Abstract
Like most positive-strand RNA viruses, hepatitis C virus (HCV) is believed to replicate its genome on the surface of rearranged membranes. We have shown previously that HCV NS4AB, but not the product NS4B, inhibits endoplasmic reticulum (ER)-to-Golgi protein traffic (K. V. Konan, T. H. Giddings, Jr., M. Ikeda, K. Li, S. M. Lemon, and K. Kirkegaard, J. Virol. 77:7843-7855). However, both NS4AB and NS4B can induce “membranous web” formation, first reported by Egger et al. (D. B Egger, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz, J. Virol. 76:5974-5984), which is also observed in HCV-infected cells (Y. Rouille, F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson, J. Virol. 80:2832-2841) and cells that bear a subgenomic NS5A-green fluorescent protein (GFP) replicon (D. Moradpour, M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice, J. Virol. 78:7400-7409). To determine the intracellular origin of the web, we examined NS4B colocalization with endogenous cellular markers in the context of the full-length or subgenomic replicon. We found that, in addition to ER markers, early endosome (EE) proteins, including Rab5, were associated with web-inducing protein NS4B. Furthermore, an immunoisolated fraction containing NS4B was found to contain both ER and EE proteins. Using fluorescence microscopy, we showed that wild-type and constitutively active Rab5 proteins were associated with NS4B. Interestingly, expression of dominant-negative Rab5 resulted in significant loss of GFP fluorescence in NS5A-GFP replicon cells. We also found that a small reduction in Rab5 protein expression decreased HCV RNA synthesis significantly. Furthermore, transfection of labeled Rab5 small interfering RNAs into NS5A-GFP replicon cells resulted in a significant decrease in GFP fluorescence. Finally, Rab5 protein was found to coimmunoprecipitate with HCV NS4B. These studies suggest that EE proteins, including Rab5, may play a role in HCV genome replication or web formation.
Hepatitis C virus (HCV) belongs to the family Flaviviridae (31, 49), which includes flaviviruses, like Kunjin virus, dengue virus, and West Nile virus, and pestiviruses, such as bovine viral diarrhea virus (BVDV). Like those of most positive-strand RNA viruses, the genomes of these viruses are translated, replicated, and packaged in the cytoplasm of infected cells. Formation of the RNA replication complexes of most of these viruses results in dramatic rearrangement of the secretory pathway of the host cell. For example, mammalian cells infected with Kunjin virus display various membrane morphologies termed “convoluted membranes” and “vesicle packets” (44, 70), whereas Vero cells infected with dengue virus contain novel membranes called “large cytoplasmic vacuoles” (44). Cells infected with BVDV display rearranged membranes called tubules and spherical vesicles that are 100 to 200 nm in diameter (24). Finally, liver biopsy specimens from HCV-infected chimpanzee or Huh7.5 cells infected with HCV display rearranged intracellular membranes termed “membranous webs” (18, 59).
To identify the putative protein(s) responsible for the induction of membrane rearrangement, proteins from positive-strand RNA viruses have been expressed singly or in combination in a variety of cells. For HCV, expression of the viral nonstructural (NS) protein NS4B (18) induces formation of the membranous webs (referred to as the web in this report). NS4B is a hydrophobic protein of approximately 27 kDa; it is an endoplasmic reticulum (ER)-associated integral membrane protein with four proposed transmembrane domains (42). NS4B has been associated in various ways with the other NS proteins (NS3, NS4A, NS5A, and NS5B), all of which are thought to be involved in HCV RNA replication (1, 2, 18, 19, 23, 30); this has led to the suggestion that NS4B associates with HCV NS proteins to form the core RNA replication complex. In this context, BVDV NS4B has been reported to interact with NS3 and NS5A, two proteins involved in BVDV genome replication (56). Taken together, these observations suggest that NS4B plays a critical role in the genome replication of the Flaviviridae family of viruses.
NS4AB is an HCV precursor protein whose function is not well understood. Like NS4B, NS4AB induces the formation of the web (33), and it is expected to be an integral membrane protein with possible association with other NS proteins involved in viral RNA replication. In addition to inducing intracellular-membrane rearrangement, NS4AB inhibits cellular protein traffic (33). These features make NS4AB a protein that may play a role both in HCV RNA replication and in the ability of the virus to temporarily evade the host immune response, because inhibiting host protein secretion should reduce the release of induced antiviral cytokines.
The intracellular membranes induced by positive-strand RNA viruses have diverse origins. For example, “convoluted membranes” and “vesicle packets” induced by Kunjin virus-infected cells contain markers from the intermediate compartment and the trans-Golgi network, respectively (32, 70). Poliovirus-infected cells accumulate membranous vesicles derived from COPII vesicles (7, 8, 60), some with characteristics similar to those of cellular autophagic vacuoles (16, 61, 67). BVDV-infected cells contain tubules and spherical vesicles that are 100 to 200 nm in diameter and are believed to originate from the rough ER (24). Several reports have shown that HCV NS4B, the mature protein responsible for web formation, colocalizes with ER markers (28, 42), suggesting that the web is derived in part from the ER compartment.
Our current hypothesis states that HCV NS4B oligomerization (46, 72) brings ER-derived vesicles together to initiate web formation. The web may be derived primarily from the ER compartment or in part from the ER compartment through selective retention of factor(s) from other intracellular membranes. Finally, the web may be a unique pooled compartment including the ER and other intracellular compartments. In the present study, we used immunofluorescence (IF) and subcellular fractionation coupled with immunoisolation to investigate the intracellular origin of the web further; we confirmed that the web is derived in part from the ER compartment. In addition, we showed that the web contains early endosome (EE) proteins, including Rab5, whose expression appears to play a functional role in HCV genome replication or web formation. The significance of this unexpected result for both the origin and other potential roles for the web in the HCV life cycle will be discussed.
MATERIALS AND METHODS
Cells.
Full-length replicon cells, C5B, have been described by Ikeda et al. (29). Cells expressing a subgenomic NS5A-green fluorescent protein (GFP) replicon were derived from a human hepatoma cell line, Huh7.5 (9); they contain a replicating, dicistronic, selectable RNA (I/5A-GFP-6) derived from genotype 1b, as described by Moradpour et al. (50). Human kidney (293T), human hepatoma (Huh7 and Huh7.5), and replicon cell lines were grown as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, nonessential amino acids, 100 units/ml penicillin, and 100 μg/ml of streptomycin at 37°C in a 5% CO2 incubator. In addition, the media for replicon cell lines contained 500 μg/ml G418 (Geneticin). All reagents were purchased from Life Technologies/GIBCO-BRL (Rockville, MD).
Construction of plasmids.
To construct plasmids that carry the DsRed-Rab5 fusion proteins, the DsRed gene was amplified by PCR from pDsRed-N1 (Clontech, Mountain View, CA). Primers were designed to introduce an NheI site at the 5′ end, a BglII site at the 3′ end, and an AUG start codon immediately upstream of the DsRed coding region. A recombinant pCR 2.1 plasmid with DsRed was cut with NheI and BglII, and the purified DsRed fragment was subcloned into NheI- and BglII-cleaved wild-type (WT) GFP-Rab5 (kindly provided by Brian Knoll, Baylor College of Medicine, Houston, TX), constitutively active (CA) GFP-Rab5, or dominant-negative (DN) GFP-Rab5 vector. To construct the bicistronic vector carrying HCV NS4B-His in the first cistron and the Rab5A gene in the second cistron, NS4B-His was amplified by PCR from NS4B-His-pIRES plasmid (33). NS4B-His primers were designed to introduce an NheI site, a SalI site, and an AUG start codon immediately upstream of the coding region and an epitope six-His tag, the stop codons UGAUAA, and an MluI site immediately downstream of the coding region. Recombinant pCR 2.1 plasmid with NS4B-His was cut with NheI and MluI, and the purified fragment was subcloned into NheI- and MluI-digested pIRES vector. To obtain the final vector containing NS4BHis-Rab5A, pIRES-Rab5A was digested with XbaI and NotI; the purified Rab5A fragment was cloned into an XbaI- and NotI-cleaved NS4BHis-pIRES vector.
