Abstract
Dengue fever, a neglected emerging disease for which no vaccine or antiviral agents exist at present, is caused by dengue virus, a member of the Flavivirus genus, which includes several important human pathogens, such as yellow fever and West Nile viruses. The NS5 protein from dengue virus is bifunctional and contains 900 amino acids. The S-adenosyl methionine transferase activity resides within its N-terminal domain, and residues 270 to 900 form the RNA-dependent RNA polymerase (RdRp) catalytic domain. Viral replication begins with the synthesis of minus-strand RNA from the dengue virus positive-strand RNA genome, which is subsequently used as a template for synthesizing additional plus-strand RNA genomes. This essential function for the production of new viral particles is catalyzed by the NS5 RdRp. Here we present a high-throughput in vitro assay partly recapitulating this activity and the crystallographic structure of an enzymatically active fragment of the dengue virus RdRp refined at 1.85-Å resolution. The NS5 nuclear localization sequences, previously thought to fold into a separate domain, form an integral part of the polymerase subdomains. The structure also reveals the presence of two zinc ion binding motifs. In the absence of a template strand, a chain-terminating nucleoside analogue binds to the priming loop site. These results should inform and accelerate the structure-based design of antiviral compounds against dengue virus.
Flaviviridae are enveloped viruses with positive-strand RNA genomes that have been grouped into three genera, Hepacivirus, Pestivirus, and Flavivirus (11, 59). Several members of the Flavivirus genus, e.g., dengue virus (DENV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus, and West Nile virus (WNV), are medically important arthropod-borne pathogens afflicting humans. DENV infects 50 to 100 million people each year, with ∼500,000 patients developing the more severe disease dengue hemorrhagic fever, leading to hospitalizations and resulting in approximately 20,000 deaths, mainly in children (24, 26, 27, 29). Based on serological studies, DENVs are further classified into four distinct serotypes, DENV 1 to 4, whose respective genomes share ∼60% sequence identity, with ∼90% sequence identity within a serotype (7, 26). The DENV RNA genome spans about 10.7 kb and contains a type I methyl guanosine cap structure at its 5′ end but is devoid of a polyadenylate tail. The genomic RNA is translated into a single polyprotein (58), which is cleaved into three structural (C-prM-E) and seven nonstructural (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) proteins by both the viral and cellular proteases (28). The viral serine protease is within the N-terminal region of NS3, and recent structural studies show that part of its catalytic site is formed by the viral cofactor NS2B upon substrate binding (18). The C-terminal region of NS3 forms the RNA helicase domain, which is thought to either separate a double-stranded RNA template into individual strands or disrupt secondary structures formed by a single-stranded RNA (ssRNA) template in order to facilitate viral genome replication by NS5 (49, 61).
With a molecular mass of 104 kDa, NS5 is the largest of the DENV proteins. Sharing a minimum of 67% amino acid sequence identity across the four DENV serotypes, NS5 is also the most conserved viral protein. Based on structural and biochemical studies, three functional domains have been identified in NS5 (Fig. 1). The N-terminal S-adenosyl methionine methyltransferase (MTase) domain, spanning amino acid residues 1 to 296, has been expressed as a soluble active enzyme, and its X-ray structure has been determined. A central S-adenosyl-methionine-dependent MTase core structure sits within a “cradle” shaped by N-terminal and C-terminal subdomains. The MTase activity of NS5 is responsible for both guanine N-7 and ribose 2′-O methylations (6, 17, 48). These two methylation events are required for proper formation of the cap structure, which is recognized by the host cell translational machinery. Yeast two-hybrid interaction studies as well as functional nuclear localization assays have mapped two nuclear localization sequences (NLS) between amino acid residues 320 and 405 of NS5 (Fig. 1). NLS signals are recognized by cellular factors, allowing protein transport to the nucleus. In the case of NS5, the precise role of this translocation event and the influence of the state of phosphorylation of NS5 on this process remain elusive (34). Interestingly, a segment of 20 amino acids that is strictly conserved in all flaviviruses also lies between amino acid residues 320 and 368 of NS5 (Fig. 1). This region of NS5, which is involved in binding β-importin (9, 33), is also thought to interact with NS3 (33). Furthermore, the association between NS5 and NS3 is expected to modulate their respective enzymatic activities. Indeed, NS5 was reported to stimulate the nucleotide triphosphatase and RNA triphosphatase activities (63) of NS3. The C-terminal region of NS5 contains five amino acid sequence motifs that form the signature of RNA-dependent RNA polymerases (RdRps) (6). In conjunction with other viral proteins and unidentified host cell proteins, the NS5 polymerase domain is responsible for synthesizing a transient double-stranded replicative RNA intermediate. This RNA duplex is composed of viral plus- and minus-strand RNAs. The newly synthesized minus strand serves in turn as a template, allowing the RdRp to synthesize additional plus-strand genomic RNA (5, 34, 44, 64). Several activity assays for the corresponding Flaviviridae RdRp domains have been described, e.g., for Kunjin virus, hepatitis C virus (HCV), and bovine viral diarrhea virus (BVDV) (25, 35, 51, 53).
FIG. 1.
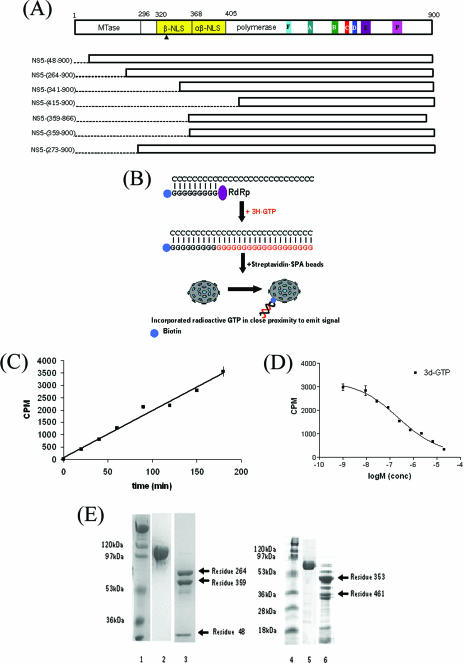
(A) Schematic representation of the distribution of domains in the DENV NS5 protein. The FL NS5 protein has three major functional domains. The N-terminal MTase spans residues 1 to 296. The NLS has been divided into αNLS (spanning residues 320 to 368) and α/βNLS (residues 369 to 405). The arrow indicates the αNLS region thought to interact with the NS3 viral helicase. The six conserved amino acid sequence motifs within the RdRp domain (residues 273 to 900) are colored as in Fig. 2C, and “P” denotes the priming loop site. Also shown are the various truncated constructs of NS5 (see the text for details). (B) Schematic representation of the polymerase assay in the SPA bead format. The newly transcribed RNAs with the incorporation of radioactive GTP were captured on streptavidin-coupled beads and detected by counting as described in Materials and Methods. (C) Steady-state kinetics of RNA synthesis by DENV 3 RdRp, using a poly(C) primer and oligo(G) template. The reaction mixture contained 1 μM enzyme, 0.25 μg poly(C), 0.0125 μg oligo(G), 4 μM total GTP, and 0.5 μCi of [3H]GTP, as described in Materials and Methods. (D) Kinetic analysis of 3′dGTP inhibition. The IC50 of 3′dGTP for DENV 3 RdRp was determined using at least two independent experiments. Assay conditions are described in Materials and Methods. (E) Limited proteolysis of the DENV 2 FL NS5 protein (left) and of the DENV 3 RdRp domain (right). Lanes 1 and 4, molecular mass markers; lanes 2 and 3, FL undigested NS5 and trypsin-digested NS5, respectively; lanes 5 and 6, undigested DENV 3 RdRp and trypsin-digested DENV 3 RdRp, respectively. The trypsin/enzyme ratio for FL NS5 was 1:100, and the reaction was carried out at room temperature for 2 h (lane 3). Similarly treated DENV 3 RdRp was completely digested (data not shown). Using a lower concentration of trypsin (1:1,000 ratio of trypsin to RdRp) for ∼30 min at room temperature yielded several smaller fragments (lane 6). The N termini of digested protein fragments were sequenced (indicated by arrows), and this information was used to derive some of the constructs shown in panel A.
