Abstract
The PML oncogenic domain (POD/ND10/PML body) is a common target of DNA viruses, which replicate their genomes in proximity to this nuclear structure. The adenovirus early protein E4 ORF3 is both necessary and sufficient to rearrange PODs from punctate bodies into track-like structures. Although multiple hypotheses exist, the precise reason for this activity has not yet been elucidated. PML, the protein responsible for nucleating PODs, is an interferon (IFN)-stimulated gene, implicating the participation of this nuclear body in an innate antiviral response. Here, we demonstrate that E4 ORF3 is critical to the replicative success of adenovirus during the IFN-induced antiviral state. When cells are pretreated with either IFN-α or IFN-γ, a mutant virus that does not express E4 ORF3 is severely compromised for replication. This result suggests the functional significance of ORF3 track formation is the inhibition of a POD-mediated, antiviral mechanism. Replication of the E4 ORF3 mutant virus can be rescued following the introduction of E4 ORF3 from evolutionarily divergent adenoviruses, suggesting a conserved function for E4 ORF3 inhibition of the IFN-induced antiviral state. Furthermore, E4 ORF3 inhibition of an IFN-induced response is unrelated to the inhibition of adenovirus replication by the Mre11-Rad50-Nbs1 DNA repair complex. We propose that the evolutionarily conserved function of the adenovirus E4 ORF3 protein is the inhibition of a host interferon response to viral infection via disruption of the PML oncogenic domain.
Adenovirus (Ad) early region 4 (E4) encodes a variety of proteins responsible for executing both disparate and essential functions within an infected cell. Among them, E4 open reading frame 3 (E4 ORF3) and E4 ORF6 are critical for efficient viral DNA replication (4, 24). E4 ORF3 and E4 ORF6 execute complementary functions in genome replication via the inhibition of the cellular Mre11-Rad50-Nbs1 (MRN) DNA repair complex. In the absence of E4 ORF3 and E4 ORF6 expression, the Ad genome induces an MRN-dependent DNA damage response that results in concatenation of viral genomes (49), thereby effectively inhibiting viral DNA replication. To counteract this host response, E4 ORF6 impedes MRN activity by targeting its components for degradation by the ubiquitin-mediated proteasome-dependent pathway (27, 42). In contrast to this strategy, E4 ORF3 inhibits MRN activity by sequestering the nucleoplasmic pool of MRN proteins into E4 ORF3-containing track-like structures (14, 27, 42, 43).
In addition to MRN complex inhibition, both E4 ORF3 and E4 ORF6 perform discrete functions (45). E4 ORF6 targets a variety of cellular proteins (e.g., p53) for degradation (22, 34) and inhibits the transport of cellular mRNAs from the nucleus (45). Distinct from those of E4 ORF6, several functions have been attributed to E4 ORF3, including the regulation of Ad mRNA splicing, cell cycle-independent virus growth, the enhancement of translation of late viral mRNAs, and the targeting of MRN to cytoplasmic aggresomes (1, 19, 32, 33, 38, 39). While the aforementioned activities occur late after infection, E4 ORF3 function is not restricted to this phase of the viral lytic cycle. In the early hours of infection, E4 ORF3 mediates the reorganization of PML oncogenic domains (PODs/ND10s/PML nuclear bodies) into track-like structures within the nucleus (6, 10) and is both necessary and sufficient to execute this process.
PML oncogenic domains are electron-dense nuclear subdomains, nucleated by promyelocytic leukemia protein, PML (3, 12). In cases of acute promyelocytic leukemia, the hematopoietic disorder for which PML was named, a reciprocal translocation event generates a fusion protein expressing the N terminus of PML and the C terminus of the retinoic acid receptor alpha (RARα). The resulting PML-RARα product disrupts the formation of discrete, punctate PODs and generates dispersed, micropunctate structures. Furthermore, the PML-RARα product stimulates uncontrolled cellular proliferation (44). Remission of the leukemic state occurs upon treatment with either all-trans retinoic acid or arsenic trioxide and is associated with restoration of the typical PML localization. As such, POD integrity is correlated with the regulation of cellular proliferation (44).
In addition to its association with proliferation and transformation, PML nuclear bodies have been implicated in a wide variety of cellular processes, including transcriptional regulation, apoptosis, and posttranslational modification (3, 23, 37, 54). This may reflect the diverse population of proteins associated with PML bodies. A subset of cellular proteins that localize to PODs are the products of IFN-stimulated genes (ISGs) (35). IFNs signal through cell surface receptors to effect the activation of specific gene transcription. IFNs are classified in two major groups (type I and type II) that bind to distinct receptors and activate overlapping and distinct pathways (35). Type I IFNs are induced directly by viral infection in most cell types, whereas a subset of lymphocytes produce type II IFN following stimulation by foreign antigen. When cells are treated with type I IFN (e.g., IFN-α) or type II IFN (IFN-γ), there is a dramatic augmentation in both the size and number of PML bodies (20, 26). This is believed to be reflective of the fact that the PML gene itself is an ISG. An IFN-stimulated response element and a gamma activation site (GAS), responsive to type I and type II IFNs, respectively, are located in the 5′ untranslated region of the PML gene (41). For this reason, PML bodies have been postulated to contribute to innate immunity by participating in the IFN-induced antiviral state (35).
Due to their association with multiple cellular processes, it is not surprising that numerous viruses target PML bodies. DNA viruses such as the herpesviruses, papovavirses, and adenoviruses undergo replication in proximity to PODs and encode proteins that associate with the nuclear bodies (16, 28). As is the case for herpes simplex virus type 1 (HSV-1) ICP0, human cytomegalovirus (HCMV) IE1, and Ad E4 ORF3, the proteins that mediate POD association also disrupt the structural integrity of these nuclear bodies (16, 28). In the case of both HSV-1 and HCMV, PML was found to play a role in mediating the antiviral effects of IFN treatment (7, 8, 15, 46). Interestingly, this interference with PML body integrity is not limited to DNA viruses, as several RNA viruses have also been found to alter POD structure (2, 9, 36).
