Abstract
It is well established that cell-free infection with human T-cell leukemia virus type 1 (HTLV-1) is less efficient than that with other retroviruses, though the specific infectivities of only a limited number of HTLV-1 isolates have been quantified. Earlier work indicated that a postentry step in the infectious cycle accounted for the poor cell-free infectivity of HTLV-1. To determine whether variations in the pol gene sequence correlated with virus infectivity, we sequenced and phenotypically tested pol genes from a variety of HTLV-1 isolates derived from primary sources, transformed cell lines, and molecular clones. The pol genes and deduced amino acid sequences from 23 proviruses were sequenced and compared with 14 previously published sequences, revealing a limited number of amino acid variations among isolates. The variations appeared to be randomly dispersed among primary isolates and proviruses from cell lines and molecular clones. In addition, there was no correlation between reverse transcriptase sequence and the disease phenotype of the original source of the virus isolate. HTLV-1 pol gene fragments encoding reverse transcriptase were amplified from a variety of isolates and were subcloned into HTLV-1 vectors for both single-cycle infection and spreading-infection assays. Vectors carrying pol genes that matched the consensus sequence had the highest titers, and those with the largest number of variations from the consensus had the lowest titers. The molecular clone from CS-1 cells had four amino acid differences from the consensus sequence and yielded infectious titers that were approximately eight times lower than those of vectors encoding a consensus reverse transcriptase.
Human T-cell leukemia virus type 1 (HTLV-1) is an oncogenic retrovirus directly associated with adult T-cell leukemia (ATL) and HTLV-associated myelopathy/tropical spastic paraparesis. An estimated 10 to 20 million people worldwide are infected with the virus, with endemic foci in southern Japan, Melanesia, central Africa, the Caribbean, and South America (2, 31, 56). Like other retroviruses, HTLV-1 depends upon the activity of its reverse transcriptase (RT) for efficient replication in target cells. However, unlike other retroviruses, primate T-cell lymphotropic viruses and the related bovine leukemia virus are known for their low genetic diversity in vivo. Interpatient nucleotide sequence variability between isolates is less than 10% worldwide and usually less than 2% within the same geographic region, while intrapatient variability is considerably less (54). This extremely low genetic variability is likely due to provirus expansion by mitotic replication (60).
HTLV-1 does not productively infect established T-cell lines because of the cytostatic effects of Tax (32, 61). This is paradoxical, because it is through the action of Tax that HTLV-1 immortalizes primary T cells. Therefore, HTLV-1 isolates are generally obtained in the form of a provirus in a chronically infected cell line. HTLV-1-transformed cell lines are obtained by coculture of peripheral blood mononuclear cells (PBMC) or cord blood leukocytes with fresh peripheral blood leukocytes (PBLs) drawn from infected patients, producing mono- or oligoclonal cell lines. Although they typically harbor defective proviruses, these HTLV-1-transformed cell lines are capable of producing infectious virions. However, the cell-free virus particles released from these cell lines are poorly infectious. In the first quantitative study of cell-free HTLV-1 infection, it was estimated that only 1 in 106 virus particles from MT-2 cells were infectious (15).
The infectious HTLV-1 molecular clones pCS-HTLV and pACH were derived from the HTLV-transformed cell lines CS-1 and CH, respectively (9, 10, 26, 28, 52), and have been widely used to study HTLV-1 transformation and infectivity in vitro. The virus particles released by cells transfected with these provirus clones appear to mimic virions produced by transformed cell lines (9), but it is unclear whether the poor replication and infectivity of these viruses are typical of HTLV-1 in nature. We previously showed that pCS-HTLV-based vectors were about 1,000-fold less infectious than human immunodeficiency virus type 1 (HIV-1) when both viruses were pseudotyped with the same envelope (9). Thus, the difference in infectivity was at a postentry step, which likely reflected a difference in viral uncoating or reverse transcription. We have begun to characterize HTLV-1 RT to determine whether or not the RT contributes to the low infectious titer of cell-free HTLV-1 (33).
To determine whether a relationship exists between RT sequence variation and infectious titer, we sequenced and phenotypically tested pol genes from a variety of sources in both single-cycle and spreading-infection assays. We found that isogenic molecular clones of HTLV-1 that contained the consensus RT gave titers that were eightfold higher than those of clones utilizing RT of the original molecular clone, pCS-HTLV.
MATERIALS AND METHODS
Cells and cell lines.
Transfected human kidney (293T), human cervical carcinoma (HeLa), and fetal rhesus lung (FRhL-B5) (12) cells were maintained in Dulbecco's modified minimum essential medium supplemented with 10% fetal bovine serum, l-glutamine, and antibiotics.
The following HTLV-1-transformed cell lines were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: C10/MJ (catalog no. 4407) from Dean Mann and Miklaus Popovic, MT-2 (catalog no. 237) and MT-4 (catalog no. 120) from Douglas Richman, and C8166 (catalog no. 404) from Robert Gallo (36, 37, 44, 48). Renu Lal and Charlene Dezzutti (Centers for Disease Control and Prevention, Atlanta, GA) generously provided the 1657, 3614, 3669, A212, EG, FS, and SP cell lines (7, 13, 16, 23, 47). The 1657, 3614, 3669, A212, C10/MJ, and C91/PL (44); C8166 and CS-1 (28); EG, FS, MT-2, MT-4, and HS-1 (30); HuT102 (43); and SP cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, l-glutamine, and antibiotics; in addition, the 1657, 3614, A212, SP, FS, and EG cultures were supplemented with interleukin-2.
Genomic DNA was extracted from the HTLV-transformed cell lines by using the QIAamp DNA mini or blood kit (QIAGEN). Genomic DNAs were isolated from PBLs of Japanese ATL patients; the isolates are designated Ptnt1, Ptnt2, Ptnt4, Ptnt5, PtntAK003, PtntAK004, PtntAK005, and PtntAK006.
Amplification and sequencing of the pol gene.