Crystal structures of 11 RdRps from several virus families have been determined either as apoenzymes or as complexes with incoming ribonucleoside triphosphates (rNTPs), primers, templates, or small-molecule inhibitors (21). These include RdRps from viruses which are members of the Flaviviridae, such as BVDV (12, 13) and HCV (2, 39, 45, 57); the Picornaviridae, including poliovirus (55), foot-and-mouth disease virus (FMDV) (20), and human rhinovirus (3); and the Caliciviridae, such as rabbit hemorrhagic disease virus (RHDV) (42) and Norwalk virus (43). In addition, structures have been reported for RdRps of the double-stranded RNA bacteriophage Φ6 (10) and for reovirus λ3 polymerase (54). In spite of a very weak overall sequence conservation, limited to the presence of a few amino acid motifs, all of these polymerases share an overall architecture composed of palm, fingers, and thumb domains (36) and a common catalytic mechanism for nucleotide incorporation that makes use of two metal ions that are coordinated by two structurally conserved aspartic acid residues (52). Nonetheless, viral RdRps have specific features that distinguish them from the basic right-handed “U-shaped” conformation adopted by DNA polymerases (21). As first seen in the HCV RdRps, the active site appears to be fully encircled by long structural elements (named “fingertips”) that connect the thumb and fingers domains. This additional structural feature gives a more spherical and compact appearance to the molecule, as opposed to the more open—and less constrained—structure for DNA polymerases. Moreover, an additional sequence motif (motif F) that was suggested to play an important role in stabilizing the nascent base pair is present only in RdRps. Because host cells are devoid of this enzymatic activity, viral polymerases represent attractive drug targets for the development of specific antiviral compounds. Indeed, selective inhibitors against the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase and the hepatitis B virus, cytomegalovirus, and herpes simplex virus polymerases have been approved as drugs for treating the associated viral infections (60). In an attempt to accelerate the search for antiviral compounds active against DENV, we have established a high-throughput screening assay based on a scintillation proximity assay format. We also report the three-dimensional structure of a catalytically active DENV RdRp domain determined to 1.85-Å resolution by X-ray crystallography. The structure of the complex between DENV RdRp and the chain-terminating nucleoside analog 3′dGTP is also discussed.
MATERIALS AND METHODS
Cloning and expression.
The catalytic domain of the DENV 3 polymerase (GenBank accession no. AY662691; nucleotides 8381 to 10264, amino acid residues 273 to 900) was amplified using forward (5′-CCACGCGTCGACAACATGGATGTCATTGGGGAAAG-3′) and reverse (5′-CCGGAATTCCTACCAAATGGCTCCCTCCGACTCCTC-3′) primers, using pDEST 14 containing the DENV 3 polymerase as the template. The underlined regions in the primer sequences correspond to SalI and EcoRI sites, respectively. The PCR fragment was cut with SalI and cloned into a pET15b plasmid that was previously cut with BamHI, blunted, and then cut with XhoI.
Escherichia coli BL21(DE3) cells (RIL; Stratagene) were transformed with the recombinant plasmid carrying the gene encoding the DENV 3 RdRp domain and were grown at 37°C in LB medium containing 100 μg ml−1 ampicillin and 50 μg ml−1 chloramphenicol until the optical density at 600 nm was 0.6 to 0.8. Protein expression was induced at 16°C by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM. After overnight growth, cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C, and the cell pellet was stored at −80°C. Protein expression for the rest of the constructs mentioned in Fig. 1A was performed similarly, and protein solubility was estimated visually by sodium dodecyl sulfate-polyacrylamide gel analysis.
Protein purification.
Cells resuspended in a lysis buffer consisting of 20 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 10 mM β-mercaptoethanol, and 10% glycerol (buffer A) supplemented with an EDTA-free protease inhibitor tablet (Roche) were lysed by sonication, and the lysate was clarified by centrifugation at 30,000 × g for 30 min at 4°C. The supernatant was purified by metal affinity, using a HisTrap HP column (GE Healthcare) equilibrated with buffer A. Unbound proteins were washed away sequentially with five column volumes each of buffer A supplemented with 25 mM and 125 mM imidazole. Proteins were eluted by using a linear gradient of imidazole from 125 to 500 mM. Fractions containing protein were pooled and dialyzed overnight against 50 mM morpholineethanesulfonic acid (MES), pH 6.5, 0.3 M NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol and were treated with thrombin to remove the hexahistidine (His6) tag.
Proteins were further purified using cation-exchange chromatography (15S) and eluted using a linear gradient ranging from 0.05 to 1.5 M NaCl in buffer B (50 mM MES, pH 6.2, 0.05 M NaCl, 5 mM β-mercaptoethanol, 1 mM EDTA). After concentration by ultrafiltration (Amicon), a final step using gel filtration chromatography was carried out (HiPrep 16/26, Superdex 200) in buffer C {20 mM Tris-HCl at pH 6.8, 0.25 M NaCl, 1 mM EDTA, 2 mM β-mercaptoethanol, and 0.1% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)}. The protein was concentrated to ∼11 mg/ml and used for crystallization. An analysis of metals bound to the protein (purified as described above, concentrated to ∼86 μM, and diluted to a final concentration of 5 μM in MilliQ water), using flame atomic absorption spectroscopy with a polarized Zeeman Z-2000 atomic absorption spectrophotometer (Hitachi), confirmed the presence of Zn2+ ions, ruling out the possibility that excess Ni2+ ions from the metal-affinity column had entered the RdRp domain during purification.
NS5 RdRp activity measured using SPA.
NS5 RdRp assays were performed in white half-area flat-bottomed 96-well plates (Corning) in a buffer containing 50 mM Tris-HCl, 10 mM KCl, 2 mM MgCl2, and 2 mM MnCl2 at pH 7.0, with 0.25 μg poly(C) annealed to 0.0125 μg biotinylated oligonucleotide G20 (Sigma) as a template and 1.75 μg of DENV 3 RdRp in a volume of 22.5 μl. Following incubation for 1 h, 2.5 μl of 4 μM of total GTP containing 0.5 μCi of [3H]GTP (6.1 Ci/mmol; GE Healthcare) was added to initiate the reaction. To measure the 50% inhibitory concentration (IC50) of the 3′dGTP inhibitor, the enzyme was preincubated with template and inhibitor for 1 h before the reaction was started by the addition of a solution containing radioactive GTP for 1 h. The reaction was stopped with EDTA containing streptavidin-coated scintillation proximity assay (SPA) beads (GE Healthcare). All reactions were carried out at room temperature.
Crystallization and data collection.
Crystals of DENV 3 RdRp were obtained using the hanging-drop vapor diffusion method at 4°C. Typically, a volume of 0.75 μl of protein solution at a concentration of 11 mg/ml in 20 mM Tris-HCl, pH 6.8, 0.25 M NaCl, 1 mM EDTA, 2 mM β-mercaptoethanol, and 0.1% (wt/vol) CHAPS was mixed with an equal volume of a reservoir solution (0.1 M Tris-HCl at pH 8.5, 0.8 M potassium-sodium tartrate, 0.5% [wt/vol] polyethylene glycol [PEG] monomethyl ether 5000). Lens-shaped crystals belonging to space group C2221, with unit-cell parameters of 160.28 Å (a), 178.77 Å (b), and 58.05 Å (c), were obtained after 3 to 4 weeks, but they diffracted very poorly. Crystal quality was markedly improved by the addition of MgSO4 or magnesium acetate at concentrations of 0.005 to 0.2 M and by subjecting crystals to a careful dehydration protocol. Prior to data collection, crystals were dehydrated in a stepwise manner by soaking them in a series of buffers containing mother liquor, with a gradual increase of the concentration of PEG 3350, from 15% to 25% to 35% (wt/vol). Crystals were then mounted in a cryo-loop and rapidly cooled to 100 K in liquid nitrogen. The Mg2+ complex structure was obtained by soaking crystals in 90 mM Tris-HCl at pH 8.5, 0.2 M MgCl2, and 25% (wt/vol) PEG 3350 for 20 h at 4°C. In order to obtain a complex with 3′dGTP, DENV 3 RdRp crystals were soaked in buffer (0.1 M Tris-HCl at pH 8.5, 0.8 M potassium-sodium tartrate, 0.5% [wt/vol] PEG monomethyl ether 5000, 40 mM MgSO4, 10 mM 3′dGTP [Trilink]) for 20 h at 4°C. Crystals were then dehydrated using a similar protocol to that described above before being flash-cooled in liquid nitrogen. Diffraction intensities were recorded on beamline X10SA at the Swiss Light Source, Switzerland, on a mar225 charge-coupled device detector, using an attenuated beam. In-house data collection was performed on a Rigaku/MSC FR-E X-ray generator with an R-AXIS IV++ imaging plate detector at the Biopolis shared-equipment facility. Integration, scaling, and merging of the intensities were carried out using programs from the CCP4 suite (4).
