Abstract
The Gryllus bimaculatus nudivirus (GbNV) infects nymphs and adults of the cricket Gryllus bimaculatus (Orthoptera: Gryllidae). GbNV and other nudiviruses such as Heliothis zea nudivirus 1 (HzNV-1) and Oryctes rhinoceros nudivirus (OrNV) were previously called “nonoccluded baculoviruses” as they share some similar structural, genomic, and replication aspects with members of the family Baculoviridae. Their relationships to each other and to baculoviruses are elucidated by the sequence of the complete genome of GbNV, which is 96,944 bp, has an AT content of 72%, and potentially contains 98 predicted protein-coding open reading frames (ORFs). Forty-one ORFs of GbNV share sequence similarities with ORFs found in OrNV, HzNV-1, baculoviruses, and bacteria. Most notably, 15 GbNV ORFs are homologous to the baculovirus core genes, which are associated with transcription (lef-8, lef-9, lef-4, vlf-1, and lef-5), replication (dnapol), structural proteins (p74, pif-1, pif-2, pif-3, vp91, and odv-e56), and proteins of unknown function (38K, ac81, and 19kda). Homologues to these baculovirus core genes have been predicted in HzNV-1 as well. Six GbNV ORFs are homologous to nonconserved baculovirus genes dnaligase, helicase 2, rr1, rr2, iap-3, and desmoplakin. However, the remaining 57 ORFs revealed no homology or poor similarities to the current gene databases. No homologous repeat (hr) sequences but fourteen short direct repeat (dr) regions were detected in the GbNV genome. Gene content and sequence similarity suggest that the nudiviruses GbNV, HzNV-1, and OrNV form a monophyletic group of nonoccluded double-stranded DNA viruses, which separated from the baculovirus lineage before this radiated into dipteran-, hymenopteran-, and lepidopteran-specific clades of occluded nucleopolyhedroviruses and granuloviruses. The accumulated information on the GbNV genome suggests that nudiviruses form a highly diverse and phylogenetically ancient sister group of the baculoviruses, which have evolved in a variety of highly divergent host orders.
The family Baculoviridae comprises viruses with large, rod-shaped, enveloped virions and covalently closed, circular, double-stranded DNA (dsDNA) genomes. They are pathogenic for insect hosts, particularly of the Lepidoptera, Hymenoptera, and Diptera, and replicate in the nuclei of infected host cells. Currently, this virus family has two genera, Nucleopolyhedrovirus (NPV) and Granulovirus (GV) (48), but their reclassification is proposed (24). The NPVs have large occlusion bodies (OBs) containing numerous virions, whereas the GVs are characterized by smaller, often ovoid OBs normally containing a single virion.
A variety of nudiviruses, previously called “nonoccluded baculoviruses,” have been reported from a wide range of host species belonging to the Coleoptera, Lepidoptera, Orthoptera, Diptera, Siphonaptera, Hymenoptera, Thysanura, Trichoptera, Neuroptera, Homoptera, Acarina, Araneina, and Crustacea (22). Similarly to the baculoviruses, these viruses also have rod-shaped virions and circular dsDNA genomes and replicate in the nuclei of the host cells. But they do not form OBs. Most of these viruses were classified based solely on morphological and very limited biological data. Whether they form a coherent group of evolutionarily related viruses or whether they are a polyphyletic assemblage of viruses with a few similar features is not yet understood.
Recently, it was shown that Heliothis zea virus 1 and Oryctes rhinoceros virus are monophyletic and should belong to the same genus, Nudivirus (52). Accordingly, renaming them to be Heliothis zea nudivirus 1 (HzNV-1) and Oryctes rhinoceros nudivirus (OrNV) was suggested and is referred to in this paper. The best-studied nudivirus is HzNV-1, with a genome 228,089 bp in size (8). HzNV-1 shares a number of homologous genes with baculoviruses, but whether these homologues derived from a common ancestor or from horizontal gene transfer remained unresolved. The partial genomic sequence analysis of OrNV, as well as a reevaluation of baculovirus-homologous genes in HzNV-1, suggested that HzNV-1 and OrNV are related to each other and might be considered as a sister group of the baculoviruses (52).
