Abstract
Secretion of interferon (IFN) by virus-infected cells is essential for activating autocrine and paracrine pathways that promote cellular transition to an antiviral state. In most mammalian cells, IFN production is initiated by the activation of constitutively expressed IFN regulatory factor 3, IRF3, which in turn leads to the induction of IRF7, the “master regulator” of IFN type I synthesis (alpha/beta IFN). Previous studies established that rotavirus NSP1 antagonizes IFN signaling by inducing IRF3 degradation. In the present study, we have determined that, in comparison to wild-type rotaviruses, rotaviruses encoding defective NSP1 grow to lower titers in some cell lines and that this poor growth phenotype is due to their failure to suppress IFN expression. Furthermore, we provide evidence that rotaviruses encoding wild-type NSP1 subvert IFN signaling by inducing the degradation of not only IRF3, but also IRF7, with both events occurring through proteasome-dependent processes that proceed with similar efficiencies. The capacity of NSP1 to induce IRF7 degradation may allow rotavirus to move across the gut barrier by enabling the virus to replicate in specialized trafficking cells (dendritic cells and macrophages) that constitutively express IRF7. Along with IRF3 and IRF7, NSP1 was found to induce the degradation of IRF5, a factor that upregulates IFN expression and that is involved in triggering apoptosis during viral infection. Our analysis suggests that NSP1 mediates the degradation of IRF3, IRF5, and IRF7 by recognizing a common element of IRF proteins, thereby allowing NSP1 to act as a broad-spectrum antagonist of IRF function.
In the immunologically naïve individual, the ability of the innate immune system to recognize and respond to a pathogen can represent a life or death event (10). Triggering of the innate immune system is based on the recognition of pathogen-associated molecular patterns (PAMP) by cellular receptor factors (5, 31). The major downstream effect of pathogen-PAMP receptor interaction is the activation of constitutively expressed cellular transcription factors that mediate the early phase of the host response (25). Included in this set of transcription factors is interferon (IFN) regulatory factor 3 (IRF3), one of the nine structurally related members of the IRF family that, as a group, exhibit a broad range of functions in regulating the immune system (14). IRF3 accumulates in its inactive monomeric form within the cytosol of the cell. PAMP recognition activates kinases that lead to the phosphorylation, dimerization, and translocation of IRF3 to the nucleus, where the factor activates genes expressing beta interferon (IFN-β) (21). Secretion of IFN-β by virus-infected cells activates the Jak/Stat pathway in neighboring uninfected cells, an event inducing the expression of transcription factors of broader action, such as IRF7. IRF7 is the primary regulator of type I IFN (IFN-α and IFN-β) production in the local and systemic reaction of the host to viral infections (30). Although cells typically contain little or no IRF7, expression of the factor is strongly upregulated in the presence of type I IFN (13). After phosphorylation and dimerization, IRF7 moves to the nucleus, where it interacts with promoters inducing the expression of IFN-α, a signaling molecule critical to triggering the expression and activation of the broad array of antiviral factors involved in establishing the antiviral state (15). Secretion of IFN-α is also essential for the efficient development of adaptive immunity (10). The fact that viruses have evolved multiple mechanisms of antagonizing activation of the innate immune system emphasizes the critical nature of this pathogen-host interface as a potential barrier to productive infection (12).
Rotaviruses, members of the family Reoviridae, contain segmented double-stranded RNA (dsRNA) genomes. Rotaviruses are an important cause of gastroenteritis in numerous species of animals, including humans (28), where these viruses represent the primary causative agent of life-threatening dehydrating diarrhea in children under the age of 5 (20). Early studies revealed that the growth of rotavirus is restricted in IFN-treated cells, but less so than other viruses, such as vesicular stomatitis virus (VSV) and reovirus (19), which are noted for their increased susceptibility to the antiviral effects of IFN. Vanden Broecke et al. found that the production of IFN was delayed in newborn calves infected with low doses of rotavirus and that this lag period was characterized by severe but transient diarrhea (35). In contrast, animals infected with high doses of virus produced IFN very early and were free of severe diarrhea. These and other studies were the first to raise the possibility that while rotaviruses are susceptible to IFN, they also may have a mechanism to suppress the IFN signaling pathway at least during early stages of infection.
Recent studies have indicated that the rotavirus nonstructural protein NSP1 interacts with IRF3 and that this interaction results in the proteasome-mediated degradation of IRF3 (4, 11). Due to the loss of IRF3, the expression of IFN-β is suppressed, thereby perturbing the establishment of the antiviral state. NSP1 is a nonessential viral protein, localizing to the cytoskeleton, that contains a highly conserved N-terminal arrangement of cysteine residues typical of RING fingers (16). The presence of similar RING fingers in E3 ubiquitin-protein ligases suggests that NSP1 may induce the degradation of IRF3 through ubiquitination activity reminiscent of E3 ligases (17). Although NSP1 exhibits a high degree of sequence variability at its C terminus, it is nonetheless through this region that NSP1 interacts with IRF3 (4, 11, 16).
IRF3 and the other members of the IRF family contain related features, including an N-terminal DNA-binding domain (34). These shared features led us to investigate the possibility that NSP1 could direct the turnover of IRF proteins in addition to IRF3. Through this analysis, we have determined that NSP1 induces the proteasome-mediated degradation of multiple members of the IRF family, including IRF3, IRF5, and IRF7. The effect of NSP1 on IRF7 is particularly significant, since this factor represents the primary regulator of type I IFN expression and is essential for the activation of IFN-α genes. By inducing the degradation of IRF5, NSP1 can downregulate the activation of genes producing proinflammatory cytokines and that initiate events leading to apoptosis. We conclude that NSP1 represents a broad-spectrum antagonist of innate immune mechanisms meant to limit virus spread.
MATERIALS AND METHODS
Cells and viruses.
The monkey kidney epithelial (MA104) cell line was grown in Medium 199 (Invitrogen) supplemented with 5% fetal bovine serum (FBS). The rhesus fetal lung (FRhL2), human colon (Caco-2), and human renal (293T) epithelial cell lines were grown in Dulbecco modified Eagle medium (BioWhittaker) supplemented with 10% FBS (complete media). Rotaviruses strains UK, BRV, and SA11-4F, -5S, -30-19, and -30-1A were propagated on MA104 cells. Virus titers were determined by plaque assay (32). The recombinant vesicular stomatitis virus, VSV-green fluorescent protein (GFP), was kindly provided John Hiscott (McGill University, Montreal, Quebec, Canada) (33). The virus, which constitutively expresses green fluorescent protein (GFP), was propagated and titers were determined on baby hamster kidney cells (6).
Expression vectors.