Structure solution, refinement, and analysis.
The structure was solved by molecular replacement, using the WNV polymerase domain structure as an initial model (PDB code 2HFZ). The refinement was carried out using programs CNS (with molecular dynamics in torsion-angle space) and REFMAC5 with the TLS (translation, liberation, screw-rotation displacement) refinement option (Table 2). The latter proved important for modeling several mobile regions of the fingers subdomains. Refinement cycles were interspersed with model rebuilding sessions with SGI computer graphics with program O. Figures 4 and 5 were drawn using the PyMOL program (14). Superpositions of structures and calculations of the root mean square (RMS) deviations were carried out using program LSQKAB from the CCP4 suite (4).
TABLE 2.
Refinement statistics
| Statistic | Value
|
|
|---|---|---|
| Mg2+ complex | 3′dGTP complex | |
| Resolution range (Å) | 20.0-1.85 | 20.0-2.6 |
| No of reflections | ||
| Completeness (%) | 99.9 | 99.7 |
| Used for refinement | 68,360 | 25,549 |
| Used for Rfree calculation | 3,646 | 1,340 |
| No. of nonhydrogen atoms (avg temp factor [Å2]) | ||
| Protein | 4,642 (36.2) | 4,621 (62.4) |
| Missing residues | 61 | 63 |
| Zn2+/Mg2+ | 2/1 (28.4, 47.0) | 2/0 (54.3)d |
| Water molecules | 506 (41.3) | 129 (64.0) |
| 3′dGTP | 13 (118.1)d | |
| Rfactora (%) | 19.6 | 21.3 |
| Rfreeb (%) | 23.1 | 28.8 |
| RMS deviations from ideality | ||
| Bond length (Å) | 0.007 | 0.010 |
| Bond angle (°) | 0.909 | 1.158 |
| Ramanchandran plot data | ||
| Residues in most favored regions (%) | 92.2 | 88.6 |
| Residues in additional allowed regions (%) | 7.0 | 10.2 |
| Residues in generously allowed regions (%) | 0.2 | 0.8 |
| Residues in disallowed regions (%) | 0.6 | 0.4 |
| Overall G factorc | 0.19 | 0.09 |
| Estimated coordinate error (Å) | 0.12 | 0.33 |
| PDB code | 2J7U | 2J7W |
Rfactor = Σ ‖Fobs| − |Fcalc‖/Σ |Fobs|.
Rfree was calculated with 5% of reflections excluded from the whole refinement procedure.
G factor is the overall measure of structural quality from PROCHECK (38).
The occupancies for 3′dGTP and Zn2+/Mg2+ were set to 1.
FIG. 4.
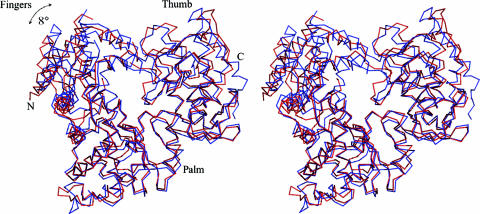
Superimposition of the α-carbon trace of DENV 3 RdRp (red) with that of WNV RdRp (blue), shown as a stereoview. The RMS deviation is 1.23 Å for 313 equivalent α-carbon atoms. The overall fold of DENV 3 RdRp is well preserved compared with that of WNV RdRp. Rotation of the fingers at an angle of ∼8° is observed between the two structures (see the text for details).
FIG. 5.
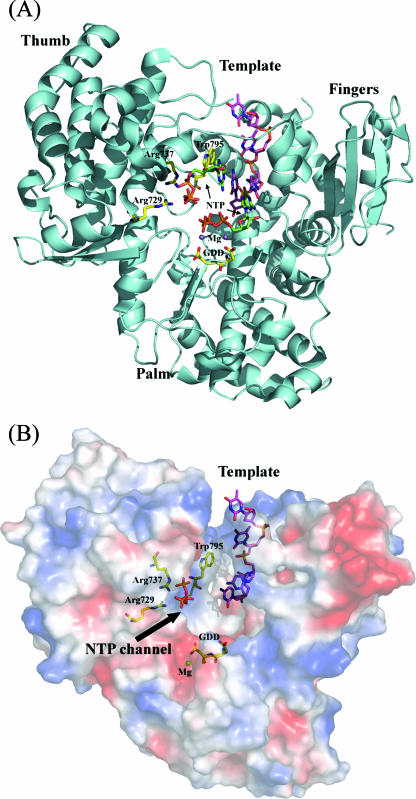
Model for the initiation complex of DENV RdRp, inspired by the Φ6 polymerase quaternary complex (10) and the HCV complex with ribonucleotides (8). (A) Ribbon representation with the priming base stacking to Trp-795 making Watson-Crick interactions with the template base. (B) Representation of the electrostatic surface of the DENV 3 RdRp viewed from the back (compared to Fig. 2A). Positive electrostatic potentials are colored blue, and negative potentials are colored red. The view illustrates a ssRNA template and provides a model for the initiation complex. The RNA template traverses the tunnel formed between the thumb and fingers subdomains, entering from the top of the figure. The NTP entry channel is at the interface between the thumb and palm subdomains, indicated by an arrow. The ssRNA template and the tP moiety are shown as sticks (the base and sugar moieties were modeled). Residues discussed in the text that play an important functional role are labeled.
Protein structure accession numbers.
The protein structure coordinates for the DENV3 NS5-Mg2+ and -3′dGTP complex structures have been submitted to the Protein Data Bank under accession numbers 2J7U and 2J7W, respectively.
RESULTS AND DISCUSSION
Protein expression.
Our attempts to crystallize the full-length (FL) NS5 protein—expressed using the baculovirus system—have so far been unsuccessful. Based on the hypothesis that the NLS region of NS5 could form a separate flexible domain, constructs starting at positions 341 (341-900) and 415 (415-900) of the NS5 protein sequence, and thus fully or partially devoid of the NLS region, were expressed, albeit mostly in an insoluble form (Fig. 1). Limited proteolysis of the FL NS5 protein identified stable fragments starting at residues 48, 264, and 359 (Fig. 1E). After screening of protein expression for the NS5 RdRps of the four DENV serotypes (data not shown), the best results were obtained with one DENV 3 polymerase truncation construct starting at amino acid 273 (50). This protein could be expressed in high quantity and purified to >95% purity, as described in Materials and Methods, typically yielding 3 mg protein from 1 liter of culture.
Enzymatic activity of RdRp and its inhibition by 3′dGTP.
To determine the activity of the recombinant protein, an SPA was set up, using homopolymer C as the template and biotin-oligo(G)20 as the primer, as outlined schematically in Fig. 1B. The addition of tritiated GTP to a primer is measured directly without the need for any wash steps since only the radioactive decay of tritium on the primer-template complex that is in close proximity to the scintillation agent via streptavidin-biotin interaction will result in light emission that can be measured. The assay was set up using FL NS5 and optimized with respect to pH and salt and divalent cation concentrations as well as other parameters, such as template, primer, substrate, and enzyme concentrations (data not shown). Figure 1C shows the activity of the truncated DENV 3 RdRp, monitored over time. Close to 1 μM DENV 3 RdRp was used to achieve signals comparable to those for FL DENV 2 NS5 (25 nM) (data not shown), implying that the domain is less active than the FL protein (see below). Nevertheless, the sensitivity and speed of the assay and the possibility to miniaturize it to a 384-well format make the assay valuable for high-throughput screening of inhibitors.
