Abstract
Recombinant adenovirus serotype 5 (rAd5) vector-based vaccines are currently being developed for both human immunodeficiency virus type 1 and other pathogens. The potential limitations associated with rAd5 vectors, however, have led to the construction of novel rAd vectors derived from rare Ad serotypes. Several rare serotype rAd vectors have already been described, but a detailed comparison of multiple rAd vectors from subgroups B and D has not previously been reported. Such a comparison is critical for selecting optimal rAd vectors for advancement into clinical trials. Here we describe the construction of three novel rAd vector systems from Ad26, Ad48, and Ad50. We report comparative seroprevalence and immunogenicity studies involving rAd11, rAd35, and rAd50 vectors from subgroup B; rAd26, rAd48, and rAd49 vectors from subgroup D; and rAd5 vectors from subgroup C. All six rAd vectors from subgroups B and D exhibited low seroprevalence in a cohort of 200 individuals from sub-Saharan Africa, and they elicited Gag-specific cellular immune responses in mice both with and without preexisting anti-Ad5 immunity. The rAd vectors from subgroup D were also evaluated using rhesus monkeys and were shown to be immunogenic after a single injection. The rAd26 vectors proved the most immunogenic among the rare serotype rAd vectors studied, although all rare serotype rAd vectors were still less potent than rAd5 vectors in the absence of anti-Ad5 immunity. These studies substantially expand the portfolio of rare serotype rAd vectors that may prove useful as vaccine vectors for the developing world.
Replication-incompetent, recombinant adenovirus serotype 5 (rAd5) vectors have been demonstrated to elicit potent antigen-specific cellular immune responses in both preclinical and clinical studies (2, 7, 25, 26, 28). In particular, rAd5 vector-based vaccines for human immunodeficiency virus type 1 (HIV-1) and other pathogens are currently being advanced into large-scale clinical studies. However, the immunogenicity and clinical utility of rAd5 vectors may be limited by the high prevalence of preexisting anti-Ad5 immunity in human populations, particularly in the developing world (13, 19, 25, 30, 31, 33, 35). Preexisting anti-Ad5 immunity has already been shown to suppress the immunogenicity of rAd5 vector-based vaccines in mice (3, 14, 15, 22, 30, 36), rhesus monkeys (6, 22), and humans (7, 25). Moreover, immunization with rAd5 vectors generates potent antivector immunity that substantially inhibits the utility of homologous vector readministration (3, 6, 24).
The generation of novel rAd vectors that circumvent anti-Ad5 immunity is therefore an important research priority. Strategies that are currently being explored include constructing hexon-chimeric rAd5 vectors (22), generating rAd vectors from nonhuman Ad serotypes (8, 11, 21, 34), and developing rAd vectors from rare human Ad serotypes (12, 14, 25, 35). Such novel rAd vectors may prove useful as vaccine vectors in populations in the developing world with high levels of preexisting anti-Ad5 immunity. Novel rAd vectors could also be utilized together with rAd5 vectors or other vectors in heterologous prime-boost regimens.
The 51 known human Ad serotypes are divided into six subgroups, A to F. A seroprevalence study showed that several Ad serotypes from subgroups B and D were rare in a Belgian population (35), suggesting that vaccine vectors derived from these serotypes may prove useful. Consistent with this hypothesis, we have recently shown that rAd11 and rAd35 vectors from subgroup B (3, 15) and rAd49 vectors from subgroup D (14) were immunogenic in mice with anti-Ad5 immunity. However, all rare serotype rAd vectors evaluated to date have proven less immunogenic than rAd5 vectors in mice and rhesus monkeys in the absence of anti-Ad5 immunity (3, 14, 15, 18, 22, 25).
Identifying rare serotype rAd vectors with improved immunogenicity would therefore be highly desirable. However, detailed comparative immunogenicity studies involving multiple rare serotype rAd vectors from subgroups B and D have not previously been reported, presumably due to the limited number of rAd vector systems available. Such studies are critical for selecting the optimal rAd vectors for advancement into clinical trials. Here we describe the initial sequencing and primary construction of three novel rAd vector systems derived from Ad26, Ad48, and Ad50. We report comparative seroprevalence and immunogenicity studies involving rAd11, rAd35, and rAd50 vectors from subgroup B; rAd26, rAd48, and rAd49 vectors from subgroup D; and rAd5 vectors from subgroup C. These studies demonstrate that a variety of rare serotype rAd vectors, and in particular rAd26, may prove useful as vaccine vectors for the developing world.
MATERIALS AND METHODS
Ad26, Ad48, and Ad50 genome sequences.
Wild-type Ad26, Ad48, and Ad50 viruses were obtained from the American Type Culture Collection (ATCC), Manassas, VA, and Jan de Jong, RIVM, The Netherlands. These viruses were propagated using PER.C6 cells and purified as described previously (35). The nucleotide sequences of wild-type Ad26, Ad48, and Ad50 were determined via shotgun sequencing (Lark Technologies Inc., Houston, TX) essentially as described previously (12).
Construction of rAd26, rAd48, and rAd50 vector systems.
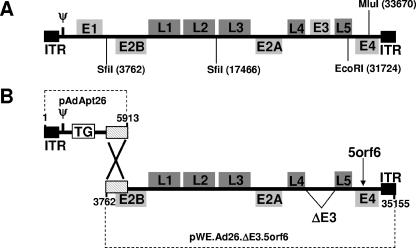
Replication-incompetent rAd26, rAd48, and rAd50 vector systems with E1/E3 deleted were constructed essentially as described previously (12, 14, 35). As shown in Fig. 1, the plasmid/cosmid systems for these vectors consist of (i) a pAdApt adaptor plasmid containing the left end of the Ad genome with a deletion of the E1 region and a transgene (TG) expression cassette and (ii) a pWE cosmid containing the majority of the Ad genome spanning from the pIX sequence to the right inverted terminal repeat (ITR) with a deletion of the E3 region and a modified E4 open reading frame 6 (E4orf6) sequence.
FIG. 1.
Novel rAd vector systems. (A) Genomic organization of replication-competent, wild-type Ad26. (B) Genomic organization of replication-incompetent rAd26 vector system with E1/E3 deleted, consisting of an adaptor plasmid (pAdApt26) and a cosmid (pWE.Ad26.ΔE3.5orf6). The E1 region was replaced by a TG cassette in the adaptor plasmid. The E3 region was deleted and the E4orf6 region was replaced by the corresponding region from Ad5 (5orf6) in the cosmid. The numbers indicate Ad26 nucleotide positions, and the region of homology that facilitates recombination between the adaptor plasmid and the cosmid is depicted. The genomic organizations and vector systems for rAd48 and rAd50 are analogous. ψ, packaging signal; E1 to E4, early genes; L1 to L5, late genes.
The pAdApt26, pAdApt48, and pAdApt50 adaptor plasmids were constructed to consist of the left ITR; the packaging signal; an expression cassette involving a cytomegalovirus promoter, a multiple cloning site for insertion of a TG, and the simian virus 40 polyadenylation transcription termination signal; and a 2.0- to 2.5-kb fragment downstream of the E1 region. This latter sequence enables homologous recombination with the pWE cosmids in E1-complementing cells such as PER.C6. Table 1 depicts the nucleotide definitions of each of these regions, and Table 2 describes the primer sequences utilized.