The plasmids pCI-NSP1 and pCI-NSP1ΔC17 contain a cytomegalovirus (CMV) immediate-early promoter that drives the expression of SA11 wild-type (wt) NSP1 and a C-terminal 17-residue truncated form of SA11 NSP1 (NSP1ΔC17), respectively (4). The pA2-SAP and the pCMVSport-IRF7H plasmids were kindly provided by Paula Pitha-Rowe (John Hopkins School of Medicine, Baltimore, MD). pA2-SAP contains an A2 IFN-α virus responsive element upstream of a thymidine kinase minimal promoter that directs the expression of secreted alkaline phosphatase (SAP) (36). pEGFP-IRF3 (20), pCMV6-IRF5 (OriGene), and pCMVSport-IRF7H (1) express human IRF3, IRF5, and IRF7, respectively, under the control of a CMV promoter. The plasmids psiRNA-hIRF3, psiRNA-hIRF7, and psiRNA-scramble have a CMV promoter that expresses GFP and an RNA polymerase III promoter that expresses a short-hairpin RNA (shRNA) that induces the silencing of IRF3 (psiRNA-hIRF3) or IRF7 (psiRNA-hIRF7) or that expresses an irrelevant shRNA (psiRNA-scramble) (InvivoGen). Plasmids were purified by isopycnic centrifugation in CsCl gradients.
VSV-GFP-based IFN detection assay.
Monolayers of FRhL2 cells grown in six-well plates were mock treated or treated with 20 μg per ml of poly(I:C) (Sigma) or were mock infected or infected with rotavirus at a multiplicity of infection (MOI) of 3. Afterward, the inoculum was removed, and trypsin-free medium containing 10% FBS was placed on the cells. The medium was collected from the cells at 24 h postinfection (p.i.), a time at which the cells displayed no cytopathic effects. One-fourth of the medium recovered from the cells at 24 h p.i. was transferred onto fresh monolayers of FRhL2 cells grown in six-well plates. After incubation for 24 h, the cells were washed with medium and either mock infected or infected with VSV-GFP at an MOI of 1. The cells were detached from plates at 24 h p.i. by incubation with a trypsin-EDTA mixture (1 part of 0.05% trypsin-0.1% EDTA solution [Quality Biological] to 3 parts of 0.2 mg per ml of EDTA solution [BioWhittaker]). The cells were collected by low-speed centrifugation, suspended in Sorter media (Quality Biological), and subjected to fluorescence-activated cell sorting using a Becton Dickinson FACScan instrument. The data were analyzed by using Flowjo software (Tree Star).
In some cases, specific anti-globulin to human IFN-β (NIAID Reference Reagent G028-501-568) or corresponding control serum globulin (G029-501-568) was included in the culture medium (dilution of 1:10,000). Plaque reduction assays indicated that this antiserum was free of SA11-4F neutralizing antibody (data not shown).
IP.
Monolayers of 293T cells grown in poly-l-lysine coated 10-cm dish plates were transfected with 12 μg in total of equivalent amounts of various plasmid DNAs by using Lipofectamine 2000 (Invitrogen). Where necessary, the pCI vector (Promega) was added to transfection mixtures to adjust total plasmid levels to 12 μg. At 48 h posttransfection, the cells were lysed in immunoprecipitation (IP) buffer (20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 5 mM MgCl2, 0.5% Triton X-100, and Complete protease inhibitor cocktail [Roche]). After sonication, the lysates were clarified by centrifugation at 10,000 × g for 10 min. IP assays were performed on the clarified lysates by using a ProFound Mammalian Co-Immunoprecipitation kit (Pierce). Briefly, the clarified lysates were precleared by incubation with the control gel component for 4 h at 4°C and then incubated with gel-immobilized anti-IRF7 antisera (SC-9083; Santa Cruz) for 16 h at 4°C. Immunoprecipitates were washed four times with the IP buffer and once with IP buffer containing 125 mM NaCl prior to elution.
Western blot assay.
Cells were scraped into cold phosphate-buffered saline containing Complete protease inhibitor cocktail, pelleted, and resuspended in radioimmunoprecipitation assay buffer (150 mM NaCl, 150 mM Tris-HCl [pH 8.0], 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, and Complete protease inhibitor cocktail). After three cycles of freeze-thawing, large debris were removed from the lysates by low-speed centrifugation. Subsequently, the proteins in the samples were resolved by electrophoresis under denaturing-reducing conditions on 10% Tris-glycine gels (Invitrogen). After the proteins were transferred from the gels onto nitrocellulose membranes, the blots were incubated with solutions of phosphate-buffered saline containing 5% milk, 0.1% Tween 20, and either rabbit antisera specific for IRF3 (1:500, Santa Cruz SC-9082), rabbit antisera specific for IRF7 (1:500, Santa Cruz SC-9083), proliferating cell nuclear antigen (PCNA; 1:1,000, Santa Cruz SC-7907), SA11 NSP1 (C19, 1:2,000) (16), mouse anti-human IRF5 monoclonal antibody (1:500, Abcam ab33478), or guinea pig antisera specific for VP6 (1:2,000). Primary antibodies were detected by using appropriate horseradish peroxidase-conjugated secondary antibody (1:10,000). Blots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce) and exposed to BioMax MR film.
Nucleofection of Caco-2 cells.
Caco-2 cells were grown to ca. 80% confluence in 150-cm2 flasks. After being washed, the cells were detached by using a trypsin-EDTA mixture, pelleted, and resuspended in Dulbecco modified Eagle medium containing 10% FBS. The viability and concentration of the cells was determined by using a Beckman Coulter Vi-CELL XR 2.03 analyzer. One million Caco-2 cells were centrifuged for 10 min at 90 × g, resuspended in 100 μl of Cell Line Nucleofector Solution T (Amaxa Biosystems), and mixed with 2 μg of equivalent amounts of various plasmid DNAs. Where necessary, the pCI vector was added to nucleofection mixtures to adjust the total plasmid levels to 2 μg. The DNA was nucleofected into the cells by using setting B-24 of an Amaxa Biosystems Nucleofector I apparatus. After incubation of the cell-DNA suspension for 5 min at room temperature, 1 ml of complete media was added, and the cells were equally distributed in three wells of a 12-well plate.
Detection of SAP activity.
At 24 h after nucleofection, Caco-2 cells containing the pA2-SAP plasmid were mock treated, treated with 20 μg per ml of poly(I:C), or infected with rotavirus at an MOI of 3. After 10 h, the cell supernatants were heated to 65°C for 20 min to selectively inactivate endogenous alkaline phosphatase (SAP is heat stable). SAP activity was detected by using a Phospha-Light SEAP reporter gene assay system (Applied Biosystems) and a Wallac Trilux 1450 Microbeta counter.
RESULTS
Loss of NSP1 function is associated with restriction of rotavirus growth in some cell lines.