The reorganization of PODs by the Ad E4 ORF3 protein is an evolutionarily conserved function among all Ad subgroups, whereas MRN relocalization by E4 ORF3 is not (43). In addition, mutational analyses of E4 ORF3 have demonstrated that PML and MRN reorganization are discrete functions of the protein (14). While POD rearrangement by E4 ORF3 is conserved, the functional consequence of this activity has not yet been delineated. Here, we demonstrate that the E4 ORF3 protein is necessary for viral DNA replication during the antiviral state induced by either IFN-α or IFN-γ. E4 ORF3-induced POD rearrangement is critical for the inhibition of an IFN-induced antiviral state, and this function is conserved among evolutionarily divergent Ad serotypes. Furthermore, E4 ORF3 inhibition of an IFN-induced response is unrelated to the inhibition of Ad replication by the Mre11-Rad50-Nbs1 DNA repair complex. We propose that the evolutionarily conserved function of the Ad E4 ORF3 protein is the inhibition of a host IFN response to viral infection.
MATERIALS AND METHODS
Cells, IFN, viruses, and infections.
Vero cells (11) were maintained in Dulbecco's modified Eagle medium supplemented with 10% bovine calf serum. IMR90 cells (ATCC) were propagated in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Cells were pretreated for 24 h with IFN (1,000 U/ml IFN-α [Hoffman LaRoche] or 2,000 U/ml IFN-γ [R&D Systems]). Following IFN pretreatment, cells were infected with different wild-type and mutant viruses. Viruses dl309 (wild-type Ad5), dl355 (E4 ORF6-minus), and E4inORF3 (E4 ORF3-minus) were described previously (21, 24). The Ad-CMV, E1 replacement viruses that express Ad5-HA-ORF3, Ad9-HA-ORF3, and Ad12-HA-ORF3, and mutant viruses dl355/D105A,L106A and dl355-N82A were described previously, (13, 14). Ad3 and Ad4 E4 ORF3 coding sequences were introduced into the E1A-CMV-HA transient expression vector (13). Virus particles were purified by CsCl equilibrium centrifugation (13). Vero cells and IMR90 cells were infected at 37°C using 200 virus particles/cell with dl309, dl355, and E4inORF3 or at 1,000 particles/cell with the E1 replacement viruses. Briefly, medium was aspirated from the cells and replaced with 1 ml of inoculum in the case of 100-mm plates or 0.5 ml in the case of coverslips. After 1 h, the viral suspension was removed and the cells washed. Dulbecco's modified Eagle medium supplemented with 10% serum was replaced and IFN added, where appropriate. The cells were then incubated at 37°C in 5% CO2.
Immunoassays.
For immunofluorescence, Vero cells and IMR90 cells were seeded onto glass coverslips and infected as described above. At 18 h after infection of Vero cells or 72 h after infection of IMR90 cells, the cells were washed with phosphate-buffered saline (PBS), fixed with −20°C methanol for 5 min, and then washed with PBS. Cells were blocked in PBS containing 10% goat serum for 1 h at room temperature. Antibodies were diluted in PBS containing 10% goat serum and incubated with fixed cells for 1 h at room temperature. The antibodies and dilutions used were as follows: 1:50 anti-DNA binding protein (DBP) DBP mouse monoclonal antibody B6-8 (from Arnold Levine, Cancer Institute of New Jersey), 1:300 anti-PML rabbit polyclonal antibody H-238 (Santa Cruz Biotechnology), 1:10 anti-ORF3 rat monoclonal 6A-11 (from Thomas Dobner, Regensburg University), 1:300 antihemagglutinin (anti-HA) rabbit polyclonal antibody Y-11 (Santa Cruz Biotechnology), and 1:2,500 anti-DBP rabbit polyclonal antibody (from P. van der Vleit, University Medical Centre Utrecht). Subsequently, the cells were washed with PBS and incubated with 1:300 dilutions of Alexa350-conjugated goat anti-mouse immunoglobulin G (IgG) (Molecular Probes), Alexa350-conjugated goat anti-rabbit IgG (Molecular Probes), fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Zymed), Texas red isothiocyanate-conjugated goat anti-rat IgG (Zymed), or Texas red isothiocyanate-conjugated goat anti-rabbit IgG (Zymed) for 45 min at room temperature in the dark. The cells were then washed with PBS, and the coverslips were mounted onto slides with Immu-Mount (Shandon). Microscopy was conducted using a Zeiss Axiovert 200 M digital deconvolution microscope, and images were captured and analyzed with Axiovision 4.5 software.
For Western blot analysis, cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (4% SDS, 120 mM Tris, pH 6.8, supplemented with protease inhibitors), and standardized quantities of protein were subjected to 12.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to Hybond-P (Amersham), and the membranes were probed with 1:10 anti-DBP mouse monoclonal antibodies A1-6 and B6-8 (from Arnold Levine) or a 1:300 anti-E1A rabbit polyclonal antibody 13S-5 (Santa Cruz Biotechnology). E1A and DBP were detected using alkaline phosphatase reagent (Promega), scanned on an ABI Storm 680 PhosphorImager, and quantified using ImageQuant software (Molecular Dynamics). To visualize γ-tubulin, 1:5,000 anti-γ-tubulin rabbit polyclonal antibody (Sigma) was utilized and detected by enhanced chemiluminescent horseradish peroxidase substrate (Millipore).