TABLE 1.
Cloning regions for the rAd26, rAd48, and rAd50 vector systems
| Adenovirus region | Nucleotide positions of:
|
||
|---|---|---|---|
| Ad26 | Ad48 | Ad50 | |
| pAdApt, left ITR and packaging signal | 1-462 | 1-462 | 1-463 |
| pAdApt, 3′Ad fragment | 3,365-5,913 | 3,362-5,910 | 3,214-5,273 |
| pBr322 subclone, middle Ad fragment | 3,762-17,466 | 3,759-20,796 | 3,383-19,351 |
| pBr322 subclone, right Ad fragment | 15,358-35,155 | 17,336-35,206 | 17,144-35,385 |
| PCR fragment A(E3) | 25,477-26,689 | 25,442-26,654 | 24,469-27,420 |
| PCR fragment B(E3) | 30,683-31,740 | 30,735-31,981 | 31,212-35,385 |
| E3 deletion | 26,690-30,682 | 26,656-30,735 | 27,420-31,212 |
| PCR fragment 1(E4-orf6) | 31,718-32,184 | 31,907-32,243 | 32,465-33,614 |
| PCR fragment 3(E4-orf6) | 33,231-33,680 | 33,290-33,739 | 33,614-35,385 |
| E4orf6 replacement | 32,185-33,230 | 32,244-33,289 | 32,465-33,614 |
TABLE 2.
Cloning primers for the rAd26, rAd48, and rAd50 vector systems
| Primer | Sequence for:
|
||
|---|---|---|---|
| Ad26 | Ad48 | Ad50 | |
| Forward primer pAdApt, left ITR and packaging signal | CCTTAATTAATCGACATCATCAATAATATACCCCAC | CAGAATTTAATTAATCGACATCATCAATAATATACCCCAC | GGCCTTAATTAACATCATCAATAATATACCTTATAG |
| Reverse primer pAdApt, left ITR and packaging signal | CGCCTAGGTCAGCTGATCTGTGACATAAAC | CAGAATCGCCTAGGTCAGCTGATCTGTGACATAAAC | CCACCTAGGCTGACACCTACGTAAAAACAC |
| Forward primer pAdApt, 3′Ad fragment | CAGAAGGGATCCAGGTAGGTTTGAGTAGTGGG | CAGAATCGGGATCCAGGTAGGTTTTGAGTAGTGGG | GCCATAAGAATGCGGCCGCTGATCAATTGACTGAGATATGACACTAAACC |
| Reverse primer pAdApt, 3′Ad fragment | CAACGCGTCGACTTAATTAATCTTGAGAGGGAATACCTAC | CAGAATACGCGTCGACTTAATTAATCTCGAGAGGGAATACCTAC | GTTCCCCGCGGGGGACCTTAATTAAGGCGATCGGCAGCGGTTCTC |
| Forward primer PCR fragment A(E3) | AAAGACTAAGGCGCGCCCAC | AAAGACTAAGGCGCGCCCAC | ATAAGAATGCGGCCGCATGTTAACCTGCCGCCCAAAG |
| Reverse primer PCR fragment A(E3) | CAGAATACTAGTGCAGTGAGTGTTGGAGACTGC | CAGAATACTAGTGCAGGTGTTGGCTACTGCTAG | CACTGCGACGCGTCGTCAATCATAGCCATCCAC |
| Forward primer PCR fragment B(E3) | CAGAATACTAGTCCATGAACTGATGTTGATTAAAAG | CAGAATACTAGTCCATGAACTGATGTTGATTAAAAC | GCCGCGACGCGTCGAAATTAATAAAAAATTACTTACTTACTTG |
| Reverse primer PCR fragment B(E3) | GATGGTAATAGAATTCCATTCTC | TCCGCCAAGGTAGACGTTAC | ATAGTTTAGCGGCCGCCATCATCAATAATATACCTTATAGATGG |
| Forward primer PCR fragment 1(E4-orf6) | GAGAATGGAATTCTATTACCATC | TCCTACTAATCCTACAACTCC | TGATTGACGCGACGCGTCGAAATTAATA |
| Reverse primer PCR fragment 1(E4-orf6) | GGGAGAAAGGACTGTTTACACTGTGAAATGG | GGGAGAAAGGACTGTGTACACTGTGAAATGG | GGGAGAAAGGACTGTGTATTCTGTTAAATG |
| Forward primer PCR fragment 2(E4-orf6) | CACAGTGTAAACAGTCCTTTCTCCCCGGCT | CACAGTGTACACAGTCCTTTCTCCCCGGCT | AACAGAATACACAGTCCTTTCTCCCCGGCT |
| Reverse primer PCR fragment 2(E4-orf6) | AGAATCCATTTCAATGACTACGTCCGGCG | AGAATCCACTACAATGACTACGTCCGGCG | CGGCAGGATAGAAATGACTACGTCCGGCG |
| Forward primer PCR fragment 3(E4-orf6) | GGACGTAGTCATTGAAATGGATTCTCTTGC | GGACGTAGTCATTGTAGTGGATTCTCTTGC | GGACGTAGTCATTTCTATCCTGCCGCTTAGT |
| Reverse primer PCR fragment 3(E4-orf6) | GATGTACGCGTAGAGCCACT | GATGTACGCGTAGAGCCACT | ATAGTTTAGCGGCCGCCATCATCAATAATATACCTTATAGATGG |
The pWE.Ad26.ΔE3.5orf6, pWE.Ad48.ΔE3.5orf6, and pWE.Ad50.ΔE3.5orf6 cosmids were constructed by first preparing two intermediate pBr322 plasmids containing subcloned partial regions of each Ad genome. The first pBr322 subclone contained the middle region of the Ad genome (see Table 1 for nucleotide definitions) flanked on both ends by linkers containing PacI and SfiI restriction sites for Ad26 and Ad48 or PacI restriction sites for Ad50. The second pBr322 subclone contained the right region of the Ad genome (see Table 1 for nucleotide definitions). The E3 region was deleted from each second pBr322 subclone by generating two PCR fragments, A(E3) and B(E3), which flanked the E3 region and contained an engineered restriction site that facilitated ligation of these fragments (see Table 1 for nucleotide definitions and Table 2 for primer sequences). The ligated PCR fragments were digested with AscI and EcoRI for Ad26, AscI and SnaBI for Ad48, or NotI and HpaI for Ad50 and ligated into similarly digested second pBr322 subclones, thereby deleting approximately 4 kb of the E3 regions.
Next, the E4orf6 region in each second pBr322 subclone was exchanged with the Ad5 E4orf6 region to enable growth in Ad5 E1-complementing cells as described previously (10). Briefly, the native E4orf6 sequence was replaced by assembling three PCR fragments (see Table 1 for nucleotide definitions and Table 2 for primer sequences). Fragment 1(E4-orf6) contained a 3′ tail homologous to the Ad5 E4orf6 sequence. Fragment 2 was obtained from the Ad5 cosmid pWE.Ad5.AflII-rITR (35) and consisted of a 1.1-kb fragment corresponding to the Ad5 sequence consisting of nt 32963 to 34077 (as in GenBank reference M73260). Fragment 3 contained a 5′ tail homologous to the Ad5 E4orf6 sequence. The three PCR fragments were combined by assembly PCR, and the total chimeric E4 region was cloned into each second pBr322 subclone using MluI and EcoRI for Ad26, MluI and SnaBI for Ad48, or MluI and NotI for Ad50.