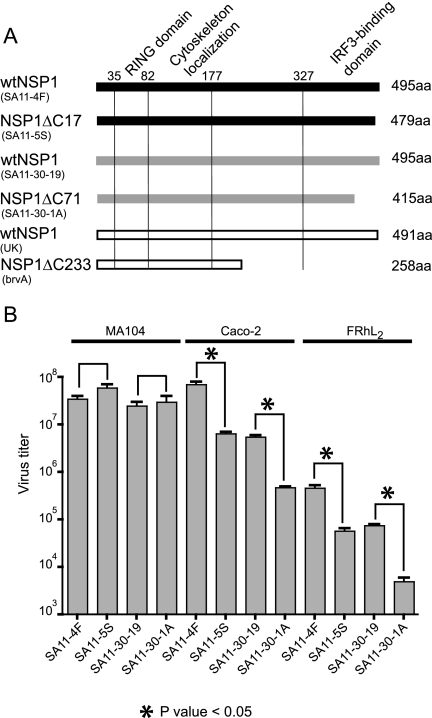
Rotavirus variants have been isolated that encode C-truncated forms of NSP1 (e.g., SA11-5S, SA11-30-19, brvA) (Fig. 1 A) (26). Unlike wt NSP1, the C-truncated NSP1 product of these variants is defective in inducing the degradation of IRF3 (4). Characteristically, such variants (NSP1-defective [NSP1-def] rotaviruses) have plaque phenotypes that are smaller than counterpart rotaviruses encoding wt NSP1 (wt rotaviruses) (26). A comparison of NSP1-def rotaviruses (SA11-5S and SA11-30-1A) and their wt counterparts (SA11-4F and SA11-30-19, respectively) also reveals that these viruses grow to different titers in vitro depending on host cell type (Fig. 1B). Most notably, while NSP1-def rotaviruses and wt rotaviruses grow to similar titers in the MA104 monkey-kidney cell line (26), wt rotaviruses grow to titers at least 1 log greater than NSP1-def rotaviruses in the Caco-2 human colon cell line and in the FRhL2 rhesus lung diploid cell line.
FIG. 1.
In vitro growth of rotaviruses encoding wt and defective forms of NSP1. (A) Representation of the NSP1 forms made by different rotavirus isolates (strain name in parentheses), including the location of putative RING and IRF3-binding domains, and cytoskeleton localization signal. The variants SA11-5S, SA11-30-1A, and brvA encode C-truncated forms of NSP1 and are sister strains of the wt viruses SA11-4F (SA11g4“O”), SA11-30-19, and bovine UK (26, 32a). GenBank accession numbers for the gene 5 sequences: AAK14071 (SA11-4F), AAK14072 (SA11-5S), AAK14069 (SA11-30-19), AAK14070 (SA11-30-1A), AAA18011 (UK), and AAA18012 (brvA). (B) Virus titers produced by infecting the indicated cell lines at an MOI of 3 with rotavirus strains encoding wt and defective NSP1. Cells were harvest at times of maximum virus yield (24 h p.i. for MA104 cells and 48 h p.i. for Caco-2 and FRhL2 cells). Titers were determined by plaque assay on MA104 cells and represent the averaged values obtained for replicate parallel infections.
Expression of IFN is suppressed in FRhL2 cells infected with wt rotaviruses but not in cells infected with NSP1-def rotaviruses.
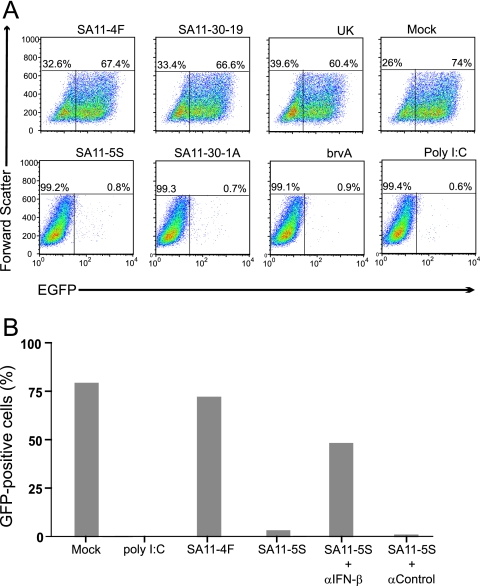
To understand the importance of NSP1 on IFN expression and, in turn, on rotavirus growth, we used a VSV-GFP-based assay system to compare the levels of IFN produced in FRhL2 cells infected with wt rotaviruses and NSP1-def rotaviruses. Similar to the Newcastle disease virus-based assay system (23), the VSV-GFP system relies on the growth sensitivity of the GFP-expressing reporter virus, VSV-GFP, to IFN as an analytical tool for detecting whether infection with a test virus (in this case, rotavirus) results in IFN expression. In our assays, monolayers of FRhL2 cells were mock infected or were infected with rotaviruses encoding wt NSP1 or C-truncated NSP1 for 1 h. Afterward, the inoculum was replaced with trypsin-free culture media containing 10% FBS, conditions inhibiting the protease-dependent activation of any released progeny virus. The culture medium was collected from the cells at 24 h p.i., a time at which the cells displayed no obvious cytopathic effect and that precedes the time of maximum virus yield in FRhL2 cells by 24 h (data not shown). Portions of the collected media were transferred onto fresh monolayers of FRhL2 cells, which were infected 24 h later with VSV-GFP. Flow cytometry showed that the percentage of GFP-positive cells in FRhL2 monolayers treated with media from wt rotavirus-infected cells (SA11-4F, SA11-30-19, and UK) reached levels nearly as high as that reached by monolayers treated with media from mock-infected cells (Fig. 2 A). In contrast, the percentage of GFP-positive cells in monolayers treated with media from NSP1-def rotavirus-infected cells (SA11-5S, SA11-30-1A, and brvA) was low, approximating the level reached by cells treated with poly(I:C) to induce IFN expression (Fig. 2A). These results show that the efficient growth of the IFN-sensitive VSV-GFP virus is correlated with the expression of wt NSP1, and not truncated NSP1, by rotavirus.
FIG. 2.
Induction of IFN associated with rotavirus infection. (A) Culture media from FRhL2 cells that were mock infected or infected with the indicated strains of rotavirus or treated with poly(I:C) were transferred onto fresh FRhL2 monolayers. After 24 h, the monolayers were infected with VSV-GFP, an IFN-sensitive virus expressing GFP. Flow cytometry was used to analyze cells for GFP fluorescence and cell size. Plots of the data indicate the percentage of cells expressing GFP (upper right quadrant) beyond the background level of fluorescence associated with uninfected control cells. (B) Same as in panel A, except that in some cases, neutralizing IFN-β antisera and control antisera were added to the culture media of SA11-5S-infected cells prior to transfer to fresh FRhL2 monolayers. The monolayers were later infected with VSV-GFP, and the percentage of cells expressing GFP was determined by flow cytometry.