The ability of 3′dGTP to block the elongation process was studied using the NS5 RdRp SPA format. In this experiment, the enzyme, template-primer complex, and inhibitor at various concentrations were incubated for 1 h prior to initiation as described above. The data presented in Fig. 1D show that the IC50 of the DENV 3 RdRp is 0.22 μM (compared with an IC50 of 5 nM for FL NS5 [data not shown]) and demonstrate that 3′dGTP can serve as a useful reference compound in the present assay format for finding new inhibitors. Furthermore, the decreased enzymatic activity of the NS5 RdRp domain is consistent with our finding that the truncated protein probably adopts a slightly more open and flexible structure than that of FL NS5, since similar amounts of trypsin resulted in more protease-accessible sites in the former than in the latter (Fig. 1E).
Protein crystallization and structure determination.
The conditions for crystallization of DENV 3 RdRp and procedures to obtain crystals diffracting to a high resolution are discussed elsewhere (62). Briefly, condition 31 from an index screen (Hampton Research) resulted in the formation of small crystals that did not diffract X-rays. The addition of divalent metal ions (MgSO4 or magnesium acetate at concentrations of 0.005 to 0.2 M) to the crystallization buffer produced larger crystals, with sizes of up to 0.1 mm, in 3 to 4 weeks, but these still diffracted weakly. Suspecting that the poor diffraction pattern obtained with the original crystals was caused by extensive dynamic properties of the enzyme, as observed previously for HIV-1 reverse transcriptase (19), we established a careful dehydration procedure, as described in Materials and Methods, in the hope that shrinkage of the unit-cell dimensions provoked by air dehydration would induce a greater stability of the protein lattice. Indeed, crystals which diffracted to 2.4-Å resolution on a rotating anode (and to >2.0-Å resolution on a third-generation synchrotron) (Table 1) could be obtained and were used for structure determination (Table 1).
TABLE 1.
Data collection and phasing statistics
| Statistic | Valuea
|
|
|---|---|---|
| Mg2+ complex | 3′dGTP complex | |
| Wavelength (Å) | 1.0 | 1.0 |
| Cell parameters (Å) for space group C2221 | ||
| a | 160.15 | 163.71 |
| b | 180.47 | 180.91 |
| c | 57.98 | 58.04 |
| Resolution range (Å) | 48-1.85 | 48-2.6 |
| No. of observed reflections | 355,151 | 131,992 |
| No. of unique reflectionsa | 72,065 (8,212) | 26,929 (3,067) |
| Completeness (%) | 99.9 (100) | 99.6 (100) |
| Multiplicity | 4.9 (4.9) | 4.9 (4.9) |
| Rmergeb | 0.09 (0.540) | 0.10 (0.627) |
| I/σ (I) | 14.62 (2.98) | 13.98 (2.54) |
| Solvent content (%) | 58.9 | 59.9 |
The numbers in parentheses refer to the last (highest) resolution shell.
Rmerge = ΣhΣi|Ihi − <Ih>|/Σh,iIhi, where Ihi is the ith observation of the reflection h, while <Ih> is its mean intensity.
Overall structure of DENV RdRp.
The distribution of domains along the DENV 3 NS5 sequence is depicted schematically in Fig. 1A. The DENV 3 RdRp has overall dimensions of approximately 65 by 60 by 40 Å (3). Its architecture assumes the canonical right-hand conformation consisting of fingers, palm, and thumb which is characteristic of known polymerase structures, but with the NLS region playing an important role in the formation of the observed structure (Fig. 2). Interestingly, the NLS domain signatures described previously (9) are distributed between the fingers and thumb subdomains. Three helices (α2 to α4) are incorporated within the thumb and the fingers (α5 to α7) subdomains, and the connection between them is realized through the segment linking helices α4 and α5. Truncations within the NLS region are thus likely to destabilize the protein structure, an observation which explains a posteriori the poor expression/solubility of constructs lacking part of this region (Fig. 1). A total of 27 α-helices (α3 is a 310 helix) and 7 β-strands are clearly defined in the current DENV 3 RdRp model, and an amino acid sequence alignment of the RdRps from several flaviviruses, including the four DENV serotypes, is shown in Fig. 3. The DENV 3 RdRp fold is analogous to that of WNV RdRp (with an RMS deviation of 1.23 Å for 313 superimposed C α). The corresponding RMS values for the three RdRp subdomains superimposed individually are 1.9, 0.8, and 1.0 Å, for the finger, palm, and thumb subdomains, respectively. Compared to the WNV RdRp, the largest variations (with RMS deviations of >1.5 Å after superimposition) are observed for residues (296 to 301, 419 to 423, 543 to 545, and 556 to 559) from the fingers subdomain. This subdomain, which is not maintained by crystal-packing contacts, is more flexible. The relative orientations between subdomains also differ, as displayed in Fig. 4. Overall, the DENV 3 RdRp assumes a more “open” conformation than the conformation observed for WNV RdRp. In the latter, the finger subdomain is rotated towards the thumb by an angle of approximately 8° (Fig. 4).
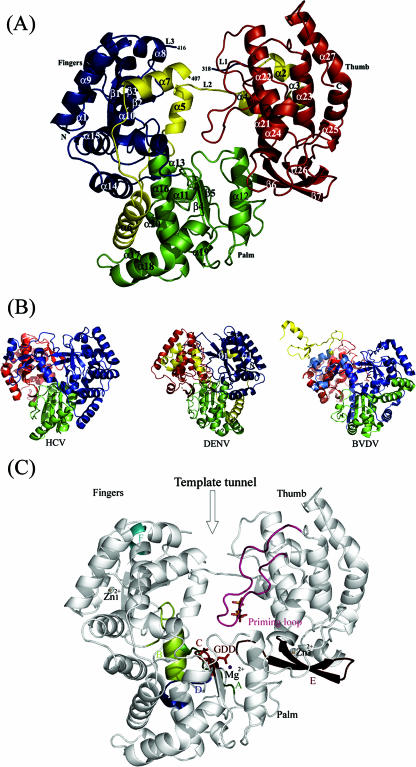
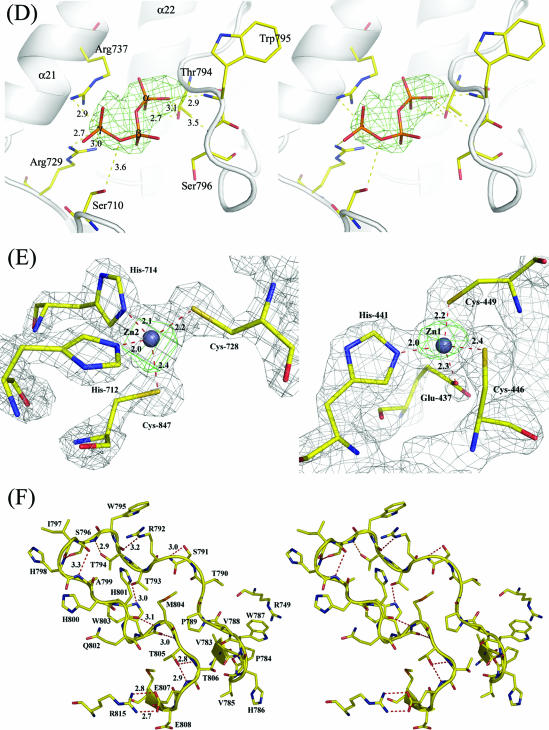
FIG. 2.