Antibodies.
Rabbit polyclonal antibody to HCV NS4B was obtained from Covance (Denver, PA). Mouse monoclonal antibody to HCV NS4B was obtained from Abcam (Cambridge, MA). HCV NS5A and NS5B antibodies were kindly provided by Craig Cameron (Pennsylvania State University, University Park, PA). Mouse monoclonal antibody to the C-terminal His tag (Penta His) was obtained from QIAGEN (Valencia, CA) and Affinity BioReagents (Golden, CO). Antibodies to EE autoantigen 1 (EEA1) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were obtained from Upstate (Lake Placid, NY) and Fitzgerald (Concord, MA), respectively. Antibodies to Rab5 isoforms and Rab4 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to calnexin and syntaxin 13 were obtained from Stressgen (Victoria, Canada). Secondary antibodies used for indirect IF were obtained from Molecular Probes (Eugene, OR). For immunoblotting, alkaline phosphatase-conjugated secondary antibodies (used in chemifluorescence) or horseradish peroxidase-conjugated secondary antibodies (used in chemiluminescence) were obtained from Vector Laboratories (Burlingame, CA).
DNA transfection.
For each experiment, human kidney 293T cells, the replicon cells (Huh7 C5B) (29), or GFP-Rep 7-2 were trypsinized and grown overnight in 100-mm tissue culture dishes to obtain 50 to 80% confluent monolayer cells. Before transfection, the cells were washed with phosphate-buffered saline (PBS) and fed with 3.5 ml OptiMEM (Invitrogen, Carlsbad, CA) for 2 h. 293T or replicon cells were transfected according to the Lipofectamine protocol as previously reported (33). In all cases, the cells were incubated for 48 h before being processed for Western blotting, IF, or immunoprecipitation.
In vitro HCV RNA transcription, small interfering RNA (siRNA) transfection, and labeling.
Full-length HCV-N plasmid DNA (29) was linearized with XbaI, phenol-chloroform extracted using Phase-Lock Gel tubes (Eppendorf, Westbury, NY), and ethanol precipitated. RNA was synthesized using the T7 RiboMAX Express Large Scale RNA Production Systems kit (Promega, Madison, WI) according to the manufacturer's instructions.
Rab5 and control siRNAs were labeled with the Silencer siRNA labeling kit-Cy3 (Ambion, Austin, TX). siRNA labeling was done according to the manufacturer's instructions. The same concentration of labeled and unlabeled siRNAs was used to transfect full-length (C5B) and NS5A-GFP subgenomic replicon cells. The cells were transfected with a combination of 25 nM, 50 nM, and 25 nM siGENOME SMARTpool siRNA (Dharmacon, Lafayette, CO) specific to RAB5A, -B, and -C, respectively, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). One hundred nanomolar functional, siCONTROL nontargeting siRNA pool (Dharmacon, Lafayette, CO) was used as a control. Transfected cells were incubated for 24 h in 10 ml of OptiMEM containing 10% fetal bovine serum and nonessential amino acids. The cells were washed, fed with Dulbecco's modified Eagle's medium containing 0.5 mg/ml of G418 (Invitrogen, Carlsbad, CA), and incubated for 24 h. The cells were then processed for immunoblotting, Northern blotting/real-time PCR, or fluorescence microscopy.
Northern blotting for HCV RNA.
Total cellular RNA was isolated using the RNeasy miniprep kit (QIAGEN, Valencia, CA) and quantified by spectrophotometry at 260 nm. Twenty micrograms of total RNA was separated by formaldehyde denaturing gel electrophoresis, transferred to a Hybond N+ nylon membrane (Amersham Biosciences, Piscataway, NJ), and hybridized with a digoxigenin-labeled (Roche, Indianapolis, IN) NS4B antisense riboprobe. The hybridized targets were detected by an anti-digoxigenin-alkaline phosphatase and visualized by chemiluminescence using the DIG Northern Starter Kit (Roche, Indianapolis, IN).
Quantitative real-time PCR.
Total cellular RNA was prepared from siRNA-transfected cells by using the RNeasy Mini Kit (QIAGEN) and was treated with RNase-free DNase (QIAGEN, Valencia, CA). First-strand cDNA was synthesized from the DNA-free RNA using random primers and the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Triplicate samples of cDNA were mixed with a Taqman probe and a set of forward and reverse primers specific for either NS4B or GAPDH, and the mixture was subjected to real-time quantitative PCR using the ABI 7300 Sequence Detection System (Applied Biosystems, Foster City, CA).
Immunoisolation of a subcellular fraction containing HCV NS4B protein.
To immunoisolate a subcellular fraction containing NS4B and associated cellular factors, parental and subgenomic NS5A-GFP replicon cells were resuspended in PBS solution containing 0.25 M sucrose and protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 1× leupeptin), and the cells were lysed with seven passages in a ball bearing homogenizer (3). The cell lysates were spun at 2,500 × g for 10 min at 4°C to pellet the cellular debris. Purified NS4B antibody (Covance, Denver, PA) was added to equal amounts of supernatants from parental and replicon cells, and the mixture was incubated overnight at 4°C with constant rotation. The resulting lysates were overlaid on a discontinuous Optiprep gradient (4 to 35%), and the gradient was centrifuged at 120,000 × g for 2 h at 4°C in an SW50.1 rotor. A total of 12 fractions were collected from top to bottom. Typically, fractions 6 to 10 contained mostly ER proteins and some Rab5; in the replicon lysate, these fractions also contained viral replicase proteins, including NS4B, NS5A, and NS5B. Fractions 1 to 5 contained Golgi and EE proteins in both the control and replicon lysates. Secondary-antibody-coated magnetic Dynabeads (M-280; Invitrogen, Carlsbad, CA) were added to the combined fractions with viral replicase proteins (6-10). After incubation at 4°C for 4 h with constant rotation, the mixture was placed in a magnetic rack (Invitrogen) for 1 min. The resulting supernatant was labeled “unbound.” NS4B-bound beads were labeled “bound.” After two washes, the “bound' fraction was resuspended in 4× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (240 mM Tris, pH 6.8, 4% SDS, 40% glycerol, 4% β-mercaptoethanol, 0.01% bromophenol blue). The “unbound” fraction was spun at 100,000 × g for 30 min at 4°C, and the resulting pellet was resuspended in 4× SDS-PAGE loading buffer. “Unbound” and “bound” fractions were separated on SDS-PAGE and examined for the presence of cellular and viral proteins.
Immunoprecipitation and immunoblot analyses.
Cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, and 2 μg/ml leupeptin), and protein concentrations were determined by Bio-Rad protein assay. For coimmunoprecipitation (co-IP) assays, equal amounts of protein from cells expressing control and test vectors were used. Cell lysates were precleared by incubating them for 1 h at 4°C with protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). The beads were pelleted by centrifugation at 3,000 rpm for 10 min at 4°C. The supernatants were recovered, mixed with primary antibody (5 μg/ml of Penta-His antibody; QIAGEN, Valencia, CA), incubated for 2 to 12 h at 4°C, mixed with protein A/G agarose, and incubated for 2 h at 4°C. Protein A/G agarose-bound immune complexes were collected by centrifugation at 13,000 rpm for 10 min and washed once with RIPA buffer, once with RIPA buffer-500 mM NaCl, and once more in RIPA buffer. For immunoblotting with crude lysate, 100 to 160 μg of protein was used. To examine the immunoprecipitates or crude lysate by SDS-PAGE, the samples were resuspended in loading buffer (34), heated at 95°C for 6 min, and centrifuged at 13,000 rpm for 2 min, and the supernatants were analyzed by 10 to 12% SDS-PAGE, followed by transfer onto an Immobilon-P membrane (polyvinylidene difluoride; Millipore, Billerica, MA). Antibody-bound proteins were detected by enhanced chemifluorescence (Amersham Pharmacia Biotech, Piscataway, NJ), visualized, and quantitated on a PhosphorImager (Typhoon 8600; Amersham Pharmacia Biotechnology Inc.-Molecular Dynamics, Piscataway, NJ). In some cases, the proteins were visualized by the enhanced chemiluminescence detection method (ECL or ECL Plus; Amersham Pharmacia Biotech, Piscataway, NJ).
Indirect IF.
Cells on coverslips were washed in PBS, fixed in 4% formaldehyde in PBS for 15 min, and permeabilized with Triton X-100 (0.05% for subgenomic NS5A-GFP replicon or Huh7.5 cells) for 5 min at room temperature. The cells were washed three times with PBS and incubated in 3% bovine serum albumin for 30 min. Primary antibodies were diluted in 3% bovine serum albumin and incubated with the cells for 1 h at room temperature. After three washes, Alexa Fluor 350 (1:1,000)-, Alexa Fluor 488 (1:1,000)-, or Alexa Fluor 594 (1:1,000)-conjugated secondary antibodies (Molecular Probes, Eugene, OR) were added to the cells for 1 h at room temperature. After being stained, the cells were washed three times in PBS, and the coverslips were mounted in Vectashield (Vector Laboratories, Inc., Burlingame, CA). Immunostained samples were analyzed by fluorescence microscopy (Zeiss Axiovert 200 M) at ×630 magnification, and digital images were taken with an Axiocam MRm charge-coupled device camera. Optical sections were deconvolved using Axiovision software to exclude out-of-focus information. All images were saved as TIFF files and imported to and processed in Adobe Photoshop. Colocalization of green (fluorescein isothiocyanate) and red (Cy3) signals produced yellow.
RESULTS
The EE protein Rab5 is associated with HCV NS4B protein.
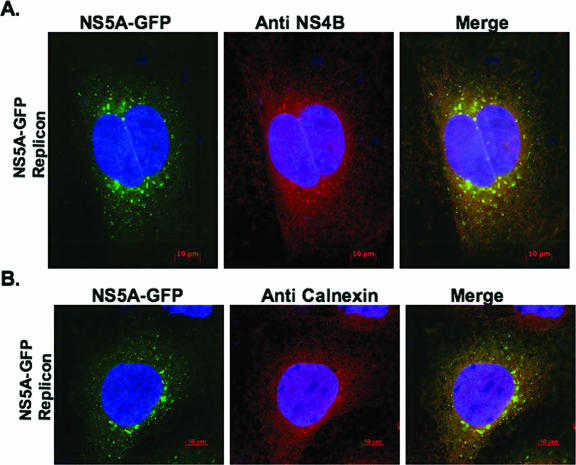
Several reports have shown that the web-inducing protein NS4B is mostly associated with ER proteins (19, 23, 28, 42), suggesting that the web is derived in part from the ER compartment. By analogy to several positive-strand RNA viruses, including poliovirus (60, 61, 67) and Kunjin virus (43, 45), we hypothesized that the HCV-induced web is derived from more than one intracellular-membrane compartment. To examine the complex intracellular origin of the web, we took advantage of two replicon-expressing cell lines: subgenomic NS5A-GFP replicon (50) and full-length C5B replicon (29) cells. As previously reported (23, 28, 42, 50), HCV NS4B displays punctate or dot-like structures in fluorescence microscopy; these dot-like structures appear as the web in electron microscopy. In this study, therefore, we used punctate fluorescence of NS5A-GFP (in subgenomic NS5A-GFP replicon cells) and NS4B (in full-length replicon cells, C5B) as an indirect assay for web detection. For cells expressing subgenomic NS5A-GFP replicon, the cells were first incubated at 37°C for 48 h, and the punctate NS5A-GFP pattern was examined using fluorescence microscopy (Fig. 1A). These cells were then stained with NS4B antibody to detect NS4B protein and to confirm its colocalization with NS5A-GFP (Fig. 1A). Colocalization of NS5A-GFP with any cellular marker was therefore used in this and subsequent studies to show colocalization of NS4B with the respective cellular marker in the context of the viral subgenomic NS5A-GFP replicon. Using this approach, we showed that NS5A-GFP and calnexin fluorescences overlapped (Fig. 1B). These results suggested that NS4B and NS5A were associated with the ER marker, calnexin, in the context of the viral genome; they are consistent with the interpretation that the ER compartment participates in web formation.
FIG. 1.
Calnexin, an ER marker, is indirectly associated with NS4B in the context of subgenomic NS5A-GFP replicon-expressing cells. Subgenomic NS5A-GFP replicon cells were grown for 48 h before being processed for IF. The cells were fixed and labeled as described in Materials and Methods. To visualize NS4B (A) or calnexin (B), labeled proteins were detected using Alexa Fluor 594-conjugated secondary antibody. Samples were observed at ×630 magnification; 0.2-μm digital sections were deconvolved with Axiovision software from Zeiss. Colocalization of green (GFP) and red (Cy3) signals produced yellow. Notice the indirect colocalization of NS4B with calnexin in the context of the replicon (B). Bars = 10 μm.
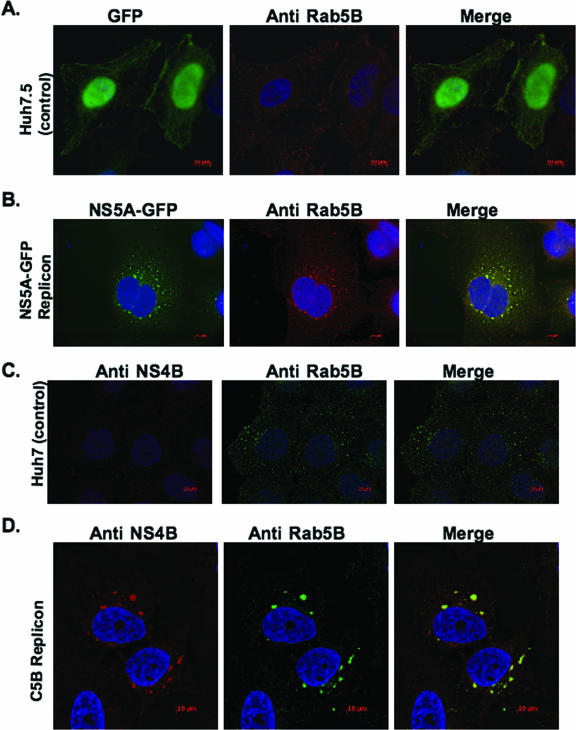
To test whether the web has a more complex origin, we examined colocalization of NS5A-GFP or NS4B with endogenous markers from other intracellular membranes, including the Golgi (golgin 97), lysosome (LAMP1), mitochondria (HSP70), and EE (Rab5). We found that NS5A-GFP or NS4B was not associated with markers from the Golgi, lysosome, and mitochondria (data not shown). However, we observed that Rab5, an EE protein, was associated with NS5A-GFP or NS4B in the context of the viral subgenomic replicon. As shown in Fig. 2B, the distribution of NS5A-GFP fluorescence in subgenomic NS5A-GFP replicon cells showed a significant overlap with endogenous Rab5B stain. Similarly, NS4B fluorescence in full-length (C5B) replicon cells showed a significant overlap with endogenous Rab5B stain (Fig. 2D). No overlap of GFP alone with Rab5B was observed when control GFP-expressing cells were examined (Fig. 2A), nor did we observe any NS4B stain in control Huh7 cells (Fig. 2C). These results suggest that endogenous Rab5B may be associated with the web induced in the context of the viral genome.
