Abstract
Cyprinid herpesvirus 3 (CyHV-3), previously designated carp interstitial nephritis and gill necrosis virus or koi herpesvirus, is the cause of a worldwide mortal disease of koi and carp. Morphologically, the virus resembles herpesviruses, yet it bears a genome of 277 to 295 kbp, which is divergent from most of the genomic sequences available in GenBank. The disease afflicts fish in the transient seasons, when the water temperature is 18 to 28°C, conditions which permit virus propagation in cultured cells. Here we report that infectious virus is preserved in cultured cells maintained for 30 days at 30°C. CyHV-3-infected vacuolated cells with deformed morphology converted to normal, and plaques disappeared following shifting up of the temperature and reappeared after transfer to the permissive temperature. Viral propagation and viral gene transcription were turned off by shifting cells to the nonpermissive temperature. Upon return of the cells to the permissive temperature, transcription of viral genes was reactivated in a sequence distinguished from that occurring in naïve cells following infection. Our results show that CyHV-3 persists in cultured cells maintained at the nonpermissive temperature and suggest that viruses could persist for long periods in the fish body, enabling a new burst of infection upon a shift to a permissive temperature.
Cyprinus carpio is a widely cultivated eurythermal fish which accounts for much of the world's aquaculture production. In recent years, this fish has been afflicted by a disease causing mass mortalities in fisheries worldwide (6, 12, 23). The sickness is seasonal, afflicting fish when the water temperature is 18 to 28°C. The agent causing the disease is a large double-stranded DNA virus designated carp interstitial nephritis and gill necrosis virus, koi herpesvirus, or cyprinid herpesvirus 3 (CyHV-3) (9, 25, 31). Morphologically, CyHV-3 resembles herpesviruses (9, 10) and shows sequence homology to two other aquatic herpesvirus-like viruses, CyHV-1 and CyHV-2, afflicting Cyprinus carpio and Carassius auratus, respectively (31). On the other hand, CyHV-3 expresses proteins resembling those of Poxviridae members and other large DNA viruses (10, 11). It is not yet known whether CyHV-3 undergoes latent infection. This question is not only important for the characterization of this unique virus but also has economic implications, since it is unclear how and where the virus is preserved between the seasons and by what means the transfer of the virus from virus-free to contaminated regions can be avoided.
CyHV-3 DNA was identified in infected fish maintained for 64 days postinfection (dpi) at the nonpermissive temperature range, but infectious virus was not found in these fish (4, 5). Recently, it was reported that infected fish maintained for ∼200 days at 12°C released infectious virus upon shift of the water temperature to a permissive temperature, i.e., 22°C (28). These reports strongly suggest that CyHV-3 persists in fish maintained at low temperatures for an extended period. In contrast, neither viral DNA nor infectious virus was found in “naturally immunized fish” (25). These fish were exposed to CyHV-3 at the permissive temperature for 2 to 3 days and then transferred to the nonpermissive temperature of 30°C for at least 3 days before being transferred to open-air ponds. These fish have been kept in open-air ponds in Israel for over 12 months and, to date, show no signs of viral infection.
In order to determine whether CyHV-3 persists in host cells maintained at a high nonpermissive temperature, we developed a tissue culture model to “follow” CyHV-3 at permissive (22°C) and nonpermissive (30°C) temperatures. We determined the length of time that CyHV-3 persists in cultured cells, as well as the sequence of events leading to suppression and reappearance of viral replication and transcription in infected cells following alteration of the temperature from permissive to nonpermissive and back. Here we demonstrate that morphologically deformed cells induced by infection of cultured common carp brain (CCB) cells at the permissive temperature (22°C) turned normal following transfer of the cell cultures to 30°C. Early transcription of viral genes is intensively inhibited at the nonpermissive temperature, and no replication of viral DNA is apparent. After establishing an infection, we tracked the down regulation of viral transcription following transfer of the cells to 30°C and the reappearance of viral mRNA after a shift to the permissive temperature. Our results show that elimination of viral mRNAs proceeds in a specific hierarchy not correlated with the reappearance of the mRNA molecules following infection. Furthermore, the establishment of the mRNA population following a temperature downshift is distinguished from that which occurred after infection of naïve cells.
MATERIALS AND METHODS
Cell cultures.
Cultures of CCB cells, generously provided by R. Riebe and S. M. Bergmann (Friedrich Loeffler Institute, Germany), and koi fin cells were prepared as previously described by Ronen et al. (25), using the methods of Hasegawa et al. (8) and Neukirch et al. (17). Cells were propagated in medium containing 75% Dulbecco's modified Eagle's medium (Sigma) and 25% Leibovitz (L-15) medium supplemented with 10% fetal calf serum and antibiotics (Biological Industries, Kibbutz Beit Haemek, Israel).
Viruses.
CyHV-3 isolated from the kidneys of sick fish was propagated in koi fin cell cultures, as previously described (21, 25). At 4 to 7 dpi, following the appearance of plaques, the culture medium was collected and cleared of cells and cell debris by centrifugation for 10 min at 10,000 × g. The viral titer was determined by a plaque assay carried out with CCB cell cultures. The virus suspension was divided into aliquots and stored at −70°C for future use. Cells were routinely propagated and infected in 50-ml flasks (Nunc) at 22°C, and the temperature shift was carried out by transferring the flasks into a 30°C incubator.
Nucleic acid extraction from cultured cells.