(A) Ribbon representation of overall structure of DENV 3 RdRp (residues 273 to 900). The various subdomains are colored as follows: fingers, blue; NLS, yellow; palm, green; and thumb, red. Secondary structure elements, as defined in Fig. 3, are labeled. The N and C termini are displayed, and the residues at the boundaries of the missing regions in the model are numbered. (B) Side-by-side views of the DENV, HCV, and BVDV RdRps, highlighting their structural differences (see the text for details). (C) The six conserved motifs present in various RdRp structures were mapped onto a ribbon representation of the DENV 3 RdRp catalytic domain and colored as follows: motif A, dark green; motif B, green; motif C, red; motif D, dark blue; motif E, magenta; motif F, light blue; and priming loop, pink. Asp-663 and Asp-664 from motif C of the catalytic site are shown in stick representation and labeled. The divalent metal ions Zn2+ and Mg2+ are represented by gold and purple spheres, respectively. An anomalous Fourier map contoured at a level of 5 σ shows two peaks at the positions corresponding to the two zinc ions. (D) Stereoviews of the interactions established by the tP moiety of the 3′dGTP inhibitor. The final difference electron density map with Fourier coefficients Fo-Fc, where the atoms from the inhibitor have been omitted from the calculation, was contoured at 3.0 σ. Residues Ser-710, Arg-729, Arg-737, Thr-794, Trp-795, and Ser-796, which are making contacts, are represented as sticks, and the distances to the α-, β-, and γ-phosphates are displayed. (E) Close-up view of the zinc binding pockets (left panel, Zn2; right panel, Zn1) located in the vicinity of motif E in the thumb and within the finger subdomains, respectively. The zinc ions are displayed as gray spheres, and residues providing the tetrahedral coordination geometry are shown as sticks. The refined electron density map (with Fourier coefficients 2Fo-Fc) is displayed at a level of 1 σ. The anomalous Fourier maps using phases from the refined final model are contoured at a level of 5 σ and displayed in green. (F) Stereoviews of the interactions of the primary loop (see text for details).
FIG. 3.
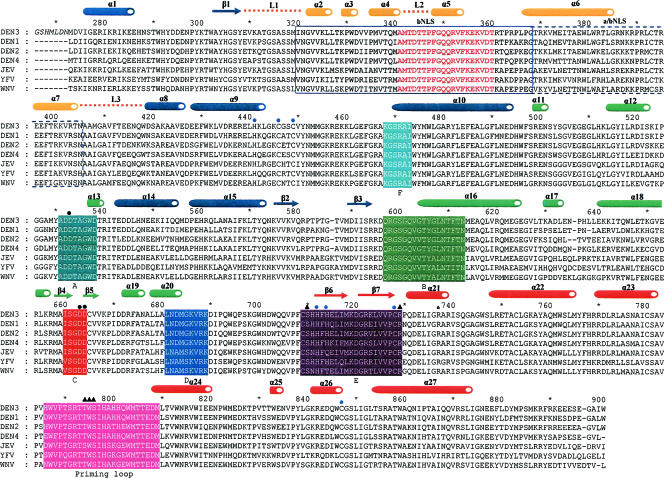
Structure-based sequence alignment of the flavivirus RdRp domains from DENV 1 to 4, JEV, YFV, and WNV. Residue numbering and secondary structure assignment are based on the DENV 3 RdRp structure, and α-helices and β-sheets are depicted as cylinders and arrows, respectively. The fingers, palm, thumb subdomains, the conserved motifs, and the priming loop are colored as in Fig. 2A and B. The βNLS and α/βNLS regions are boxed. Catalytic aspartate residues are marked by black circles. Residues interacting with tP moieties of 3′dGTP in the DENV 3 RdRp complex are marked by triangles, and residues coordinating the zinc ion are indicated with blue circles. The three loops, L1 to L3, discussed in the text are depicted as dotted lines and labeled.
NLS region.
The amino-terminal region comprising the βNLS and α/βNLS sequences (residues 316 to 415) forms a peculiar feature of DENV RdRp, and such sequences are not present in either the HCV or BVDV polymerase (Fig. 2B). The structure adopted by the βNLS (residues 316 to 368) includes a helix-turn-helix motif that lies on top of the thumb domain, and helices α7 and α6 of the α/βNLS (residues 369 to 415) are buried in the fingers and between the fingers and the palm domain, respectively (Fig. 2). Previous biochemical pull-down assays have shown that the βNLS region of NS5 from DENV 3 interacts with the NS3 helicase, probably via a well-conserved stretch of 20 amino acids. This segment (341-AMTDTTPFGQQRVFKEKVDT) within the βNLS (9, 35) (Fig. 1 to 3) forms part of a mobile region of the thumb subdomain. In the present structure, residues 350 to 360, which contain the 310 helix α3, are properly oriented to interact with the importin β site, with residues Arg-352, Phe-354, Glu-356, and Lys-357 positioned to interact with the binding partner. Together, the βNLS and the α/βNLS (9, 35) form part of a functional import system bringing flaviviral NS5 to the nucleus of the host cell, where it contributes to a significant proportion (ca. 20%) of the total RdRp activity from cells infected with WNV, JEV, and DENV (56). Furthermore, a link to the observed dengue pathogenesis was suggested by the nuclear location of NS5 in a report showing that the chemokine interleukin-8 is induced after DENV 2 infection, presumably via a direct interaction with a CAAT/enhancer binding protein (41).
Fingers subdomain.
The finger subdomain comprises residues 273 to 315, 416 to 496, and 543 to 600. As shown by a TLS analysis (which models pseudo-rigid-body displacements of the various polymerase subdomains within the crystal lattice), the fingers subdomain, especially the “fingertip region” (located at the top of the RdRp molecule in Fig. 2), appears to be more mobile than the other palm and thumb subdomains. Residues 311 to 317, 408 to 415, and 454 to 466 (preceding motif F) were not observed in the electron density map and were thus not included in the present model. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis of dissolved protein crystals ruled out proteolytic cleavage, indicating the high mobility of these segments.
In spite of low sequence similarity, the extended “fingertip” feature is similar to that in other viral RdRps, including primer-independent RdRps, such as those of BVDV, HCV, and Φ6 (PDB codes 1S48, 1GX6, and 1HHS, respectively) (8, 10, 13), and primer-dependent RdRps, such as those of FMDV and RHDV (PDB codes 1WNE and 1KHV, respectively) (3, 22). The base of the fingers domain forms a concave surface shaped by the solvent-exposed residues of helices (α6, α14, and α15) near the N terminus of the protein (Fig. 2). This concave surface could be involved in accommodating the N-terminal MTase domain. A more precise map of the interaction between the MTase and the RdRp domains was provided by Malet et al. for the WNV structure (40a).
Compared to the RdRps of HCV and BVDV, several differences can be noted in the DENV RdRp molecule. In DENV 3 RdRp, an additional N-terminal segment of 35 amino acids (absent in HCV and BVDV) folds into helix α1 and strand β1, connecting to helix α2 of the NLS, which is deeply buried in the thumb domain. Two flexible loops, L1 and L2 (residues 309 to 321 and 342 to 347, respectively), linking the fingers and the thumb domains are likely to transmit conformational changes between the two domains and/or restrict their individual mobility. Furthermore, the location of the template tunnel beneath these connecting loops suggests that any transition from a “closed” to an “open” conformation of the RdRp during replication could be modulated by these loops (2, 39). The additional β1 strand belongs to a three-stranded antiparallel β-sheet that is surrounded by helices α1 and α5. The corresponding region in the HCV polymerase is occupied by a five-stranded antiparallel β-sheet, and such formation of β-sheets is not observed in the BVDV RdRp. The L3 loop (residues 405 to 418) connecting α7 and α8 is highly mobile. This loop is also rightly positioned to regulate access of the ssRNA substrate at the entrance of the template tunnel. In contrast, the fingertips of the corresponding loops in BVDV and HCV twist away from the active site towards the fingers domain.
Palm domain and catalytic active site.