The pol gene was PCR amplified in two overlapping segments by using HotStarTaq DNA polymerase (QIAGEN). Nucleotides (nt) 2473 to 3856 (1,384 bp) comprised the 5′ segment, and nt 3243 to 4341 (1,099 bp) comprised the 3′ segment. At least two independent PCR amplifications of the target sequence were performed. PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen), and a minimum of two insert-containing TOPO vectors from each PCR amplification were sequenced. In some instances, direct sequencing of the patient sample PCR product was also performed. The HTLV-1 virus expression vector, pCS-HTLV, was directly sequenced as a reference for all constructs currently used in our lab. The pACH infectious molecular clone was also sequenced for comparison to the published pol gene sequence of the CH isolate (40, 46). The region between nt 2493 and 4319 of the provirus (1,827 bp) was sequenced in all isolates. In addition, the region between nt 4319 and 5127 was sequenced in the MT-2 and C10/MJ isolates.
Phylogenetic analysis.
Nucleotide sequences were aligned with the CLUSTAL W program. Phylogenetic trees were constructed by the neighbor-joining method and rooted with the pol sequence of the HTLV-1c Mel5 isolate as an outlier. The reliability of each branch on the neighbor-joining tree was estimated by bootstrap analysis of 1,000 samplings of the original sequence alignments. Pairwise genetic distances were estimated on each sampling by the Kimura two-parameter method.
Plasmids.
The cloning and construction of the pCS-HTLV plasmid vector, which contains a full-length provirus from the CS-1 cell line, has been described previously (10). The CS-1 cell line was obtained by the cocultivation of cord blood leukocytes with irradiated HTLV-1-infected HS-1 cells. The PstI-to-SstII fragment of pCS-HTLV, containing the pX region, was replaced with the homologous fragment from MT-2 to produce the pX1MT infectious clone (11). The NotI-to-SphI fragment of pX1MT was replaced with the homologous region of pCMVHT-1Δenv (MT-2 RT), described below, to create the pX1MT-M infectious molecular clone. The initial pCMVHT-1Δenv packaging plasmid was derived from the pCS-HTLV infectious molecular clone by replacing the 5′ long terminal repeat promoter with a cytomegalovirus (CMV) promoter linked to a fragment (positions 439 to 567) of the R region (9). The infectious molecular clone pACH was provided by Lee Ratner (26).
To analyze the phenotypic effects of HTLV-1 isolate RT variations on viral genomic replication and infectivity, we replaced portions of the pCMVHT-1Δenv pol gene with the homologous region of selected isolates. The 2,366-bp BglII-to-SphI sequence (nt 2762 to 5127) of pACH was ligated into pCMVHT-1Δenv to create pCMVHT-1Δenv (CH RT). The 1,876-bp KpnI-to-SphI region (nt 3252 to 5127) of pCMVHT-1Δenv was replaced with the homologous regions of C10/MJ and MT-2 to create pCMVHT-1Δenv (C10/MJ RT) and pCMVHT-1Δenv (MT-2 RT), respectively. It is worthwhile to note that the RT polypeptide sequence encoded by pCMVHT-1Δenv (C10/MJ RT) is identical to the MT-2 RT sequence (see Fig. 2). The KpnI-to-XbaI region (nt 3252 to 4081) of pCMVHT-1Δenv (C10/MJ RT) was replaced with the homologous fragments from PCR-amplified EG, Ptnt1, and Ptnt4 DNAs to construct the pCMVHT-1Δenv (EG RT), pCMVHT-1Δenv (Ptnt1 RT), and pCMVHT-1Δenv (Ptnt4 RT) plasmid vectors. The 1,046-bp XbaI-to-SphI fragment (nt 4081 to 5127) of pCMVHT-1Δenv was replaced with the homologous region of MT-2 to create pCMVHT-1Δenv (3614 RT). Site-directed mutagenesis was used to construct pCMVHT-1Δenv RT Q463R. The sequences of all plasmid constructs were confirmed by sequencing.
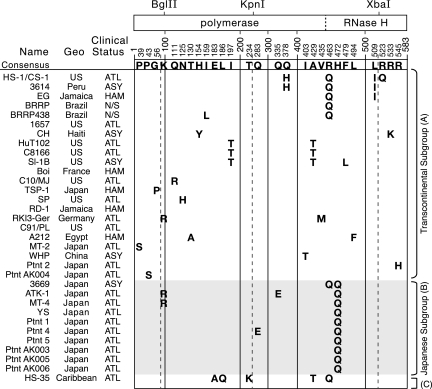
FIG. 2.
Amino acid variations in the RTs of HTLV-1a isolates are mostly random. An alignment of RT amino acid sequences from various HTLV-1a isolates is shown. Each column indicates a site of amino acid variation. The HTLV-1a consensus amino acid is shown at the top of the column, and isolates with variations are presented underneath. Numbering is based on Gly166 of the pro open reading frame as the N-terminal residue of RT (20) and Leu583 as the approximate C-terminal residue (33). The locations of restriction endonuclease sites in the pol gene used for cloning and the junction between the polymerase and RNase H domain are indicated relative to the sites of variation. The sequence of the FS isolate was not included because it contains three stop codons. Isolates are organized by their Cosmopolitan lineage subgroup: Transcontinental (A), Japanese (B), and West African/Caribbean (C).
Single-cycle replication and spreading-infection assays.
Infectivity was measured by means of a single-cycle replication assay that has been described previously (9). In brief, vesicular stomatitis virus G protein (VSV-G)-pseudotyped virus-like particles were generated by cotransfection of 293T cells with a pCMVHT-1Δenv-derived packaging vector, a VSV-G Env expression vector (pCMV-VSV-G), and a reporter vector (pHTC-GFPluc). The reporter vector expresses a surrogate HTLV-1 genomic mRNA containing an internal CMV promoter and luciferase reporter gene that is packaged into VSV-G-pseudotyped virions. Virus-containing supernatants were collected 40 h after transfection and cleared by low-speed centrifugation and filtration through 0.45-μm-pore-size low-protein-binding filters (Millipore). Gag protein in the virus-containing supernatants was quantified using an HTLV-1 p19 (MA) enzyme-linked immunosorbent assay (ELISA) kit (Zeptometrix). The surrogate HTLV genome, containing the luciferase reporter gene, was transduced into HeLa cells by infection with 1.0 ml of the virus-containing supernatant supplemented with 5 μg/ml Polybrene. Three days after infection, HeLa cells were lysed in GLO lysis buffer (Promega). An equal volume of the cleared cell lysate was mixed with Bright GLO luciferase assay reagent (Promega), and luciferase activity was measured from triplicate readings on a luminometer. Viral infectivity was measured as the relative light units per picogram of p19 antigen present in the virus supernatant used for infection.