The extraction procedure for total DNA varied according to our requirements. For PCR analyses, cultured cells were pelleted and washed twice in phosphate-buffered saline. Pellets were then resuspended in Tris-EDTA (pH 7.6) supplemented with 0.5% sodium dodecyl sulfate and incubated for 10 min at 37°C, and subsequently, DNA was purified by phenol-chloroform and ethanol precipitation. For pulsed-field gel electrophoresis (PFGE), cells were embedded in agarose plugs by using standard methods (27). In brief, washed cell pellets were resuspended in buffer L (0.01 M Tris-Cl, pH 7.6, 0.1 M EDTA, 0.02 M NaCl) at a concentration of 1 × 107 cells/ml, and an equal amount of molten 1% low-melting-temperature SeaPlaque agarose (BMA) was added to the cell mixture and allowed to set in a 1-ml syringe barrel. Plugs were then transferred to 5 volumes of buffer L supplemented with 0.1 mg/ml proteinase K (Sigma-Aldrich) and 1% (wt/vol) Sarkosyl (Sigma-Aldrich) and incubated for 16 h at 50°C. Following incubation, plugs were repeatedly washed in Tris-EDTA containing 40 μg/ml phenylmethylsulfonyl fluoride (Sigma-Aldrich) and then stored at 4°C. Total cellular RNA was extracted from CCB cells by using an EZ-RNA kit (Biological Industries, Kibbutz Beit Haemek, Israel) according to the manufacturer's instructions.
Quantitative PCR (qPCR) analysis.
Primers NHRT-F (5′-CCAGATCCACCAGCTGCTGT-3′) and NHRT-R (5′-AAGATGGGATCTCTCGGAGG-3′) were designed to amplify a 200-bp amplicon of the CyHV-3 B22R homologue (GenBank accession no. AY661550) by PrimerExpress (Applied Biosystems), using default settings. Reaction mixtures containing 250 nM of each primer, 12.5 μl of 2× SYBR green master mix (Applied Biosystems), and 2.5 μl of sample in a final volume of 25 μl were run on an ABI PRISM 7700 instrument (Applied Biosystems). CyHV-3 DNA from sucrose gradient-purified virions (10) was used to generate a standard linear curve at a range of 5 ng to 0.25 fg (R = 0.99). DNA samples were assayed in quadruplicate.
RT-PCR.
For the reverse transcriptase (RT) reaction, total RNA preparations extracted from infected and noninfected cells were treated with DNA-free (Ambion) according to the manufacturer's protocol. cDNAs were generated from 1 μg of total RNA by using SuperScript RNase H RT according to the manufacturer's protocol (Invitrogen), using a final volume of 20 μl. Following reverse transcription, 2 μl of the cDNA was amplified using HotStart Taq DNA polymerase (Roche) with a specific primer set for each gene (Table 1). The amplification protocol included 1 cycle of initial activation at 95°C for 10 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min; and 1 cycle of terminal extension at 72°C for 10 min. Ten microliters of the PCR product was run in a 1% agarose gel and stained with ethidium bromide. The primers listed in Table 1 represent the 20 viral open reading frames (ORFs) published thus far in GenBank and comprise approximately 14% of the viral transcriptome. Amplification of carp β-actin with primers 5′-GCCAACACAGTGCTGTCTGG-3′ (forward) and 5′-GCTGATCCACATCTGCTGG-3′ (reverse) was used as a control.
TABLE 1.
Oligonucleotides used as primers to identified CyHV-3 mRNAs
| CyHV-3 gene | Accession no. | PCR product size (bp) | Primers (5′-3′)a |
|---|---|---|---|
| DNA polymerase (DNAP) | AY939862 | 303 | F, CTC-TGA-GGA-CCA-AGC-TGG-AG; R, GTC-GCC-GTA-CAC-GAC-GGT-CA |
| Thymidine kinase (TK) | AJ535112 | 650 | F, GGA-TCC-ATG-GCT-ATG-CTG-GAA-CTG-GT; R, GGA-TCC-GGC-GCA-CCC-AGT-AGA-TTA-TG |
| Thymidylate kinase (TmpK) | DQ118125 | 400 | F, GGA-TCC-ACA-AGA-GCG-ATC-TGG-ACG-AT; R, GGA-TCC-GAG-GAT-GTC-ACC-ATG-GAC-CT |
| Helicase | AY939857 | 401 | F, GGA-TCC-GTT-TCG-ACA-CCA-TCC-AGT-CC; R, GGA-TCC-GAA-CCT-GTC-GCA-GTT-GTT-GA |
| Ribonucleotide reductase (RNR) | AY208989 | 700 | F, CCA-ACT-CGG-AGG-AGG-TGG-CCG; R, TGC-GCA-TGT-TAT-CGT-TCC |
| Major capsid protein (MCP) | AY787402 | 200 | F, GGA-TCC-CCA-GGC-GTA-CTT-CAT-GTC-CT; R, GGA-TCC-ACG-ATG-GGC-ACC-AAC-TTT-AG |
| Intercapsomeric triplex protein (ITP) | AY939859 | 303 | F, GGA-TCC-TCC-CTG-GAC-GTG-ATC-TTC-TC; R, GGA-TCC-TCC-GCT-GTA-AGC-GTT-GTA-GA |
| Major Envelope protein (Orf4) | AB178324 | 200 | F, GGA-TCC-CCA-ACG-ATC-CCA-ACT-TTG-AT; R, GGA-TCC-TAG-ATG-ACC-CCG-ATC-AGG-AA |
| Membrane protein (Orf2) | AB178324 | 200 | F, GGA-TCC-TGG-TCT-TAT-TCG-GGC-TTG-TC; R, GGA-TCC-GTG-TCG-TTG-ACG-ATG-CTC-TC |
| Membrane protein (ORF3) | AB178324 | 200 | F, GGA-TCC-ACA-ACA-GGT-TGA-GGG-ACG-AC; R, GGA-TCC-ACA-AAC-ACG-CTG-ATG-GTC-AC |
| Glycoprotein (GP) | AY660950 | 200 | F, GGA-TCC-CCC-GTG-TGT-CCG-TAC-TTT-GT; R, GGA-TCC-CTC-CGA-CCT-GAA-TTG-GTC-AG |
| Clone D (B22Rh exon 1) | AY661550 | 600 | F, GGA-TCC-CGC-CTT-CTC-TCT-AGC-ACA-GC; R, GGA-TCC-AAG-AGG-CAC-ATT-CCC-ATG-TC |
| Clone E (B22Rh exon 2) | AY661550 | 517 | F, GGA-TCC-AGA-CGG-TGA-CGG-TCA-CCC; R, GCC-CAG-AGT-CAC-TTC-CAG-CTT-CG |
| Clone A | AY208988 | 701 | F, CCC-ATG-AGG-CTG-AGG-AAC-GCC-G; R, GCA-CCC-CCG-TGA-TGG-TCT-TGC |
| Clone Y | AY873800 | 493 | F, GGA-TCC-GCA-CCT-TTT-CCT-TCA-TGC-AC; R, GGA-TCC-GTG-CCA-AGG-CCT-ATG-ATG-TT |
| Hypothetical protein (Orf1) | AB178324 | 200 | F, GGA-TCC-AAC-GTG-AAG-GGC-AAC-AAC-AT; R, GGA-TCC-GTT-CAG-AGG-GTG-ATC-GTG-GT |
| Hypothetical protein (Orf5) | AB178324 | 200 | F, GGA-TCC-CAA-CGT-CCA-CAG-CTA-CAT-CG; R, GGA-TCC-CAG-GAG-TTT-TCG-CAG-GTC-TC |
| Hypothetical protein 1 (HP-1) | AY858588 | 500 | F, GGA-TCC-TGC-CAC-ACG-TTG-AAG-AGT-TC; R, GGA-TCC-TCA-AGC-AGC-TCA-CCA-ACA-TC |
| Replicon 9/5 | AF411803 | 484 | F, GAC-GAC-GCC-GGA-GAC-CTT-GTG; R, CAC-AAG-TTC-AGT-CTG-TTC-CTC-AAC |
| Gray SphI | AY568950 | 290 | F, GAC-ACC-ACA-TCT-GCA-AGG-AG; R, GAC-ACA-TGT-TAC-AAT-GGT-GGC |
F, forward; R, reverse.