The palm domain consists of residues 497 to 542 and 601 to 705 and is composed of a small antiparallel β-strand platform, β4 and β5, surrounded by eight helices (α11 to α13 and α16 to α20) (Fig. 2). Overall, the palm domain appears to be the most structurally conserved among all known polymerases, reflecting the preservation of the architecture of the catalytic site during evolution. Indeed, the catalytic domain of DENV RdRp shows good superimposition with those of other RdRps (e.g., that of BVDV or HCV), with RMS deviations of ∼1.7 Å and with the active site residues superimposing closely with the equivalent residues from other RdRps.
A comparative analysis of RNA polymerases from positive-strand viruses identified four of six conserved sequence motifs responsible for NTP binding and catalysis, located in the palm domain (Fig. 3) (15, 46). The GDD catalytic active site (motif C, comprising Asp-663 and Asp-664) is located in the turn between strands β4 and β5. The antiparallel β-sheet that houses the active site has shorter strands of approximately 10 Å (like in WNV RdRp), in contrast to the equivalent β-strands found in BVDV and HCV, which are about 20 Å long. In addition, the connection between motifs B and C appears to be more elaborate in RdRps from flaviviruses (comprising helices α17 and α18) than in RdRps from viruses in other genera, which have only a short α-helix directly attached to the β-sheet of the active site. These differences constitute interesting signatures differentiating RdRps of flaviviruses from those of pestiviruses and hepaciviruses that occurred during the divergent evolution of these viruses. As in the case of HCV, the active site of the RdRp from DENV 3 is encircled by several loops that contribute to shaping the tunnel into which the template RNA can gain access to the catalytic site.
Crystals soaked with 0.2 M MgCl2 (form I) clearly revealed the presence of one hydrated Mg2+ ion close to the expected catalytic position, hexa-coordinated by Asp-533 (motif A) via a water molecule and by Asp-664 (motif C) (Fig. 2 and 3).
Thumb domain.
The thumb domain (residues 706 to 900), which forms the C-terminal end of the RdRp of DENV, is the most structurally variable among known polymerase structures. It contains two conserved sequence motifs. Motif E forms an antiparallel β-sheet wedged between the palm domain and several α-helices of the thumb domain. Interestingly, the loop that links helices α21 and α22 in RdRp of DENV 3 (residues 740 to 747) adopts a different conformation from that in HCV and BVDV, where it forms a β-hairpin. Together with the fingertips, the path adopted by this loop contributes to shape the RNA template tunnel. A second loop spanning amino acids 782 to 809 forms the priming loop that partially occludes the active site (Fig. 2). The path followed by this long and potentially flexible loop is clearly defined in the electron density map. It is stabilized by several “internal” (intraloop) interactions, including hydrogen bonds formed between Thr-794 and Ser-796 and a salt bridge between Glu-807 and Arg-815 that projects from helix α24, as well as stacking interactions between the δ-guanido group of Arg-749 and the indole ring of Trp-787. In addition, hydrogen bonds formed by residues from this loop with residues projecting from helix α21 contribute to maintain its orientation with respect to the protein (Fig. 2F). Together with loop L3, the priming loop forms the upper part of the RNA template tunnel and is thus likely to regulate access of and exit into the active site (Fig. 2). Replacement of the corresponding β-hairpin with a short turn in the RdRp from HCV converts the protein from a solely primer-independent enzyme to a primer-dependent one that can use double-stranded RNA as a substrate (30). Notably, RdRps from HCV, Norwalk virus, and Φ6 also possess unique C-terminal regions that fold back into the active site cleft (1, 43). Collectively, these specific structural features differentiate primer-independent RdRps (e.g., those from BVDV, HCV, and DENV) from primer-dependent RdRps (e.g., those from FMDV and RHDV). Accordingly, RdRps of poliovirus and calicivirus, which lack such features, initiate RNA transcription by using a template-primer duplex as a substrate for polynucleotide incorporation (42). In the DENV RdRp, the distance between the last residue (Met-883) that is visible in the electron density map and GDD motif C is 39 Å, and a more precise assessment of the role played by C-terminal residues in regulating access to the active site requires further structural studies.
Zinc binding sites.
Unexpectedly, our crystal structure revealed two zinc binding pockets, Zn1 and Zn2, in the fingers and thumb subdomains, respectively (Fig. 2). Both zinc ligands are buried within the protein and have tetrahedral coordination geometry. Since no zinc ions were included in the crystallization conditions or during protein purification, they likely originated from the bacterial growth medium. One zinc atom (Zn2) is coordinated by His-712, His-714, Cys-728 of motif E, and Cys-847 of helix α26. Strong peaks were observed in the residual difference electron density (at a contour level of 18 σ) and anomalous (at 8 σ) maps, and this site seems to be fully occupied in the protein, with a low-temperature factor of 24 Å2 (Fig. 2E). The residues involved in coordinating the zinc ion Zn2 are conserved in the four DENV serotypes and also in the RdRp from YFV. Residue His-714 is replaced by a threonine in the RdRp from WNV, and a disulfide bridge linking the two evolutionarily conserved cysteine residues is observed instead (PDB code 2HFZ). The second structural zinc (Zn1) located in the fingers subdomains is coordinated by Cys-446, Cys-449, His-441, and the carboxylate group of Glu-437. This zinc binding site is conserved in the WNV RdRp molecule (Fig. 2C). This site seems less occupied, as shown by a higher temperature factor and an anomalous signal visible at a contour level of 5 σ. The presence of zinc atoms bound to the protein was independently confirmed using flame atomic spectrometric analysis. Interestingly, the HIV reverse transcriptase assumes a sharp turn after motif E that serves as a pivot point for movements of the thumb subdomain upon template-primer binding (32). The zinc ion Zn2 likely contributes to the structural stability of the region near motif E of the DENV polymerase. Interestingly, this pocket is also located near the functionally important residues Ser-710 and Arg-729, which bind to the incoming rNTP. The Zn2 ion could therefore also play a role in regulating conformational switches within the thumb subdomain that occur along the reaction pathway. Further mutagenesis experiments are needed to address the exact functional significance, if any, of these two zinc binding sites.
The priming loop and its role in the initiation complex.
A high concentration of GTP is required for de novo initiation by the DENV RdRp, regardless of the precise nucleotide sequence at the 3′ end of the RNA template (44). The structure of the complex between DENV RdRp and the 3′dGTP nucleoside analog should shed light on the GTP binding site(s) and its effect on de novo initiation. No large conformational change of DEN RdRp occurs upon binding of 3′dGTP. The electron density map obtained for a crystal soaked with 3′dGTP and magnesium ions revealed a clear extra density corresponding to the triphosphate (tP) moieties of 3′dGTP near the priming loop, about 7 Å away from the catalytic site (Fig. 2C). No magnesium ions could be seen in the 3′dGTP complex, possibly because of their chelation by sodium-potassium tartrate that was included in the soaking buffer. Only very weak electron density was present for the guanosine base and the sugar moiety, indicating their mobility in the absence of a template. No metal ion was observed to coordinate the tP moiety. Comparisons with the RdRps from BVDV and HCV in complex with nucleosides reveal the presence of an rNTP substrate positioned similarly in the vicinity of the priming loop (P site). Binding of ribonucleotides in the catalytic (C) site of the HCV RdRp (8) is mediated by divalent ions which bridge the carboxylic groups from the aspartic acid residues of the active site with the phosphate groups of the nucleotide (8). In crystal form, the absence of Mg2+ ions due to their chelation by sodium-potassium tartrate is the most likely explanation for the fact that the C site is not occupied. Further work is in progress to substitute the sodium-potassium tartrate component for other buffers suitable for divalent metal soaking experiments.