Spreading-infection assays were initiated by transfection of FRhL-B5 cells by calcium phosphate coprecipitation and glycerol shock (15%, vol/vol). Cells were diluted 1:5 at 3- to 4-day intervals after transfection until the cytopathic effects of virus infection were observed (12). To measure virus production, cell supernatants were collected at each passage for HTLV-1 p19 ELISA.
Nucleotide sequence accession numbers.
The accession numbers of the isolates used in this study are as follows: 1657, EF076680, AMB66539, and AMB66540; 3614, EF076681, AMB66541, and AMB66542; 3669, EF076682, AMB66543, and AMB66544; A212, EF076683, AMB66545, and AMB66546; EG, EF076688, AMB66555, and AMB66556; FS, EF076689, ABM66557, and ABM66558; SP, EF076702, ABM66583, and ABM66584; HuT102, EF076691, ABM66561, and ABM66562; C8166, EF076686, AMB66551, and AMB66552; MT-2, EF076692, ABM66563, and ABM66564; MT-4, EF076693, ABM66565, and ABM66566; C10/MJ, EF076684, AMB66547, and AMB66548; C91/PL, EF076685, AMB66549, and AMB66550; CS-1, EF076687, AMB66553, and AMB66554; HS-1, EF076690, ABM66559, and ABM66560; Ptnt1, EF076694, ABM66567, and ABM66568; Ptnt2, EF076695, ABM66569, and ABM66570; Ptnt4, EF076696, ABM66571, and ABM66572; Ptnt5, EF076697, ABM66573, and ABM66574; PtntAK003, EF076698, ABM66575, and ABM66576; PtntAK004, EF076699, ABM66577, and ABM66578; PtntAK005, EF076700, ABM66579, and ABM66580; PtntAK006, EF076701, ABM66581, and ABM66582; RKI3-Ger, AF042071; SI-1 B, AF139170; WHP, AF259264; HS-35, D13784; TSP-1, M86840; ATK-1, J02029; RD-1, L10341; Boi, L36905; EL, S74562; YS, U19949; BRRP, AY563953; BRRP438, AY563954; and Mel5, L05234. The sequence of HTLV-1CH is not present in GenBank and was transcribed from reference 46.
RESULTS
Nucleotide sequence analysis.
Proviral pol gene sequences were amplified and sequenced from genomic DNAs isolated from 14 HTLV-1-transformed cell lines and from PBLs of eight ATL patients. The pol regions of the infectious molecular clones pCS-HTLV and pACH were also sequenced. HTLV-1 pol gene sequences available in GenBank were included in the sequence analysis; these previously published isolates were from primary sources (Boi, RKI3-Ger, YS, ATK-1, BRRP, BRRP438, WHP, and Mel5) (4, 6, 14, 50, 51) and cell cultures (SI-1 B, HS-35, CH, RD-1, EL, Mel5, and TSP-1) (1, 19, 22, 24, 39, 50, 59). The WHP, BRRP, and BRRP438 sequences are presumed to be from uncultured isolates, but this information was not specified in their GenBank entries. In addition, uncultured PBMC, cultured PBMC, and a transformed T-cell line are all listed as sources of Mel5 genomic DNA, but it is not clear which source provided the template for pol sequencing. The published Mel5 pol gene includes two missense mutations as well as a premature stop codon in the pol reading frame, which would clearly result in the synthesis of a defective RT. These mutations could represent sequencing errors or could reflect somatic or reverse transcription errors.
The pol gene sequence of pCS-HTLV was identical to the homologous provirus sequence in CS-1 and HS-1 cells (Fig. 1). The CS-1 cell line was derived by coculture of cord blood lymphocytes with HS-1 cells (28). Likewise, the pACH pol sequence was identical to the previously published sequence of the pCH molecular clone (46) from which it was derived (26), indicating that no mutations of this sequence occurred during construction of the clones. No genetic differences were observed when we compared the pol sequence of HuT102 with that of C8166, both of which originated from the same patient. The HuT102 T-lymphoblast cell line was established in 1977 from the tumor cells of an ATL patient (16, 42). Two years later, PBLs from this patient were used to establish the HTLV-transformed cell line, CTCL-3 (42, 43), and a clone of cells, CR-M2, isolated from CTCL-3 cells was cocultivated with cord blood leukocytes to establish C8166 (also called C63/CRII-2 or C81-66-45) (48). Together, these results agree with other observations of low sequence diversity between HTLV-1 isolates separated in time by in vivo passage (17).
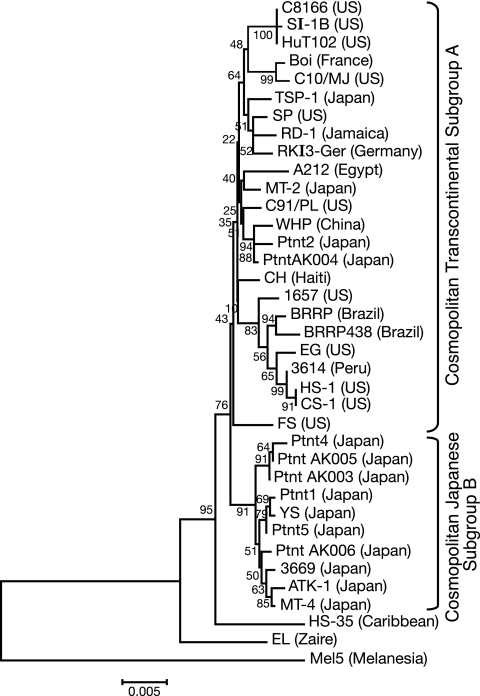
FIG. 1.