PFGE and Southern blotting.
Agarose-embedded samples were resolved in a 1% Seakem Gold agarose (Cambrex) gel by PFGE via a counterclamped homogenous electric field (CHEF-DRII; Bio-Rad). Running conditions were as follows: 6 V/cm at 14°C for 22 h, with switch times ramped from 50 to 90 s. After being transferred to a Hybond-N nylon membrane (Amersham), viral DNA was detected using a DIG DNA labeling and detection kit (Roche). The digoxigenin-labeled probe was generated by PCR, using the primers for clone A (Table 1).
RESULTS
Stability of CyHV-3 in culture medium.
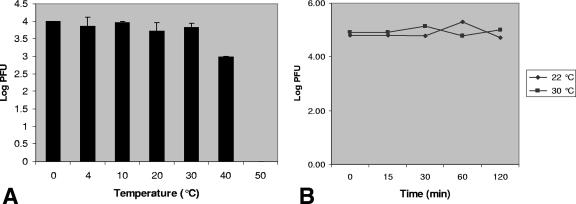
Fish succumb to CyHV-3 infection at a distinct temperature range of 18 to 28°C. In order to determine whether this phenomenon is due to virion instability outside the permissive temperature range, we measured the residual infectivity of CyHV-3 incubated at 0 to 50°C. Medium containing the virus was thawed on ice, dispensed into aliquots, and incubated for different times and temperature intervals. Following incubation, the titer of the residual infectious virus was determined by plaque assay. Figure 1A shows that incubation of the medium containing virus for 30 min at 50°C was sufficient to neutralize it. However, the virus was quite stable in medium maintained at a temperature range of permissive and both high and low nonpermissive temperatures. Incubation at 30°C for 2 h showed no decline in the residual amounts of infectious virus compared to incubation at the permissive temperature (Fig. 1B). These results indicate that CyHV-3 is stable during the time frame used for infecting cultured cells in the following experiments.
FIG. 1.
Residual viral infectivity following incubation at permissive and nonpermissive temperatures. Medium containing virus was thawed on ice, dispensed into aliquots, and (A) incubated at temperatures ranging from 0 to 50°C for 30 min or (B) incubated at 22°C and 30°C for the designated times. Following incubation, residual infectivity was measured by plaque assay on CCB cells. Results shown are mean values for three independent assays.
CPE induced by CyHV-3 is temperature dependent.
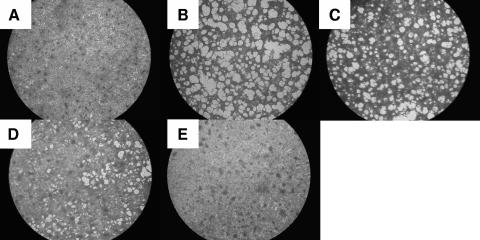
We traced the appearance of the cytopathic effect (CPE) in cells infected at the permissive temperature and then transferred to the nonpermissive temperature. Figure 2 demonstrates that infected cells kept at the nonpermissive temperature show no sign of CPE (Fig. 2E) and are indistinguishable from noninfected control cells (Fig. 2A). Plaques began forming at the permissive temperature at 3 to 4 dpi, and on day 9 extensive CPE was evident in the infected culture (Fig. 2B). Yet once infected cells were shifted to the permissive temperature, the onset of CPE was evident, in concord with the time cells spent under nonpermissive conditions (Fig. 2C and D).
FIG. 2.
Temperature shift assay. CCB cells were infected with wt CyHV-3. Following infection, the cells were either kept at 22°C (B) or shifted up to 30°C. At 24 hpi (C) or 48 hpi (D), the cells were returned to 22°C. At 9 dpi, the cells were fixed, stained, and photographed. (A) Noninfected control. (E) Infected cells kept at 30°C after infection. Magnification, ×20.
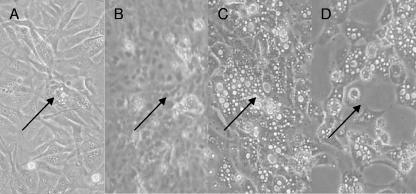
A temperature shift to nonpermissive conditions reverted an already evident plaque to its normal morphological appearance. Vacuolated cells appeared in CCB cell cultures at 3 dpi, as shown in Fig. 3A. These cells shed the virus to neighboring cells to produce plaques. Such an early plaque was labeled by drawing a circle on the bottom of the flask, enabling us to follow the same region following transfer of the flask to a 30°C incubator. The CPE disappeared, and 3 days later the cell morphology seemed normal (Fig. 3B). This normal appearance of cell morphology was maintained as long as cells were kept at the nonpermissive temperature. However, upon downshifting of the culture temperature to 22°C, the CPE reappeared after 7 days (Fig. 3C) and developed into plaques 10 days later (Fig. 3D).