The complete pWE.Ad26.ΔE3.5orf6, pWE.Ad48.ΔE3.5orf6, and pWE.Ad50.ΔE3.5orf6 cosmids were then produced by utilizing the two pBr322 subclones and Genehogs bacteria (Invitrogen, Breda, The Netherlands). For Ad26 and Ad48, the first and second pBr322 subclones were digested with PacI to free the adenoviral sequences from the pBr322 backbone and with SrfI (Ad26; nucleotide 15433) or FseI (Ad48; nucleotide 20757) to facilitate ligation of the two sequences. For Ad50, the first pBr322 subclone was digested with PacI and the second pBr322 subclone was digested with NotI to free the adenoviral sequences, and the NheI restriction site (Ad50; nucleotide 19351) was used to facilitate ligation of the two sequences. The pWE backbone was digested with PacI for Ad26 and Ad48 or with PacI and NotI for Ad50 to allow ligation of the two adenoviral sequences into the backbone.
Vector production.
The rAd5, rAd11, rAd35, rAd50, rAd26, rAd48, and rAd49 vectors with E1/E3 deleted (summarized in Table 3) were produced by homologous recombination of the adaptor plasmid expressing a TG of interest with the relevant cosmid in adherent PER.C6 packaging cells as previously described (12, 14, 35). Adaptor plasmids were produced containing enhanced green fluorescent protein (eGFP), luciferase (Luc), or simian immunodeficiency virus SIVmac239 Gag (SIV Gag) TGs. The plasmids were linearized prior to transfection of PER.C6 cells that utilized lipofectamine in T25 flasks. Cells were passaged into T75 flasks after 48 h and maintained until virus cytopathic effect was observed. The vectors were plaque-purified, analyzed for TG expression, amplified in 24 triple-layer T175 flasks, purified by double CsCl gradient ultracentrifugation, and dialyzed into phosphate-buffered saline (PBS) containing 5% sucrose. Purified rAd vectors were stored at −80°C. Virus particle (vp) titers were determined by spectrophotometry (17). Specific infectivity was assessed by PFU assays.
TABLE 3.
Adenovirus serotypes
| Ad subgroup | Serotype |
|---|---|
| B | Ad11 (rare) |
| Ad35 (rare) | |
| Ad50 (rare) | |
| C | Ad5 (common) |
| D | Ad26 (rare) |
| Ad48 (rare) | |
| Ad49 (rare) |
Human serum samples.
Two hundred serum samples from healthy adults were obtained from various countries in sub-Saharan Africa. These countries included Angola, Botswana, Cameroon, Central African Republic, Congo, Ethiopia, Ghana, Ivory Coast, Kenya, Liberia, Nigeria, Rwanda, Senegal, Sierra Leone, Somalia, South Africa, Sudan, Uganda, Zaire, and Zambia. Seroprevalence studies utilizing human serum samples were approved by our Institutional Review Boards (IRB).
Virus neutralization assays.
Ad-specific neutralizing antibody (NAb) titers using human or mouse serum samples were assessed by Luc-based virus neutralization assays as described previously (27). A549 human lung carcinoma cells were plated at a density of 1 × 104 cells per well in 96-well plates and infected with replication-incompetent rAd-Luc reporter constructs at a multiplicity of infection of 500 with 2-fold serial dilutions of serum in 200-μl reaction mixture volumes. Following a 24-hour incubation, Luc activity in the cells was measured using the Steady-Glo Luc reagent system (Promega, Madison, WI) with a Victor 1420 multilabel counter (Perkin Elmer, Wellesley, MA). Neutralization titers were defined as the maximum serum dilution that neutralized 90% of Luc activity.
Animals and immunizations.
Six- to eight-week-old C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA) or Taconic (Hudson, NY). Mice were injected intramuscularly (i.m.) with various doses of the replication-incompetent rAd5, rAd11, rAd35, rAd50, rAd26, rAd48, or rAd49 vectors or with a plasmid DNA vaccine expressing SIV Gag in 100 μl sterile PBS in both quadriceps muscles. The plasmid DNA vaccine was provided by Gary Nabel, Vaccine Research Center, National Institutes of Health, Bethesda, MD. To induce active antivector immunity, mice were preimmunized twice, separated by a 4-week interval, i.m. with 1010 vp rAd5-Empty in 100 μl sterile PBS.
Adult rhesus monkeys (Macaca mulatta) that did not express the major histocompatibility complex class I allele Mamu-A*01 were housed at New England Primate Research Center, Southborough, MA. Monkeys were immunized i.m. with 1011 vp replication-incompetent rAd5, rAd26, rAd48, or rAd49 vectors expressing SIV Gag in 1 ml sterile PBS containing 5% sucrose in both quadriceps muscles. All animal studies were approved by our Institutional Animal Care and Use Committees (IACUC).
Tetramer binding assays.
Tetrameric H-2Db complexes folded around the immunodominant SIV Gag AL11 epitope (AAVKNWMTQTL) (3) were prepared and utilized to stain peptide-specific CD8+ T lymphocytes from C57BL/6 mice as described previously (1, 3). Mouse blood was collected in RPMI 1640 containing 40 U/ml heparin. Following lysis of red blood cells, 0.1 μg of phycoerythrin-labeled Db/AL11 tetramer in conjunction with allophycocyanin-labeled anti-CD8α monoclonal Ab (Ly-2; Caltag, San Francisco, CA) was utilized to stain AL11-specific CD8+ T lymphocytes. The cells were washed in PBS containing 2% fetal bovine serum (FBS) and fixed in 0.5 ml PBS containing 1.5% paraformaldehyde. Samples were analyzed by two-color flow cytometry on a fluorescence-activated cell sorter array (BD Pharmingen, San Diego, CA). Gated CD8+ T lymphocytes were examined for staining with the Db/AL11 tetramer. CD8+ T lymphocytes from naïve mice were utilized as negative controls and exhibited <0.1% tetramer staining.
ELISPOTs.
Gag-specific cellular immune responses in vaccinated mice or rhesus monkeys were assessed by gamma interferon (IFN-γ) enzyme-linked immunospot assays (ELISPOTs) as described previously (3, 18). Overlapping 15-amino-acid peptides spanning the SIV Gag protein were obtained from the NIH AIDS Research and Reference Reagent Program. Ninety-six-well multiscreen plates (Millipore, Bedford, MA) were coated overnight with 100 μl/well of 10 μg/ml anti-mouse or anti-human IFN-γ (BD Pharmingen, San Diego, CA) in endotoxin-free Dulbecco's PBS (D-PBS). The plates were then washed three times with D-PBS containing 0.25% Tween 20 (D-PBS-Tween), blocked for 2 h with D-PBS containing 5% FBS at 37°C, washed three times with D-PBS-Tween, rinsed with RPMI 1640 containing 10% FBS to remove the Tween 20, and incubated with 2 μg/ml of each peptide and 5 × 105 murine splenocytes or 2 × 105 rhesus monkey peripheral blood mononuclear cells (PBMC) in triplicate in 100-μl reaction mixture volumes. Following an 18-h incubation at 37°C, the plates were washed nine times with PBS-Tween and once with distilled water. The plates were then incubated with 2 μg/ml biotinylated anti-mouse or anti-human IFN-γ (BD Pharmingen, San Diego, CA) for 2 h at room temperature, washed six times with PBS-Tween, and incubated for 2 h with a 1:500 dilution of streptavidin-alkaline phosphatase (Southern Biotechnology Associates, Birmingham, AL). Following five washes with PBS-Tween and one with PBS, the plates were developed with nitro blue tetrazolium-5-bromo-4-chloro-3-indolyl-phosphate chromogen (Pierce, Rockford, IL), stopped by washing with tap water, air-dried, and read using an ELISPOT reader (Cellular Technology Ltd., Cleveland, OH). The numbers of spot-forming cells (SFC) per 106 cells were calculated. Media backgrounds were consistently <15 SFC per 106 cells.