FIG. 2.
Rab5, a marker for EE, is associated with HCV NS4B. Subgenomic NS5A-GFP replicon cells (B) and full-length replicon (C5B) cells (D) were grown and processed for fluorescence as described in Materials and Methods. Endogenous Rab5B was detected using Alexa Fluor 594-conjugated secondary antibody. Notice the colocalization of NS5A-GFP (B) and NS4B (D) with endogenous Rab5B; a control GFP vector (A) shows the specificity of NS5A-GFP colocalization with Rab5. Also, notice the redistribution of Rab5B in cells with the replicon. Bars = 10 μm.
HCV NS4B interacts with Rab5 protein.
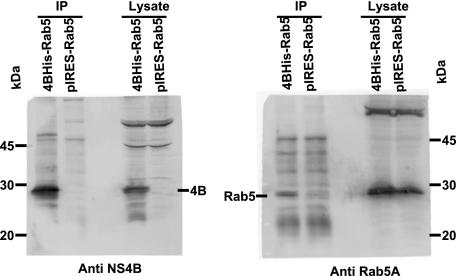
The findings that the ER-derived web also contains EE protein, Rab5 (11, 21, 36, 52), prompted us to determine how this marker might be recruited into the web. We hypothesized that Rab5 is recruited into the web through its interaction with NS4B, since NS4B colocalizes with endogenous Rab5 when expressed alone (data not shown). To determine whether NS4B and Rab5A interact, we performed a co-IP assay. Human 293T cells were transfected with a bicistronic construct to ensure that each cell expressing NS4B protein (with a C-terminal six-His tag) also expressed Rab5A, in addition to endogenous Rab5 protein. Controls for this experiment included the vector expressing Rab5A alone. At 48 h posttransfection, the cells were lysed and the homogenate was immunoprecipitated with Penta His antibody, followed by immunoblotting with anti-NS4B and anti-Rab5A antibodies. As shown in Fig. 3, immunoprecipitation of NS4B resulted in co-IP of Rab5A in the bicistronic vector containing both NS4B and Rab5A, but not in the control vector with Rab5A alone. These results suggest that HCV NS4B interacts with Rab5A protein.
FIG. 3.
HCV NS4B interacts with Rab5. 293T cells were transfected with a bicistronic vector expressing NS4B-His and Rab5A, and the cells were collected at 48 h posttransfection. Cell lysates were immunoprecipitated with Penta His monoclonal antibody. Immunoprecipitates were separated on 13% SDS-PAGE and subjected to immunoblotting with anti-NS4B and anti-Rab5A antibodies. Notice that immunoprecipitation (IP) of NS4B results in co-IP of Rab5A.
Other EE proteins are associated with the web.
The previous findings suggested that the web might include the ER and at least one EE marker, Rab5. To determine whether additional EE proteins are associated with the web, we examined the colocalization of other EE proteins with NS4B in the context of a full-length genomic replicon, C5B. These EE proteins included two Rab5 effectors, EEA1 (63, 64, 66), rabaptin 5 (15, 27, 41), and Rab4 (6, 17, 48), a protein found in both the EE and the recycling endosome. Full-length replicon cells were seeded on coverslips and incubated at 37°C for 48 h; the cells were stained with antibody specific to EEA1, rabaptin 5, or Rab4 protein. As shown in Fig. 4B, D, and F, NS4B fluorescence from the full-length replicon overlapped with the fluorescence from EEA1, rabaptin 5, or Rab4, respectively. These results suggest that several EE proteins may be associated with the NS4B-induced web.
FIG. 4.
Three EE markers, EEA1 (A and B), rabaptin 5 (C and D), and Rab4 (E and F), are associated with HCV NS4B. Full-length (C5B) replicon cells were grown for 48 h and processed for endogenous EEA1, rabaptin 5, or Rab4 fluorescence as described in Materials and Methods. EEA1, rabaptin 5, and Rab4 were stained with their respective antibodies and detected with Alexa Fluor 594-conjugated secondary antibody to EEA1, rabaptin 5, and Rab4. Notice the direct colocalization of NS4B with EEA1 (B), rabaptin-5 (D), or Rab4 (F), as well as the redistribution of these markers in cells with the replicon. Bars = 10 μm.
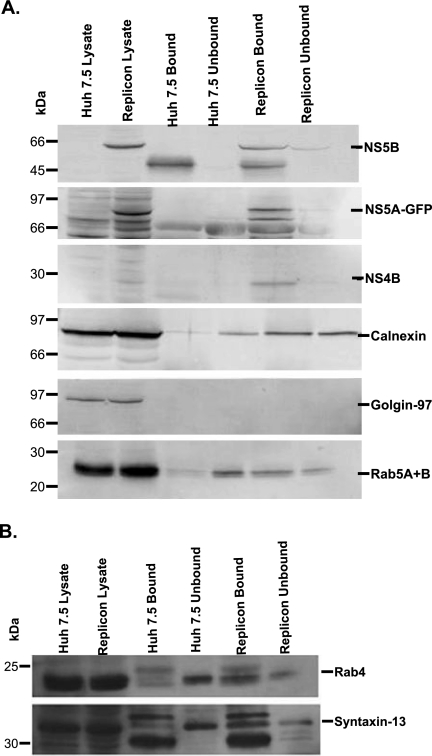
The colocalization of ER and several EE proteins with the web-inducing protein NS4B led us to hypothesize that a subcellular fraction containing NS4B would also include the ER and EE proteins. To test this hypothesis, we used rabbit NS4B antibody coupled with subcellular fractionation and magnetic-bead-conjugated secondary anti-rabbit antibody to immunoisolate the intracellular fraction containing NS4B from parental and NS5A-GFP replicon cell homogenates, as described in Materials and Methods. Using this protocol, we were successful in isolating a fraction containing not only NS4B, but also several replicase complex proteins, including NS5A and NS5B (Fig. 5A). NS4B, NS5A, and NS5B were mostly found in the bound fraction from replicon lysate, which is the fraction containing the magnetic-bead-conjugated secondary antibody to NS4B antibody. No band equal in size to HCV NS4B, NS5A, or NS5B was found in the bound fraction from parental cells (Fig. 5A), indicating specificity of the antibodies used in the current studies. When calnexin, an ER protein, was examined, it was found only in the unbound fraction in the parental cells. However, we found calnexin in both bound and unbound fractions from NS5A-GFP replicon cells, indicating that the immunoisolated fraction with replicase complex proteins also contained ER membrane-bound protein (Fig. 5A). We also tested for the presence of several EE proteins, including Rab5 (11, 21, 36, 52), syntaxin 13 (25, 47, 53), and Rab4 (6, 17), in the immunoisolated fraction. These EE proteins were predominantly found in the unbound fraction from parental cells (Fig. 5A and B). Interestingly, we found Rab5 (Fig. 5A), syntaxin 13, and Rab4 (Fig. 5B) in both bound and unbound fractions from NS5A-GFP replicon cells. However, most of these proteins were enriched in the bound fraction from replicon cells, consistent with the interpretation that EE proteins are indirectly associated with the web-inducing protein NS4B (Fig. 5A and B). As expected, we did not find golgin 97 (35), a trans-Golgi protein, in immunoisolated fractions from either the parental or replicon cells (Fig. 5A), suggesting specificity for cellular proteins coimmunoisolated with HCV NS4B protein.