FIG. 3.
Deactivation and reactivation of CyHV-3 by temperature shifts. CCB cells grown at 22°C were infected with wt CyHV-3. At 3 dpi, plaques began forming (A), and the cells were transferred to 30°C. Three days after the upshift, cells returned to their normal morphological state (B) and were shifted down to 22°C. Seven days after the downshift, the onset of CPE was evident (C). Thirteen days after the downshift, cells detached from the flask and the culture died (D). Arrowheads point to the reference marker on the flask. Magnification, ×20.
Viral replication at permissive and nonpermissive temperatures.
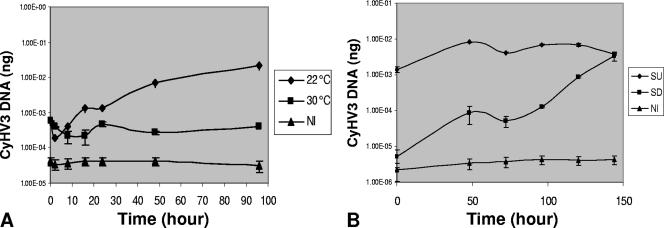
We initially determined whether CyHV-3 replicates at 30°C. CCB cells were infected at a multiplicity of infection (MOI) of 0.01 for 1 h at 22°C, washed, and then either kept at the permissive temperature or shifted to the nonpermissive temperature of 30°C. Total DNA was extracted from the infected cells at time intervals, and the amounts of viral DNA were measured using qPCR (Fig. 4). Cells harvested at 1 hpi (T0) showed the amounts of viral DNA in the initial inoculums. At the permissive temperature, replication of viral DNA commenced as early as 8 to 16 hpi and proceeded for 96 hpi, while at the nonpermissive temperature no viral DNA multiplication was apparent.
FIG. 4.
Replication of viral DNA at permissive and nonpermissive temperatures, measured by qPCR. CCB cells were infected (MOI, 3) at 22°C for 1 h. Cells were washed and incubated at either 22°C or 30°C. (A) At 0, 2, 8, 16, 24, 48, and 96 hpi, total DNA was extracted from a cell culture maintained at each temperature, and viral DNA was measured by using qPCR. (B) At 96 hpi, cells grown at 22°C were transferred to 30°C (SU) and those at 30°C were shifted to 22°C (SD). Viral DNA was quantified at 0, 48, 72, 96, 120, and 144 h postshift. NI, total DNA assayed from noninfected CCB cells.
In order to determine whether the virus maintained at 30°C would reactivate upon shifting of the cells to the permissive temperature, we monitored viral DNA replication in cultures swapped from nonpermissive to permissive temperatures and vice versa. Figure 4B shows that cultures reallocated to 22°C recommenced viral DNA replication as early as 48 h postshift. Interestingly, no change in the amount of viral DNA was evident in cells moved to the nonpermissive temperature for a period of 144 h postshift.
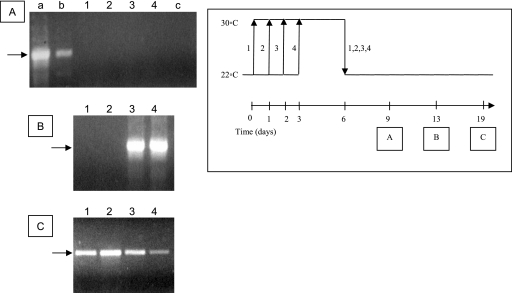
We were interested to determine, firstly, how soon after the downshift of temperature the infected cells recommenced virus production, and secondly, whether the delay in virus production was dependent on the duration that the infected cells were maintained at the permissive temperature. To this end, we infected CCB cells with CyHV-3 at an MOI of 0.1 and transferred cultures at 1-day intervals to 30°C (Fig. 5). At 6 dpi, all cultures were transferred to 22°C, and the newly synthesized virus released into the medium was identified by PCR amplification. No viral particles were released into the medium on the third day after the shift (9 dpi) (Fig. 5A). However, at 13 dpi (a week following temperature downshift), cells which were moved to 30°C at 3 and 4 dpi released virus into the medium (Fig. 5B, lanes 3 and 4), while cells which remained at 22°C for only 24 and 48 hpi produced viruses as late as 19 dpi, after 2 weeks at the permissive temperature (Fig. 5C). These results indicate that a lag of at least 3 to 7 days at the permissive temperature is required for CyHV-3-infected CCB cells to release virus progenies into the culture medium.
FIG. 5.
Identification of viral progeny of temperature-deactivated/reactivated virus by PCR analysis. The scheme on the right describes the assay protocol. CCB cells were infected at 22°C and either kept at the permissive temperature (arrows 2, 3, and 4) or shifted up to 30°C (arrow 1). At time intervals of 24 h, flasks 2, 3, and 4 were shifted to 30°C. At 6 dpi, cells were downshifted to 22°C. At 9 (A), 13 (B), and 19 (C) dpi, medium was collected from each cell culture and assayed for the presence of viral DNA by PCR. Lane a, purified CyHV-3 DNA used as a template; lane b, medium collected from infected cells at 22°C at 3 dpi; lane c, medium collected from noninfected control cells. Arrows to the left of the gels represent a 700-bp DNA marker.