ELISA.
SIV Gag-specific serum antibody titers from immunized monkeys were measured by a direct enzyme-linked immunosorbent assay (ELISA) as described previously (3). Ninety-six-well plates coated overnight with 100 μl/well of 1 μg/ml recombinant SIV Gag p27 (ImmunoDiagnostics, Woburn, MA) in PBS were blocked for 2 h with PBS containing 2% bovine serum albumin and 0.05% Tween 20. Sera were then added in serial dilutions and incubated for 1 h. The plates were washed three times with PBS containing 0.05% Tween 20 and incubated for 1 h with a 1:2,000 dilution of a peroxidase-conjugated, affinity-purified rabbit anti-human secondary antibody (Jackson Laboratories, Bar Harbor, ME). The plates were washed three times, developed with tetramethylbenzidine (Kirkegaard & Perry Laboratories, Gaithersburg, MD), stopped with 1% HCl, and analyzed at 450 nm with a Dynatech MR5000 ELISA plate reader.
Statistical analyses.
Statistical analyses were performed with GraphPad Prism version 4.01 (GraphPad Software, Inc., 2004). Immune responses among groups of mice are presented as means with standard errors. Comparisons of mean immune responses were performed by analysis of variance. Comparisons of vector infectivity in nontransfected versus transfected cell lines were performed using the t test. In all cases, P values of less than 0.05 were considered significant.
Nucleotide sequence accession numbers.
The total genome sequences for wild-type Ad26, Ad48, and Ad50 were deposited in GenBank under accession number EF153474 for Ad26, EF153473 for Ad48, and AY737798 for Ad50.
RESULTS
Construction of rAd26, rAd48, and rAd50 vector systems.
To expand the diversity of rAd vectors available for comparative seroprevalence and immunogenicity studies, we first constructed three novel rAd vector systems derived from Ad26 (subgroup D), Ad48 (subgroup D), and Ad50 (subgroup B). We determined the complete genome sequences for these viruses by shotgun sequencing. As depicted for Ad26 in Fig. 1A, the genome organization of these viruses proved comparable to that of previously sequenced adenoviruses (35). Replication-incompetent rAd26, rAd48, and rAd50 vector systems with E1/E3 deleted were then constructed as detailed in Materials and Methods essentially as we have previously described (12, 14, 35).
As depicted for rAd26 in Fig. 1B, the rAd26, rAd48, and rAd50 vector systems consisted of an adaptor plasmid and a cosmid. The adaptor plasmid contained the left end of the Ad genome with a TG expression cassette in place of the E1 region. The cosmid contained the majority of the Ad genome, spanning the pIX sequence to the right ITR, with a deletion of the E3 region and with the E4orf6 sequence replaced by the corresponding sequence from Ad5 to allow growth in Ad5 E1-complementing cell lines. Homologous recombination of the adaptor plasmid and cosmid resulted in the generation of rAd26, rAd48, and rAd50 vectors, which grew to high titers and could be purified using standard cesium chloride gradient purification protocols (35). Final yields of rAd26, rAd48, and rAd50 vectors expressing eGFP, Luc, or SIV Gag were approximately 5 × 1012 to 5 × 1013 vp from research-scale productions involving 24 triple-layer T175 flasks.
Receptor binding studies.
We next investigated cellular receptor usage by these novel vectors. Ad5 from subgroup C has been shown to utilize the coxsackievirus and adenovirus receptor (CAR) for facilitating entry into cells (5, 23), whereas Ad11 and Ad35 from subgroup B have been reported to utilize CD46 as their primary receptor (9). In contrast, receptors utilized by Ads from subgroup D are less well defined. We therefore assessed receptor usage by the rAd5 vector from subgroup C; the rAd11, rAd35, and rAd50 vectors from subgroup B; and the rAd26, rAd48, and rAd49 vectors from subgroup D.
Cells stably transfected with CAR or CD46 as well as untransfected control cells were infected with rAd vectors expressing eGFP in triplicate at a multiplicity of infection of 1,000 for 2 h. Cells were washed to remove unbound virus and incubated for 2 days before virus infectivity was assessed by flow cytometry. As shown in Fig. 2A, the CHO-CAR cells were transduced efficiently only by the rAd5-eGFP vector (P <0.005 for comparison of the infectivity of CHO-CAR versus CHO-k1 cells using the t test). In contrast, as depicted in Fig. 2B, the B16F10-CD46 cells were transduced effectively by the rAd11, rAd35, rAd50, rAd26, rAd48, and rAd49 vectors expressing eGFP (P <0.005 for comparison of the infectivity of B16F10-CD46 versus B16F10 cells using the t test). These data suggest that a variety of rAd vectors from both subgroup B and D can utilize CD46 to facilitate cellular entry. The rAd vectors from subgroup D, however, appeared less efficient than those from subgroup B at transducing B16F10-CD46 cells. As a result, we cannot exclude the possibility that the rAd vectors from subgroup D may also utilize other receptors in addition to CD46.
FIG. 2.
Receptor binding studies. Receptor usage was determined by assessing the abilities of rAd5, rAd11, rAd35, rAd50, rAd26, rAd48, and rAd49 vectors expressing eGFP to transduce (A) CHO cells stably transfected with CAR or (B) B16F10 cells stably transfected with CD46. Untransfected cells were utilized as negative controls. TG expression was assessed by flow cytometry. *, P < 0.005.
Comparative seroprevalence studies in sub-Saharan Africa.
We evaluated adenovirus seroprevalence in a cohort of 200 adults from various regions in sub-Saharan Africa using rAd5, rAd11, rAd35, rAd50, rAd26, rAd48, and rAd49 vectors expressing Luc. Luc-based virus neutralization assays were performed using serum samples essentially as described previously (27). As shown in Fig. 3, Ad5 seroprevalence was nearly universal in these samples, and 48% of these individuals had high Ad5-specific NAb titers of >1,000 as expected. In contrast, the six rAd vectors from subgroups B and D had markedly lower seroprevalence. The Ad11 seroprevalence was 28%, and 9% of these samples had Ad11-specific NAb titers of >1,000. Ad35, Ad50, Ad26, and Ad49 seroprevalences were lower, at 17 to 22%, and 0 to 2% of these individuals had NAb titers of >1,000. Ad48 seroprevalence was the lowest among all the serotypes in this study, at 3%, and no subjects had Ad48-specific NAb titers of >1,000. These data demonstrate that a variety of rAd vectors from subgroups B and D have a low seroprevalence in populations in sub-Saharan Africa. The slightly higher seroprevalence of Ad11 as compared with the other serotypes is consistent with our previous findings (12, 33). The seroprevalence of these Ads appeared to be independently distributed and without a clearly detectable geographic pattern among these samples (data not shown). These data suggest the potential utility of these rAd vectors for vaccine development for populations in sub-Saharan Africa.