In the DENV 3 RdRp molecule, the tP moiety of 3′dGTP is coordinated by Ser-710, Arg-729, and Arg-737 (Fig. 2D). These three residues are strictly conserved across positive-strand RNA viruses known to initiate replication using a de novo mechanism (Fig. 3), including DENV 1 to 4, YFV, JEV, and WNV. In several de novo RdRp initiation complexes reported, the base of the ribonucleotide bound at the P site stacks against a tyrosine residue (Tyr-630 for the Φ6 bacteriophage polymerase, Tyr-448 for HCV polymerase, and Tyr-581 for BVDV polymerase) (8, 10, 13). The orientation of the tP moiety in our crystal structure (which was inferred from superimpositions with the RdRp structures mentioned above) places the base at the right distance to form stacking interactions with Trp-795. The side chain of Trp-795 could thus help position the base such that it can make Watson-Crick hydrogen bonds with the base located at the 3′ end of the template. Additional interactions between the polymerase and the 3′dGTP α-phosphate include hydrogen bonds with Thr-794 and Ser-796. Residue Arg-737, projecting from helix α21, makes a salt bridge with the β-phosphate, and Arg-729 (motif E) makes a salt bridge with the γ-phosphate. Residue Ser-710 (from motif E) makes a hydrogen bond with the γ-phosphate. Those residues thus provide the platform for de novo RNA initiation by the flavivirus polymerase domain. In a separate study, residues Ser-498 and Arg-517 in the BVDV polymerase (corresponding to Ser-710 and Arg-729 in DENV RdRp) were mutated individually to alanine in order to test their contribution to an elongating (primer-dependent) versus de novo (primer-independent) mode of RNA synthesis (37). The de novo mode of RNA synthesis was almost completely abolished by these single mutations, while RNA elongation was reduced only approximately two- to ninefold. Thus, the mutation of these residues from the BVDV RdRp is able to confer specificity for a primer-dependent mechanism, as opposed to a de novo mechanism, of RNA synthesis. The structural conservation of these residues within motif E with the BVDV RdRp suggests a similar phenotype when Ser-710 and Arg-729 are mutated in the DENV RdRp enzyme. A comparable study of the HCV RdRp demonstrated a similar role played by Arg-386 and Arg-394 (also leading to a severe decrease in de novo initiation). These two residues are structurally equivalent to Arg-729 and Arg-737 in the DENV 3 RdRp (47). Taken together, these data suggest an essential role for this GTP-binding site for de novo initiation of RNA synthesis by the DENV RdRp. Interestingly, one zinc atom (Zn2) located between the antiparallel β-sheets β6 and β7 and helix α26 (Fig. 2) is found in the vicinity of Arg-729 and could thus play a role in modulating de novo initiation.
Model for RNA synthesis.
We built a model for a ssRNA template bound to the DENV 3 RdRp (Fig. 5), based on the experimental complex observed between the RdRp of bacteriophage Φ6 and a template RNA strand (PDB code 1HI0). The ensuing position and orientation of the Φ6 RNA template strand introduce minimal steric clashes with DENV 3 RdRp, suggesting that the mode of binding could be essentially preserved between the two enzymes. The sugar-phosphate backbone of the template is oriented towards the solvent, pointing away from the binding groove of the fingers domain. The positive charge at the surface of the fingers subdomain provides an electrostatic interaction with the ssRNA template, stabilizing its 3′ end at the active site cavity. Together, loops L1, L2, and L3, the linker α21-α22, and the priming loop encircle the catalytic active site and shape the template tunnel. The tunnel is approximately 30 Å deep and 24 Å wide. These dimensions are compatible with the presence of only a ssRNA chain of about five to seven nucleotides, not a duplex, in the active site. Therefore, several loops are likely to adopt a different conformation that would allow the translocation of a duplex formed by the newly synthesized RNA strand and template. The fact that the tunnel observed in our structure has dimensions that would permit access to the active site only for a ssRNA has implications for the dynamics of the flavivirus polymerase. A motion of helices in the fingers domain and a compression (or closing) of the active site around a bound primer/template have been observed in a number of ternary complexes of DNA polymerases, RNA polymerases, and reverse transcriptases (16, 31, 40). For example, the ternary complex formed between HIV reverse transcriptase and a template-primer-dNTP showed an inward rotation of the fingers subdomain towards the palm (by an angle of ∼20°) and the repacking of β-strands β3 and β4 (31) (or of helices in the fingers subdomain for the bacteriophage T7 polymerase), leading to a closed form of the active site in the catalytic complex. In the case of the HCV RdRp template complex (PDB code 1NB7) (45), the HCV RdRp apoenzyme structure appears to adopt an energetically favorable closed conformation, and no major conformational changes of the fingers subdomain upon soaking short RNA templates in pregrown crystals were observed. The high temperature factors for the fingers domains of DENV 3 RdRp suggest that the binding of a short RNA could stabilize this domain and trigger a transition to a more closed form. Compared to the WNV RdRp, the DENV 3 RdRp displays an outward rotation, with an angle of 8° of the fingers with respect to the palm subdomain. The precise assessment of the importance of conformational changes for the formation of a catalytic complex by flavivirus RdRps requires further structural studies. It is possible that the polymerase domain expressed separately is more flexible in the absence of the N-terminal MTase region. Alternatively, the flavivirus polymerases might represent a separate group (compared to the hepacivirus polymerases) where the catalytically competent site is not essentially preformed but instead requires movements of the fingers that would close in the ternary complex. These movements of subdomains are likely to be accompanied by disorder-to-order transitions in the segments linking the fingers and thumb domains (e.g., in the linker between helices α4 and α5). Further structural and mutagenesis work is needed to clarify these issues. Interestingly, Gamarnik and collaborators recently identified a 5′ minimal element of 70 bases forming stem-loop A (SLA), which appears to play an essential role for DENV polymerase RNA binding, leading to template replication (23).
In conclusion, the first high-resolution view of the DENV RdRp catalytic domain described here should facilitate structure-based efforts for the design of antiviral compounds against DENV, and molecules interfering with the enzyme's dynamics could hold great promise as specific inhibitors.
Acknowledgments
James Tam is thanked for encouraging the strong interaction between the structural virology groups led by J.L. and the Novartis Institute for Tropical Diseases. Lee Wan Yen and Daying Wen are thanked for their help with protein expression and purification. We are grateful to the Canard Laboratory (supported by VIZIER integrated project LSHG-CT-2004-511960 of the European Union 6th Framework Programme [FP6] as well as the Conseil Régional de la Région Provence-Alpes-Côte d'Azur) for very helpful discussions and sharing the coordinates of WNV RdRp. We are very grateful to Clemens Scheufler, Sandra Jacob, and Hans Widmer for help with data collection for this work that was performed at beamline X10SA (PXII) of the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland. We also acknowledge the excellent beamline time provided by the ESRF (Grenoble, France), where some early data were collected for this work.
J.L.'s laboratory is supported by grants from NTU (RG 29/05 and RG119/05) and the Singapore Biomedical Research Council (05/1/22/19/405 and 02/1/22/17/043).
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 3.Appleby, T. C., H. Luecke, J. H. Shim, J. Z. Wu, I. W. Cheney, W. Zhong, L. Vogeley, Z. Hong, and N. Yao. 2005. Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer. J. Virol. 79:277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, S. 1994. The Ccp4 Suite—programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Bartholomeusz, A., and P. Thompson. 1999. Flaviviridae polymerase and RNA replication. J. Viral Hepat. 6:261-270. [DOI] [PubMed] [Google Scholar]
- 6.Bartholomeusz, A. I., and P. J. Wright. 1993. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch. Virol. 128:111-121. [DOI] [PubMed] [Google Scholar]
- 7.Blok, J. 1985. Genetic relationships of the dengue virus serotypes. J. Gen. Virol. 66:1323-1325. [DOI] [PubMed] [Google Scholar]
- 8.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks, A. J., M. Johansson, A. V. John, Y. B. Xu, D. A. Jans, and S. G. Vasudevan. 2002. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J. Biol. Chem. 277:36399-36407. [DOI] [PubMed] [Google Scholar]
- 10.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. L. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 12.Choi, K. H., A. Gallei, P. Becher, and M. G. Rossmann. 2006. The structure of bovine viral diarrhea virus RNA-dependent RNA polymerase and its amino-terminal domain. Structure 14:1107-1113. [DOI] [PubMed] [Google Scholar]
- 13.Choi, K. H., J. M. Groarke, D. C. Young, R. J. Kuhn, J. L. Smith, D. C. Pevear, and M. G. Rossmann. 2004. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. USA 101:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delano, W. L. 2004. The PYMOL user's manual. DeLano Scientific, San Carlos, CA.