Phylogenetic tree of HTLV-1 pol. The phylogenetic tree was constructed by the neighbor-joining method, based on the 1,827-bp RT-encoding regions of the pol gene. The tree was rooted by assuming the pol gene sequence of the HTLV-1c Mel5 isolate as the outgroup. The bootstrap test of phylogeny was used to determine tree branch reliability; values are indicated at each node.
Phylogenetic analyses have been useful for studying worldwide dissemination of the virus and categorizing HTLV-1 isolates into the three major lineages: Cosmopolitan (HTLV-1a), Central African (HTLV-1b), and Melanesian (HTLV-1c) (3, 5, 18, 19, 29, 34, 35, 49, 57). In addition, the Cosmopolitan group has been further subdivided into five currently accepted subgroups: Transcontinental (A), Japanese (B), West African/Caribbean (C), North African (D), and Black Peruvian (E) (38, 54, 58). Although the level of nucleotide sequence diversity among the HTLV-1 pol genes used in our analysis was typically low, it was sufficient to construct a phylogenetic tree capable of resolving subgroups within the Cosmopolitan lineage. Our phylogenetic analysis of pol nucleotide sequences from the aforementioned isolates showed that they all belonged to the Cosmopolitan lineage (HTLV-1a). Within this lineage, the transformed cell line isolates 1657, 3614, A212, C10/MJ, C91/PL, EG, FS, HS-1/CS-1, and HuT102/C8166 and patient sample isolates Ptnt2 and PtntAK004 belonged to the Transcontinental subgroup (HTLV-1aA), while the HTLV-1 provirus of the 3669 cell line and patient isolates Ptnt1, Ptnt4, Ptnt5, PtntAK003, PtntAK005, and PtntAK006 belonged to the Japanese subgroup (HTLV-1aB).
All of the Cosmopolitan nucleotide sequences analyzed (including those already available in GenBank) were very similar to one another in the RT-coding region, differing from one another by less than 2.5%. There were 140 sites of variation among the Cosmopolitan isolates, with 55 positions variable in more than one isolate; i.e., 85 variations were unique to one HTLV-1a isolate. Nucleotide variations favored transitions between guanine (G) and adenine (A) (39%) about as much as between cytosine (C) and thymidine (T) (39%). These are consistent with the previously noted error bias of HTLV-1 RT (6, 8, 27). The sequence context of these substitutions does not immediately suggest the activity of any known host restriction factor, such as APOBEC3G. Most of the observed genetic changes were synonymous substitutions, which suggests that positive selection of the Pol amino acid sequence occurs.
Deduced RT amino acid sequences.
The consensus HTLV-1a RT amino acid sequence was generated from 23 Transcontinental isolates, 10 Japanese isolates, and 1 West African/Caribbean isolate, which gave it a Transcontinental character. No amino acid variations specific to either cell culture or primary HTLV-1 isolates were observed. As noted in previous studies that examined the env or LTR sequences, there was no correlation between isolate genotype and specific HTLV-associated pathologies (27). Instead, sequence similarity is generally seen between isolates of a common geographical origin, regardless of the patient's clinical status.
The most common variation from the consensus sequence was exclusive to Japanese subgroup isolates, which could be distinguished from Transcontinental isolates by the presence of a glutamine at position 472 of RT (Fig. 2). Of the less frequently seen variations, a histidine residue at position 378 in place of the consensus glutamine and an isoleucine at position 509 instead of leucine were variations observed in HS-1/CS-1 (United States) and 3614 (Peru). Likewise, a threonine in place of the consensus isoleucine at amino acid residue 197 was present in U.S. isolates SI-1 B and HuT102/C8166. Several other variations were seen in more than one Cosmopolitan subgroup. A few Transcontinental and Japanese isolates, besides those sequenced here, have arginine at position 100 of RT (45, 46), though lysine is the consensus residue at this position. A glutamine at residue 463 of RT was prevalent in HTLV-1aA isolates from the Caribbean region and South America (BRRP, BRRP438, 3614, EG, and HS-35) but was also observed in Japanese subgroup isolate 3669 and in the HTLV-1c isolate Mel5 (not shown). The threonine residue variation at position 429 of RT was seen in SI-1 B and HuT102/C8166 of the Transcontinental subgroup, as well as in the HTLV-1aC subgroup isolate HS-35.
Phenotypic effects of variation on viral infectivity.
To determine whether certain amino acid variations in RT correlated with virus infectivity, we used a sensitive, quantitative single-cycle infection assay to measure reporter gene transduction (9). To ensure a similar viral context in which to compare the effects of RT variations, the pol gene in the CS-1-derived vector pCMVHT-1Δenv (9) was replaced with pol gene fragments from the MT-2, C10/MJ, CH, 3614, Ptnt1, Ptnt4, and EG proviruses. These isolates were selected because they either encoded the consensus RT sequence (MT-2 and C10/MJ), encoded the RT of the other common HTLV-1 infectious molecular clone (CH), represented the consensus Japanese subgroup RT (Ptnt1) or a variant thereof (Ptnt4), or clustered with CS-1 on the phylogenetic tree (3614 and EG).
Vectors encoding an RT that closely matched the consensus RT sequence, such as MT-2, C10/MJ, and the prototypical Japanese subgroup sequence Ptnt1, displayed the highest titers in a single-cycle infection assay (Table 1). In general, variations from the consensus sequence resulted in decreased viral replication. The presence of a glutamate at amino acid 283 of the Ptnt4 RT significantly decreased viral replication in comparison to the activity of either the Ptnt1 or consensus RT. One or both of the consensus sequence variations present in the CH isolate, Tyr154 and Lys533, reduced viral infectivity nearly threefold relative to the consensus RT. The variations seen in a cluster of U.S., Caribbean, and South American isolates had a significant effect on viral replication. The relative infectivity of these isolates decreased from 36% (EG) to almost 14% (CS-1) of that of the MT-2 RT isolate activity as a result of variations at a combination of positions in the connection and RNase H domains. A derivative of CS-1, where the glutamine at position 463 was mutated to the consensus arginine (Q463R), had a twofold-higher relative infectivity than the CS-1 clone, in agreement with our observation that RT sequences closer to the consensus sequence generally display a higher infectivity.
TABLE 1.