To determine whether rotavirus encoding wt NSP1 promoted the growth of VSV-GFP by subverting IFN expression, FRhL2 cells were mock infected or were infected with wt rotavirus (SA11-4F) or NSP1-def rotavirus (SA11-5S). Culture media collected from these cells were either directly transferred onto fresh FRhL2 monolayers or transferred after first mixing them with IFN-β neutralizing antisera or control antisera lacking such neutralizing antibodies. After infection with VSV-GFP, the percentage of GFP-positive cells in the monolayers was determined by flow cytometry (Fig. 2B). Consistent with the results presented above (Fig. 2A), a high percentage of GFP-positive cells was detected in monolayers treated with media from mock-infected cells and from cells infected with wt rotavirus (SA11-4F). Although culture media from cells infected with NSP1-def rotavirus (SA11-5S) was associated with only a low percentage of GFP-positive cells in FRhL2 monolayers (Fig. 2B), the addition of neutralizing IFN-β antisera to this media increased the percentage of GFP-positive cells to a point that it was ∼60% of monolayers treated with media from mock-infected cells. These data indicate that wt rotaviruses promote the growth of VSV-GFP through a mechanism mediated by the capacity of wt NSP1 to suppress IFN-β expression.
wt NSP1 expression is correlated with reduced levels of IRF7 accumulation.
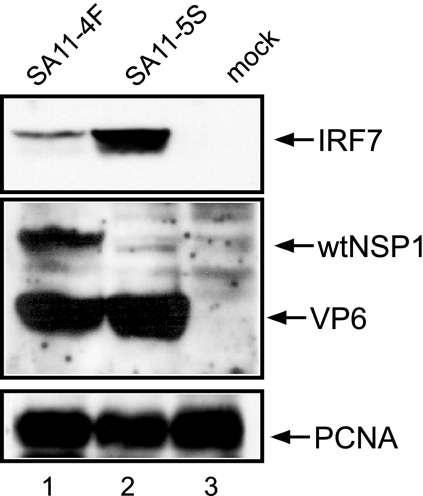
Although IRF3 contributes to type I IFN induction, its effect is largely targeted toward inducing IFN-β expression. In comparison, IRF7 has broader effects since this factor not only stimulates further IFN-β expression but also triggers the expression of IFN-α. The importance of IRF7 to the IFN signaling pathway raises the possibility that viruses may have evolved mechanisms of inactivating IRF7 in addition to IRF3. Because IRF3 and IRF7 sequences show extensive similarity, we postulated that rotaviruses might direct the degradation of IRF7 through the same NSP1-dependent mechanism that it uses to degrade IRF3. To probe this possibility, we mock infected or infected Caco-2 cells with wt rotavirus (SA11-4F) or NSP1-def rotavirus (SA11-5S) and then analyzed the levels of IRF7 in the cells at 18 h p.i. by using a Western blot assay. The cells were also examined for wt NSP1, the viral capsid protein VP6, and PCNA (Fig. 3). The results showed that, in contrast to mock-infected cells, in which no IRF7 was detected, high levels of IRF7 were present in cells infected with the NSP1-def virus. Thus, as demonstrated using SA11-5S, rotavirus infection has the potential for strongly upregulating expression of IRF7, the primary inducer of type I IFN. However, as demonstrated using SA11-4F, rotaviruses expressing wt NSP1 subvert pathways that lead to IRF7 induction.
FIG. 3.
IRF7 levels in rotavirus-infected cells. Lysates prepared at 18 h p.i. from Caco-2 cells infected with SA11-4F or SA11-5S were examined for IRF7, full-length NSP1, VP6, and PCNA by Western blot assay. The SA11 NSP1(C19) antiserum was prepared against a peptide representing the last 19 amino acids of wt NSP1 (15). As a result, the antiserum does not recognize the C-truncated NSP1 (NSP1ΔC17) encoded by SA11-5S.
NSP1 expression induces proteasome-mediated degradation of IRF7.
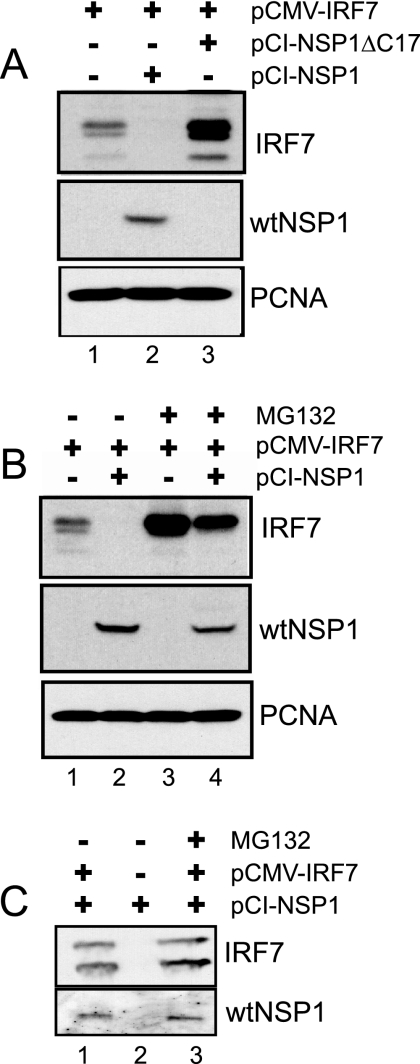
The decreased levels of IRF7 in wt rotavirus-infected cells may result from two alternative actions of NSP1. (i) NSP1 could induce the proteasome-mediated degradation of IRF7, as it does IRF3, or (ii) by inducing IRF3 degradation, NSP1 could suppress the expression of IFN-β, an activator of the IRF7 gene. To address these possibilities, 293T cells were transfected with vectors encoding IRF7 (pCMV-IRF7) and either wt NSP1 (pCI-NSP1) or the defective form of NSP1 made by SA11-5S (pCI-NSP1ΔC17). Lysates prepared from the cells at 48 h posttransfection were analyzed by Western blot assay for IRF7, wt NSP1, and PCNA. The results show that coexpression with wt NSP1 reduced IRF7 accumulation to barely detectable levels (Fig. 4 A). In contrast, IRF7 accumulated to levels higher when coexpressed with defective NSP1 than occurred upon expression of IRF7 by itself. These results indicate that wt NSP1, and not the defective form of NSP1, induces the degradation of IRF7. The mechanism by which the defective form of NSP1 causes an increase in IRF7 levels is not clear, but the effect is reproducible. One possible explanation is that the defective protein interacts with and ties up cellular proteins involved in ubiquitination and/or proteasome function, thereby inhibiting the degradation of IRF7, a protein typically with a short half-life (27).
FIG. 4.