FIG. 5.
Identification of viral and cellular proteins in the purified subcellular fraction. Huh7.5 (control) and NS5A-GFP replicon cells (1.5 × 106 cells/100-mm dish) were grown for 48 h. The cells were then harvested, lysed with a ball bearing homogenizer, and incubated overnight with NS4B antibody. The homogenate was layered onto an Optiprep gradient and fractionated as described in Materials and Methods. Secondary-antibody-coated magnetic Dynabeads were added to the pooled fraction and subjected to magnetic immunoisolation as described in Materials and Methods. The bound and unbound fractions were separated on SDS-PAGE, followed by immunoblotting with the respective antibody. Crude lysate was used as a control. Proteins were visualized using chemifluorescence (A) or chemiluminescence (B) substrate, as described in Materials and Methods.
Rab5A, -B, and -C siRNAs or DN Rab5 expression results in reduced web fluorescence.
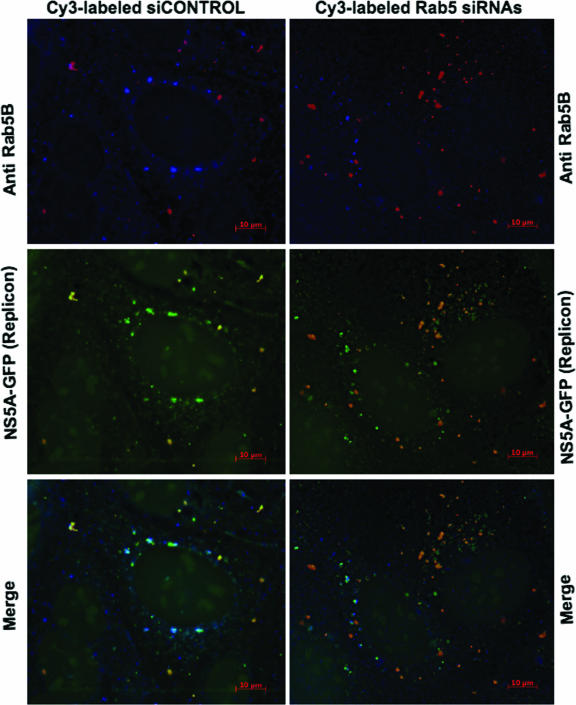
The findings that Rab5, a well-characterized EE marker, is associated with web-inducing NS4B, led us to hypothesize that Rab5 silencing could reduce web formation in replicon cells. To test this hypothesis, we transfected labeled siCONTROL or pooled Rab5A, -B, and -C siRNAs into subgenomic NS5A-GFP replicon-expressing cells and examined web formation, defined as the punctate GFP fluorescence from the replicon. As shown in Fig. 6, Cy3-labeled siCONTROL did not appear to affect Rab5B or GFP replicon fluorescence. However, the presence of Cy3-labeled Rab5A, -B, and -C siRNAs in the replicon cells affected Rab5B and GFP fluorescence. Cells with more Rab5 siRNAs appeared to show a drastic decrease in Rab5B fluorescence; in most cases, it was below detection levels in comparison to siCONTROL-transfected cells. We also observed that cells with substantial Rab5 siRNA presence appeared to show reduced and somewhat diffuse GFP fluorescence, whereas cells with fewer Rab5 siRNAs had GFP fluorescence sometimes comparable to that of siCONTROL-transfected cells. These results suggest that Rab5 may play a role in web formation.
FIG. 6.
Silencing of human Rab5 expression results in a decrease in GFP fluorescence from the subgenomic NS5A-GFP replicon. NS5A-GFP replicon cells were transfected with labeled siCONTROL (red) or Rab5A, -B, and -C siRNAs (red) as described in Materials and Methods. At 48 h posttransfection, endogenous Rab5B protein (blue) was stained with rabbit polyclonal antibody and detected with Alexa Fluor 355-conjugated secondary antibody. Rab5B fluorescence was reduced overall in cells with Cy3-labeled Rab5 siRNAs (red). Notice the overall decrease in GFP fluorescence from the NS5A-GFP replicon in cells with substantial Rab5 siRNA presence. Cy3-labeled siCONTROL (red) does not affect Rab5B or GFP fluorescence from the replicon. Bars = 10 μm.
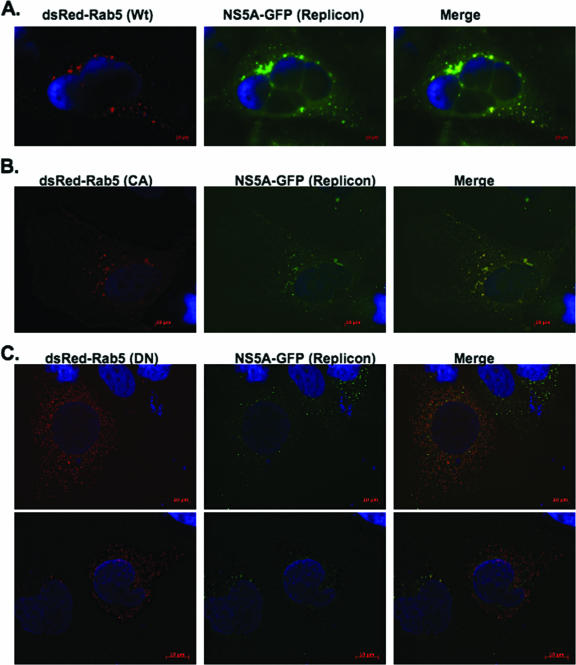
We also examined the effects of different mutant forms of Rab5 on web formation in the subgenomic NS5A-GFP replicon-expressing cells. WT Rab5 cycles between the active, GTP-bound, membrane-associated form and the inactive, GDP-bound, cytoplasmic form (11, 26). CA Rab5 (GTP bound), a GTPase-defective mutant (40), and DN Rab5, locked in the GDP conformation (36), were used in these studies, along with WT Rab5. CA Rab5 is membrane associated, whereas DN Rab5 is mostly cytoplasmic (55). DsRed-fused WT, CA, and DN Rab5 vectors were expressed in the subgenomic NS5A-GFP replicon-expressing cells. If the different forms of DsRed-Rab5A are associated with the web, we anticipated that the merged DsRed and GFP fluorescence would result in yellow fluorescence in each case. As shown in Fig. 7A and B, WT Rab5A or CA Rab5A fluorescence overlapped with GFP fluorescence from the NS5A-GFP replicon, resulting in yellow fluorescence. In addition, expression of WT Rab5 and CA Rab5A did not significantly affect the subgenomic GFP fluorescence intensity. However, the results from DN Rab5A expression (Fig. 7C) in the subgenomic NS5A-GFP replicon-expressing cells were strikingly different. As expected, we found DN Rab5A fluorescence to be cytoplasmic, consistent with Rab5-GDP cytoplasmic localization in the cell (13, 21). Moreover, the punctate GFP fluorescence from the subgenomic NS5A-GFP replicon was greatly reduced in cells expressing DN Rab5A in comparison to cells with WT or CA Rab5A (compare Fig. 7C to A and B). These results suggest that Rab5 might play a role in web formation.
FIG. 7.