Single-cycle infection assay of HTLV-1 vectors encoding various RTs
| Isolatea | Amino acid variation from consensus RT sequence:
|
Relative infectivity (%)b | |||||||
|---|---|---|---|---|---|---|---|---|---|
| H-154 | Q-283 | Q-378 | R-463 | H-472 | L-509 | R-523 | R-533 | ||
| MT-2 | 100 | ||||||||
| C10/MJ | 84 | ||||||||
| Ptnt1 | Q | 135 | |||||||
| Ptnt4 | E | Q | 37 | ||||||
| CH | Y | K | 42 | ||||||
| EG | Q | I | 36 | ||||||
| 3614 | H | Q | I | 37 | |||||
| CS-1 (Q463R) | H | I | Q | 24 | |||||
| CS-1 | H | Q | I | Q | 13 | ||||
The RT-encoding pol region of the indicated HTLV-1 isolates and the CS-1 Q463R mutant were cloned into pCMVHT-1Δenv.
Normalized to MT-2. The mean of results from duplicate infections was determined for each isolate. CS-1 and MT-2 were tested 10 times. Most other isolates were tested a minimum of three times.
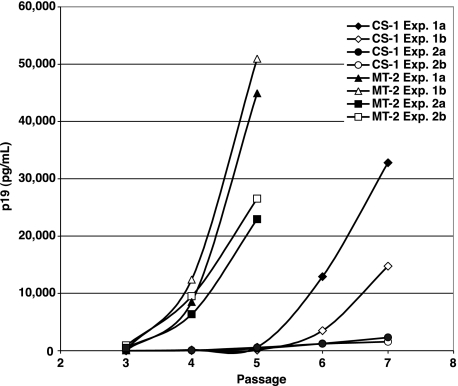
The difference in replication between virus with the consensus sequence RT, represented by MT-2, and the CS-1 variant was further confirmed in a spreading-infection assay. FRhL-B5 cells were transfected with pX1MT or pX1MT-M, encoding the CS-1 and MT-2 RT sequences, respectively, and virus production was monitored by p19 ELISA. As seen in Fig. 3, virus production in cell cultures transfected with pX1MT typically peaked at around 3 weeks after transfection, while peak virus production in cultures transfected with pX1MT-M consistently occurred after only 2 weeks.
FIG. 3.
Replication kinetics in spreading-infection assays correlate with RT sequence. A comparison of the virus infection kinetics between the HTLV-1 infectious molecular clones encoding the CS-1 and MT-2 isolate RT sequences is shown. For each experiment, infections were initiated in duplicate by transfection of FRhL-B5 cells with cloned provirus. Cells were passaged at a 1:5 dilution every 3 to 4 days. At each passage, virus expression was monitored by HTLV-1 p19 ELISA of the culture supernatant. The clone containing MT-2 RT consistently achieved peak virus production two passages before the CS-1 RT clone.
DISCUSSION
To determine if the low infectivity of HTLV-1 virions in vitro is strictly a feature of viruses produced by HTLV-transformed cell lines and the infectious molecular clones derived from these cell lines, we performed a genotypic and phenotypic comparison of the HTLV-1 pol genes from primary isolates, transformed cells, and molecular clones. A phylogenetic analysis of these isolates showed that they belonged to the Cosmopolitan lineage and varied from other HTLV-1a isolates by less than 2.5%. A few of these nucleotide sequence variations also translated into changes in the amino acid sequence of RT, suggesting positive selection for RT function. Some variations from the consensus sequence were shared among several isolates, most notably the glutamine at position 472 in all Japanese subgroup isolates, but most of the variations were unique to a single HTLV-1 isolate. Amino acid variations did not correlate with virus isolates from transformed cell lines versus primary sources, nor did variations correlate with disease status. Finally, no mutations in the pol gene sequence occurred during construction of the molecular clones. Therefore, the pol gene sequences of HTLV-transformed cell lines and infectious molecular clones appear to accurately represent primary HTLV-1 isolates.
The relationship between Pol amino acid sequence and HTLV-1 infectious titer was examined by inserting pol gene sequences from different virus isolates into the packaging plasmid pCMVHT-1Δenv for single-cycle infection assays or into the infectious molecular clone pX1MT for spreading infections. Recombinant viruses whose pol genes most closely matched the consensus sequences, such as MT-2, C10/MJ, and Ptnt1, gave the highest titers. Viral titers decreased roughly in proportion to the number of amino acid variations from the consensus sequence that were present. These data suggest that it is unlikely that a very-high-titer variant of HTLV-1 with a unique pol gene exists, analogous to the high-titer beta strain of RSV (55). The differences in titers for HTLV-1 clones with variant pol genes is reminiscent of the difference in viral titers observed for highly pathogenic SIVmne027 and slow-replicating SIVmneCl8, which differ functionally as the result of a single K412E mutation in the connection domain of RT (25, 41; Jason Kimata, personal communication).
There have been only two reports that made an attempt to quantify infectious titers for cell-free HTLV-1 particles. In the first, Fan et al. used virions produced from MT-2 cells, and measured infection by PCR of nascent reverse transcription products (minus-strand strong-stop DNA) (15). Those authors calculated that one in 106 particles were infectious, a value that is at least 1,000-fold lower than the specific infectivity of HIV-1 particles. However, the specific infectivity for HTLV-1 that is based on MT-2 virions is likely to underestimate the actual value by about 100-fold, because MT-2 cells contain seven defective proviruses and one full-length provirus. Thus, assuming that all proviruses are expressed and packaged, which appears to be likely from RNA analyses (21, 53), only one in 64 particles would contain a dimer of full-length genomic RNA. This would put the actual specific infectivity of HTLV-1 particles at closer to one in 104. In a later study, we used VSV-G-pseudotyped viral vectors to show that HTLV-1 particles gave about 1,000-fold-lower titers than HIV-1 particles (9). These calculations were based on HTLV-1 vectors that contain the CS-1 RT sequence, which give about 10-fold-lower titers than the vectors containing a consensus RT. Calculations based on results from the latter HTLV-1 vectors indicate that HTLV-1 particles are only 100-fold less infectious than HIV-1 particles in cell-free infection experiments. These data indicate that the lower relative titers for pseudotyped HTLV-1 particles are manifested at a postentry step and are consistent with the results of Fan et al. (15), which suggested that this reflects a difference in reverse transcription. The data presented here suggest that cell-free infection in vitro with HTLV-1 is not as low as previously thought. The new HTLV-1 vectors, which encode a consensus RT, increase the sensitivity of in vitro infection assays by 10 times, and this should significantly enhance future studies with HTLV-1 particles containing HTLV-1 Env.