Analysis of viral DNA replication.
To determine the extent of viral DNA replication, high-molecular-weight DNA was prepared from nuclei of infected cells isolated at 18 h after infection, as described elsewhere (13). The DNA was diluted 1:50, denatured in 0.3 N NaOH for 1 h at 65°C, and then neutralized in 2 M ammonium acetate. Samples were applied to Hybond N+ (Amersham) with a slot blot apparatus. The DNAs were cross-linked using a Stratalinker (Stratagene) and then probed by Southern hybridization. An Ad5 whole-genome probe labeled with [32P]dATP by random priming was utilized (13). The results were visualized on an ABI Storm 680 PhosphorImager and quantified using ImageQuant software (Molecular Dynamics).
RESULTS
E4 ORF3 is necessary for efficient viral DNA replication following IFN treatment.
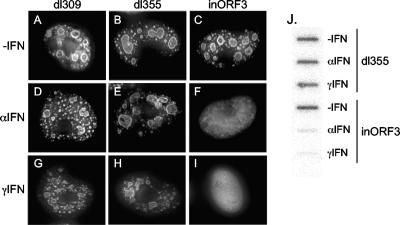
Vero cells were utilized in these analyses, as they respond well to IFN treatment and lack the endogenous type I IFN locus, preventing autocrine activation (11, 50). Cells were pretreated with either IFN-α or IFN-γ for 24 h or left untreated. Subsequently, cells were infected with dl309 (wild-type Ad5), dl355 (E4 ORF6-minus), or E4inORF3 (E4 ORF3-minus). After 18 h, the cells were fixed and immunostained with an antibody against the viral DBP (Fig. 1). The presence of circular, DBP-positive viral replication centers in the nucleus directly correlates with active viral DNA replication (48) and was apparent in untreated cells infected with all three viruses (Fig. 1A to C), signifying the replication competence of the viruses in Vero cells under standard culture conditions. dl309 and dl355 were capable of significant viral DNA replication following infection of cells treated with either IFN-α or IFN-γ (Fig. 1D, E, G, and H). In contrast, only diffuse nuclear DBP staining was evident in cells infected with mutant E4inORF3 and treated with either IFN-α or IFN-γ (Fig. 1F and I), indicative of a replication defect (48).
FIG. 1.
E4 ORF3 is required for efficient Ad DNA replication following IFN treatment. Vero cells were grown on coverslips and were untreated (±IFN) (A to C) or pretreated with IFN-α (D to F) or IFN-γ (G to I) for 24 h. Subsequently, the cells were infected with dl309 (ORF3+/ORF6+) (A, D, and G), dl355 (ORF3+/ORF6−) (B, D, and H), or E4inORF3 (ORF3−/ORF6+) (C, E, and I). At 18 h after infection, cells were immunostained for Ad DBP. The images represent compressed, deconvolved Z-stacks. J. Vero cells were untreated (−IFN) or pretreated with IFN-α or IFN-γ for 24 h. Subsequently, the cells were infected with dl355 (ORF3+/ORF6−) or E4inORF3 (ORF3−/ORF6+). Total nuclear DNA was isolated at 18 h after infection, slot blotted, and probed by Southern hybridization.
To confirm this observation biochemically, a viral DNA replication assay was performed to quantify the levels of replication. Cells were pretreated with either IFN-α or IFN-γ for 24 h, or left untreated, and infected with dl355 or E4inORF3. Total nuclear DNA was isolated 18 h after infection, slot blotted, and probed by Southern blot analysis. Viral DNA at the 18-h time point corresponds to newly replicated DNA. These results recapitulated the immunofluorescence data and demonstrated that mutant E4inORF3 exhibited a marked reduction in viral DNA replication in cells treated with either IFN-α or IFN-γ (Fig. 1J). Mutant E4inORF3 replication was reduced 15- to 20-fold in the presence of IFN. In contrast, dl355 replicated efficiently either in the presence or absence of IFN treatment. These results demonstrate that E4 ORF3 is necessary for efficient Ad DNA replication during an IFN response and contributes a unique function to the process, which is not provided by other Ad5 proteins, including E4 ORF6.
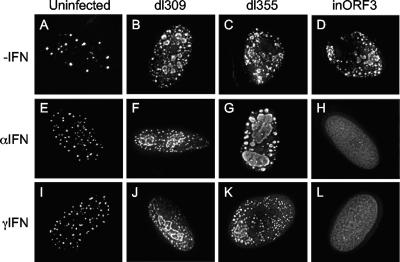
To more thoroughly investigate the functional significance of E4 ORF3 during the IFN-induced antiviral state, we examined the effect of IFN treatment following infection of primary human diploid fibroblasts, IMR90 cells. Here, cells were pretreated with either IFN-α or IFN-γ for 24 h and subsequently infected with dl309, dl355, or E4inORF3 or were mock infected (Fig. 2). The cells were fixed and immunostained with antibodies against either PML or Ad DBP at 72 h postinfection. PML bodies became more numerous in both IFN-α- and IFN-γ-stimulated cells, signifying that IMR90 cells are responsive to IFN (Fig. 2A, E, and I). Upon infection under standard tissue culture conditions, staining for Ad DBP revealed the presence of numerous viral replication centers in cells infected with dl309, dl355, and E4inORF3 (Fig. 2B to D). As observed in Vero cells, both dl309 and dl355 were capable of replicating in cells that had been subjected to either IFN-α or IFN-γ pretreatment (Fig. 2F, G, J, and K). In contrast, E4inORF3 did not form viral replication centers in cells stimulated with either IFN-α or IFN-γ, as indicated by diffuse DBP staining (Fig. 2H and L). These data further strengthen the assertion that E4 ORF3 is necessary for efficient Ad viral DNA replication during the IFN-induced antiviral state and demonstrate its functional significance in human cells.