Proteasome-dependent effect of NSP1 on IRF7 levels. (A) 293T cells were cotransfected with equal amounts of pCMV-IRF7 and pCI-NSP1ΔC17 or pCI-NSP1. Where necessary, the empty pCI vector was also added to normalize amounts of plasmid DNA contained in transfection mixtures. Lysates prepared from the cells were examined at 48 h posttransfection for IRF7, wt NSP1, and PCNA by Western blot assay. (B) Same as in panel A, except that at 48 h posttransfection the culture media over some monolayers was adjusted to 10 μM MG132, an inhibitor of proteasome function. Lysates were prepared 12 h later and analyzed by Western blot assay. (C) Same as in panel B, except that immunoprecipitates were recovered by using IRF7-antibody linked to agarose beads from cellular lysates prepared at 24 posttransfection. The immunoprecipitates were analyzed for IRF7 and wt NSP1 by Western blot assay.
The potential involvement of the proteasomal degradation system in the failure of IRF7 to accumulate in cells containing wt NSP1 was evaluated using the proteasome inhibitor MG132. The analysis showed that the addition of MG132 to 293T cells coexpressing IRF7 and wt NSP1 resulted in the accumulation of significant levels of IRF7 (Fig. 4B, lane 4). In contrast, cells expressing these same proteins but in the absence of MG132 contained barely detectable levels of IRF7 (lane 2). We conclude from these data that wt NSP1 induces the proteasome-mediated degradation of IRF7 and thus likely uses the same mechanism to degrade IRF7 that it does IRF3.
To address the possibility that the effect of NSP1 on IRF7 involved a direct interaction between the proteins, wt NSP1 was expressed alone or in combination with IRF7 in 293T cells maintained in the presence or absence of MG132. Clarified lysates were prepared from the cells at 24 h posttransfection, a point prior to the 48 h posttransfection time when IRF7 is no longer detectable in cells coexpressing wt NSP1 and IRF7 (Fig. 4A). The lysates were incubated with IRF7-specific antisera, and the immunoprecipitates recovered from the samples were analyzed for IRF7 and wt NSP1 by Western blot assay. The analysis showed that wt NSP1 was present in the IRF7 immunoprecipitates regardless of whether or not the coexpressing cells were maintained in MG132 (Fig. 4C, lanes 1 and 3). Wild-type NSP1 was not recovered in immunoprecipitates prepared from cell lysates not expressing IRF7 (lane 2). These results indicate that the mechanism by which NSP1 induces IRF7 degradation may involve the physical interaction of the proteins. The similarity in amounts of IRF7 recovered from cellular lysates containing NSP1 and IRF7, with or without MG132 (Fig. 4C, lanes 1 and 3), results from the saturation of the IRF7 antibody added to the immunoprecipitation assays.
IRF3 and IRF7 expression restricts the growth of NSP1-defective rotaviruses.
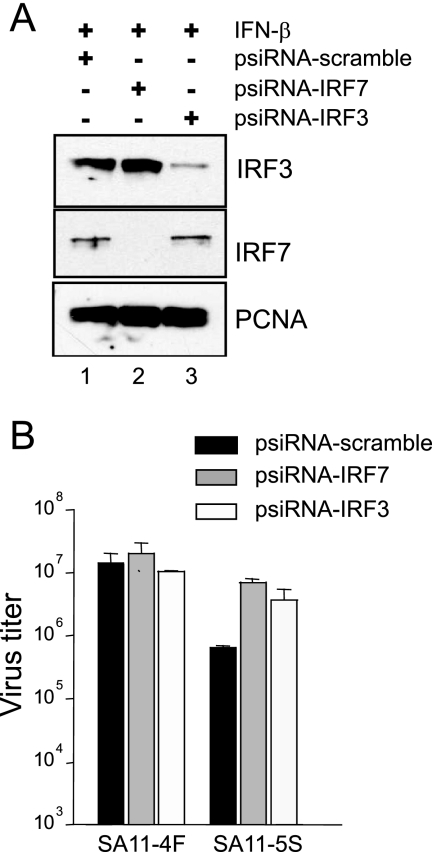
Our results suggest that wt rotaviruses grow to higher titers than NSP1-def rotaviruses in Caco-2 and FRhL2 cell lines because wt NSP1, unlike defective NSP1, can subvert IFN production by inducing IRF3 and IRF7 degradation. To further validate this possibility, we analyzed rotavirus growth in Caco-2 cells in which the expression of IRF3 or IRF7 was suppressed by treatment with appropriate gene-specific small interfering RNAs (siRNAs). As shown in Fig. 5 A, the introduction of vectors producing IRF7- and IRF3-specific siRNAs into Caco-2 cells was effective in suppressing the expression of IRF7 and IRF3, respectively, upon subsequent exposure of the cells to the triggering factor, IFN-β.
FIG. 5.
Importance of IRF3 and IRF7 expression on rotavirus growth. (A) Vectors producing IRF7- or IRF3-specific siRNAs (psiRNA-IRF7 and -IRF3, respectively) or a control irrelevant siRNA (psiRNA-scramble) were nucleofected into Caco-2 cells. After incubation with 103 U of IFN-β per ml for 10 h, the cells were analyzed for IRF3, IRF7, and PCNA by Western blot assay. (B) Vectors producing IRF7- or IRF3-specific siRNAs or a control irrelevant siRNA were nucleofected into Caco-2 cells. After 24 h, the cells were infected with SA11-4F or SA11-5S at an MOI of 3. The infected Caco-2 cells were harvested at 48 h p.i., a time by which SA11-4F has reached maximum titers in this cell line. Virus titers were determined by plaque assay on MA104 cells. The titers shown reflect the averaged results of two separate experiments, with plaque assays performed in duplicate.
Analysis of Caco-2 cells expressing either IRF3- or IRF7-specific siRNAs showed that the cells supported growth of wt rotaviruses to titers indistinguishable from that reached in cells expressing a control (scramble) siRNA (Fig. 5B). In contrast, Caco-2 cells expressing either IRF3- or IRF7-specific siRNAs supported the growth of NSP1-def rotaviruses to a titer close to 1 log higher than cells expressing the control siRNA. These results indicate that both IRF3 and IRF7 can trigger antiviral effects in rotavirus-infected cells that limit virus growth and that wt NSP1, but not defective NSP1, has activities that help the virus overcome these limitations. The results also indicate that the growth restriction observed for NSP1-def rotaviruses in Caco-2 cells can be assigned, at least in part, to the inability of these viruses to induce the degradation of IRF3 or IRF7.
Effect of NSP1 on the transactivation of IRF7-dependent promoter elements.