FIG. 3.
Seroprevalence studies. Serum samples from 200 adults from sub-Saharan Africa were evaluated for Ad5-, Ad11-, Ad35-, Ad50-, Ad26-, Ad48-, and Ad49-specific NAbs. NAb titers were arbitrarily divided into the following categories: <16 (negative), 16 to 200, 200 to 1,000, and >1,000.
Comparative immunogenicity studies using mice.
We next compared the immunogenicity of rAd5 vectors from subgroup C; rAd11, rAd35, and rAd50 vectors from subgroup B; and rAd26, rAd48, and rAd49 vectors from subgroup D. Naïve C57BL/6 mice (n = 4/group) were immunized i.m. with a single injection of 109 vp, 108 vp, or 107 vp of each vector expressing SIV Gag. Vaccine-elicited CD8+ T-lymphocyte responses specific for the dominant AL11 epitope (AAVKNWMTQTL) were monitored by Db/AL11 tetramer binding assays at multiple time points following immunization (3). As shown in Fig. 4A, all rAd vectors were highly immunogenic at the high dose of 109 vp. At the intermediate dose of 108 vp, the rAd5 and rAd26 vectors proved significantly more immunogenic than the rAd11, rAd35, rAd50, rAd48, and rAd49 vectors (P of <0.01 for comparison of responses on day 28 using analysis of variance). At the low dose of 107 vp, the rAd5 vectors still elicited Gag-specific cellular immune responses, whereas none of the rare serotype rAd vectors elicited detectable responses. These data suggest a relative hierarchy of vector immunogenicity in mice. In C57BL/6 mice without anti-Ad5 immunity, rAd5-Gag was consistently the most immunogenic vector among those studied. Among the six rare serotype rAd vectors, rAd26-Gag appeared the most immunogenic. This hierarchy was confirmed by functional IFN-γ ELISPOT assays performed on day 35 using a Gag peptide pool, the CD8+ T-lymphocyte epitopes AL11 (AAVKNWMTQTL) and KV9 (KSLYNTVCV) (3), and the CD4+ T-lymphocyte epitope DD13 (DRFYKSLRAEQTD) (16) shown in Fig. 4B.
FIG. 4.
Immunogenicity studies using mice. (A, B) Naïve C57BL/6 mice (n = 4/group) were immunized i.m. with 109, 108, or 107 vp rAd5, rAd11, rAd35, rAd50, rAd26, rAd48, or rAd49 expressing SIV Gag. (C, D) C57BL/6 mice with anti-Ad5 immunity were similarly immunized with 109 vp of each vector. Gag-specific cellular immune responses were assessed by Db/AL11 tetramer binding assays at multiple time points following immunization (A, C) and by IFN-γ ELISPOT assays in response to a Gag peptide pool as well as the CD8+ T-lymphocyte epitopes AL11 and KV9 and the CD4+ T-lymphocyte epitope DD13 on day 35 (B, D). Mean responses with standard errors are shown.
Since the majority of individuals in the developing world have high levels of preexisting anti-Ad5 immunity, we repeated this study using mice with anti-Ad5 immunity. C57BL/6 mice (n = 4/group) were preimmunized with two injections of 1010 vp rAd5-Empty separated by a 4-week interval. This regimen induced Ad5-specific NAb titers of 8,192 to 16,384 (data not shown) (3, 22), which models the upper 20% of Ad5-specific NAb titers found in individuals in sub-Saharan Africa. Four weeks later, these mice were primed with 109 vp of each vector, and vaccine-elicited cellular immune responses were evaluated by Db/AL11 tetramer binding assays and IFN-γ ELISPOT assays. As shown in Fig. 4C and D, the immunogenicity of rAd5-Gag was essentially ablated by high levels of anti-Ad5 immunity. In contrast, the immunogenicity of the six rare serotype rAd-Gag vectors was not detectably suppressed in mice with anti-Ad5 immunity as compared with their immunogenicity in naïve mice (Fig. 4A and B). Importantly, all rare serotype rAd-Gag vectors proved significantly more immunogenic than rAd5-Gag in the presence of preexisting anti-Ad5 immunity (P < 0.001).
Heterologous prime-boost regimens in mice.
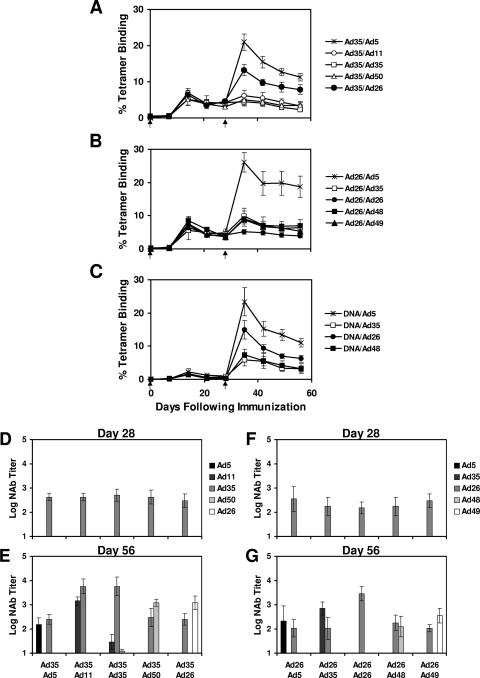
All rAd vectors may be limited by the rapid generation of potent antivector immunity following immunization, which substantially inhibits readministration of the homologous vector (3, 6, 15, 24, 32). Heterologous prime-boost regimens consisting of two rAd vectors derived from different serotypes may overcome this problem. As shown in Fig. 5A, naïve C57BL/6 mice (n = 4/group) were primed i.m. on day 0 with 109 vp rAd35-Gag and boosted on day 28 with 109 vp rAd5-Gag, rAd11-Gag, rAd35-Gag, rAd50-Gag, or rAd26-Gag. Boosting with rAd5-Gag expanded mean tetramer+ CD8+ T-lymphocyte responses to 21.0% on day 35, and these responses declined to 11.3% on day 56. Boosting with rAd26-Gag also expanded tetramer+ CD8+ T-lymphocyte responses, although to a lesser degree (13.3% on day 35; 7.8% on day 56). In contrast, boosting with rAd11-Gag, rAd35-Gag, and rAd50-Gag proved substantially less effective, consistent with our recent finding that rAd11 and rAd35 vectors from subgroup B elicit cross-reactive vector-specific NAbs (15, 32). These data suggest that immune responses primed by rAd35 vectors from subgroup B are boosted more effectively by rAd vectors from subgroups C or D than by other rAd vectors from subgroup B.
FIG. 5.
Heterologous prime-boost studies with mice. Naïve mice (n = 4/group) were primed on day 0 with (A) 109 vp rAd35-Gag, (B) 109 vp rAd26-Gag, or (C) 50 μg DNA-Gag and then boosted on day 28 with 109 vp of the rAd-Gag vectors shown. Arrows indicate immunizations. Gag-specific cellular immune responses were assessed by Db/AL11 tetramer binding assays at multiple time points following immunization. (D, E) Serum Ad5-, Ad11-, Ad35-, Ad50-, and Ad26-specific NAb titers were determined using the mice in the study whose results are depicted in panel (A). (F, G) Serum Ad5-, Ad35-, Ad26-, Ad48-, and Ad49-specific NAb titers were determined using the mice in the study whose results are depicted in panel (B). Mean responses with standard errors are shown.