- 15.Dolja, V. V., and E. V. Koonin. 1991. Phylogeny of capsid proteins of small icosahedral RNA plant-viruses. J. Gen. Virol. 72:1481-1486. [DOI] [PubMed] [Google Scholar]
- 16.Doublie, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 angstrom resolution. Nature 391:251-258. [DOI] [PubMed] [Google Scholar]
- 17.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erbel, P., N. Schiering, A. D'Arcy, M. Renatus, M. Kroemer, S. P. Lim, Z. Yin, T. H. Keller, S. G. Vasudevan, and U. Hommel. 2006. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 13:372-373. [DOI] [PubMed] [Google Scholar]
- 19.Esnouf, R. M., J. S. Ren, E. F. Garman, D. O. Somers, C. K. Ross, E. Y. Jones, D. K. Stammers, and D. I. Stuart. 1998. Continuous and discontinuous changes in the unit cell of HIV-1 reverse transcriptase crystals on dehydration. Acta Crystallogr. D 54:938-953. [DOI] [PubMed] [Google Scholar]
- 20.Ferrer-Orta, C., A. Arias, R. Agudo, R. Perez-Luque, C. Escarmis, E. Domingo, and N. Verdaguer. 2006. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 25:880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrer-Orta, C., A. Arias, C. Escarmis, and N. Verdaguer. 2006. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16:27-34. [DOI] [PubMed] [Google Scholar]
- 22.Ferrer-Orta, C., A. Arias, R. Perez-Luque, C. Escarmis, E. Domingo, and N. Verdaguer. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279:47212-47221. [DOI] [PubMed] [Google Scholar]
- 23.Filomatori, C. V., M. F. Lodeiro, D. E. Alvarez, M. M. Samsa, L. Pietrasanta, and A. V. Gamarnik. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 20:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 26.Guzman, M. G., and G. Kouri. 2002. Dengue: an update. Lancet Infect. Dis. 2:33-42. [DOI] [PubMed] [Google Scholar]
- 27.Halstead, S. B. 1988. Pathogenesis of dengue—challenges to molecular biology. Science 239:476-481. [DOI] [PubMed] [Google Scholar]
- 28.Halstead, S. B. 2002. Dengue. Curr. Opin. Infect. Dis. 15:471-476. [DOI] [PubMed] [Google Scholar]
- 29.Henchal, E. A., and J. R. Putnak. 1990. The dengue viruses. Clin. Microbiol. Rev. 3:376-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. H. Yao, J. Y. N. Lau, and W. D. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 31.Huang, H. F., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 32.Jager, J., S. J. Smerdon, J. M. Wang, D. C. Boisvert, and T. A. Steitz. 1994. Comparison of 3 different crystal forms shows HIV-1 reverse-transcriptase displays an internal swivel motion. Structure 2:869-876. [DOI] [PubMed] [Google Scholar]
- 33.Johansson, M., A. J. Brooks, D. A. Jans, and S. G. Vasudevan. 2001. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-ss and the viral helicase, NS3. J. Gen. Virol. 82:735-745. [DOI] [PubMed] [Google Scholar]
- 34.Kapoor, M., L. W. Zhang, M. Ramachandra, J. Kusukawa, K. E. Ebner, and R. Padmanabhan. 1995. Association between NS3 and NS5 proteins of dengue virus type-2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270:19100-19106. [DOI] [PubMed] [Google Scholar]
- 35.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal-structure at 3.5 angstrom resolution of HIV-1 reverse-transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 37.Lai, V. C. H., C. C. Kao, E. Ferrari, J. Park, A. S. Uss, J. Wright-Minogue, Z. Hong, and J. Y. N. Lau. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski, R. A., M. W. Macarthur, D. S. Moss, and J. M. Thornton. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 39.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 40.Li, Y., S. Korolev, and G. Waksman. 1998. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17:7514-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Malet, H., M. P. Egloff, B. Selisko, R. E. Butcher, P. J. Wright, M. Roberts, A. Gruez, G. Sulzenbacher, C. Vonrhein, G. Bricogne, J. M. Mackenzie, A. A. Khromykh, A. D. Davidson, and B. Canard. 7 February 2007, posting date. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. [Epub ahead of print]. [DOI] [PubMed]
- 41.Medin, C. L., K. A. Fitzgerald, and A. L. Rothman. 2005. Dengue virus nonstructural protein NS5 induces interleukin-8 transcription and secretion. J. Virol. 79:11053-11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng, K. K., M. M. Cherney, A. L. Vazquez, A. Machin, J. M. Alonso, F. Parra, and M. N. James. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277:1381-1387. [DOI] [PubMed] [Google Scholar]
- 43.Ng, K. K., N. Pendas-Franco, J. Rojo, J. A. Boga, A. Machin, J. M. Alonso, and F. Parra. 2004. Crystal structure of Norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279:16638-16645. [DOI] [PubMed] [Google Scholar]
- 44.Nomaguchi, M., M. Ackermann, C. Yon, S. You, and R. Padmanabhan. 2003. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J. Virol. 77:8831-8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 46.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of 4 conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 48.Ray, D., A. Shah, M. Tilgner, Y. Guo, Y. Zhao, H. Dong, T. S. Deas, Y. Zhou, H. Li, and P. Y. Shi. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sampath, A., T. Xu, A. Chao, D. H. Luo, J. Lescar, and S. G. Vasudevan. 2006. Structure-based mutational analysis of the NS3 helicase from dengue virus. J. Virol. 80:6686-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selisko, B., H. Dutartre, J. C. Guillemot, C. Debarnot, D. Benarroch, A. Khromykh, P. Despres, M. P. Egloff, and B. Canard. 2006. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 351:145-158. [DOI] [PubMed] [Google Scholar]
- 51.Steffens, S., H. J. Thiel, and S. E. Behrens. 1999. The RNA-dependent RNA polymerases of different members of the family Flaviviridae exhibit similar properties in vitro. J. Gen. Virol. 80:2583-2590. [DOI] [PubMed] [Google Scholar]
- 52.Steitz, T. A. 1998. Structural biology—a mechanism for all polymerases. Nature 391:231-232. [DOI] [PubMed] [Google Scholar]
- 53.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 54.Tao, Y. Z., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage—structural studies of reovirus polymerase lambda 3. Cell 111:733-745. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, A. A., and O. B. Peersen. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 23:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchil, P. D., A. V. A. Kumar, and V. Satchidanandam. 2006. Nuclear localization of flavivirus RNA synthesis in infected cells. J. Virol. 80:5451-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 58.Wengler, G., and G. Wengler. 1981. Terminal sequences of the genome and replicative-form RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology 113:544-555. [DOI] [PubMed] [Google Scholar]
- 59.Westaway, E. G. 1987. Flavivirus replication strategy. Adv. Virus Res. 33:45-90. [DOI] [PubMed] [Google Scholar]
- 60.Wu, J. Z., N. H. Yao, M. Walker, and Z. Hong. 2005. Recent advances in discovery and development of promising therapeutics against hepatitis C virus NS5B RNA-dependent RNA polymerase. Mini Rev. Med. Chem. 5:1103-1112. [DOI] [PubMed] [Google Scholar]
- 61.Xu, T., A. Sampath, A. Chao, D. Y. Wen, M. Nanao, P. Chene, S. G. Vasudevan, and J. Lescar. 2005. Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 angstrom. J. Virol. 79:10278-10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yap, T. L., Y. L. Chen, T. Xu, D. Wen, S. G. Vasudevan, and J. Lescar. 2007. A multi-step strategy to obtain crystals of the dengue virus RNA-dependent RNA polymerase that diffract to high resolution. Acta Crystallogr. F 63:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yon, C., T. Teramoto, N. Mueller, J. Phelan, V. K. Ganesh, K. H. M. Murthy, and R. Padmanabhan. 2005. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 280:27412-27419. [DOI] [PubMed] [Google Scholar]
- 64.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]