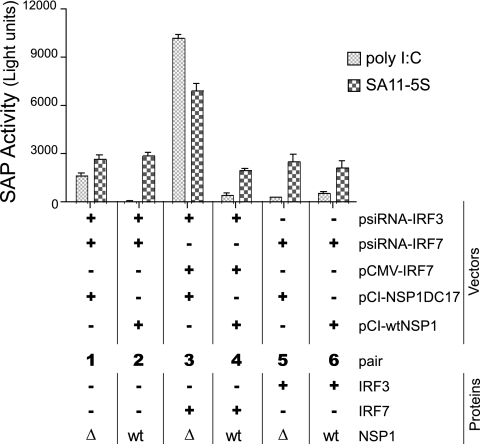
Activated IRF7 dimerizes and translocates to the nuclei, where it becomes part of a multiprotein complex that interacts with the promoter of the IFN-α gene, triggering its expression (29). To probe the effect of NSP1 on the activation of the IFN-α gene, Caco-2 cells were transfected with pA2-SAP, a reporter plasmid that contains an IRF7-activated promoter element that controls the expression of SAP. Along with pA2-SAP, the cells were cotransfected with various combinations of vectors, some producing siRNAs that suppressed the expression of endogenous IRF3 (psiRNA-hIRF3) or IRF7 (psiRNA-hIRF7), one directing the expression of exogenous IRF7 (pCMVSport-IRF7H), and others directing the expression of wt NSP1 (pCI-NSP1) or C-truncated NSP1 (pCI-NSP1ΔC17) (Fig. 6). To stimulate IFN expression, the transfected cells were subsequently treated with poly(I:C) or infected with NSP1-def rotavirus (SA11-5S) and then assayed 10 h later for levels of SAP. The results showed that the activity of the IFN-α promoter element was greatest in stimulated cells expressing both IRF7 and NSP1ΔC17 (Fig. 6, bar pair 3). In contrast, expression of IRF7 with wt NSP1 (bar pair 4) reduced the activity of the IFN-α promoter element to levels similar to those in stimulated cells that lacked IRF3 and IRF7 due to siRNA knockdown (bar pairs 1 and 2). Thus, wt NSP1 is effective in preventing the activation of the IRF7-dependent IFN-α promoter.
FIG. 6.
Effect of NSP1 on the activation of the IFN-α promoter element. Caco-2 cells were nucleofected with combinations of vectors expressing IRF3- or IRF7-specific siRNAs, wt NSP1 or NSP1ΔC17, IRF7, and pA2-SAP, a reporter plasmid containing an IFN-α response element that drives the expression of SAP. After 24 h, the cells were treated with poly(I:C) or infected with SA11-5S to stimulate IFN expression. Subsequently, cell supernatants were analyzed for the presence of SAP activity. Relevant proteins present in the cells are indicated below the row defining bar pairs 1 to 6. wt, wild-type NSP1; Δ, NSP1ΔC17.
IRF3 expression did not increase the activity of the IFN-α promoter element in stimulated cells expressing wt NSP1 or NSP1ΔC17 (Fig. 6, bar pairs 5 and 6) beyond that of cells that lacked both IRF3 and IRF7 (pair 1 and 2). This result is expected given that activation of the IFN-α promoter is driven by IRF7 and not by IRF3.
Basal levels of IFN-α promoter activity were detected even in stimulated cells transfected with vectors producing siRNAs designed to knockdown IRF3 and IRF7 expression (Fig. 6, bar pair 1). This is mostly likely due to transfection efficiencies being <100%, leaving some percentage of the cells within the population capable of producing IRF3 and IRF7 upon treatment with poly(I:C) or infection with SA11-5S. We also observed that infection of cells with SA11-5S generally resulted in greater activity of the IFN-α promoter element than occurred upon treating the cells with poly(I:C) (Fig. 6, bar pairs 1, 2, 5, and 6). This may best be explained by considering that poly(I:C) is probably only able to induce IFN expression through a single dsRNA-detector, TLR3, whereas viral infection is likely to induce IFN expression through multiple dsRNA detectors and receptors (e.g., TLR3, RIG-I, and MDA5), thereby making viral infection a stronger stimulus of IRF7 expression and SAP activity.
NSP1-dependent degradation of multiple members of the IRF family, including IRF5.
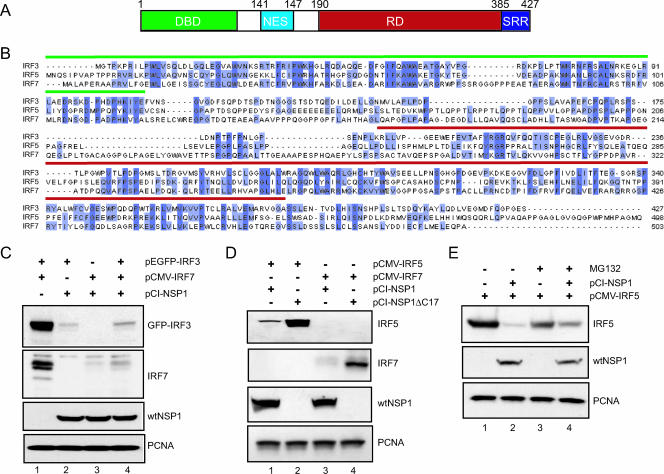
Cotransfection of 293T cells with vectors expressing IRF3 (pEGFP-IRF3), IRF7 (pCMV-IRF7), and wt NSP1 (pCI-NSP1) indicates that NSP1 can simultaneously and, without obvious preference for one factor over the other, induce the intracellular degradation of IRF3 and IRF7 (Fig. 7 C). Given the overall similarity in the organization and sequence of the members of the IRF family (Fig. 7A and B) and the fact that NSP1 has been shown to target two of the members of the family, it could be reasoned that NSP1 is a broad-spectrum antagonist of the innate immune system, targeting not just IRF3 and IRF7 but also multiple IRF members. To examine this hypothesis, we tested whether wt NSP1 could induce the degradation of IRF5, an IRF family member that is constitutively expressed in B lymphocytes and dendritic cells and that plays a role not only in the expression of type I IFN but also in cell cycle regulation and apoptosis (2, 3). As shown in Fig. 7D, cotransfection of 293T cells with vectors encoding IRF5 and wt NSP1 (pCI-NSP1) or C-truncated NSP1 (pCI-NSP1ΔC17) showed that the expression of wt NSP1, relative to C-truncated NSP1, was correlated with decreased IRF5 accumulation. The effect of wt versus truncated NSP1 on IRF5 levels mirrors the effect of these NSP1 species on IRF7 and IRF3 levels (Fig. 7C), indicating that NSP1 subverts the innate immune response by targeting multiple members of the IRF family for degradation. Addition of the proteasome inhibitor, MG132, to 293T cells coexpressing wt NSP1 and IRF5 resulted in a partial recovery in IRF5 accumulation in comparison to similar cells not treated with MG132 (Fig. 7E, lanes 2 and 4). This result implies that NSP1 uses the same proteasome-dependent mechanism to induce the degradation of IRF5 that it uses in inducing the degradation of IRF3 and IRF7.
FIG. 7.