FIG. 2.
E4 ORF3 is required for efficient Ad DNA replication following IFN treatment in primary human diploid fibroblasts. IMR90 cells were grown on coverslips and were untreated (−IFN) (A to D) or pretreated with IFN-α (E to H) or IFN-γ (I to L) for 24 h. Subsequently, the cells were infected with dl309 (ORF3+/ORF6+) (B, F, and J), dl355 (ORF3+/ORF6−) (C, G, and K), or E4inORF3 (ORF3−/ORF6+) (D, H, and L) or were mock infected (A, E, and I). At 72 h after infection, cells were immunostained for either PML (A, E, and I) or Ad DBP (B to D, F to H, and J to L). The images represent compressed, deconvolved Z-stacks.
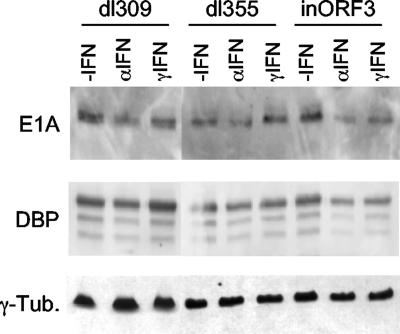
To determine if this replication defect was fully attributable to the absence of E4 ORF3 and not a consequence of reduced synthesis of other early viral proteins essential to viral DNA replication, expression of early viral genes was examined. Vero cells were pretreated with IFN-α, IFN-γ, or left untreated, and infected with dl309, dl355, or E4inORF3. Cell lysates were prepared, and E1A and DBP levels were measured by Western blot analysis (Fig. 3). γ-Tubulin levels were assayed as a loading control. All three viruses, with or without IFN treatment, synthesized similar levels of these viral proteins. When the results were quantified, modest decreases (twofold or less) in E1A and DBP levels were observed in cells infected with dl309 and dl355 following IFN treatment, which was slightly greater than that in IFN-treated cells infected with E4inORF3 (two- to fourfold decreases). Such modest differences in E1A and DBP levels between dl309, dl355, and E4inORF3 are not likely to explain the replication defect observed in IFN-treated cells infected with mutant E4inORF3 and indicate that this defect does not occur as a consequence of reduced synthesis of these early viral proteins.
FIG. 3.
IFN treatment does not significantly reduce the expression of Ad early proteins. Vero cells were untreated (−IFN) or pretreated with IFN-α or IFN-γ for 24 h. Subsequently, the cells were infected with dl309 (ORF3+/ORF6+), dl355 (ORF3+/ORF6−), or E4inORF3 (ORF3−/ORF6+). Total cell lysates were prepared 18 h after infection and analyzed by Western blotting using antibodies against E1A, DBP, and γ-tubulin.
E4 ORF3 rearranges PML bodies into tracks following IFN treatment.
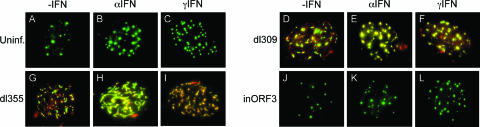
PML is an IFN-stimulated gene, and both the size and number of PODs are augmented by IFN treatment (20, 26, 41). As such, this nuclear subdomain has been implicated in cellular antiviral activities (35). For this reason, we examined if E4 ORF3 was capable of rearranging PODs into track-like structures in the cellular environment of increased PML synthesis associated with IFN stimulation. Vero cells were pretreated with IFN-α or IFN-γ for 24 h or were left untreated. Subsequently, the cells were infected with either dl309, dl355, E4inORF3, or mock infected. The cells were then fixed and immunostained with antibodies against PML and E4 ORF3 (Fig. 4). As previously reported (20, 26, 41), both the size and number of PML bodies were increased as a result of treatment with either IFN-α or IFN-γ (Fig. 4A to C). Both dl309 and dl355 were fully capable of rearranging PML into characteristic track-like structures that contain E4 ORF3 following IFN treatment in a manner indistinguishable from PML body rearrangement in virus-infected, untreated cells (Fig. 4D to I). Rearrangement of IFN-induced PML was not observed when cells were infected with mutant E4inORF3 (Fig. 4J to L). Because E4 ORF3 is known to be both necessary and sufficient for PML rearrangement (6, 10) and the deletion of E4 ORF3 elicits a dramatic replication defect during an IFN response (Fig. 1 and 2), these data indicate that E4 ORF3-induced PML track formation during an IFN response inhibits an IFN-induced, antiviral function of the POD.
FIG. 4.
E4 ORF3 rearranges PML during an IFN response. Vero cells seeded onto coverslips were untreated (A, D, G, and J) or pretreated with IFN-α (B, E, H, and K) or IFN-γ (C, F, I, and L) for 24 h. Subsequently, the cells were left uninfected (A to C) or infected with dl309 (ORF3+/ORF6+) (D to F), dl355 (ORF3+/ORF6−) (G to I), or E4inORF3 (ORF3−/ORF6+) (J to L). At 18 h after infection, the cells were immunostained for PML (monoclonal antibody PG-M3, fluorescein isothiocyanate-coupled secondary antibody) and ORF3 (monoclonal antibody 6A11, tetramethyl rhodamine isocyanate (TRITC)-coupled secondary antibody). The images represent a merge of compressed, deconvolved Z-stacks.
Inhibition of an IFN response is a conserved function of E4 ORF3 among evolutionarily divergent Ad serotypes.