DN Rab5 expression affects web formation in subgenomic NS5A-GFP replicon cells. NS5A-GFP replicon cells were transfected with DsRed-fused WT, CA, or DN Rab5. The cells were processed at 48 h posttransfection for fluorescence microscopy. Notice that DN Rab5 expression drastically reduces GFP fluorescence from the replicon, whereas WT or CA Rab5 has little or no effect on GFP fluorescence.
Rab5 protein plays a functional role in HCV RNA synthesis.
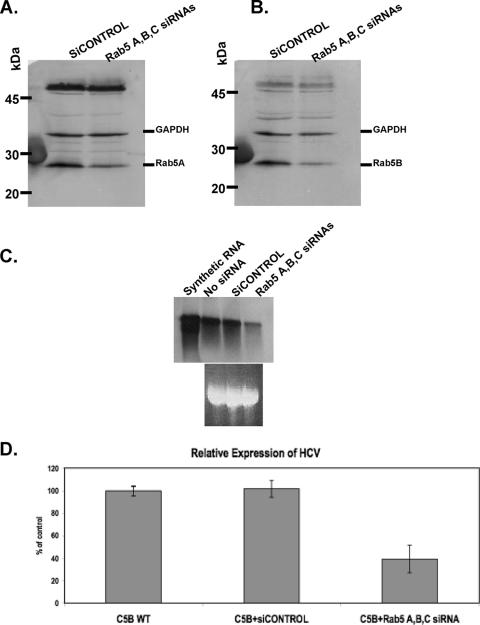
The presence of an EE marker, Rab5, in the web has raised the possibility that this protein plays a role in the replication of the viral genome. To test this hypothesis, siRNA was used to silence Rab5A, -B, and -C isoforms in the genome-length replicon-expressing C5B cells. The results shown below are representative of three separate transfection experiments. Transfection of C5B cells with Rab5A, -B, and -C siRNAs resulted in a 25 to 32% decrease in Rab5A and -B expression in comparison to siCONTROL-transfected cells (Fig. 8A and B). Rab5C silencing results were not included in this report because the endogenous levels of Rab5C were barely detectable in the siCONTROL-transfected cells or in mock-transfected cells. To determine whether the reduced accumulation of cellular Rab5 had any effect on HCV RNA synthesis, total RNA was collected from the same transfection experiments, and viral RNA levels were detected using both Northern blot analysis and real-time PCR. As shown in Fig. 8C, partial silencing of Rab5 resulted in a significant reduction of replicon RNA accumulation in comparison to siCONTROL-transfected cells. Indeed, real-time PCR experiments performed on these RNA samples indicated that replicon cells with Rab5 siRNAs had an approximately 60% decrease in HCV RNA levels in comparison to mock- or siCONTROL-transfected cells (Fig. 8D). These results suggest that Rab5 activity might play a functional role in HCV genome replication.
FIG. 8.
Silencing of human Rab5 expression results in a significant decrease in HCV RNA synthesis. Full-length replicon cells (C5B) were transfected with siCONTROL or Rab5A, -B, and -C siRNAs. Immunoblot analyses were performed using anti-Rab5A (A) or anti-Rab5B (B) antibodies. GAPDH was used as an internal control. (C) Northern blot analysis of HCV RNA in extracts from mock-, siCONTROL-, or siRNA-transfected cells. The negative-sense RNA riboprobe is complementary to the NS4B sequence of the HCV-N strain. (D) Total cellular RNAs used in Northern blot analysis were also used to determine the level of HCV RNA by quantitative real-time PCR. Values from three independent transfection experiments are presented as percentages of the mock-transfected replicon cells. The results were normalized to GAPDH. The error bars indicate standard deviations.
DISCUSSION
Like most positive-strand RNA viruses, HCV is expected to replicate its genome on the surface of rearranged intracellular membranes. Ultrastructural evidence for such membranes has been reported in liver biopsy specimens from chimpanzees infected with HCV (18), in HCV-infected Huh7.5 cells (59), and in cells expressing either the subgenomic replicon (23, 50), the entire HCV polyprotein (18), NS4AB (33), or NS4B protein alone (18, 33). Recently, the role of the web in viral-RNA replication has been further strengthened by the localization of nascent RNA on this novel membrane (23).
Very little is known about the mechanism of web formation following HCV infection. Using IF, various reports have shown that most of the HCV NS proteins involved in viral RNA replication are associated with the ER membrane (19, 28, 42, 71), suggesting that the web is derived in part from the ER compartment. In this report, we used IF coupled with immunoisolation of a subcellular fraction containing HCV NS4B to examine the possible contributions of other intracellular compartments in forming the web (18, 33). We have confirmed that ER markers, including calnexin (42) (Fig. 1B and 5A), are associated with the web-inducing protein NS4B. These observations, together with previous reports (19, 28, 42, 71), clearly suggest that the web is derived in part from the ER compartment.
It is not clear whether the web is derived from a single intracellular compartment, from a single compartment incorporating markers from other intracellular membranes, or from a unique pooled compartment. For instance, the poliovirus-induced double membrane vesicles have been reported to derive from the ER by a mechanism that selectively excludes ER-resident proteins (67); interestingly, these membrane vesicles also appear to contain markers specific for the Golgi and the lysosomal compartments (61). Kunjin virus, a flavivirus closely related to HCV, induces membranes derived in part from the ER and the trans-Golgi network (70). We did not find that the web contains markers associated with the Golgi, lysosomal, or mitochondrial membranes (data not shown), nor was Golgi-associated protein, golgin 97 (35), immunoisolated with HCV NS4B protein (Fig. 5A). These results suggest that the web is not derived in part from these compartments, but we cannot rule out the possibility that a marker from one of these membranes is selectively incorporated into the web.
Using IF and immunoisolation approaches, we found that Rab5, an EE protein, is associated with the web-inducing protein NS4B (Fig. 2B and D and 5A). To determine the extent of involvement of the EE membrane in web formation, we examined colocalization between NS4B and Rab5 effectors, including EEA1 (63, 64, 66) and rabaptin 5 (27). Syntaxin 13 (10, 25, 47), an EE integral membrane protein, and Rab4 (39, 65, 69), a protein transiently associated with the EE, were also examined for their association with NS4B. All these markers, especially those tested (EEA1 and Rab4), were found to costain with endogenous Rab5B protein under IF conditions (data not shown). Interestingly, as reported by Lundin et al. (42), we have also found that NS4B does not always colocalize with EEA1 when expressed alone (data not shown). However, EEA1, rabaptin 5, and Rab4 were found to be associated with NS4B in the context of the full-length and subgenomic NS5A-GFP replicons. Furthermore, like Rab5, most of these EE proteins were immunoisolated, along with NS4B (Fig. 5A and B). These results are consistent with our current hypothesis stating that the web is formed in part from the ER compartment through selective retention of a factor(s) from other intracellular membranes or from a unique pooled compartment including the ER and other intracellular compartments. However, it is also possible that the web actually consists of a heterogeneous population of membranes. The current results do not allow us to distinguish between these possibilities. Further biochemical characterization of the immunoisolated fraction, coupled with IF and electron microscopy, will help to determine the origin of the web.
