endosomes (8, 18). On the other hand, the catalytic activity of ICAM-1 allows infection of HRV14, HRV3, and HRV89 even in the presence of endosomotropic agents like Baf that increase the endosomal pH to neutrality (1, 19, 35). The ability of HRV54 to infect RD cells lacking ICAM-1 might indicate that it has to be highly acid labile, allowing the low endosomal pH alone to trigger uncoating. We thus determined virus inactivation as a function of the pH. HRVs at 107 TCID50 were incubated in sodium acetate buffer adjusted to pH 7, 6.5, 6.0, 5.6, 5.2, 4.8, and 4.4 for 30 min at room temperature. After reneutralization with phosphate buffer, the infectivity was determined. HRV2 and HRV89 were used as controls representing each receptor group. As depicted in Fig. 7, HRV54 and HRV2 were inactivated at below pH 5.6, while HRV89 remained infective even at pH 4.4, although at substantially reduced titer. The pH thresholds for inactivation, as determined in this experimental setup, are all somewhat below the pH encountered in late endosomes, the site of uncoating; nevertheless, they indicate a similar acid lability of HRV54 and HRV2. This suggests that usage of a receptor different from ICAM-1 requires the virus to be unstable at a pH that prevails in the endosomal system. This correlates well with the previous data on the HRV89 mutants; acquiring affinity for HS went hand in hand with the virus becoming less acid stable than the wt (35).
FIG. 7.
HRV54 is more sensitive to low pH than HRV89, another major-group virus. HRVs were incubated in buffer at the pH indicated for 30 min at room temperature. The solutions were neutralized, and virus infectivity was determined. HRV2 and HRV89 were used as representatives for each receptor group. Values are means from three independent experiments ± standard deviations.
HRV54 uncoating in the absence of ICAM-1 depends on low endosomal pH.
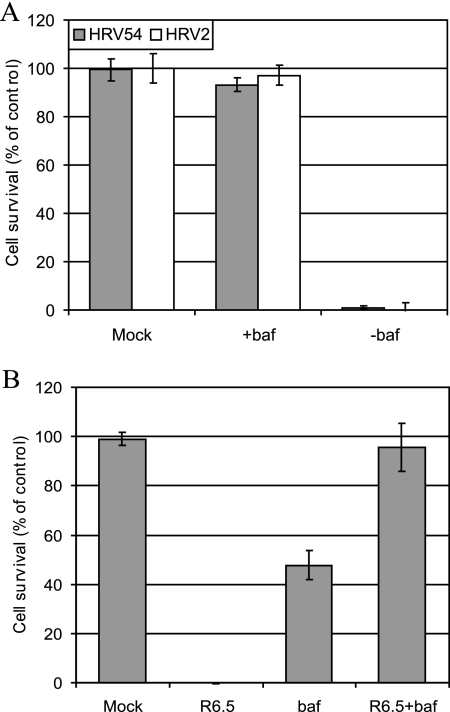
Baf is well established in investigating the role of the low endosomal pH in virus uncoating. This drug inhibits vesicular H+-ATPases and has been widely employed in investigations of the influence of the endosomal pH on HRV2 and HRV14 infection (1, 22, 35). For example, HRV2 infection is completely blocked by Baf, while infection with HRV14 proceeds, albeit with lower efficiency (1, 19). This is in agreement with the in vitro experiments indicating that ICAM-1-catalyzed uncoating occurs at neutral pH (7). We thus conducted experiments to investigate the pH dependency of HRV54 infection. RD cells were preincubated with Baf and infected with HRV54, and cell damage was monitored at 24 h p.i. As shown in Fig. 8A, virtually 100% of the cells survived in the presence of the drug; thus, in RD cells, HRV54 behaves as HRV2 does with respect to strict dependence on the low endosomal pH for uncoating. When the same experiment was carried out using RD-ICAM cells, substantial cell damage occurred in the presence of Baf, making it clear that the low pH is not required for HRV54 infection if ICAM-1 is present (Fig. 8B). However, when ICAM-1 was blocked with MAb R6.5, the RD-ICAM cells behaved like wt RD cells.
FIG. 8.
In the absence of ICAM-1, HRV54 infection depends on the low endosomal pH. RD cells (A) and RD-ICAM cells (B) were preincubated with Baf and infected with HRV54 (and also with HRV2 in the case of RD cells). Where indicated, MAb R6.5 was also present during the entire experiment. Cell survival was monitored after 24 h as described for Fig. 1. Values are means from three independent experiments ± standard deviations.