As shown in Fig. 5B, additional groups of naïve C57BL/6 mice (n = 4/group) were primed i.m. on day 0 with 109 vp rAd26-Gag and boosted on day 28 with 109 vp rAd5-Gag, rAd35-Gag, rAd26-Gag, rAd48-Gag, or rAd49-Gag. Boosting with rAd5-Gag expanded the mean tetramer+ CD8+ T-lymphocyte responses to 26.1% on day 35, and these responses declined to 18.7% on day 56. Boosting with rAd35-Gag, rAd48-Gag, and rAd49-Gag also expanded tetramer+ CD8+ T-lymphocyte responses, although to a lesser degree (8.8 to 9.9% on day 35; 5.3 to 7.0% on day 56). As expected, boosting with the homologous rAd26-Gag vector did not detectably enhance responses. These data suggest that immune responses primed by the rAd26 vectors from subgroup D are boosted effectively by the rAd5 vectors from subgroup C, but they can also be boosted by heterologous rAd vectors from subgroup B as well as subgroup D.
Priming with plasmid DNA vaccines and boosting with rAd5 vectors has been shown by a number of laboratories to elicit particularly potent antigen-specific immune responses (4, 26, 29). To explore the relative capacities of rare serotype rAd vectors to boost responses primed by DNA vaccines, we immunized C57BL/6 mice (n = 4/group) i.m. on day 0 with 50 μg plasmid DNA expressing SIV Gag and boosted them on day 28 with 109 vp rAd5-Gag, rAd35-Gag, rAd26-Gag, or rAd48-Gag. As shown in Fig. 5C, boosting with rAd5-Gag expanded the mean tetramer+ CD8+ T-lymphocyte responses to 23.4% on day 35, and these responses declined to 11.0% on day 56. Boosting with rAd26-Gag expanded the mean tetramer+ CD8+ T-lymphocyte responses to 14.9% on day 35, and these responses declined to 6.2% on day 56. In contrast, boosting with rAd35-Gag and rAd48-Gag proved less effective. These data are consistent with the relative hierarchy of rAd vector immunogenicity observed in the results presented in Fig. 4A and B. These data also suggest that prime-boost regimens consisting of two heterologous rAd vectors are comparably immunogenic to regimens consisting of DNA priming and rAd boosting.
We assessed the vector-specific NAb titers for the mice whose results are depicted in Fig. 5A and B after both the priming immunization and the boost immunization. As shown in Fig. 5D, all mice primed with rAd35 generated Ad35-specific NAbs, as expected. As depicted in Fig. 5E, rAd35-primed mice boosted with rAd11 developed anamnestic Ad35-specific NAb responses, and mice that received two injections of rAd35 developed cross-reactive Ad11-specific NAbs. These cross-reactive Ad11/Ad35-specific NAbs are consistent with our previous results (15, 32). In contrast with these observations, we were unable to detect cross-reactive vector-specific NAbs in mice immunized with rAd26, rAd48, and rAd49 vectors from subgroup D. As shown in Fig. 5F and G, boosting rAd26-primed mice with rAd26, rAd48, or rAd49 resulted in NAbs specific for both the priming and boosting vectors but no detectable cross-reactive NAbs. These data are consistent with the immunogenicity study results shown in Fig. 5B that suggest that the rAd26, rAd48, and rAd49 vectors did not elicit functionally relevant cross-reactive NAbs. The vector-specific NAbs of the mice whose results are depicted in Fig. 5C demonstrated responses to the homologous serotypes at titers comparable to those observed in the results shown in Fig. 5D and F (data not shown).
Comparative immunogenicity studies using rhesus monkeys.
We next evaluated the immunogenicity of the novel rAd vectors from subgroup D in a pilot study of nonhuman primates to confirm the results of the immunogenicity studies with mice, since mice lack the optimal CD46 receptor. Twelve adult rhesus monkeys (n = 3/group) that did not express the Mamu-A*01 class I allele were immunized once i.m. with 1011 vp rAd5-Gag, rAd26-Gag, rAd48-Gag, or rAd49-Gag. Gag-specific cellular and humoral immune responses were assessed by pooled peptide IFN-γ ELISPOT assays and ELISAs at multiple time points following immunization. As shown in Fig. 6A, all rAd vectors elicited detectable Gag-specific cellular immune responses in all animals by week 2 following immunization. As expected, rAd5-Gag elicited high-frequency mean ELISPOT responses of 638 SFC/106 PBMC at week 12. The rAd26-Gag vector elicited slightly lower mean ELISPOT responses of 482 SFC/106 PBMC at week 12. In contrast, rAd48-Gag and rAd49-Gag elicited severalfold lower mean ELISPOT responses, which were comparable in magnitude to those elicited by rAd35-Gag in our previous studies of rhesus monkeys (18). As depicted in Fig. 6B, rAd5-Gag also elicited high-titer Gag-specific antibodies by ELISA, whereas rAd26-Gag elicited lower antibody responses. In contrast, rAd48-Gag and rAd49-Gag elicited marginal to undetectable Gag-specific ELISA responses. These data suggest that the hierarchy of vector immunogenicity observed for nonhuman primates is similar to that observed for mice (Fig. 4A and B and 5C).
FIG. 6.
Immunogenicity studies using rhesus monkeys. Adult rhesus monkeys (n = 3/group) were immunized with a single injection of 1011 vp rAd5-Gag, rAd26-Gag, rAd48-Gag, or rAd49-Gag. (A) IFN-γ ELISPOT assays in response to a Gag peptide pool and (B) Gag-specific ELISAs were performed at multiple time points following immunization. Mean responses with standard errors are shown.
DISCUSSION
The utility of rAd5 vector-based vaccines for HIV-1 and other pathogens may be limited by the high prevalence of anti-Ad5 immunity in the developing world (2, 25). It is therefore important to identify novel serotype rAd vectors that circumvent anti-Ad5 immunity and that are also highly immunogenic. In this study, we compared the seroprevalence and immunogenicity of rAd11, rAd35, and rAd50 vectors from subgroup B and rAd26, rAd48, and rAd49 vectors from subgroup D. The seroprevalence of these rAd vectors was substantially lower than that of rAd5 vectors in a cohort of individuals from sub-Saharan Africa. All six rare serotype rAd vectors also proved immunogenic in mice both with and without anti-Ad5 immunity. The immunogenicity of the rAd26 vectors appeared particularly promising in both mice and rhesus monkeys, suggesting the potential of rAd26 as a vaccine vector for the developing world.
Previous studies have assessed the utility of several novel rAd vectors, including capsid chimeric rAd vectors (18, 22), nonhuman serotype rAd vectors (8, 11, 21), and rare human serotype rAd vectors (12, 14, 35). These studies, however, each evaluated only limited numbers of novel rAd vectors. The present study extends these prior studies by developing three novel rAd vector systems (rAd26, rAd48, and rAd50) and presenting comparative assessments of six rare serotype rAd vectors in which we attempted to minimize experimental variables to the extent possible. For example, the seroprevalence studies utilized the same set of serum samples and standardized virus neutralization assays, and the murine and primate immunogenicity studies utilized vectors expressing the same SIV Gag antigen and standardized tetramer and ELISPOT assays. Such controlled comparative studies are critical for selecting optimal rAd vectors for advancement into clinical trials.