Members of the IRF family as targets of NSP1. (A) Schematic representation of the structural domains of the IRF3 protein: DBD DNA-binding domain; NES, nuclear export signal; RD, regulatory domain; and SRR, serine-rich region. (B) Sequence alignment of human IRF3 (accession number Q14653), IRF5 (Q92985), and IRF7 (NM_032643). The DNA-binding domains and regulatory domains are overlined in green and red, respectively. Conserved residues are highlighted in dark blue. Residues present in two of the three sequences are shown in light blue. (C) EGFP-IRF3 and IRF7 were transiently expressed individually or in combination with NSP1 in 293T cells by transfection with the indicated plasmids. The levels of EGFP-IRF3, IRF7, wt NSP1, and PCNA were determined by Western blot assay. After 48 h, the cells were examined for levels of IRF3, IRF7, wt NSP1, and PCNA by Western blot assay. (D) IRF5 and IRF7 were individually expressed with wt NSP1 or NSP1ΔC17 in 293T cells by transfection with the indicated plasmids. After 48 h, the cells were examined for levels of IRF5, IRF7, wt NSP1, and PCNA by Western blot assay. (E) 293T cells were transfected with pCMV-IRF5 alone or in combination with pCI-NSP1. At 48 h posttransfection, the culture medium was adjusted to 10 μM MG132. Cell lysates were prepared 12 h later and then analyzed by Western blot assay for IRF5, wt NSP1, and PCNA.
DISCUSSION
Serial passage of rotavirus at a high MOI in cell culture relieves the pressure on the virus to spread efficiently from one cell to another. Under such passage conditions, variants appear in the rotavirus population with aberrant genotypes due to intragenic sequence rearrangements occurring within the gene 5 RNA (26). Such rearrangements typically interrupt the NSP1 open reading frame in the gene 5 RNA, creating rotaviruses that produce C-truncated forms of NSP1. Previous studies have shown that these NSP1-def rotaviruses have a small plaque phenotype in comparison to their wt rotavirus counterparts (13). As described here, we have also found that rotaviruses encoding defective NSP1 grow to lower titers in some cell types (FRhL2 and Caco-2) than rotaviruses encoding wt NSP1. Based on IFN detection assays performed with VSV-GFP, the relatively poor growth characteristics of NSP1-def rotaviruses was linked to the failure of these variant viruses to prevent IFN expression. These results provide direct experimental evidence that NSP1 plays a critical role in preventing IFN expression in rotavirus-infected cell and that this function is dependent upon the C-terminal region of NSP1.
In an earlier study, we showed that infection with wt rotavirus leads to a rapid and nearly complete loss of the constitutively expressed transcription factor IRF3 (4). Here we showed that infection with wt rotavirus also prevents the accumulation of the inducible transcription factor IRF7. Given the role of IRF3 as the initial trigger of IFN-β expression and IRF7 as the “master regulator” of type I IFN (15), infection with wt rotavirus would thus interfere with the ability of the host to activate IFN-dependent autocrine and paracrine pathways required to establish an antiviral state. Our analysis indicates that wt NSP1 can interact with both IRF3 and IRF7 in the infected cell and that this interaction results in the degradation of these factors through a proteasome-dependent process. Unlike wt NSP1, C-truncated forms of NSP1 (e.g., NSP1ΔC17) cannot induce the degradation of IRF3 and IRF7. This inactivity of the C-truncated forms is consistent with the results of previous yeast two-hybrid experiments, which indicated that the C terminus of NSP1 is required for the interaction with IRF3 (11). Our analysis indicates that NSP1 is equally effective in inducing the degradation of IRF3 and IRF7. In toto these data indicate that NSP1 uses a similar mechanism to induce the turnover of IRF3 and IRF7.
In most mammalian cells, activation of IRF3 leads to the expression of IFN-β, which in turn induces the expression of IRF7. By targeting IRF3 for degradation, NSP1 removes an upstream transcription factor needed for IRF7 expression, thereby subverting the production of type I IFN. Thus, the low levels of IRF7 present in some rotavirus-infected cells (e.g., Caco-2) likely reflects not only the ability of NSP1 to induce IRF7 degradation but also the ability of NSP1 to induce the degradation of IRF3. The dual mechanism of NSP1 in subverting IRF7 function may be crucial for successful virus replication in the host at later stages of infection, where the virus may be challenged with replicating in cells that have transitioned from a naive to an antiviral status due to exposure to cytokines or debris from neighboring infected cells. In such activated cells, successful rotavirus replication may rely largely on the capacity of NSP1 to induce the degradation of preexisting IRF7. For rotaviruses encoding defective NSP1, infection results not only in the induction of IRF3 and IRF7 but also in the activation of IFN-β- and IFN-α-dependent promoter elements (4) (Fig. 6). Hence, infection with NSP1-def rotaviruses leads to the expression of both IFN-β and IFN-α and, as a consequence, activation of the complete IFN signaling pathway.
Recent reports have established that rotavirus infection can lead to a viremic phase in the host, with a number of organs becoming positive for viral antigen and RNA (8, 9). The spread of rotavirus from the gut to the circulatory system may be mediated by the trafficking of dendritic cells and macrophages, cell types indicated to be capable of supporting rotavirus replication (24). Unlike most cells, IRF7 is maintained at high levels in dendritic cells and macrophages due to its constitutive expression (1). The ability of NSP1 to target IRF7 for degradation may be essential for the growth of rotavirus in such trafficking cells and therefore for the movement of the virus across the gut barrier to produce a viremia.
Comparative analysis of the NSP1 sequences of rotaviruses recovered from different animal species shows evidence of a highly conserved N-terminal RING finger-like motif. However, the C-terminal half of the protein, including the IRF-interactive domain, is remarkably poorly conserved. The fact that the NSP1 sequence is much more conserved among virus strains originating from common animal species suggests that the protein has evolved such that it may be somewhat species specific in its function. Hence, different varieties of NSP1 may show considerable variability in the capacity to induce the degradation of any one IRF, depending on how close that IRF comes to the natural target of the NSP1 protein. Such species specificity of NSP1 function may explain the variation seen in the ability of different rotaviruses to grow and spread in animal model systems.
The observation that NSP1 can induce the degradation of IRF3 and IRF7 led us to investigate the possibility that NSP1 targeted multiple members of the IRF family for degradation. Indeed, our analysis indicates that NSP1 can induce the degradation of IRF5, a transcription factor that is constitutively expressed in dendritic cells and B lymphocytes, that stimulates the expression of subtypes of IFN-α, and that is regulated by type I IFN (2, 22). Evidence exists that IRF5 also has roles in stimulating the expression of cytokines and chemokines that cause the recruitment of T lymphocytes, regulating the cell cycle, triggering apoptosis, and acting as a p53-controlled tumor suppressor (3, 7). The ability of NSP1 to induce the degradation of IRF3, IRF5, and IRF7 suggests that NSP1 recognizes a common element in all three factors, with the likeliest being the conserved N-terminal DNA-binding domain or one or more shared islands of sequence conservation located in its regulatory domain (Fig. 7A).