In order to gain a better understanding of the possible origin, evolution, and divergence of baculoviruses and the evolutionarily related nudiviruses, genomic comparisons and phylogenetic analyses of putatively ancient members of these viruses are supposed to form a straightforward and powerful approach (53). Therefore, we have completely sequenced the genome of a putative nudivirus infecting Gryllus bimaculatus. We refer to this virus as Gryllus bimaculatus nudivirus (GbNV). GbNV was identified in diseased nymphs and adults of the field cricket, Gryllus bimaculatus (Orthoptera: Gryllidae) (21). It also infects other cricket species, such as G. campestris, Teleogryllus oceanicus, and T. commodus (21). GbNV replicates in the nuclei of the fat body cells and shows rod-shaped and enveloped virions (21). Its cricket hosts belong to the hemimetabolous Orthoptera, which evolutionarily diverged much earlier than other known insect host orders of baculoviruses and nudiviruses. Hence, the genomic sequence of GbNV should in principle provide valuable insight into the origin and early evolution of baculoviruses and baculovirus-related nudiviruses. Here, we report the complete genomic sequence of GbNV and compare it to the genomes of baculoviruses HzNV-1 and OrNV.
MATERIALS AND METHODS
Viruses, DNA purification, cloning, and sequencing.
Viruses were isolated from laboratory-maintained G. bimaculatus, which were infected with GbNV as described by Huger (21). Briefly, the diseased crickets were homogenized, and the debris was removed by low-speed centrifugation. Purified virions were obtained after 30 to 60% (wt/vol) sucrose-gradient ultra-centrifugation. Genomic DNA was extracted from the viruses by using the standard phenol-chloroform method. Five hundred to 600 ng of the viral DNA was incubated at 37°C for 2 h with 10 U of BamHI, EcoRI, or HindIII. Subsequently, the digested DNAs were loaded onto a 0.7% agarose gel, which was electrophoresed at 80 V for 3 h. For cloning, viral DNA was sheared to fragments of 1.5 to 2.5 kb and ligated into SmaI-cut, dephosphorylated pUC19 vectors (GENterprise, Mainz, Germany). Sequencing was performed using an ABI 3730 automated 48 capillary sequencer (Applied Biosystems) with the ABI prism BigDye Terminator cycle sequencing ready reaction kit version 3.1 by GENterprise (Mainz, Germany) (http://www.genterprise.de/). The genomic DNA of GbNV was sequenced to 9.2-fold coverage. To close sequencing gaps, primer walking was performed using GbNV sequence-specific oligonucleotides.
DNA sequence analysis.
Sequences were assembled by using SeqMan (Lasergene 5.0 software; DNAStar, Inc.). The trace files were checked by eye, and minor mistakes were corrected as necessary. The simulated restriction digestion of the sequence was done with the GeneQuest program integrated in Lasergene software. Methionine-initiated open reading frames (ORFs) encoding 50 amino acids (aa) or more and showing minimum overlap (<60 bp) were found using the ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and GeneQuest program. ORFs with less than 50 aa were only considered as putative genes in cases of clear homology to ORFs in other dsDNA viruses. Sequence comparisons of all predicted ORFs to public databases were done by using the BLASTP, TBLASTN, and PSI-BLAST programs (1, 46). A local database containing all up-to-date completed genomic sequences of baculoviruses as well as HzNV-1 and OrNV was also included in BLAST searches. The following criteria were considered for assigning a putative homologue to the GbNV ORFs: (i) a BLASTP search showed a dsDNA virus match with an E value of 0.1 or less, (ii) amino acid identity to a dsDNA virus homologue was 20% or greater based on MegAlign ClustalW analysis of entire ORFs, or (iii) a conserved domain was found. The putative coding regions were numbered as GbNV ORFs. All ORFs were investigated for characteristic sequence signatures using the InterProScan (http://www.ebi.ac.uk/InterProScan/) and PROSITE (http://au.expasy.org/prosite/) programs. Repeated and palindromic sequences were identified using the REPuter program (http://bibiserv.techfak.uni-bielefeld.de/reputer/submission.html), Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.submit.options.html), and the GeneQuest program (Lasergene 5.0).
Phylogenetic analysis.
Neighbor-joining (NJ), minimum evolution (ME), and maximum parsimony (MP) phylogenetic analyses were performed using MEGA 3.1 (30).
Nucleotide sequence accession number.