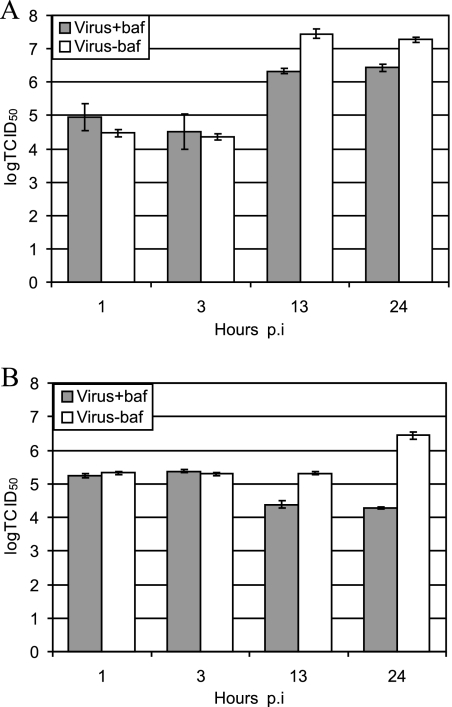
We further confirmed the dependency of the virus uncoating on the low endosomal pH by determining de novo virus production in the presence or absence of the drug. Virus replication took place in the presence of Baf in RD-ICAM cells (Fig. 9A) but not in wt RD cells (Fig. 9B). These results further support the strict requirement of HRV54 for a low-pH environment for uncoating and infection when using HS as a receptor in the absence of ICAM-1.
FIG. 9.
Effect of Baf on viral replication in RD-ICAM (A) and RD (B) cells. Cells were preincubated with or without Baf and infected with HRV54. After the incubation times indicated, the titer of the virus was determined. Note that the decrease in virus titer in the presence of Baf at 13 and 24 h p.i. (B) is due to degradation, which is not counterbalanced by de novo viral synthesis. Values are means from three parallel experiments ± standard deviations.
DISCUSSION
Although phylogenetically very similar and causing the same disease, the many rhinovirus serotypes use two structurally and functionally unrelated receptors for infection and follow distinct pathways for uncoating (2, 26). In particular, a single lysine residue in the HI loop of VP1 of minor-group HRVs is essential in the interaction with members of the LDLR family (34) that bind close to the fivefold-symmetry axis, whereas ICAM-1 binds within the viral canyon of major-group HRVs (21). This lysine residue is strictly conserved in all 12 minor-group HRVs and is also present in nine serotypes of the major group of rhinoviruses, yet neither of these latter groups is able to use LDLRs for infection. This indicates that the lysine is necessary but not sufficient (36). Reexamination of the receptor specificity of these K-type HRVs at a high multiplicity of infection (50 TCID50/cell) revealed, to our surprise, that HRV54 was able to kill RD cells that lack ICAM-1 expression. Since infection by this serotype was not inhibited by MBP-V33333, a recombinant concatemer of repeat 3 of VLDLR that strongly neutralizes all minor-group HRVs (34, 36), involvement of LDLRs in cell attachment was excluded. We thus investigated cell surface HS as a likely candidate receptor. Indeed, heparin as well as HS strongly inhibited HRV54-induced cytopathic effect in RD cells, but with different efficiencies. The more sulfated heparin completely protected the cells already at concentrations of ≥1 mg/ml. This compares well with HS-binding echoviruses, which require between 125 μg/ml and 2 mg/ml of heparin to completely prevent infection (6). The involvement of HS in virus binding was further supported by treatment of the cells with heparinase 1 and the use of CHO mutants with defects in glycan synthesis; virus attachment was drastically reduced upon enzymatic removal of HS, and CHO pgsA-745 and pgsD-677 cells showed only background binding.
The interaction between HRV54 and HS is relatively specific, since DS and CS were only marginally effective in preventing virus binding. Furthermore, sulfate modification of the glycan is required, since growth of the cells in the presence of chlorate substantially reduced attachment of HRV54.
The type of interaction between HS and various ligands, including viruses, is mostly of an ionic nature. The binding site is often formed by basic residues that are far from each other in the sequence but come close together in the three-dimensional structure. However, the HS binding motives BBXB and BBBXXB (with B being a basic residue and X any other) have also been characterized in some cases (32). Scanning of VP1 of HRV54, the only capsid protein with an available sequence, using ScanProsite (http://www.expasy.ch/cgi-bin/prosite/PSScan.cgi) revealed the presence of such a pattern (HHFK) at the BC loop. Interestingly, a similar pattern was also found in HRV62, -65, -83, and -98 at a comparable position. Since the three-dimensional structures of all these serotypes are not available, it is not known whether this motif is sufficiently accessible for binding. Even more, the presence of an HS binding motif does not necessarily mean that it is being used (5), and there are some viruses which lack such a sequence pattern altogether and yet bind HS (4, 24, 35).