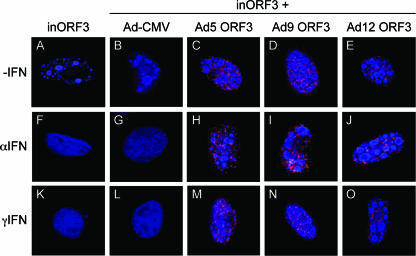
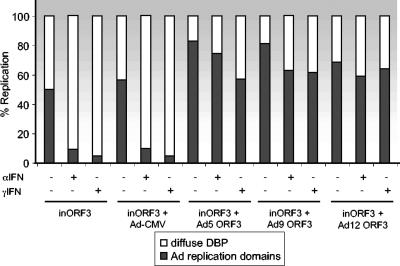
The ability of E4 ORF3 to rearrange PML bodies from punctate nuclear domains into track-like structures is a common function of E4 ORF3 of all Ad serotypes examined to date (43). This observation implies that E4 ORF3-mediated rearrangement of PML performs an essential function; however, such a function has not yet been elucidated. Due to POD association with innate immune responses, PML rearrangement could potentially relate to Ad replicative success during the IFN-induced antiviral state. To examine if the E4 ORF3 proteins of different Ad serotypes are capable of blocking the effect of IFN on Ad replication, Vero cells were pretreated with IFN-α, IFN-γ, or left untreated and then coinfected with two viruses: mutant E4inORF3 and one of several E1-replacement viruses that express E4 ORF3 proteins of different Ad serotypes. The E1-replacement viruses carry a deletion of E4 open reading frames 1 to 3, and so they do not express E4 ORF3 from the natural E4 context. Mutant E4inORF3 expresses the E1A and E1B proteins missing in the E1 replacement viruses, and the E1 replacement viruses express E4 ORF3 proteins missing in mutant E4inORF3. Ad DBP and E4 ORF3 were visualized after infection via immunofluorescence (Fig. 5). The presence of DBP-positive viral replication centers directly correlates with active viral DNA replication, whereas diffuse DBP staining is indicative of a replication defect (48) (Fig. 1). As described above, mutant E4inORF3 replicated in the absence of IFN treatment, but not following treatment of cells with either IFN-α or IFN-γ (Fig. 5A, F, and K). No E4 ORF3 expression was observed, as expected, due to mutation of the E4 ORF3 protein. The same results were observed in cells coinfected with E4inORF3 and an E1 replacement virus that does not express the E4 ORF3 protein (Ad-CMV) (Fig. 5B, G, and L). Coinfection of IFN-treated cells with mutant E4inORF3 and E1 replacement viruses that express E4 ORF3 proteins of Ad5 (Fig. 5C, H, and M), Ad9 (Fig. 5D, I, and N), or Ad12 (Fig. 5E, J, and O) resulted in the restoration of viral replication centers following IFN pretreatment to levels comparable to that of mutant E4inORF3 without IFN treatment. Similarly, E4inORF3 infection of IFN-treated cells in conjunction with transfection of vectors for the expression of E4 ORF3 of Ad3 or Ad4 yielded comparable results (data not shown). E4 ORF3 was evident in track-like structures with all five serotypes. Replication was not detected in any cells infected with the E1 replacement viruses alone (data not shown). The quantification of these results is shown in Fig. 6. In the absence of IFN treatment, Ad replication domains were observed in >50% of cells infected with mutant E4inORF3. Similar results were observed in cells coinfected with E4inORF3 and Ad-CMV. In both cases, IFN treatment reduced the number of cells showing replication domains to <10%. When cells were coinfected with mutant E4inORF3 and E1-replacement viruses expressing E4 ORF3 of Ad9 or Ad12 without IFN treatment, the number of cells exhibiting the formation of replication domains increased to 70 to 80%. This result is consistent with the increase in virus growth observed when E4 ORF3 is expressed under ordinary culture conditions (4, 24). When cells were treated with either IFN-α or IFN-γ, the expression of E4 ORF3 by the E1 replacement viruses rescued the replication defect observed with mutant E4inORF3 such that ≥60% of cells expressing HA-ORF3 tracks exhibited viral DNA replication domains, in contrast to the <10% of cells infected with mutant E4inORF3 alone or E4inORF3 in conjunction with Ad-CMV. These results demonstrate that the inhibition of the IFN-induced antiviral state by the E4 ORF3 protein is a function that is conserved among Ad serotypes.
FIG. 5.
E4 ORF3 proteins of different Ad serotypes block the effect of IFN on Ad replication. Vero cells were seeded onto coverslips and were untreated (A to E) or pretreated with IFN-α (F to J) or IFN-γ (K to O) for 24 h. Subsequently, the cells were infected with mutant E4inORF3 alone (A, F, and K) or coinfected with E4inORF3 and an E1 replacement virus that does not express any E4 ORF3 protein (Ad-CMV) (B, G, and L) or E1 replacement viruses that express E4 ORF3 proteins of Ad5 (C, H, and M), Ad9 (D, I, and N), and Ad12 (E, J, and O). At 18 h after infection, cells were immunostained for DBP (monoclonal antibody B6-8, Alexa350-coupled secondary antibody) and HA (rabbit antibody Y-11, TRITC-coupled secondary antibody). The images represent a merge of compressed, deconvolved Z-stacks.
FIG. 6.
Quantification of Ad replication centers in coinfection experiments with E4 ORF3 proteins of different Ad serotypes. The percentage of cells exhibiting viral replication centers is presented in cells, with or without IFN pretreatment, infected with mutant E4inORF3 alone or coinfected with E1 replacment viruses that express no E4 ORF3 protein (Ad-CMV) or E4 ORF3 proteins of Ad5, Ad9, and Ad12. Replication was quantified by counting cells that exhibited viral replication centers in comparison to cells that displayed diffuse nuclear DBP staining. The results represent the average of three independent experiments where >100 infected cells were quantified per virus infection.