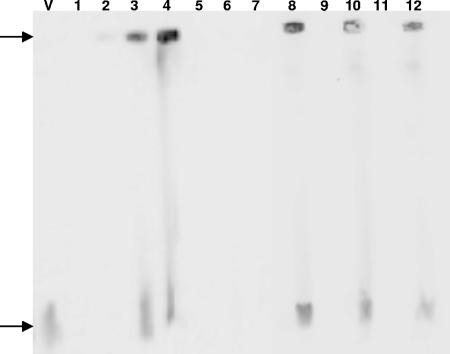
In order to determine the state of the viral DNA upon infection, we performed PFGE and Southern blot analysis. At the permissive temperature, we were able to detect high-molecular-weight (“well DNA”) viral DNA as early as 8 hpi (Fig. 6, lane 2), and at 12, 24, and 72 hpi, linear forms appeared (Fig. 6, lanes 3, 4, and 8). These results suggest that the synthesis of viral DNA initiates in the form of “endless” DNA (30), which fails to migrate from sample wells during PFGE (20). Shifting the cells to the nonpermissive temperature immediately following infection inhibited viral replication (Fig. 6, lanes 5 to 7, 9, and 11), and neither the high-molecular-weight nor the linear form was detected. Once viral DNA was allowed to replicate, being kept at 22°C for 24 hpi prior to the temperature upshift, both forms of viral DNA were apparent in the infected cells maintained under nonpermissive conditions.
FIG. 6.
Southern blot analysis of CyHV-3 DNA at permissive versus nonpermissive temperatures. CCB cells were infected (MOI, 10) at 22°C and either kept at the permissive temperature or shifted to 30°C. DNA samples were separated by PFGE (100-μl plug/slot), and viral DNA was detected by Southern hybridization using a digoxigenin-labeled clone A probe. Lanes: V, purified viral DNA; 1, cells immediately postinfection; 2, cells at 8 hpi at 22°C; 3, cells at 12 hpi at 22°C; 4, cells at 24 hpi at 22°C; 5, cells at 8 hpi at 30°C; 6, cells at 12 hpi at 30°C; 7, cells at 24 hpi at 30°C; 8, cells at 48 hpi at 22°C; 9, cells at 48 hpi at 30°C; 10, cells infected for 24 h at 22°C and maintained at 30°C for 48 h; 11, cells at 72 hpi at 30°C; 12, cells infected for 24 h at 22°C and maintained at 30°C for 72 h.
Elimination of viral mRNA molecules from CyHV-3-infected cells following temperature upshift.
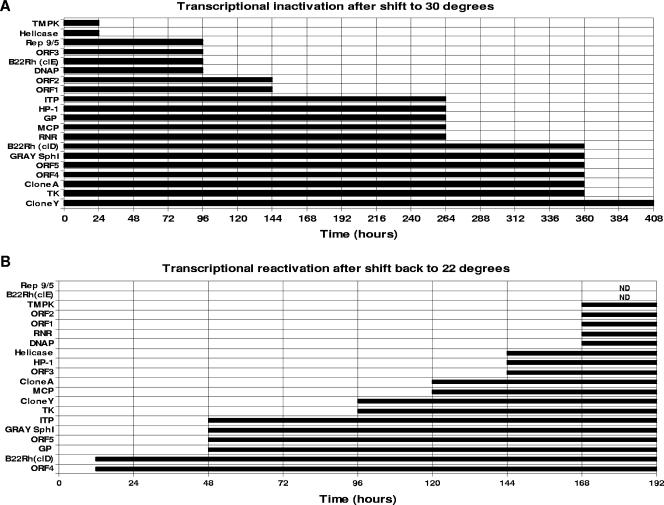
CCB cells were infected with CyHV-3 at an MOI of 0.1, and at 3 dpi culture cells were split (1:2) to maximize the number of infected cells in the cultures. Two days later (at 5 dpi), the infected CCB cells transcribed all the viral mRNAs tested and released viral progenies into the medium (Fig. 7A, zero time). In order to follow the kinetics of mRNA elimination from the infected cells following the temperature upshift, cell cultures were transferred to a 30°C incubator, and cell cultures harvested daily were used to extract the total RNA molecules. The viral mRNA molecules were identified by RT-PCR. To verify that no DNA molecules were amplified by RT-PCR, RNA samples used as a control were amplified without preexposure to RT. No DNA products were apparent without synthesizing cDNA by reverse transcription (not shown). Amplification of endogenous β-actin mRNA was used to ensure the fidelity of the RT-PCR throughout the experiments (not shown). Figure 7A shows that the viral mRNAs involved in DNA synthesis, namely, those for TmpK and helicase, were eliminated from the cells as early as 24 h following the temperature upshift. On the other hand, ribonucleotide reductase (RNR) and thymidine kinase (TK) mRNAs, which are also involved in nucleotide preparation for DNA synthesis, disappeared as late as 11 and 15 days after the cells were shifted to 30°C. The mRNA encoding DNA polymerase was eliminated 4 days following the temperature rise. Interestingly, some of the viral mRNAs persisted for as long as 15 or even 17 days following the temperature upshift. It should be emphasized here that this experiment reveals the presence of viral mRNAs in the cells without distinguishing between newly synthesized mRNAs and molecules characterized by a long half-life in cells.
FIG. 7.
Inactivation and reactivation of viral transcription following temperature shift. (A) Infected CCB cells (5 dpi) were shifted to 30°C, and total RNA was extracted daily for 17 days. (B) On day 20, cells were shifted back to 22°C, and RNAs were extracted for 10 days. RT-PCR using the primers listed in Table 1 was used to monitor viral transcription. Endogenous carp β-actin was used as a positive control for the RT reaction (not shown). ND, not detected.
Reappearance of viral mRNA molecules in CyHV-3-infected cells following temperature downshift.