It is intriguing that Rab5 or any other EE marker is an intrinsic component of the web. Rab5 belongs to the Ras superfamily of small GTPases; it is involved in the regulation of membrane fusion in the early endocytic pathway (11, 12, 52). Rab5 has three isoforms, Rab5A, -B, and -C (11, 14, 22). These isoforms localize to the EE, regulate endocytosis (11), and share close to 85% amino acid sequence identity. Rab5 activity has been linked to the H-Ras signal transduction pathway, and expression of oncogenic H-Ras is known to stimulate endocytosis (5). Activation of the H-Ras signal transduction pathway, including phosphatidylinositol 3-kinase (PI3K) and protein kinase B/Akt, appears to be directly linked to Rab5 activation and endocytosis, since DN Rab5 blocks endocytosis by the H-Ras pathway (4, 37, 38). Interestingly, activation of the N-Ras-PI3K-Akt pathway has been reported in Huh7 cells expressing HCV replicon (46). These findings, coupled with the report that PI3K interacts directly with both Ras and Rab5 (58, 62), suggest that HCV might target the signal transduction pathway between Ras and Rab5 activation of endocytosis. Several pathogens are known to target Rab5 function. For example, phagosomes containing Mycobacterium tuberculosis are reported to bind tightly to Rab5, leading to delayed phagolysosome maturation (54). The SpoE protein of Salmonella acts as a Rab5-specific nucleotide exchange factor; it recruits nonprenylated Rab5 on Salmonella-containing phagosomes to promote fusion with EE and prevents phagolysosome maturation (51). Aside from the role of Rab5 in adenovirus cell entry (57), there is no known report of direct involvement of Rab5 in viral-genome replication. Rotavirus VP4 spike protein, for example, has been reported to interact with Rab5 protein (20), but the role of Rab5 in the virus life cycle has not been elucidated yet. Replication of the coronavirus mouse hepatitis virus has been reported to occur on late endosomal membranes (68), but not on the EE. Thus, HCV appears to be the first reported virus whose replication may require EE-derived membranes.
We tested the hypothesis that Rab5 plays a role in the formation of the web and perhaps in HCV genome replication by using siRNAs specific to Rab5 isoforms. We found that endogenous Rab5 can be partially silenced without any noticeable effect on cell viability (data not shown); this silencing resulted in a decrease in viral RNA synthesis. Using labeled Rab5 siRNAs, we examined the relationship between viral-RNA synthesis and web formation. The presence of labeled Rab5 siRNAs resulted in a substantial decrease in Rab5B and GFP fluorescence from the subgenomic NS5A-GFP replicon. In addition, using a WT, CA, or DN form of Rab5A, we have found that DN Rab5A expression results in a drastic decrease in GFP fluorescence from the subgenomic NS5A-GFP replicon. These results suggest that Rab5 may play a direct role in web formation. However, we cannot rule out the possibility that a decrease in GFP fluorescence is the result of a decrease in HCV RNA synthesis, nor can we rule out the possibility that the effect of Rab5 silencing and DN Rab5A on HCV synthesis could be indirect, due to interference with the formation of EEs.
In an attempt to determine how EE proteins are retained in the web, we examined possible interaction between Rab5 (the first EE protein shown to colocalize with NS4B) and HCV NS4B protein. Using a co-IP assay, we have shown that Rab5A interacts with HCV NS4B when expressed alone (Fig. 3), but not in the context of the full-length replicon (data not shown). We did not observe interaction between NS4B and Rab4 or NS4B and EEA1 (data not shown). However, by combining subcellular fractionation with NS4B immunoisolation, we have shown that a purified fraction containing HCV replicase proteins, including NS4B, also contains the EE proteins Rab5, syntaxin 13, and Rab4. Taken together, these results suggest that Rab5 is retained in the web through its direct interaction with NS4B or indirectly through its association with the EE. Studies are under way to examine in more detail the significance of Rab5 presence in the web and in the HCV life cycle.
Acknowledgments
We are grateful to Karla Kirkegaard, Stanford University, for allowing Kouacou Konan to start this work in her laboratory and for her valuable advice and to Avery August, Leslie Parent, and Anthony Schmitt, Penn State, University Park, for critical reading of the manuscript.
This work was supported by K01 award CA95086-04 from the National Institutes of Health and by a gift from Intermune, Inc.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, M. J. 1997. Epidemiology of hepatitis C. Hepatology 26:62S-65S. [DOI] [PubMed] [Google Scholar]
- 3.Balch, W. E., and J. E. Rothman. 1985. Characterization of protein transport between successive compartments of the Golgi apparatus: asymmetric properties of donor and acceptor activities in a cell-free system. Arch. Biochem. Biophys. 240:413-425. [DOI] [PubMed] [Google Scholar]
- 4.Barbieri, M. A., A. D. Kohn, R. A. Roth, and P. D. Stahl. 1998. Protein kinase B/akt and rab5 mediate Ras activation of endocytosis. J. Biol. Chem. 273:19367-19370. [DOI] [PubMed] [Google Scholar]
- 5.Bar-Sagi, D., and J. R. Feramisco. 1986. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 233:1061-1068. [DOI] [PubMed] [Google Scholar]
- 6.Bielli, A., P. O. Thornqvist, A. G. Hendrick, R. Finn, K. Fitzgerald, and M. W. McCaffrey. 2001. The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1. Biochem. Biophys. Res. Commun. 281:1141-1153. [DOI] [PubMed] [Google Scholar]
- 7.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39-48. [DOI] [PubMed] [Google Scholar]
- 9.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandhorst, D., D. Zwilling, S. O. Rizzoli, U. Lippert, T. Lang, and R. Jahn. 2006. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl. Acad. Sci. USA 103:2701-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bucci, C., A. Lutcke, O. Steele-Mortimer, V. M. Olkkonen, P. Dupree, M. Chiariello, C. B. Bruni, K. Simons, and M. Zerial. 1995. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 366:65-71. [DOI] [PubMed] [Google Scholar]
- 12.Bucci, C., R. G. Parton, I. H. Mather, H. Stunnenberg, K. Simons, B. Hoflack, and M. Zerial. 1992. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70:715-728. [DOI] [PubMed] [Google Scholar]
- 13.Calero, M., C. Z. Chen, W. Zhu, N. Winand, K. A. Havas, P. M. Gilbert, C. G. Burd, and R. N. Collins. 2003. Dual prenylation is required for Rab protein localization and function. Mol. Biol. Cell. 14:1852-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavrier, P., R. G. Parton, H. P. Hauri, K. Simons, and M. Zerial. 1990. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62:317-329. [DOI] [PubMed] [Google Scholar]
- 15.Cosulich, S. C., H. Horiuchi, M. Zerial, P. R. Clarke, and P. G. Woodman. 1997. Cleavage of rabaptin-5 blocks endosome fusion during apoptosis. EMBO J. 16:6182-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dales, S., H. J. Eggers, I. Tamm, and G. E. Palade. 1965. Electron microscopic study of the formation of poliovirus. Virology 26:379-389. [DOI] [PubMed] [Google Scholar]
- 17.Daro, E., P. van der Sluijs, T. Galli, and I. Mellman. 1996. Rab4 and cellubrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc. Natl. Acad. Sci. USA 93:9559-9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 20.Enouf, V., S. Chwetzoff, G. Trugnan, and J. Cohen. 2003. Interactions of rotavirus VP4 spike protein with the endosomal protein Rab5 and the prenylated Rab acceptor PRA1. J. Virol. 77:7041-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes, A. Q., B. R. Ali, J. S. Ramalho, R. F. Godfrey, D. C. Barral, A. N. Hume, and M. C. Seabra. 2003. Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell. 14:1882-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorvel, J. P., P. Chavrier, M. Zerial, and J. Gruenberg. 1991. Rab5 controls early endosome fusion in vitro. Cell 64:915-925. [DOI] [PubMed] [Google Scholar]