The GbNV genomic sequence has been deposited in GenBank under accession no. EF203088.
RESULTS AND DISCUSSION
Sequence analysis.
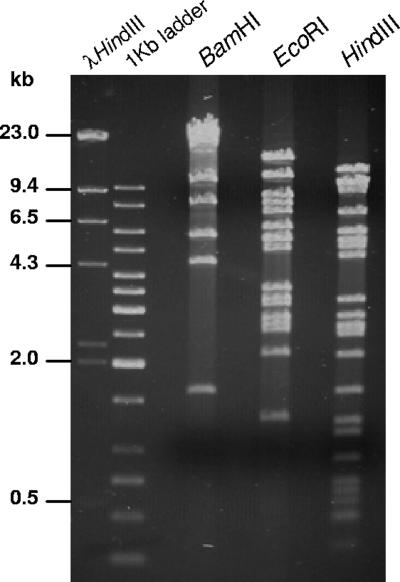
The GbNV genome is 96,944 bp in length, which is in good agreement with the estimation of 95 kbp based on DNA restriction endonuclease profiles of the GbNV genomic DNA determined in an agarose gel (Fig. 1). These were identical to those predicted from the genomic sequence (data not shown), indicating that the cloning, sequencing, and sequence assembling were accurate. The AT content of the GbNV genome is 72%, which is one of the highest of any sequenced dsDNA virus isolated from insects. For example, the AT contents of currently sequenced baculoviruses range from 42 to 67%, while the predicted AT contents of some GVs appear to be up to 76% (25); OrNV and HzNV-1 have AT contents of 57 and 58%, respectively (8, 41, 52).
FIG. 1.
Restriction endonuclease profiles of GbNV DNA for the restriction endonucleases as shown above each lane. Molecular sizes are shown on the left.
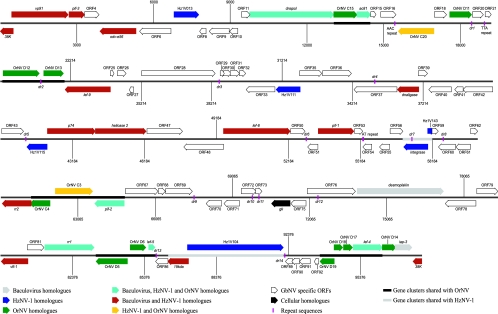
In agreement with the genome orientation of HzNV-1 (8), the first GbNV ORF was defined as the first ORF located in the largest XhoI fragment of the GbNV genome. The first nucleotide of this XhoI segment was considered to be number 1, and the successive nucleotides were numbered in the clockwise direction of the GbNV ORF1 (Fig. 2). A total of 98 methionine-initiated ORFs with 50 or more amino acids and minimal overlap with adjacent ORFs were predicted, of which 57 were in a clockwise orientation and 41 were in a counterclockwise orientation (Fig. 2). As is typical for most large dsDNA viruses, the genome of GbNV is densely packed with ORFs, which account for 93.6% of the total genome and represent a gene density of 0.93 kbp per ORF.
FIG. 2.
Linear map of the GbNV genome. ORFs and their transcriptional directions are indicated as arrows.
Gene content analysis.
Forty of the predicted 98 GbNV ORFs were considered to be homologous (amino acid identity of encoded proteins of 20 to 46%) to the known genes from other large dsDNA viruses, particularly those from OrNV, HzNV-1, and the baculoviruses; one ORF (ORF74) is homologous to a cellular gene (Table 1; Fig. 2). However, the remaining 57 ORFs revealed no homology or poor similarities to any gene in the current gene databases. GbNV shares 29 homologues with the complete genome of HzNV-1 and 19 homologues with the partial genome sequence of OrNV (Table 1; Fig. 2). Since only one-third of the OrNV genome is determined (52), it can be expected that the number of genes shared with OrNV will substantially increase when its complete genome sequence is available. Most notably, 15 ORFs are homologous to the baculovirus core genes (Table 2). So far, 29 so-called baculovirus core genes have been identified in all sequenced baculovirus genomes (18, 24). These genes play crucial roles in infection by baculoviruses, and their products have been classified into five functional categories: mRNA transcription, DNA replication, structural proteins, auxiliary proteins, and proteins of unknown function. The genes have also been used to infer the phylogenetic history of baculoviruses and for identification and classification purposes (17, 33).