The chronic CyHV-3-infected cells incubated at 30°C (described above) were used to study the kinetics of viral mRNA reappearance upon moving the cells to the permissive temperature (Fig. 7B). The experiment commenced at 21 dpi, when no viral mRNAs could be detected by RT-PCR. Incubation of the chronically infected cultured cells for 8 days at 22°C was sufficient for the reemergence of all the viral mRNA molecules tested in this experiment. The B22R homologue and Orf4 genes were retranscribed as early as 12 to 24 h, and the Orf5 and Gray Sph1 genes reappeared after 2 days of incubation at the permissive temperature. Interestingly, these four gene transcripts were abrogated quite late from the infected cells following temperature upshift (Fig. 7A). Importantly, the viral DNA polymerase, RNR, and TmpK gene transcripts, which are required for DNA replication, as well as Orf1 and Orf2 transcripts, were synthesized long after shifting of the cells to the permissive temperature, probably when all the machinery of viral replication was ready and after the reinitiation of viral DNA synthesis (Fig. 7B). However, the results presented in Fig. 7A and B do not reveal a close correlation between the elimination and reappearance of many viral mRNAs after moving the chronically infected cells from a permissive to nonpermissive temperature and back to 22°C.
Prolonged incubation of cells under nonpermissive conditions revealed that CyHV-3 persistence in cultured cells is limited. Infected cells kept at 30°C for 30 days were positive for viral DNA by PCR, and 14 days following transfer of these cells to 22°C, plaques began forming. Infected cells incubated for 70 days at 30°C were negative for the presence of viral DNA by PCR and showed no sign of viral reactivation after being shifted back to the permissive temperature (not shown).
Early inhibition of gene transcription following temperature upshift.
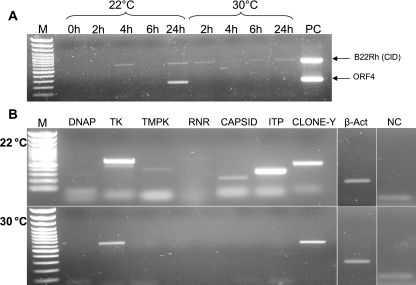
We tested whether a temperature shift immediately following infection inhibits the transcription of viral genes, which may account for the inhibition of viral DNA replication (Fig. 4B). Cells were exposed to the virus at 22°C for an hour of absorption and then immediately shifted to 30°C. At short intervals of 2 h, total RNA was extracted and RT-PCR analysis was performed on CyHV-3 gene transcripts. Figure 8A shows that while Orf4 and B22Rh were both transcribed at 22°C, only the latter appeared at 30°C. Further analysis showed that at 24 hpi, all of the known viral transcripts appeared at the permissive temperature (not shown), yet only the clone Y, TK, intercapsomeric triplex protein (ITP), and B22Rh genes (Fig. 8A and B) were transcribed at 30°C.
FIG. 8.
Early gene transcription following temperature shift. CCB cells were infected (MOI, 3) at 22°C and either kept at 22°C or shifted to 30°C immediately p.i. (A) Transcription kinetics of B22Rh and Orf4 at 22°C versus 30°C. At the times indicated, RT-PCR analysis was performed using primers for the designated viral replicons. (B) RT-PCR at 24 hpi at 22°C versus 30°C, using primers for the genes designated in the header. M, 100-bp DNA ladder (PeqLab); PC, CyHV-3 DNA; β-Act, endogenous carp β-actin control; NC, no-RT template control using β-actin primers.
DISCUSSION
Carp and koi disease afflicts hatcheries when the water temperature is 18 to 28°C; in Israel, the disease appears in the spring and fall, while in Europe it appears once a year, in the summer. It is still unclear how the virus is preserved during the cold and hot seasons. One possibility is that infectious virus remains in the fish dropping sediment on the pond bottom (3). However, the possibility that the virus persists in the host cells cannot be excluded. Previously, it was shown that viral DNA and infectious virus persist for a long period (approximately 200 days) in infected fish maintained at nonpermissive low temperatures (28). Gilad et al. (5) reported the presence of very few viral genomic copies in fish exposed to the pathogenic virus at permissive temperatures at 62 dpi. However, to date, there is no solid evidence indicating the presence of infectious virus in fish surviving the disease (4) or in so-called “naturally immunized” carps, as well as fish immunized with the attenuated virus, maintained under permissive conditions (18, 25, 26). It is conceivable that at permissive and high temperatures, but not at low temperatures, the specific immune antibodies efficiently remove the virus and the infected cells expressing viral antigens from the fish (15, 16).
In order to resolve the question of whether CyHV-3 persists in host cells, we used the following two different approaches: cultured cells were used rather than live fish, and infected carp cells were maintained at a nonpermissive 30°C rather than at low temperatures. Studying persistence in cultured cells is advantageous, as the system is not affected by host immune responses, and because most of the cells bear the virus, it facilitates molecular studies. The results of this study clearly show that CyHV-3 persists in cultured cells kept at 30°C for at least 30 dpi, as transferring the cells to permissive conditions at 30 but not at 70 dpi reactivates the virus. Since the cells were continually passaged over the incubation period at the nonpermissive temperature, the virus's inability to reactivate at 70 dpi may be due to its dilution in the culture. Whether these results indicate that the virus persists in the fish body for such a long period at high temperatures is not yet known, and resolving this question will require additional intensive investigations.
Here we report that CyHV-3 is stable in culture medium and that virus incubated at 22°C or 30°C for 2 h remains active. This observation correlates with our previous results showing that the virus remains active for 4 h in a water tank (19). The foamy vacuolated cytoplasm and the morphology deformation induced by the virus (21) are completely dependent on viral gene expression, as holding back the viral mRNA and/or viral DNA synthesis by elevating the temperature is concomitant with converting the cell morphology to normal.
Early viral gene transcription, with the exception of that of the TK, B22Rh, ITP, and clone Y genes, was completely abolished when cells were transferred to 30°C immediately following infection (Fig. 8). Interestingly, the four genes transcribed at the nonpermissive temperature were the last genes to be shut off when cells were shifted to the nonpermissive temperature (Fig. 7A), suggesting that unlike the case for other viral genes, their transcription is not temperature dependent. Whether these genes share common regulatory motifs or play a role in persistence of CyHV-3 is a matter of further study.