- 23.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray, E. W., and P. F. Nettleton. 1987. The ultrastructure of cell cultures infected with border disease and bovine virus diarrhoea viruses. J. Gen. Virol. 68:2339-2346. [DOI] [PubMed] [Google Scholar]
- 25.Grosshans, B. L., D. Ortiz, and P. Novick. 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103:11821-11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffenberg, S., J. C. Sanford, S. Liu, D. S. Daniel, M. Tuvin, B. J. Knoll, M. Wessling-Resnick, and B. F. Dickey. 1995. Biochemical and functional characterization of a recombinant GTPase, Rab5, and two of its mutants. J. Biol. Chem. 270:5048-5056. [DOI] [PubMed] [Google Scholar]
- 27.Horiuchi, H., R. Lippe, H. M. McBride, M. Rubino, P. Woodman, H. Stenmark, V. Rybin, M. Wilm, K. Ashman, M. Mann, and M. Zerial. 1997. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90:1149-1159. [DOI] [PubMed] [Google Scholar]
- 28.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 31.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konan, K. V., T. H. Giddings, Jr., M. Ikeda, K. Li, S. M. Lemon, and K. Kirkegaard. 2003. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 77:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 35.Larocca, M. C., R. A. Shanks, L. Tian, D. L. Nelson, D. M. Stewart, and J. R. Goldenring. 2004. AKAP350 interaction with cdc42 interacting protein 4 at the Golgi apparatus. Mol. Biol. Cell. 15:2771-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, G., M. A. Barbieri, M. I. Colombo, and P. D. Stahl. 1994. Structural features of the GTP-binding defective Rab5 mutants required for their inhibitory activity on endocytosis. J. Biol. Chem. 269:14631-14635. [PubMed] [Google Scholar]
- 37.Li, G., C. D'Souza-Schorey, M. A. Barbieri, J. A. Cooper, and P. D. Stahl. 1997. Uncoupling of membrane ruffling and pinocytosis during Ras signal transduction. J. Biol. Chem. 272:10337-10340. [PubMed] [Google Scholar]
- 38.Li, G., C. D'Souza-Schorey, M. A. Barbieri, R. L. Roberts, A. Klippel, L. T. Williams, and P. D. Stahl. 1995. Evidence for phosphatidylinositol 3-kinase as a regulator of endocytosis via activation of Rab5. Proc. Natl. Acad. Sci. USA 92:10207-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, G., and P. D. Stahl. 1993. Post-translational processing and membrane association of the two early endosome-associated rab GTP-binding proteins (rab4 and rab5). Arch. Biochem. Biophys. 304:471-478. [DOI] [PubMed] [Google Scholar]
- 40.Li, G., and P. D. Stahl. 1993. Structure-function relationship of the small GTPase rab5. J. Biol. Chem. 268:24475-24480. [PubMed] [Google Scholar]
- 41.Lippe, R., H. Horiuchi, A. Runge, and M. Zerial. 2001. Expression, purification, and characterization of Rab5 effector complex, rabaptin-5/rabex-5. Methods Enzymol. 329:132-145. [DOI] [PubMed] [Google Scholar]
- 42.Lundin, M., M. Monne, A. Widell, G. Von Heijne, and M. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Improved membrane preservation of flavivirus-infected cells with cryosectioning. J. Virol. Methods 56:67-75. [DOI] [PubMed] [Google Scholar]
- 45.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 46.Mannova, P., and L. Beretta. 2005. Activation of the N-Ras-PI3K-Akt-mTOR pathway by hepatitis C virus: control of cell survival and viral replication. J. Virol. 79:8742-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride, H. M., V. Rybin, C. Murphy, A. Giner, R. Teasdale, and M. Zerial. 1999. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell 98:377-386. [DOI] [PubMed] [Google Scholar]
- 48.McCaffrey, M. W., A. Bielli, G. Cantalupo, S. Mora, V. Roberti, M. Santillo, F. Drummond, and C. Bucci. 2001. Rab4 affects both recycling and degradative endosomal trafficking. FEBS Lett. 495:21-30. [DOI] [PubMed] [Google Scholar]
- 49.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee, K., S. Parashuraman, M. Raje, and A. Mukhopadhyay. 2001. SpoE acts as an Rab5-specific nucleotide exchange factor and recruits non-prenylated Rab5 on Salmonella-containing phagosomes to promote fusion with early endosomes. J. Biol. Chem. 276:23607-23615. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen, E., F. Severin, J. M. Backer, A. A. Hyman, and M. Zerial. 1999. Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol. 1:376-382. [DOI] [PubMed] [Google Scholar]
- 53.Novick, P., M. Medkova, G. Dong, A. Hutagalung, K. Reinisch, and B. Grosshans. 2006. Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem. Soc. Trans. 34:683-686. [DOI] [PubMed] [Google Scholar]
- 54.Perskvist, N., K. Roberg, A. Kulyte, and O. Stendahl. 2002. Rab5a GTPase regulates fusion between pathogen-containing phagosomes and cytoplasmic organelles in human neutrophils. J. Cell Sci. 115:1321-1330. [DOI] [PubMed] [Google Scholar]
- 55.Prada-Delgado, A., E. Carrasco-Marin, C. Pena-Macarro, E. Del Cerro-Vadillo, M. Fresno-Escudero, F. Leyva-Cobian, and C. Alvarez-Dominguez. 2005. Inhibition of Rab5a exchange activity is a key step for Listeria monocytogenes survival. Traffic 6:252-265. [DOI] [PubMed] [Google Scholar]
- 56.Qu, L., L. K. McMullan, and C. M. Rice. 2001. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in viral cytopathogenicity. J. Virol. 75:10651-10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rauma, T., J. Tuukkanen, J. M. Bergelson, G. Denning, and T. Hautala. 1999. Rab5 GTPase regulates adenovirus endocytosis. J. Virol. 73:9664-9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez-Viciana, P., P. H. Warne, R. Dhand, B. Vanhaesebroeck, I. Gout, M. J. Fry, M. D. Waterfield, and J. Downward. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370:527-532. [DOI] [PubMed] [Google Scholar]
- 59.Rouille, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin, H. W., M. Hayashi, S. Christoforidis, S. Lacas-Gervais, S. Hoepfner, M. R. Wenk, J. Modregger, S. Uttenweiler-Joseph, M. Wilm, A. Nystuen, W. N. Frankel, M. Solimena, P. De Camilli, and M. Zerial. 2005. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170:607-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simonsen, A., J. M. Gaullier, A. D'Arrigo, and H. Stenmark. 1999. The Rab5 effector EEA1 interacts directly with syntaxin-6. J. Biol. Chem. 274:28857-28860. [DOI] [PubMed] [Google Scholar]
- 64.Simonsen, A., R. Lippe, S. Christoforidis, J. M. Gaullier, A. Brech, J. Callaghan, B. H. Toh, C. Murphy, M. Zerial, and H. Stenmark. 1998. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394:494-498. [DOI] [PubMed] [Google Scholar]
- 65.Sonnichsen, B., S. De Renzis, E. Nielsen, J. Rietdorf, and M. Zerial. 2000. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149:901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stenmark, H., R. Aasland, B. H. Toh, and A. D'Arrigo. 1996. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 271:24048-24054. [DOI] [PubMed] [Google Scholar]
- 67.Suhy, D. A., T. H. Giddings, and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vitale, G., V. Rybin, S. Christoforidis, P. Thornqvist, M. McCaffrey, H. Stenmark, and M. Zerial. 1998. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 17:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Westaway, E. G., A. A. Khromykh, and J. M. Mackenzie. 1999. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology 258:108-117. [DOI] [PubMed] [Google Scholar]
- 71.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, G. Y., K. J. Lee, L. Gao, and M. M. Lai. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80:6013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]