Acknowledgments
We are sincerely grateful to Eugenio Ramirez for generously providing patient sample genomic DNA and especially thankful to his patients, though we were unable to present the sequencing data from his patients in this study. We also thank Mary Kearney for taking time to explain methods in retroviral phylogenetic analysis, for constructing the phylogenetic tree, and for proofreading relevant sections of the manuscript. We thank Stephen Hughes, Paul Boyer, John Julias, Frank Maldarelli, and John Mellors for helpful discussions.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Andreani, T., R. Modigliani, Y. le Charpentier, A. Galian, J. C. Brouet, M. Liance, J. R. Lachance, B. Messing, and B. Vernisse. 1983. Acquired immunodeficiency with intestinal cryptosporidiosis: possible transmission by Haitian whole blood. Lancet i:1187-1191. [DOI] [PubMed] [Google Scholar]
- 2.Bangham, C. R. 2000. HTLV-1 infections. J. Clin. Pathol. 53:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastian, I., J. Gardner, D. Webb, and I. Gardner. 1993. Isolation of a human T-lymphotropic virus type I strain from Australian aboriginals. J. Virol. 67:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazarbachi, A., M. Huang, A. Gessain, F. Saal, A. Saib, J. Peries, H. De The, and F. Galibert. 1995. Human T-cell-leukemia virus type I in post-transfusional spastic paraparesis: complete proviral sequence from uncultured blood cells. Int. J. Cancer 63:494-499. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., L. Zekeng, M. Yamashita, J. Takehisa, T. Miura, E. Ido, I. Mboudjeka, J. M. Tsague, M. Hayami, and L. Kaptue. 1995. HTLV type I isolated from a Pygmy in Cameroon is related to but distinct from the known central African type. AIDS Res. Hum. Retrovir. 11:1529-1531. [DOI] [PubMed] [Google Scholar]
- 6.Chou, K. S., A. Okayama, N. Tachibana, T. H. Lee, and M. Essex. 1995. Nucleotide sequence analysis of a full-length human T-cell leukemia virus type I from adult T-cell leukemia cells: a prematurely terminated PX open reading frame II. Int. J. Cancer 60:701-706. [DOI] [PubMed] [Google Scholar]
- 7.Constantine, N. T., D. A. Scott, M. Kamal, C. R. Roberts, A. Rolfs, H. C. Schumacher, and P. Marx. 1992. Tropical spastic paraparesis associated with HTLV-1 in Egypt. Trans. R. Soc. Trop. Med. Hyg. 86:300-301. [DOI] [PubMed] [Google Scholar]
- 8.Dekaban, G. A., E. E. King, D. Waters, and G. P. Rice. 1992. Nucleotide sequence analysis of an HTLV-I isolate from a Chilean patient with HAM/TSP. AIDS Res. Hum. Retrovir. 8:1201-1207. [DOI] [PubMed] [Google Scholar]
- 9.Derse, D., S. A. Hill, P. A. Lloyd, H. Chung, and B. A. Morse. 2001. Examining human T-lymphotropic virus type 1 infection and replication by cell-free infection with recombinant virus vectors. J. Virol. 75:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derse, D., J. Mikovits, M. Polianova, B. K. Felber, and F. Ruscetti. 1995. Virions released from cells transfected with a molecular clone of human T-cell leukemia virus type I give rise to primary and secondary infections of T cells. J. Virol. 69:1907-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derse, D., J. Mikovits, and F. Ruscetti. 1997. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology 237:123-128. [DOI] [PubMed] [Google Scholar]
- 12.Derse, D., J. Mikovits, D. Waters, S. Brining, and F. Ruscetti. 1996. Examining the molecular genetics of HTLV-I with an infectious molecular clone of the virus and permissive cell culture systems. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:1-5. [DOI] [PubMed] [Google Scholar]
- 13.Dezzutti, C. S., D. L. Rudolph, C. R. Roberts, and R. B. Lal. 1993. Characterization of human T-lymphotropic virus type I- and II-infected T-cell lines: antigenic, phenotypic, and genotypic analysis. Virus Res. 29:59-70. [DOI] [PubMed] [Google Scholar]
- 14.Ellerbrok, H., C. Fleischer, M. Salemi, P. Reinhardt, W. D. Ludwig, A. M. Vandamme, and G. Pauli. 1998. Sequence analysis of the first HTLV-I infection in Germany without relations to endemic areas. AIDS Res. Hum. Retrovir. 14:1199-203. [DOI] [PubMed] [Google Scholar]
- 15.Fan, N., J. Gavalchin, B. Paul, K. H. Wells, M. J. Lane, and B. J. Poiesz. 1992. Infection of peripheral blood mononuclear cells and cell lines by cell-free human T-cell lymphoma/leukemia virus type I. J. Clin. Microbiol. 30:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazdar, A. F., D. N. Carney, P. A. Bunn, E. K. Russell, E. S. Jaffe, G. P. Schechter, and J. G. Guccion. 1980. Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood 55:409-417. [PubMed] [Google Scholar]
- 17.Gessain, A., R. C. Gallo, and G. Franchini. 1992. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J. Virol. 66:2288-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gessain, A., R. Yanagihara, G. Franchini, R. M. Garruto, C. L. Jenkins, A. B. Ajdukiewicz, R. C. Gallo, and D. C. Gajdusek. 1991. Highly divergent molecular variants of human T-lymphotropic virus type I from isolated populations in Papua New Guinea and the Solomon Islands. Proc. Natl. Acad. Sci. USA 88:7694-7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, B. H., G. M. Shaw, M. Popovic, A. Lo Monico, R. C. Gallo, and F. Wong-Staal. 1984. Molecular cloning and analysis of a new variant of human T-cell leukemia virus (HTLV-ib) from an African patient with adult T-cell leukemia-lymphoma. Int. J. Cancer 34:613-618. [DOI] [PubMed] [Google Scholar]