Viral DNA synthesis is completely restrained at nonpermissive temperatures, and viral DNA replication resumes within 48 h following cell transfer to permissive temperatures (Fig. 4). However, maturation and release of infectious virus into the culture medium are time-consuming and occur 7 to 19 days after transfer of the cells to 22°C (Fig. 5), suggesting that DNA synthesis precedes the expression of many other viral genes required for virus maturation. The finding that 5 of 20 genes tested in our study reappeared as late as a week after moving the infected cells to the permissive temperature supports this assumption.
A correlation exists between the qPCR and Southern blot experiments showing that no viral replication occurs, implying that CyHV-3 is inhibited at early stages of infection when shifted to 30°C. When cells were infected at the permissive temperature, the viral DNA assumed a high-molecular-weight state, as observed in herpes simplex virus type 1 (HSV-1) (13, 22, 29), the prototype virus of the family Herpesviridae, prior to active lytic replication. Although the cells were infected at a high MOI, we were unable to detect the state of the viral DNA when they were shifted to 30°C immediately following infection. Once viral replication initiated at the permissive temperature, both the high-molecular-weight (“endless”) and linear forms were maintained when cells were shifted to 30°C for up to 72 h. Whether the high-molecular-weight latency-associated form of the viral DNA persists for longer periods under nonpermissive conditions is a matter for further study.
While viral DNA replication is suppressed upon shifting to 30°C, viral mRNA transcripts are present in cells for up to 17 days under nonpermissive conditions. This may explain the “naturally immunized carp” phenomenon (25). In carps, low environmental temperature enhances nonspecific cytotoxic cell activity and decreases antibody production. In contrast, a high environmental temperature has no effect on these parameters compared to the standard temperature (15). Elevating the temperature gives the fish immune system an advantage over the virus. While the pathogenic virus does not replicate in these fish, it continues to produce viral antigens, triggering a specific immune response which is fully active under these conditions.
Moving the infected cells from the permissive temperature to 30°C immediately halted viral DNA synthesis, while the DNA polymerase gene mRNA was eliminated 4 days later. In case the cellular translation machinery is not affected by the temperature upshift, it is conceivable that the viral DNA polymerase is a temperature-sensitive enzyme. This mechanism limits the synthesis of viral DNA to the environmental conditions appropriate for virus propagation.
The question of whether CyHV-3 undergoes latent infection is economically and epidemiologically important. The experiments described here and those previously reported by St.-Hilaire et al. and Gilad et al. (4, 5, 28) showing that infectious virus and/or viral information persists in fish maintained at a low temperature support the assumption that CyHV-3 undergoes latent infection. These experiments inform us that CyHV-3 replication and disease induction are restrained under nonpermissive conditions. Converting the environmental conditions to permissive is sufficient to reactivate the virus.
Unique viral genes control latent infection of HSV-1, for example, which takes place under normal permissive conditions, and alteration of the physiological status of the host cells causes the reactivation of the virus (24). Several tissue culture models were developed to study the infection, repression, and reactivation of HSV-1. These systems usually employ viral replication inhibitors and/or mutant viruses (7, 14, 32, 33) or infecting wild-type (wt) virus outside its natural host (1). A recent publication by Bringhurst and Schaffer shows a correlation of stress induced by temperature with the ability of a mutant HSV-1 to propagate in a tissue culture model (2).
Whether the molecular mechanisms regulating CyHV-3 and other herpesviruses, such as HSV-1, are parallel remains to be determined. Our model, which studies a wt virus in its natural host, may facilitate this endeavor.
The conditional quiescence in which CyHV-3 persists in poikilothermic vertebrate host cells maintained under nonpermissive conditions enables it to survive under nonpermissive conditions and provides an evolutionary selective advantage. It is plausible that this genetic characteristic of the poikilothermic vertebrate viruses could be developed into genes responsible for latency in large DNA viruses of homoeothermic vertebrate organisms.
Acknowledgments
We thank R. Riebe and S. M. Bergmann, NRL for Fish Diseases, Friedrich-Loeffler-Institute, Greifswald-Insel Riems, Germany, for supplying the cultured CCB cells. We thank S. Amir for editing the manuscript.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Barreca, C., and P. O'Hare. 2006. Characterization of a potent refractory state and persistence of herpes simplex virus 1 in cell culture. J. Virol. 80:9171-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bringhurst, R. M., and P. A. Schaffer. 2006. Cellular stress rather than stage of the cell cycle enhances the replication and plating efficiencies of herpes simplex virus type 1 ICP0− viruses. J. Virol. 80:4528-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dishon, A., A. Perelberg, J. Bishara-Shieban, M. Ilouze, M. Davidovich, S. Werker, and M. Kotler. 2005. Detection of carp interstitial nephritis and gill necrosis virus in fish droppings. Appl. Environ. Microbiol. 71:7285-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilad, O., S. Yun, M. A. Adkison, K. Way, N. H. Willits, H. Bercovier, and R. P. Hedrick. 2003. Molecular comparison of isolates of an emerging fish pathogen, koi herpesvirus, and the effect of water temperature on mortality of experimentally infected koi. J. Gen. Virol. 84:2661-2667. [DOI] [PubMed] [Google Scholar]