Preincubation of HRV54 with heparin (to block its heparin binding sites) together with blocking of ICAM-1 on the cell surface with MAb R6.5 completely prevented infection, indicating that receptor usage is limited to ICAM-1 and HS. This makes the involvement of an additional receptor, including LDLRs, unlikely. Interestingly, MAb R6.5 did not appreciably diminish binding of radiolabeled virus to RD-ICAM cells, whereas heparin reduced virus attachment by about 40%. Apparently, the ICAM-1 molecules are limiting in virus binding whereas HS molecules are not.
ICAM-1 has a catalytic ability facilitating uncoating, presumably by stabilizing an intermediate conformation of the capsid. Furthermore, RNA release also occurs in the presence of lysosomotropic agents and drugs that neutralize the endosomal pH (19). Since LDLRs lack such an activity, the minor-group virus HRV2, and most probably all members of the minor group, are strictly dependent on the low-pH environment for uncoating. We thus asked how HVR54 is uncoated in the absence of ICAM-1 and determined its pH stability. Comparison with HRV2 and the major-group HRV89 revealed that it exhibits a low acid stability similar to that of HRV2. Is the conserved lysine in the HI loop of VP1 responsible for this property? To address this question we also tested the pH sensitivities of some of the other K-type HRVs. Interestingly, we observed that the other major-group K-type viruses tested (HRV8, -18, and -24) were substantially more stable at low pH (data not shown). This makes a contribution of the lysine in pH lability unlikely. The threshold pH for HRV2 inactivation was somewhat lower than the one previously determined (8), which might be due to different experimental setups. Nevertheless, it is evident that HRV2 and HRV54 exhibit similar acid sensitivity profiles. Accordingly, like in the case of HRV2, infection by HRV54 was indeed completely blocked by the vesicular H+-ATPase inhibitor Baf in the absence of ICAM-1; a slight decrease in viral titer at 13 and 24 h p.i. is most probably due to inactivation of the input virus (Fig. 9B).
A number of enteroviruses have been shown to use HS as an alternate receptor for cell entry; since most of them normally attach to members of the immunoglobulin superfamily that bind within the viral canyon and aid in uncoating, it is not clear how the RNAs of these acid-stable viruses become released within the cell. Are there additional factors catalyzing uncoating? It would be interesting to examine the HS-binding enteroviruses for their pH stability. They might be more labile than those that exclusively bind ICAM-1, the poliovirus receptor, or the coxsackie-adenovirus receptor.
Finally, we compared the efficiencies of infection via the two different receptors. The virus yield was consistently by about 1 order of magnitude lower when ICAM-1-negative cells were infected, and appreciable viral de novo synthesis was clearly seen only at 24 h p.i., whereas it was virtually finished within 13 h in RD-ICAM cells. This went hand in hand with cell death; no RD-ICAM cells were left at 13 h, but some few intact RD cells were still found at 24 h p.i. This fact can be tentatively explained by assuming that the uncoating either is less efficient or occurs at an unfavorable site within the endocytic pathway when ICAM-1 is absent.
In summary, our results demonstrate that HRV54, a major-group HRV, is able to use HS in addition to ICAM-1 for productive infection. In contrast to HRV89, which required extensive adaptation (32 blind passages in HEp-2 cells that express ICAM-1 only at a very low level, each followed by a boost in HeLa cells) to acquire HS binding (24, 35), this property seems to be intrinsic to wt HRV54. Some other picornaviruses have been shown to acquire HS binding on tissue culture propagation. Since natural HRV54 isolates were not available for comparison, we cannot exclude that a few passages in tissue culture suffice for adaptation. However, the lack of HS binding by HRV2 and HRV14, which have been serially passaged many times in our laboratory, makes this unlikely and demonstrates that the phenomenon is not a general one.