Our results provide evidence that rotavirus NSP1 represents a broad-spectrum antagonist of the function of IRFs. Based on these results, we can predict that the origin and nature of NSP1 expressed by any particular vaccine strain of rotavirus may have a bearing on the behavior and efficacy of the vaccine within the vaccinee. Instead of relying on reassortants formed by animal and human rotaviruses to generate vaccine strains, mutation of the NSP1 gene of human rotavirus strains may give rise to attenuated vaccine candidates that are more suitable in inducing relevant and broadly protective B- and T-cell responses in the vaccinee (18).
Acknowledgments
We thank John Hiscott (McGill University, Montreal, Quebec, Canada) and Paula Pitha-Rowe (Johns Hopkins School of Medicine, Baltimore, MD).
This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Au, W. C., P. A. Moore, D. W. LaFleur, B. Tombal, and P. M. Pitha. 1998. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J. Biol. Chem. 273:29210-29217. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, B. J., M. J. Kellum, K. E. Pinder, J. A. Frisancho, and P. M. Pitha. 2003. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 63:6424-6431. [PubMed] [Google Scholar]
- 4.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowie, A. G., and I. R. Haga. 2005. The role of Toll-like receptors in the host response to viruses. Mol. Immunol. 42:859-867. [DOI] [PubMed] [Google Scholar]
- 6.Cave, D. R., F. M. Hendrickson, and A. S. Huang. 1985. Defective interfering virus particles modulate virulence. J. Virol. 55:366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, T. F., S. Brzostek, O. Ando, S. Van Scoy, K. P. Kumar, and N. C. Reich. 2006. Differential activation of IFN regulatory factor (IRF)-3 and IRF-5 transcription factors during viral infection. J. Immunol. 176:7462-7470. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, S. E., D. G. Patel, E. Cheng, Z. Berkova, J. M. Hyser, M. Ciarlet, M. J. Finegold, M. E. Conner, and M. K. Estes. 2006. Rotavirus viremia and extraintestinal viral infection in the neonatal rat model. J. Virol. 80:4820-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenaux, M., M. A. Cuadras, N. Feng, M. Jaimes, and H. B. Greenberg. 2006. Extraintestinal spread and replication of a homologous EC rotavirus strain and a heterologous rhesus rotavirus in BALB/c mice. J. Virol. 80:5219-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firth, M. A., P. E. Shewen, and D. C. Hodgins. 2005. Passive and active components of neonatal innate immune defenses. Anim. Health Res. Rev. 6:143-158. [DOI] [PubMed] [Google Scholar]
- 11.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. L. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hata, N., M. Sato, A. Takaoka, M. Asagiri, N. Tanaka, and T. Taniguchi. 2001. Constitutive IFN-alpha/beta signal for efficient IFN-alpha/beta gene induction by virus. Biochem. Biophys. Res. Commun. 285:518-525. [DOI] [PubMed] [Google Scholar]
- 14.Honda, K., and T. Taniguchi. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644-658. [DOI] [PubMed] [Google Scholar]
- 15.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 16.Hua, J., X. Chen, and J. T. Patton. 1994. Deletion mapping of the rotavirus metalloprotein NS53 (NSP1): the conserved cysteine-rich region is essential for virus-specific RNA binding. J. Virol. 68:3990-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 18.Kearney, K., D. Chen, Z. F. Taraporewala, P. Vende, Y. Hoshino, M. A. Tortorici, M. Barro, and J. T. Patton. 2004. Cell-line-induced mutation of the rotavirus genome alters expression of an IRF3-interacting protein. EMBO J. 23:4072-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Bonnardiere, C., C. de Vaureix, R. L'Haridon, and R. Scherrer. 1980. Weak susceptibility of rotavirus to bovine interferon in calf kidney cells. Arch. Virol. 64:167-170. [DOI] [PubMed] [Google Scholar]
- 20.Leung, A. K., J. D. Kellner, and H. D. Davies. 2005. Rotavirus gastroenteritis. Adv. Ther. 22:476-487. [DOI] [PubMed] [Google Scholar]
- 21.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancl, M. E., G. Hu, N. Sangster-Guity, S. L. Olshalsky, K. Hoops, P. Fitzgerald-Bocarsly, P. M. Pitha, K. Pinder, and B. J. Barnes. 2005. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J. Biol. Chem. 280:21078-21090. [DOI] [PubMed] [Google Scholar]
- 23.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narvaez, C. F., J. Angel, and M. A. Franco. 2005. Interaction of rotavirus with human myeloid dendritic cells. J. Virol. 79:14526-14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill, L. A. 2006. How Toll-like receptors signal: what we know and what we don't know. Curr. Opin. Immunol. 18:3-9. [DOI] [PubMed] [Google Scholar]
- 26.Patton, J. T., Z. Taraporewala, D. Chen, V. Chizhikov, M. Jones, A. Elhelu, M. Collins, K. Kearney, M. Wagner, Y. Hoshino, and V. Gouvea. 2001. Effect of intragenic rearrangement and changes in the 3′ consensus sequence on NSP1 expression and rotavirus replication. J. Virol. 75:2076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prakash, A., and D. E. Levy. 2006. Regulation of IRF7 through cell type-specific protein stability. Biochem. Biophys. Res. Commun. 342:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saif, L. J., and F. M. Fernandez. 1996. Group A rotavirus veterinary vaccines. J. Infect. Dis. 174(Suppl. 1):S98-S106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 30.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 31.Seth, R. B., L. Sun, and Z. J. Chen. 2006. Antiviral innate immunity pathways. Cell Res. 16:141-147. [DOI] [PubMed] [Google Scholar]
- 32.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Small, C., M. Barro, T. L. Brown, and J. T. Patton. 2007. Genome heterogeneity of SA11 rotavirus due to reassortment with “O” agent. Virology 359:415-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojdl, D. F., B. D. Lichty, B. R. tenOever, J. M. Paterson, A. T. Power, S. Knowles, R. Marius, J. Reynard, L. Poliquin, H. Atkins, E. G. Brown, R. K. Durbin, J. E. Durbin, J. Hiscott, and J. C. Bell. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263-275. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 35.Vanden Broecke, C., A. Schwers, L. Dagenais, A. Goossens, M. Maenhoudt, P. P. Pastoret, and J. Werenne. 1984. Interferon response in colostrum-deprived newborn calves infected with bovine rotavirus: its possible role in the control of the pathogenicity. Ann. Rech. Vet. 15:29-34. [PubMed] [Google Scholar]
- 36.Yeow, W. S., W. C. Au, Y. T. Juang, C. D. Fields, C. L. Dent, D. R. Gewert, and P. M. Pitha. 2000. Reconstitution of virus-mediated expression of interferon alpha genes in human fibroblast cells by ectopic interferon regulatory factor-7. J. Biol. Chem. 275:6313-6320. [DOI] [PubMed] [Google Scholar]