- 5.Gilad, O., S. Yun, F. J. Zagmutt-Vergara, C. M. Leutenegger, H. Bercovier, and R. P. Hedrick. 2004. Concentrations of a koi herpesvirus (KHV) in tissues of experimentally infected Cyprinus carpio koi as assessed by real-time TaqMan PCR. Dis. Aquat. Organ. 60:179-187. [DOI] [PubMed] [Google Scholar]
- 6.Haenen, O. L. M., K. Way, S. M. Bergmann, and E. Ariel. 2004. The emergence of koi herpesvirus and its significance to European aquaculture. Bull. Eur. Assoc. Fish Pathol. 24:293-307. [Google Scholar]
- 7.Harris, R. A., and C. M. Preston. 1991. Establishment of latency in vitro by the herpes simplex virus type 1 mutant in 1814. J. Gen. Virol. 72:907-913. [DOI] [PubMed] [Google Scholar]
- 8.Hasegawa, S., T. Somamoto, C. Nakayasu, T. Nakanishi, and N. Okamoto. 1997. A cell line (CFK) from fin of isogeneic ginbuna crusian carp. Fish Pathol. 32:127-128. [Google Scholar]
- 9.Hedrick, R. P., O. Gilad, S. Yun, J. Spangenberg, G. Marty, R. Nordhausen, M. Kebus, H. Bercovier, and A. Eldar. 2000. A herpesvirus associated with mass mortality of juvenile and adult koi, a strain of common carp. J. Aquat. Anim. Health 12:44-55. [DOI] [PubMed] [Google Scholar]
- 10.Hutoran, M., A. Ronen, A. Perelberg, M. Ilouze, A. Dishon, I. Bejerano, N. Chen, and M. Kotler. 2005. Description of an as yet unclassified DNA virus from diseased Cyprinus carpio species. J. Virol. 79:1983-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilouze, M., A. Dishon, T. Kahan, and M. Kotler. 2006. Cyprinid herpes virus-3 (CyHV-3) bears genes of genetically distant large DNA viruses. FEBS Lett. 580:4473-4478. [DOI] [PubMed] [Google Scholar]
- 12.Ilouze, M., A. Dishon, and M. Kotler. 2006. Characterization of a novel virus causing a lethal disease in carp and koi. Microbiol. Mol. Biol. Rev. 70:147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamieson, D. R., L. H. Robinson, J. I. Daksis, M. J. Nicholl, and C. M. Preston. 1995. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus type 1 Vmw65 mutants. J. Gen. Virol. 76:1417-1431. [DOI] [PubMed] [Google Scholar]
- 15.Le Morvan, C., P. Deschaux, and D. Troutaud. 1996. Effects and mechanisms of environmental temperature on carp (Cyprinus carpio) anti-DNP antibody response and non-specific cytotoxic cell activity: a kinetic study. Dev. Comp. Immunol. 20:331-340. [DOI] [PubMed] [Google Scholar]
- 16.Le Morvan, C., D. Troutaud, and P. Deschaux. 1998. Differential effects of temperature on specific and nonspecific immune defences in fish. J. Exp. Biol. 201:165-168. [DOI] [PubMed] [Google Scholar]
- 17.Neukirch, M., K. Bottcher, and S. Bunnajirakul. 1999. Isolation of a virus from koi with altered gills. Bull. Eur. Assoc. Fish Pathol. 19:221-224. [Google Scholar]
- 18.Perelberg, A., A. Ronen, M. Hutoran, Y. Smith, and M. Kotler. 2005. Protection of cultured Cyprinus carpio against a lethal viral disease by an attenuated virus vaccine. Vaccine 23:3396-3403. [DOI] [PubMed] [Google Scholar]
- 19.Perelberg, A., M. Smirnov, M. Hutoran, A. Diamant, I. Bejerano, and M. Kotler. 2003. Epidemiological description of a new viral disease afflicting cultured Cyprinus carpio in Israel. Isr. J. Aquacult. 55:5-12. [Google Scholar]
- 20.Petrillo-Peixoto, M. L., and S. M. Beverley. 1988. Amplified DNAs in laboratory stocks of Leishmania tarentolae: extrachromosomal circles structurally and functionally similar to the inverted-H-region amplification of methotrexate-resistant Leishmania major. Mol. Cell. Biol. 8:5188-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pikarsky, E., A. Ronen, J. Abramowitz, B. Levavi-Sivan, M. Hutoran, Y. Shapira, M. Steinitz, A. Perelberg, D. Soffer, and M. Kotler. 2004. Pathogenesis of acute viral disease induced in fish by carp interstitial nephritis and gill necrosis virus. J. Virol. 78:9544-9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poffenberger, K. L., and B. Roizman. 1985. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 53:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pokorova, D., T. Vesely, V. Piackova, S. Reschova, and J. Hulova. 2005. Current knowledge on koi herpesvirus (KHV): a review. Vet. Med. Czech. 50:139-147. [Google Scholar]
- 24.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 25.Ronen, A., A. Perelberg, J. Abramowitz, M. Hutoran, S. Tinman, I. Bejerano, M. Steinitz, and M. Kotler. 2003. Efficient vaccine against the virus causing a lethal disease in cultured Cyprinus carpio. Vaccine 21:4677-4684. [DOI] [PubMed] [Google Scholar]
- 26.Ronen, A., A. Perelberg, M. Hutoran, Y. Shapira, M. Steinitz, B. Levavi-Sivan, E. Pikarsky, and M. Kotler. 2005. Prevention of a mortal disease of carps induced by the carp interstitial nephritis and gill necrosis virus (CNGV) in Israel. Bull. Fish. Res. Agency 2005(Suppl. 2):9-11. [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.St.-Hilaire, S., N. Beevers, K. Way, R. M. Le Deuff, P. Martin, and C. Joiner. 2005. Reactivation of koi herpesvirus infections in common carp Cyprinus carpio. Dis. Aquat. Organ. 67:15-23. [DOI] [PubMed] [Google Scholar]
- 29.Strang, B. L., and N. D. Stow. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su, Y. H., M. J. Moxley, A. K. Ng, J. Lin, R. Jordan, N. W. Fraser, and T. M. Block. 2002. Stability and circularization of herpes simplex virus type 1 genomes in quiescently infected PC12 cultures. J. Gen. Virol. 83:2943-2950. [DOI] [PubMed] [Google Scholar]
- 31.Waltzek, T. B., G. O. Kelley, D. M. Stone, K. Way, L. Hanson, H. Fukuda, I. Hirono, T. Aoki, A. J. Davison, and R. P. Hedrick. 2005. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J. Gen. Virol. 86:1659-1667. [DOI] [PubMed] [Google Scholar]
- 32.Wigdahl, B., A. C. Scheck, R. J. Ziegler, E. De Clercq, and F. Rapp. 1984. Analysis of the herpes simplex virus genome during in vitro latency in human diploid fibroblasts and rat sensory neurons. J. Virol. 49:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, N., S. C. Watkins, P. A. Schaffer, and N. A. DeLuca. 1996. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J. Virol. 70:6358-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]