- 20.Heidecker, G., S. Hill, P. A. Lloyd, and D. Derse. 2002. A novel protease processing site in the transframe protein of human T-cell leukemia virus type 1 PR76(gag-pro) defines the N terminus of reverse transcriptase. J. Virol. 76:13101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, S. A., M. Shuh, and D. Derse. 1999. Comparisons of defective HTLV-I proviruses predict the mode of origin and coding potential of internally deleted genomes. Virology 263:273-281. [DOI] [PubMed] [Google Scholar]
- 22.Hirose, S., Y. Uemura, M. Fujishita, T. Kitagawa, M. Yamashita, J. Imamura, Y. Ohtsuki, H. Taguchi, and I. Miyoshi. 1986. Isolation of HTLV-I from cerebrospinal fluid of a patient with myelopathy. Lancet ii:397-398. [DOI] [PubMed] [Google Scholar]
- 23.Janssen, R. S., J. E. Kaplan, R. F. Khabbaz, R. Hammond, R. Lechtenberg, M. Lairmore, M. A. Chiasson, A. Punsalang, B. Roberts, R. R. McKendall, et al. 1991. HTLV-I-associated myelopathy/tropical spastic paraparesis in the United States. Neurology 41:1355-1357. [DOI] [PubMed] [Google Scholar]
- 24.Karpas, A., K. Malik, and J. Lida. 1987. Studies of human retroviruses in relation to adult T-cell leukaemia, acquired immune deficiency syndrome, and multiple sclerosis. Arch. Virol. 95:237-249. [DOI] [PubMed] [Google Scholar]
- 25.Kimata, J. T., J. M. Wilson, and P. G. Patel. 2004. The increased replicative capacity of a late-stage simian immunodeficiency virus mne variant is evident in macrophage- or dendritic cell-T-cell cocultures. Virology 327:307-317. [DOI] [PubMed] [Google Scholar]
- 26.Kimata, J. T., F. H. Wong, J. J. Wang, and L. Ratner. 1994. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology 204:656-664. [DOI] [PubMed] [Google Scholar]
- 27.Komurian, F., F. Pelloquin, and G. de The. 1991. In vivo genomic variability of human T-cell leukemia virus type I depends more upon geography than upon pathologies. J. Virol. 65:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longo, D. L., E. P. Gelmann, J. Cossman, R. A. Young, R. C. Gallo, S. J. O'Brien, and L. A. Matis. 1984. Isolation of HTLV-transformed B-lymphocyte clone from a patient with HTLV-associated adult T-cell leukaemia. Nature 310:505-506. [DOI] [PubMed] [Google Scholar]
- 29.Mahieux, R., F. Ibrahim, P. Mauclere, V. Herve, P. Michel, F. Tekaia, C. Chappey, B. Garin, E. Van Der Ryst, B. Guillemain, E. Ledru, E. Delaporte, G. de The, and A. Gessain. 1997. Molecular epidemiology of 58 new African human T-cell leukemia virus type 1 (HTLV-1) strains: identification of a new and distinct HTLV-1 molecular subtype in Central Africa and in Pygmies. J. Virol. 71:1317-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann, D. L., J. Clark, M. Clarke, M. Reitz, M. Popovic, G. Franchini, C. D. Trainor, D. M. Strong, W. A. Blattner, and R. C. Gallo. 1984. Identification of the human T cell lymphoma virus in B cell lines established from patients with adult T cell leukemia. J. Clin. Investig. 74:56-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manns, A., M. Hisada, and L. La Grenade. 1999. Human T-lymphotropic virus type I infection. Lancet 353:1951-1958. [DOI] [PubMed] [Google Scholar]
- 32.Marriott, S. J., and O. J. Semmes. 2005. Impact of HTLV-I Tax on cell cycle progression and the cellular DNA damage repair response. Oncogene 24:5986-5995. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, M. S., J. Tozser, G. Princler, P. A. Lloyd, A. Auth, and D. Derse. 2006. Synthesis, processing, and composition of the virion-associated HTLV-1 reverse transcriptase. J. Biol. Chem. 281:3964-3971. [DOI] [PubMed] [Google Scholar]
- 34.Miura, T., T. Fukunaga, T. Igarashi, M. Yamashita, E. Ido, S. Funahashi, T. Ishida, K. Washio, S. Ueda, K. Hashimoto, et al. 1994. Phylogenetic subtypes of human T-lymphotropic virus type I and their relations to the anthropological background. Proc. Natl. Acad. Sci. USA 91:1124-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miura, T., M. Yamashita, V. Zaninovic, L. Cartier, J. Takehisa, T. Igarashi, E. Ido, T. Fujiyoshi, S. Sonoda, K. Tajima, and M. Hayami. 1997. Molecular phylogeny of human T-cell leukemia virus type I and II of Amerindians in Colombia and Chile. J. Mol. Evol. 44(Suppl. 1):S76-S82. [DOI] [PubMed] [Google Scholar]
- 36.Miyoshi, I., I. Kubonishi, S. Yoshimoto, T. Akagi, Y. Ohtsuki, Y. Shiraishi, K. Nagata, and Y. Hinuma. 1981. Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature 294:770-771. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi, I., H. Taguchi, I. Kubonishi, S. Yoshimoto, Y. Ohtsuki, Y. Shiraishi, and T. Akagi. 1982. Type C virus-producing cell lines derived from adult T-cell leukemia. Gann Monogr. Cancer Res. 28:219-228. [Google Scholar]
- 38.Moynet, D., J. Y. Cosnefroy, I. Bedjabaga, G. Roelants, M. C. Georges-Courbot, and B. Guillemain. 1995. Identification of new genetic subtypes of human T cell leukemia virus type I in Gabon from encoding sequence of surface envelope glycoprotein. AIDS Res. Hum. Retrovir. 11:1407-1411. [DOI] [PubMed] [Google Scholar]