The MRN complex is not responsible for the inhibition of Ad replication during an IFN response.
The E4 ORF3 proteins of subgroup C adenoviruses are capable of rearranging the MRN complex into track-like structures, but E4 ORF3 proteins produced by adenoviruses of other subgroups are not (43). This function is hypothesized to protect the linear double-stranded DNA genomes from nucleolytic cleavage by the endo- and exonuclease activities of Mre11 (52). While Mre11, Rad50, and Nbs1 do not appear to be ISGs, as assessed by protein levels in the presence or absence of IFN treatment (data not shown), the possibility remained that the enzymatic activities of MRN are enhanced due to IFN treatment. To determine if the MRN complex is involved in the IFN-induced replication defect of mutant E4inORF3, Vero cells were coinfected with mutant E4inORF3 and one of two mutant viruses carrying specific point mutants in the E4 ORF3 reading frame in the natural E4 context: dl355-D105A/L106A or dl355-N82A. The D105A/L106A mutations in E4 ORF3 prevent rearrangement of the MRN complex into track-like structures but do not affect the ability of the mutant protein to rearrange PML (14), thereby segregating the ability of E4 ORF3 to rearrange PML from its ability to rearrange the MRN complex. The N82A mutation in E4 ORF3 blocks both PML and MRN reorganization (42). Both E4 ORF3 mutants are in the genomic background of dl355, a virus that does not express the E4 ORF6 protein. Both of these mutant viruses are completely defective for viral growth due to the mutations in the E4 ORF3 and E4 ORF6 proteins. Thus, in these coinfection experiments, functional E4 ORF6 is provided by the virus E4inORF3 and the D105A/L106A or N82A mutant E4 ORF3 proteins are provided by the viruses dl355-D105A/L106A or dl355-N82A.
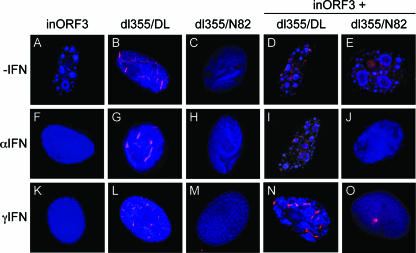
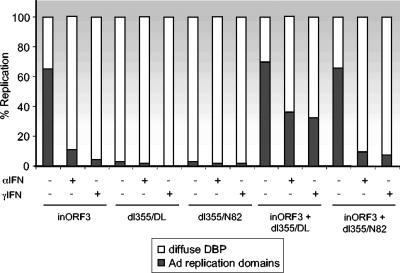
Vero cells were pretreated with either IFN-α or IFN-γ, or left untreated, and infected individually or in combination with these viruses. Viral replication centers and the E4 ORF3 proteins were visualized after infection via immunofluorescence (Fig. 7). Mutant E4inORF3 exhibited a significant number of viral replication centers in the absence of IFN treatment, as described above. In single infections, mutants dl355-D105A/L106A and dl355-N82A were defective for replication due to the mutations in the E4 ORF3 and E4 ORF6 proteins (Fig. 7A to C), as previously described (42). None of these three viruses replicated in single infections when cells were pretreated with IFN-α or IFN-γ (Fig. 7F to H and K to M). In the absence of IFN treatment, efficient viral replication was evident in cells coinfected with E4inORF3 and either dl355-D105A/L106A or dl355-N82A, as indicated by the formation of numerous viral replication centers (Fig. 7D and E), as observed with E4inORF3 infection alone (Fig. 7A). When cells were pretreated with either IFN-α or IFN-γ, coinfection of cells with E4inORF3 and dl355-D105A/L106A rescued the replication defect observed with E4inORF3 infection alone in the presence of IFNs (Fig. 7F and K versus I and N). In contrast, coinfection of cells with E4inORF3 and dl355-N82A did not rescue the replication defect when cells were pretreated with either IFN-α or IFN-γ, as indicated by diffuse DBP nuclear staining (Fig. 7J and O). Quantification of these results is presented in Fig. 8. Because the E4 ORF3 mutant D105A/L106A is capable of inhibiting the IFN-induced antiviral state and is able to reorganize PML but not MRN, we conclude that the IFN-induced replication defect does not relate to inhibition of the MRN complex. This conclusion is consistent with the replication defect observed in IFN-treated cells infected with mutant E4inORF3 alone where the E1B 55K-E4 ORF6 complex is present and would be expected to inhibit MRN activity by degradation, yet IFN treatment still inhibited viral DNA replication.
FIG. 7.
The MRN complex does not contribute the effect of IFN on Ad replication. Vero cells were seeded onto coverslips and were untreated (−IFN) (A to E) or pretreated with IFN-α (F to J) or IFN-γ (K to O) for 24 h. Subsequently, the cells were infected with mutant E4inORF3 alone (A, F, and K), dl355-D105A/L106A alone (B, G, and L), dl355-N82A alone (C, H, and M), or coinfected with E4inORF3 and dl355-D105A/L106A (D, I, and N) or dl355-N82A (E, J, and O). At 18 h after infection, the cells were immunostained DBP (monoclonal antibody B6-8, Alexa350-coupled secondary antibody) and E4 ORF3 (monoclonal antibody 6A11, TRITC-coupled secondary antibody). The images represent a merge of compressed, deconvolved Z-stacks.
FIG. 8.
Quantification of Ad replication centers in coinfection experiments with E4 ORF3 mutant viruses. The percentage of cells exhibiting viral replication centers was quantified as described for Fig. 6.