Although there is abundant HS on the cell surface and attachment appears to be difficult to saturate, infection via proteoglycan is much less efficient than infection via ICAM-1, which is present at much lower concentrations. This might point to differences in virus uptake, uncoating, and/or routing to the site most efficient for RNA release and might explain why the ubiquitously and strongly expressed HS is not a good receptor for efficient infection. Whereas uptake by ICAM-1 occurs via the clathrin-coated pit pathway (3), linkage of the glycosaminoglycans to glycosyl-phosphatidylinositol-anchored proteins might direct the virus to lipid rafts (31). Further work will be aimed at differentiating the pathways followed by this virus when taken up by ICAM-1 compared to HS.
Acknowledgments
We thank I. Goesler for virus production and titer determination. We thank Bohringer Ingelheim for the kind gift of MAb R6.5.
This work was supported by Austrian Science Foundation grant P17516-B10. A.G.K. is the recipient of a fellowship of the Higher Education Commission of Pakistan.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Bayer, N., E. Prchla, M. Schwab, D. Blaas, and R. Fuchs. 1999. Human rhinovirus HRV14 uncoats from early endosomes in the presence of bafilomycin. FEBS Lett. 463:175-178. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, N., D. Schober, E. Prchla, R. F. Murphy, D. Blaas, and R. Fuchs. 1998. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 72:9645-9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeTulleo, L., and T. Kirchhausen. 1998. The clathrin endocytic pathway in viral infection. EMBO J. 17:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escribano-Romero, E., M. A. Jimenez-Clavero, P. Gomes, J. A. Garcia-Ranea, and V. Ley. 2004. Heparan sulphate mediates swine vesicular disease virus attachment to the host cell. J. Gen. Virol. 85:653-663. [DOI] [PubMed] [Google Scholar]
- 5.Fry, E. E., J. W. Newman, S. Curry, S. Najjam, T. Jackson, W. Blakemore, S. M. Lea, L. Miller, A. Burman, A. M. King, and D. I. Stuart. 2005. Structure of foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation. J. Gen. Virol. 86:1909-1920. [DOI] [PubMed] [Google Scholar]
- 6.Goodfellow, I. G., A. B. Sioofy, R. M. Powell, and D. J. Evans. 2001. Echoviruses bind heparan sulfate at the cell surface. J. Virol. 75:4918-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greve, J. M., C. P. Forte, C. W. Marlor, A. M. Meyer, H. Hooverlitty, D. Wunderlich, and A. McClelland. 1991. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol. 65:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruenberger, M., D. Pevear, G. D. Diana, E. Kuechler, and D. Blaas. 1991. Stabilization of human rhinovirus serotype-2 against pH-induced conformational change by antiviral compounds. J. Gen. Virol. 72:431-433. [DOI] [PubMed] [Google Scholar]
- 9.Herdy, B., L. Snyers, M. Reithmayer, P. Hinterdorfer, and D. Blaas. 2004. Identification of the human rhinovirus serotype 1A binding site on the murine low-density lipoprotein receptor by using human-mouse receptor chimeras. J. Virol. 78:6766-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes, J. H., D. C. Thomas, V. V. Hamparian, and H. G. Cramblett. 1973. Acid lability of rhinovirus type 14: effect of pH, time, and temperature. Proc. Soc. Exp. Biol. Med. 144:555-560. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, T., F. M. Ellard, R. A. Ghazaleh, S. M. Brookes, W. E. Blakemore, A. H. Corteyn, D. I. Stuart, J. W. Newman, and A. M. Q. King. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 70:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez-Clavero, M. A., E. Escribano-Romero, A. J. Douglas, and V. Ley. 2001. The N-terminal region of the VP1 protein of swine vesicular disease virus contains a neutralization site that arises upon cell attachment and is involved in viral entry. J. Virol. 75:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson, E. S., L. Xing, R. H. Cheng, and D. R. Shafren. 2004. Enhanced cellular receptor usage by a bioselected variant of coxsackievirus a21. J. Virol. 78:12603-12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laine, P., S. Blomqvist, C. Savolainen, K. Andries, and T. Hovi. 2006. Alignment of capsid protein VP1 sequences of all human rhinovirus prototype strains: conserved motifs and functional domains. J. Gen. Virol. 87:129-138. [DOI] [PubMed] [Google Scholar]
- 15.Laine, P., C. Savolainen, S. Blomqvist, and T. Hovi. 2005. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J. Gen. Virol. 86:697-706. [DOI] [PubMed] [Google Scholar]