- 39.Nerurkar, V. R., K. J. Song, R. R. Melland, and R. Yanagihara. 1994. Genetic and phylogenetic analyses of human T-cell lymphotropic virus type I variants from Melanesians with and without spastic myelopathy. Mol. Neurobiol. 8:155-173. [DOI] [PubMed] [Google Scholar]
- 40.Paine, E., J. Garcia, T. C. Philpott, G. Shaw, and L. Ratner. 1991. Limited sequence variation in human T-lymphotropic virus type 1 isolates from North American and African patients. Virology 182:111-123. [DOI] [PubMed] [Google Scholar]
- 41.Patel, P. G., M. T. Yu Kimata, J. E. Biggins, J. M. Wilson, and J. T. Kimata. 2002. Highly pathogenic simian immunodeficiency virus mne variants that emerge during the course of infection evolve enhanced infectivity and the ability to downregulate CD4 but not class I major histocompatibility complex antigens. J. Virol. 76:6425-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poiesz, B. J., F. W. Ruscetti, J. W. Mier, A. M. Woods, and R. C. Gallo. 1980. T-cell lines established from human T-lymphocytic neoplasias by direct response to T-cell growth factor. Proc. Natl. Acad. Sci. USA 77:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popovic, M., P. S. Sarin, M. Robert-Gurroff, V. S. Kalyanaraman, D. Mann, J. Minowada, and R. C. Gallo. 1983. Isolation and transmission of human retrovirus (human T-cell leukemia virus). Science 219:856-859. [DOI] [PubMed] [Google Scholar]
- 45.Ramirez, E., L. Cartier, C. Villota, and J. Fernandez. 2002. Genetic characterization and phylogeny of human T-cell lymphotropic virus type I from Chile. Virus Res. 84:135-149. [DOI] [PubMed] [Google Scholar]
- 46.Ratner, L., T. Philpott, and D. B. Trowbridge. 1991. Nucleotide sequence analysis of isolates of human T-lymphotropic virus type 1 of diverse geographical origins. AIDS Res. Hum. Retrovir. 7:923-941. [DOI] [PubMed] [Google Scholar]
- 47.Ratner, L., N. Vander Heyden, E. Paine, D. Frei-Lahr, R. Brown, P. Petruska, S. Reddy, and M. D. Lairmore. 1990. Familial adult T-cell leukemia/lymphoma. Am. J. Hematol. 34:215-222. [DOI] [PubMed] [Google Scholar]
- 48.Salahuddin, S. Z., P. D. Markham, F. Wong-Staal, G. Franchini, V. S. Kalyanaraman, and R. C. Gallo. 1983. Restricted expression of human T-cell leukemia-lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 129:51-64. [DOI] [PubMed] [Google Scholar]
- 49.Salemi, M., S. Van Dooren, E. Audenaert, E. Delaporte, P. Goubau, J. Desmyter, and A. M. Vandamme. 1998. Two new human T-lymphotropic virus type I phylogenetic subtypes in seroindeterminates, a Mbuti pygmy and a Gabonese, have closest relatives among African STLV-I strains. Virology 246:277-287. [DOI] [PubMed] [Google Scholar]
- 50.Sarin, P. S., P. Rodgers-Johnson, D. K. Sun, A. H. Thornton, O. S. Morgan, W. N. Gibbs, C. Mora, G. McKhann II, D. C. Gajdusek, and C. J. Gibbs, Jr. 1989. Comparison of a human T-cell lymphotropic virus type I strain from cerebrospinal fluid of a Jamaican patient with tropical spastic paraparesis with a prototype human T-cell lymphotropic virus type I. Proc. Natl. Acad. Sci. USA 86:2021-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw, G. M., B. H. Hahn, M. Popovic, A. Lo Monico, F. Wong-Staal, and R. Gallo. 1984. Acquired immune deficiency syndrome: proceedings of a Schering Corp.-UCLA symposium held in Park City, Utah, February 5-10, 1984. A.R. Liss, New York, NY.
- 53.Shuh, M., S. A. Hill, and D. Derse. 1999. Defective and wild-type human T-cell leukemia virus type I proviruses: characterization of gene products and trans-interactions between proviruses. Virology 262:442-451. [DOI] [PubMed] [Google Scholar]
- 54.Slattery, J. P., G. Franchini, and A. Gessain. 1999. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 9:525-540. [PubMed] [Google Scholar]
- 55.Sudol, M., T. L. Lerner, and H. Hanafusa. 1986. Polymerase-defective mutant of the Bryan high-titer strain of Rous sarcoma virus. Nucleic Acids Res. 14:2391-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchiyama, T. 1997. Human T cell leukemia virus type I (HTLV-I) and human diseases. Annu. Rev. Immunol. 15:15-37. [DOI] [PubMed] [Google Scholar]
- 57.Vandamme, A. M., H. F. Liu, P. Goubau, and J. Desmyter. 1994. Primate T-lymphotropic virus type I LTR sequence variation and its phylogenetic analysis: compatibility with an African origin of PTLV-I. Virology 202:212-223. [DOI] [PubMed] [Google Scholar]
- 58.Van Dooren, S., E. Gotuzzo, M. Salemi, D. Watts, E. Audenaert, S. Duwe, H. Ellerbrok, R. Grassmann, E. Hagelberg, J. Desmyter, and A. M. Vandamme. 1998. Evidence for a post-Columbian introduction of human T-cell lymphotropic virus [type I] [corrected] in Latin America. J. Gen. Virol. 79:2695-708. [DOI] [PubMed] [Google Scholar]
- 59.Waziri, A., S. S. Soldan, M. D. Graf, J. Nagle, and S. Jacobson. 2000. Characterization and sequencing of prototypic human T-lymphotropic virus type 1 (HTLV-1) from an HTLV-1/2 seroindeterminate patient. J. Virol. 74:2178-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wodarz, D., and C. R. Bangham. 2000. Evolutionary dynamics of HTLV-I. J. Mol. Evol. 50:448-455. [DOI] [PubMed] [Google Scholar]
- 61.Yoshida, M. 2001. Multiple viral strategies of HTLV-1 for dysregulation of cell growth control. Annu. Rev. Immunol. 19:475-496. [DOI] [PubMed] [Google Scholar]