DISCUSSION
PML nuclear bodies have been implicated in the innate immune response due to the dramatic amplification of both POD size and number, which occurs upon stimulation with either type I or type II IFN. This likely is attributable to the fact that a variety of proteins known to localize at the POD, including PML itself, are ISGs (35). Reorganization of PML bodies into track-like structures is a conserved function of the Ad E4 ORF3 protein (43), yet the consequence(s) of this activity previously has not been elucidated. For this reason, we examined the potential function of E4 ORF3-mediated POD rearrangement during the IFN-induced antiviral state.
We have demonstrated that when cells have mounted either a type I or type II IFN response, the E4 ORF3 protein is required for efficient Ad DNA replication. This phenomenon is illustrated by the significant replication defect exhibited by cells that have been prestimulated with either IFN-α or IFN-γ and subsequently infected with E4inORF3, a mutant virus that does not express E4 ORF3 (Fig. 1 and 2). Significantly, this IFN-induced replication defect mimics the phenotype associated with deletion of the entire E4 region. Here, viral DNA replication is severely compromised (21, 51); however, under standard culture conditions, restoration of either E4 ORF3 or E4 ORF6 rescues viral genome replication to levels comparable to that of wild-type virus (4, 24). In contrast, this IFN-induced replication defect is not abrogated by the presence of E4 ORF6. These results demonstrate that E4 ORF3 is necessary for efficient viral DNA replication during the IFN-induced antiviral state and define a novel function for this viral protein.
To ensure that E4 ORF3 is directly responsible for permitting viral DNA replication in IFN-treated cells, we verified that loss of E4 ORF3 does not impede production of the Ad gene products required for viral DNA replication. Six viral early proteins are essential for Ad DNA replication in the absence of E4 ORF3: E1A, E1B 55K, DBP, Ad pol, Ad pTP, and E4 ORF6. We quantified the levels of E1A, the immediate early gene required for the onset of the Ad transcriptional program, and DBP, one of the essential E2 gene products, by Western blot analysis (Fig. 3). Similar levels of these proteins were synthesized by wild-type, ORF6-deleted, and ORF3-deleted viruses irrespective of IFN prestimulation. Thus, IFN treatment did not significantly alter Ad early gene expression in the absence of E4 ORF3 expression.
Substantial colocalization of E4 ORF3 with PML in track-like structures was evident following pretreatment of cells with either IFN-α or IFN-γ (Fig. 4). E4 ORF3 elicited PML body rearrangement despite the increased PML synthesis associated with IFN treatment (41). This characteristic POD reorganization by E4 ORF3 is a conserved function among all Ad serotypes investigated, whereas the reorganization of the MRN complex is not (43). Our results demonstrate that the E4 ORF3 proteins of evolutionarily divergent Ad serotypes are capable of inhibiting the IFN response, thereby facilitating efficient viral DNA replication (Fig. 5 and 6 and data not shown). Furthermore, an Ad5 E4 ORF3 mutant (D105A/L106A) that is competent for PML, but not MRN rearrangement, rescued the inhibitory effect of IFN treatment (Fig. 7 and 8). Together, these data strongly indicate that MRN complex activity does not participate in the IFN-mediated inhibition of Ad DNA replication in the absence of E4 ORF3 expression. This conclusion is consistent with the fact that the E1B 55K and E4 ORF6 proteins are expressed with mutant E4inORF3 and would be expected to inhibit MRN activity via proteasome-dependent degradation (27, 42).
A variety of viruses express proteins that compromise POD integrity. Herpesviruses such as HSV-1, HCMV, and Epstein-Barr virus encode gene products capable of disrupting PML bodies (15, 16). Similarly, RNA viruses such as lymphocytic choriomeningitis virus, rabies virus, and vesicular stomatitis virus have been documented to produce analogous proteins (35). In several instances, the viral gene products responsible for mediating POD disruption inhibit IFN-stimulated antiviral activities. In the case of HSV-1, the ICP0 protein directs PML protein degradation (15, 16), and an ICP0 mutant virus is sensitive to the effects of IFN treatment in a PML-dependent manner (7, 30). Further, reduction in PML protein levels enhances the growth properties of an ICP0 mutant virus (18). These results support both the antiviral role of PML under normal growth conditions and the amplification of such activity in response to IFN. This idea is consistent with the observation that failure to disrupt PML bodies in HSV-1-infected cells inhibits viral DNA replication (5). Similar results have also been described for HCMV IE1 mutant viruses (15, 16, 46).
In the case of both HSV-1 and HCMV, the major antiviral effects of PML and IFN involve down egulation of immediate early gene expression and are reversed upon inhibition of PML activity (7, 18, 30, 31, 46, 47, 53). With respect to the aforementioned RNA viruses, the antiviral effects of PML are executed via inhibition of viral RNA and protein synthesis (35). Of particular interest is the fact that the role of the Ad E4 ORF3 in the inhibition of an IFN response appears to be distinct from the previous illustrations, since in the absence of E4 ORF3 expression, we did not observe a significant reduction in early gene expression following IFN treatment (Fig. 3).