Using both mice and rhesus monkeys, we observed a similar relative hierarchy of vector immunogenicity among these rAd serotypes. In the absence of anti-Ad5 immunity, rAd5 vectors were consistently the most immunogenic among the vectors that we studied. The reasons underlying the potency of rAd5, however, are not yet fully understood, although we speculate that it may reflect efficient CAR-mediated entry into cells or effective triggering of innate immunity that amplifies adaptive immune responses. Among the rare serotype rAd vectors evaluated in this study, the rAd26 vectors consistently appeared to be the most immunogenic (Fig. 4A and B, 5C, and 6A and B). Moreover, in the presence of high levels of anti-Ad5 immunity, all six rare serotype rAd vectors proved significantly more immunogenic than rAd5 vectors (Fig. 4C and D).
Priming with a DNA vaccine and boosting with an rAd5 vector has been shown by a number of laboratories to generate high-frequency immune responses in a variety of experimental systems (26, 29). In contrast, heterologous prime-boost regimens consisting of two serologically distinct rAd vectors have not been studied as extensively. As shown in Fig. 5, priming with a rare serotype rAd vector such as rAd26 or rAd35 followed by boosting with rAd5 proved remarkably potent. These observations extend prior reports exploring the immunogenicity of heterologous rAd prime-boost regimens (15, 20, 32). In the present study, the highest magnitude and most durable responses were generated using the rAd26 prime, rAd5 boost regimen. In fact, the rAd26 prime, rAd5 boost regimen appeared as immunogenic as the DNA prime, rAd5 boost regimen in these studies and may also offer practical advantages over regimens that require DNA priming.
Our data indicate that rAd26 vectors have several characteristics that are attractive for vaccine development. First, Ad26 seroprevalence is low in populations in sub-Saharan Africa. Second, rAd26 vectors are highly immunogenic both as a single vector modality and as a component of heterologous prime-boost regimens. Third, the rAd26 vectors grow to high titers in currently available Ad5 E1-complementing cell lines, thus making large-scale clinical manufacturing of rAd26 vectors potentially feasible. These findings suggest that rAd26 should be evaluated further as a candidate vaccine vector for advancement into clinical trials for HIV-1 and other pathogens.
Acknowledgments
We thank Jan de Jong, Ben Berkhout, Maria Grazia Pau, Jerome Custers, Brad Spiller, Gary Nabel, Faye Stephens, Norman Letvin, Michelle Lifton, and Andrew Bates for generous advice, assistance, and reagents. The SIV Gag overlapping peptides were obtained from the NIH AIDS Research and Reference Reagent Program.
We acknowledge support from NIH grants AI066305 (D.H.B.), AI066924 (D.H.B.), AI058727 (D.H.B.), P30 AI060354, and P51 RR00168.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Altman, J. D., P. A. Moss, P. J. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 2.Barouch, D. H., and G. J. Nabel. 2005. Adenovirus vector-based vaccines for human immunodeficiency virus type 1. Hum. Gene Ther. 16:149-156. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., M. G. Pau, J. H. Custers, W. Koudstaal, S. Kostense, M. J. Havenga, D. M. Truitt, S. M. Sumida, M. G. Kishko, J. C. Arthur, B. Korioth-Schmitz, M. H. Newberg, D. A. Gorgone, M. A. Lifton, D. L. Panicali, G. J. Nabel, N. L. Letvin, and J. Goudsmit. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 172:6290-6297. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., Z. Y. Yang, W. P. Kong, B. Korioth-Schmitz, S. M. Sumida, D. M. Truitt, M. G. Kishko, J. C. Arthur, A. Miura, J. R. Mascola, N. L. Letvin, and G. J. Nabel. 2005. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 79:8828-8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 6.Casimiro, D. R., L. Chen, T. M. Fu, R. K. Evans, M. J. Caulfield, M. E. Davies, A. Tang, M. Chen, L. Huang, V. Harris, D. C. Freed, K. A. Wilson, S. Dubey, D. M. Zhu, D. Nawrocki, H. Mach, R. Troutman, L. Isopi, D. Williams, W. Hurni, Z. Xu, J. G. Smith, S. Wang, X. Liu, L. Guan, R. Long, W. Trigona, G. J. Heidecker, H. C. Perry, N. Persaud, T. J. Toner, Q. Su, X. Liang, R. Youil, M. Chastain, A. J. Bett, D. B. Volkin, E. A. Emini, and J. W. Shiver. 2003. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J. Virol. 77:6305-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catanzaro, A. T., R. A. Koup, M. Roederer, R. T. Bailer, M. E. Enama, Z. Moodie, L. Gu, J. E. Martin, L. Novik, B. K. Chakrabarti, B. T. Butman, J. G. Gall, C. R. King, C. A. Andrews, R. Sheets, P. L. Gomez, J. R. Mascola, G. J. Nabel, and B. S. Graham. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J. Infect. Dis. 194:1638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farina, S. F., G. P. Gao, Z. Q. Xiang, J. J. Rux, R. M. Burnett, M. R. Alvira, J. Marsh, H. C. Ertl, and J. M. Wilson. 2001. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 75:11603-11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 10.Havenga, M., R. Vogels, D. Zuijdgeest, K. Radosevic, S. Mueller, M. Sieuwerts, F. Weichold, I. Damen, J. Kaspers, A. Lemckert, M. van Meerendonk, R. van der Vlugt, L. Holterman, D. Hone, Y. Skeiky, R. Mintardjo, G. Gillissen, D. Barouch, J. Sadoff, and J. Goudsmit. 2006. Novel replication-incompetent adenoviral B-group vectors: high vector stability and yield in PER.C6 cells. J. Gen. Virol. 87:2135-2143. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann, C., P. Loser, G. Cichon, W. Arnold, G. W. Both, and M. Strauss. 1999. Ovine adenovirus vectors overcome preexisting humoral immunity against human adenoviruses in vivo. J. Virol. 73:6930-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holterman, L., R. Vogels, R. van der Vlugt, M. Sieuwerts, J. Grimbergen, J. Kaspers, E. Geelen, E. van der Helm, A. Lemckert, G. Gillissen, S. Verhaagh, J. Custers, D. Zuijdgeest, B. Berkhout, M. Bakker, P. Quax, J. Goudsmit, and M. Havenga. 2004. Novel replication-incompetent vector derived from adenovirus type 11 (Ad11) for vaccination and gene therapy: low seroprevalence and noncross-reactivity with Ad5. J. Virol. 78:13207-13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostense, S., W. Koudstaal, M. Sprangers, G. J. Weverling, G. Penders, N. Helmus, R. Vogels, M. Bakker, B. Berkhout, M. Havenga, and J. Goudsmit. 2004. Adenovirus types 5 and 35 seroprevalence in AIDS risk groups supports type 35 as a vaccine vector. Aids 18:1213-1216. [DOI] [PubMed] [Google Scholar]