- 16.Ledford, R. M., N. R. Patel, T. M. Demenczuk, A. Watanyar, T. Herbertz, M. S. Collett, and D. C. Pevear. 2004. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J. Virol. 78:3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, J., and S. C. Thorp. 2002. Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 22:1-25. [DOI] [PubMed] [Google Scholar]
- 18.Neubauer, C., L. Frasel, E. Kuechler, and D. Blaas. 1987. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology 158:255-258. [DOI] [PubMed] [Google Scholar]
- 19.Nurani, G., B. Lindqvist, and J. M. Casasnovas. 2003. Receptor priming of major group human rhinoviruses for uncoating and entry at mild low-pH environments. J. Virol. 77:11985-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nykjaer, A., and T. E. Willnow. 2002. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol. 12:273-280. [DOI] [PubMed] [Google Scholar]
- 21.Olson, N. H., P. R. Kolatkar, M. A. Oliveira, R. H. Cheng, J. M. Greve, A. McClelland, T. S. Baker, and M. G. Rossmann. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA. 90:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prchla, E., E. Kuechler, D. Blaas, and R. Fuchs. 1994. Uncoating of human rhinovirus serotype 2 from late endosomes. J. Virol. 68:3713-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddi, H. V., and H. L. Lipton. 2002. Heparan sulfate mediates infection of high-neurovirulence Theiler's viruses. J. Virol. 76:8400-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reischl, A., M. Reithmayer, G. Winsauer, R. Moser, I. Gosler, and D. Blaas. 2001. Viral evolution toward change in receptor usage: adaptation of a major group human rhinovirus to grow in ICAM-1-negative cells. J. Virol. 75:9312-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reithmayer, M., A. Reischl, L. Snyers, and D. Blaas. 2002. Species-specific receptor recognition by a minor-group human rhinovirus (HRV): HRV serotype 1A distinguishes between the murine and the human low-density lipoprotein receptor. J. Virol. 76:6957-6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schober, D., P. Kronenberger, E. Prchla, D. Blaas, and R. Fuchs. 1998. Major- and minor-receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J. Virol. 72:1354-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semler, B. L., and E. Wimmer. 2002. Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 28.Shafren, D. R. 1998. Viral cell entry induced by cross-linked decay- accelerating factor. J. Virol. 72:9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staunton, D. E., V. J. Merluzzi, R. Rothlein, R. Barton, S. D. Marlin, and T. A. Springer. 1989. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 56:849-853. [DOI] [PubMed] [Google Scholar]
- 30.Stott, E. J., and R. A. Killington. 1972. Rhinoviruses. Annu. Rev. Microbiol. 26:503-524. [DOI] [PubMed] [Google Scholar]
- 31.Tkachenko, E., E. Lutgens, R. V. Stan, and M. Simons. 2004. Fibroblast growth factor 2 endocytosis in endothelial cells proceed via syndecan-4-dependent activation of Rac1 and a Cdc42-dependent macropinocytic pathway. J. Cell Sci. 117:3189-3199. [DOI] [PubMed] [Google Scholar]
- 32.Trybala, E., T. Bergstrom, D. Spillmann, B. Svennerholm, S. J. Flynn, and P. Ryan. 1998. Interaction between pseudorabies virus and heparin/heparan sulfate. Pseudorabies virus mutants differ in their interaction with heparin/heparan sulfate when altered for specific glycoprotein C heparin-binding domain. J. Biol. Chem. 273:5047-5052. [DOI] [PubMed] [Google Scholar]
- 33.Tumova, S., A. Woods, and J. R. Couchman. 2000. Heparan sulfate proteoglycans on the cell surface: versatile coordinators of cellular functions. Int. J. Biochem. Cell Biol. 32:269-288. [DOI] [PubMed] [Google Scholar]
- 34.Verdaguer, N., I. Fita, M. Reithmayer, R. Moser, and D. Blaas. 2004. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11:429-434. [DOI] [PubMed] [Google Scholar]
- 35.Vlasak, M., I. Goesler, and D. Blaas. 2005. Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J. Virol. 79:5963-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlasak, M., M. Roivainen, M. Reithmayer, I. Goesler, P. Laine, L. Snyers, T. Hovi, and D. Blaas. 2005. The minor receptor group of human rhinovirus (HRV) includes HRV23 and HRV25, but the presence of a lysine in the VP1 HI loop is not sufficient for receptor binding. J. Virol. 79:7389-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zautner, A. E., U. Korner, A. Henke, C. Badorff, and M. Schmidtke. 2003. Heparan sulfates and coxsackievirus-adenovirus receptor: each one mediates coxsackievirus B3 PD infection. J. Virol. 77:10071-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]