We believe that PML body rearrangement by E4 ORF3 inhibits the deleterious effects of IFN on Ad replication. Clearly, the E4 ORF3 protein is capable of circumventing aspects of the IFN-induced antiviral state to permit successful viral genome replication. It is, therefore, unlikely that E4 ORF3 interferes with the induction of upstream IFN signaling pathways. Rather, it is intuitive that E4 ORF3 counteracts the antiviral effect of PML overexpression following IFN stimulation and/or the function of a distinct IFN-stimulated gene product(s). It is plausible that PML is required for the function of a yet-uncharacterized ISG whose synthesis is upregulated during an IFN response. This could potentially affect a critical aspect of Ad viral DNA replication, such as terminal protein priming or Ad DNA polymerase activity. Alternatively, IFN-mediated inhibition of Ad replication in the absence of E4 ORF3 may reflect the intrinsic, antiviral activity of the POD itself. It is well established that the genomes of DNA viruses localize in proximity to PODs early after infection (25, 28, 29). Indeed, recent results have demonstrated that de novo PML body assembly occurs at the location of infecting viral genomes (17) and that viral genomes that successfully enter the DNA replication program are primarily those that localized at PODs following infection (40). The DNA virus strategy of PML body rearrangement facilitates viral genome replication by an unknown mechanism. Conversely, inhibition of POD reorganization diminishes the process. It remains to be determined how precisely the Ad E4 ORF3 protein combats the antiviral state induced by IFNs, but these results underscore the complex interplay that has evolved between host cell defense and viral success.
Acknowledgments
We thank several colleagues for the generous gifts of antibodies, including Thomas Dobner for antibody against E4 ORF3 and Arnold Levine and Peter van der Vleit for antibodies against DBP. We acknowledge the excellent technical assistance of Mary Anderson and Sarah Van Scoy. We thank Chris Gordon for advice on microscopy and members of our laboratory for informed discussions.
This work was supported by NIH grant CA028146 to P.H. and N.C.R. A.J.U. is supported by NIH training grant CA009176.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondel, D., T. Regad, N. Poisson, B. Pavie, F. Harper, P. P. Pandolfi, H. De The, and M. K. Chelbi-Alix. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957-7970. [DOI] [PubMed] [Google Scholar]
- 3.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkham, J., D. M. Coen, C. B. Hwang, and S. K. Weller. 2001. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75:2353-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, A. V., and B. Roizman. 2004. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J. Virol. 78:4185-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djavani, M., J. Rodas, I. S. Lukashevich, D. Horejsh, P. P. Pandolfi, K. L. Borden, and M. S. Salvato. 2001. Role of the promyelocytic leukemia protein PML in the interferon sensitivity of lymphocytic choriomeningitis virus. J. Virol. 75:6204-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 11.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 12.Eskiw, C. H., and D. P. Bazett-Jones. 2002. The promyelocytic leukemia nuclear body: sites of activity? Biochem. Cell Biol. 80:301-310. [DOI] [PubMed] [Google Scholar]
- 13.Evans, J. D., and P. Hearing. 2003. Distinct roles of the Adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365-374. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 17.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrum, F. D., and D. A. Ornelles. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 73:7474-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grotzinger, T., T. Sternsdorf, K. Jensen, and H. Will. 1996. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML). Eur. J. Biochem. 238:554-560. [DOI] [PubMed] [Google Scholar]
- 21.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann, T. G., and H. Will. 2003. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 10:1290-1299. [DOI] [PubMed] [Google Scholar]
- 24.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871-876. [PubMed] [Google Scholar]
- 27.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79:14004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 29.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 30.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA. 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordqvist, K., K. Ohman, and G. Akusjarvi. 1994. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohman, K., K. Nordqvist, and G. Akusjarvi. 1993. Two adenovirus proteins with redundant activities in virus growth facilitates tripartite leader mRNA accumulation. Virology 194:50-58. [DOI] [PubMed] [Google Scholar]
- 34.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 36.Regad, T., A. Saib, V. Lallemand-Breitenbach, P. P. Pandolfi, H. de The, and M. K. Chelbi-Alix. 2001. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20:3495-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salomoni, P., and P. P. Pandolfi. 2002. The role of PML in tumor suppression. Cell 108:165-170. [DOI] [PubMed] [Google Scholar]
- 38.Shepard, R. N., and D. A. Ornelles. 2004. Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background. J. Virol. 78:9924-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shepard, R. N., and D. A. Ornelles. 2003. E4orf3 is necessary for enhanced S-phase replication of cell cycle-restricted subgroup C adenoviruses. J. Virol. 77:8593-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sourvinos, G., and R. D. Everett. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 21:4989-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadler, M., M. K. Chelbi-Alix, M. H. Koken, L. Venturini, C. Lee, A. Saib, F. Quignon, L. Pelicano, M. C. Guillemin, C. Schindler, et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565-2573. [PubMed] [Google Scholar]
- 42.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 43.Stracker, T. H., D. V. Lee, C. T. Carson, F. D. Araujo, D. A. Ornelles, and M. D. Weitzman. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strudwick, S., and K. L. Borden. 2002. Finding a role for PML in APL pathogenesis: a critical assessment of potential PML activities. Leukemia 16:1906-1917. [DOI] [PubMed] [Google Scholar]
- 45.Tauber, B., and T. Dobner. 2001. Adenovirus early E4 genes in viral oncogenesis. Oncogene 20:7847-7854. [DOI] [PubMed] [Google Scholar]
- 46.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, J. L., D. Unverrich, W. J. O'Brien, and K. W. Wilcox. 2000. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J. Interferon Cytokine Res. 20:805-815. [DOI] [PubMed] [Google Scholar]
- 48.Voelkerding, K., and D. F. Klessig. 1986. Identification of two nuclear subclasses of the adenovirus type 5-encoded DNA-binding protein. J. Virol. 60:353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg, D. H., and G. Ketner. 1983. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc. Natl. Acad. Sci. USA 80:5383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weitzman, M. D., C. T. Carson, R. A. Schwartz, and C. E. Lilley. 2004. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amsterdam) 3:1165-1173. [DOI] [PubMed] [Google Scholar]
- 53.Xu, Y., J. H. Ahn, M. Cheng, C. M. apRhys, C. J. Chiou, J. Zong, M. J. Matunis, and G. S. Hayward. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75:10683-10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]