- 14.Lemckert, A. A., J. Grimbergen, S. Smits, E. Hartkoorn, L. Holterman, B. Berkhout, D. H. Barouch, R. Vogels, P. Quax, J. Goudsmit, and M. J. Havenga. 2006. Generation of a novel replication-incompetent adenoviral vector derived from human adenovirus type 49: manufacture on PER.C6 cells, tropism and immunogenicity. J. Gen. Virol. 87:2891-2899. [DOI] [PubMed] [Google Scholar]
- 15.Lemckert, A. A., S. M. Sumida, L. Holterman, R. Vogels, D. M. Truitt, D. M. Lynch, A. Nanda, B. A. Ewald, D. A. Gorgone, M. A. Lifton, J. Goudsmit, M. J. Havenga, and D. H. Barouch. 2005. Immunogenicity of heterologous prime-boost regimens involving recombinant adenovirus serotype 11 (Ad11) and Ad35 vaccine vectors in the presence of anti-ad5 immunity. J. Virol. 79:9694-9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, J., B. A. Ewald, D. M. Lynch, A. Nanda, S. M. Sumida, and D. H. Barouch. 2006. Modulation of DNA vaccine-elicited CD8+ T-lymphocyte epitope immunodominance hierarchies. J. Virol. 80:11991-11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115-125. [DOI] [PubMed] [Google Scholar]
- 18.Nanda, A., D. M. Lynch, J. Goudsmit, A. A. Lemckert, B. A. Ewald, S. M. Sumida, D. M. Truitt, P. Abbink, M. G. Kishko, D. A. Gorgone, M. A. Lifton, L. Shen, A. Carville, K. G. Mansfield, M. J. Havenga, and D. H. Barouch. 2005. Immunogenicity of recombinant fiber-chimeric adenovirus serotype 35 vector-based vaccines in mice and rhesus monkeys. J. Virol. 79:14161-14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nwanegbo, E., E. Vardas, W. Gao, H. Whittle, H. Sun, D. Rowe, P. D. Robbins, and A. Gambotto. 2004. Prevalence of neutralizing antibodies to adenoviral serotypes 5 and 35 in the adult populations of The Gambia, South Africa, and the United States. Clin. Diagn. Lab. Immunol. 11:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinto, A. R., J. C. Fitzgerald, W. Giles-Davis, G. P. Gao, J. M. Wilson, and H. C. Ertl. 2003. Induction of CD8+ T cells to an HIV-1 antigen through a prime boost regimen with heterologous E1-deleted adenoviral vaccine carriers. J. Immunol. 171:6774-6779. [DOI] [PubMed] [Google Scholar]
- 21.Reddy, P. S., N. Idamakanti, Y. Chen, T. Whale, L. A. Babiuk, M. Mehtali, and S. K. Tikoo. 1999. Replication-defective bovine adenovirus type 3 as an expression vector. J. Virol. 73:9137-9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts, D. M., A. Nanda, M. J. Havenga, P. Abbink, D. M. Lynch, B. A. Ewald, J. Liu, A. R. Thorner, P. E. Swanson, D. A. Gorgone, M. A. Lifton, A. A. Lemckert, L. Holterman, B. Chen, A. Dilraj, A. Carville, K. G. Mansfield, J. Goudsmit, and D. H. Barouch. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441:239-243. [DOI] [PubMed] [Google Scholar]
- 23.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santra, S., M. S. Seaman, L. Xu, D. H. Barouch, C. I. Lord, M. A. Lifton, D. A. Gorgone, K. R. Beaudry, K. Svehla, B. Welcher, B. K. Chakrabarti, Y. Huang, Z. Y. Yang, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Replication-defective adenovirus serotype 5 vectors elicit durable cellular and humoral immune responses in nonhuman primates. J. Virol. 79:6516-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiver, J. W., and E. A. Emini. 2004. Recent advances in the development of HIV-1 vaccines using replication-incompetent adenovirus vectors. Annu. Rev. Med. 55:355-372. [DOI] [PubMed] [Google Scholar]
- 26.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 27.Sprangers, M. C., W. Lakhai, W. Koudstaal, M. Verhoeven, B. F. Koel, R. Vogels, J. Goudsmit, M. J. Havenga, and S. Kostense. 2003. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing preexisting immunity to vaccine and gene therapy vectors. J. Clin. Microbiol. 41:5046-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan, N. J., T. W. Geisbert, J. B. Geisbert, L. Xu, Z. Y. Yang, M. Roederer, R. A. Koup, P. B. Jahrling, and G. J. Nabel. 2003. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 30.Sumida, S. M., D. M. Truitt, M. G. Kishko, J. C. Arthur, S. S. Jackson, D. A. Gorgone, M. A. Lifton, W. Koudstaal, M. G. Pau, S. Kostense, M. J. Havenga, J. Goudsmit, N. L. Letvin, and D. H. Barouch. 2004. Neutralizing antibodies and CD8+ T lymphocytes both contribute to immunity to adenovirus serotype 5 vaccine vectors. J. Virol. 78:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sumida, S. M., D. M. Truitt, A. A. Lemckert, R. Vogels, J. H. Custers, M. M. Addo, S. Lockman, T. Peter, F. W. Peyerl, M. G. Kishko, S. S. Jackson, D. A. Gorgone, M. A. Lifton, M. Essex, B. D. Walker, J. Goudsmit, M. J. Havenga, and D. H. Barouch. 2005. Neutralizing antibodies to adenovirus serotype 5 vaccine vectors are directed primarily against the adenovirus hexon protein. J. Immunol. 174:7179-7185. [DOI] [PubMed] [Google Scholar]
- 32.Thorner, A. R., A. A. Lemckert, J. Goudsmit, D. M. Lynch, B. A. Ewald, M. Denholtz, M. J. Havenga, and D. H. Barouch. 2006. Immunogenicity of heterologous recombinant adenovirus prime-boost vaccine regimens is enhanced by circumventing vector cross-reactivity. J. Virol. 80:12009-12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorner, A. R., R. Vogels, J. Kaspers, G. J. Weverling, L. Holterman, A. A. Lemckert, A. Dilraj, L. M. McNally, P. M. Jeena, S. Jepsen, P. Abbink, A. Nanda, P. E. Swanson, A. T. Bates, K. L. O'Brien, M. J. Havenga, J. Goudsmit, and D. H. Barouch. 2006. Age dependence of adenovirus-specific neutralizing antibody titers in individuals from sub-Saharan Africa. J. Clin. Microbiol. 44:3781-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuboly, T., and E. Nagy. 2001. Construction and characterization of recombinant porcine adenovirus serotype 5 expressing the transmissible gastroenteritis virus spike gene. J. Gen. Virol. 82:183-190. [DOI] [PubMed] [Google Scholar]
- 35.Vogels, R., D. Zuijdgeest, R. van Rijnsoever, E. Hartkoorn, I. Damen, M. P. de Bethune, S. Kostense, G. Penders, N. Helmus, W. Koudstaal, M. Cecchini, A. Wetterwald, M. Sprangers, A. Lemckert, O. Ophorst, B. Koel, M. van Meerendonk, P. Quax, L. Panitti, J. Grimbergen, A. Bout, J. Goudsmit, and M. Havenga. 2003. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 77:8263-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Z. Y., L. S. Wyatt, W. P. Kong, Z. Moodie, B. Moss, and G. J. Nabel. 2003. Overcoming immunity to a viral vaccine by DNA priming before vector boosting. J. Virol. 77:799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]