TABLE 1.
ORFs and their proteins predicted in GbNV
| ORF | Stranda | Position
|
Length
|
Intergenic distance (bp)b | % A+T | Best BLAST match
|
Signaturec | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | Nucleotides | aa | ORF, protein encoded, or mass | Species | BLASTP score | aa identity (% match identity) | E value | |||||
| 1 | − | 505 | 96583 | 867 | 288 | 53 | 71.3 | 38K (AhNPV ORF74) | Adoxophyes honmai nucleopolyhedrovirus | 48.1 | 34/79 (43) | 8e-04 | SP |
| 2 | + | 450 | 2669 | 2,220 | 739 | −56 | 66.3 | VP91 capsid protein | HzNV-1 | 91.7 | 81/357 (22) | 2e-16 | Chitin binding peritrophin-A domain; SP; TM |
| 3 | + | 2692 | 3294 | 603 | 200 | 22 | 66.8 | Per os infectivity factor 3 (Hz1V088) | HzNV-1 | 75.9 | 57/198 (28) | 2e-12 | SP; TM Bipartite nuclear targeting sequence |
| 4 | + | 3303 | 3869 | 567 | 188 | 8 | 75.5 | ||||||
| 5 | − | 5310 | 3907 | 1,404 | 467 | 37 | 64.0 | ODV envelope-56 protein (Hz1V076) | HzNV-1 | 80.5 | 93/397 (23) | 3e-13 | TM |
| 6 | − | 6685 | 5456 | 1,230 | 409 | 145 | 70.4 | Leucine zipper pattern | |||||
| 7 | + | 6807 | 7769 | 963 | 320 | 121 | 72.7 | Hz1V013 | HzNV-1 | 42.4 | 20/51 (39) | 5e-02 | FAD-dependent thiol oxidase; TM |
| 8 | − | 8052 | 7843 | 210 | 69 | 73 | 78.6 | ||||||
| 9 | − | 8955 | 8179 | 777 | 258 | 126 | 71.2 | ||||||
| 10 | − | 9299 | 8982 | 318 | 105 | 26 | 72.3 | Microbody C-terminal targeting signal; TM | |||||
| 11 | + | 9438 | 9773 | 336 | 111 | 138 | 68.2 | ||||||
| 12 | + | 9760 | 13056 | 3,297 | 1,098 | −14 | 73.1 | DNA polymerase B | OrNV | 311.0 | 215/671 (32) | 1e-85 | DNA_pol_B; DNA_pol_B_exo |
| 13 | + | 13061 | 13960 | 900 | 299 | 4 | 69.3 | ORFC15 | OrNV | 105.0 | 67/228 (29) | 4e-24 | |
| 14 | + | 13963 | 14502 | 540 | 179 | 2 | 69.3 | Ac81 | OrNV | 135.0 | 63/144 (43) | 1e-33 | Leucine zipper pattern; TM |
| 15 | + | 14499 | 14714 | 216 | 71 | −4 | 73.2 | ||||||
| 16 | + | 14882 | 15493 | 612 | 203 | 167 | 66.2 | Cell attachment sequence; threonine-rich region; SP | |||||
| 17 | − | 16979 | 15588 | 1,392 | 463 | 94 | 70.8 | ORFC20 | OrNV | 127.0 | 89/296 (30) | 1e-30 | ATP/GTP-binding site motif A (P-loop) |
| 18 | + | 16997 | 17473 | 477 | 158 | 17 | 66.7 | ||||||
| 19 | + | 17590 | 18447 | 858 | 285 | 116 | 71.5 | ORFD11 | OrNV | 161.0 | 98/255 (38) | 4e-41 | Alpha/beta-hydrolases; lipolytic enzymes; SP; TM |
| 20 | + | 18491 | 18910 | 420 | 139 | 43 | 70.5 | ||||||
| 21 | + | 18990 | 19214 | 225 | 74 | 79 | 56.4 | ||||||
| 22 | + | 19356 | 20879 | 1,524 | 507 | 141 | 69.9 | ORFD12 | OrNV | 79.3 | 104/443 (23) | 4e-16 | Bipartite nuclear targeting sequence |
| 23 | + | 21075 | 21902 | 828 | 275 | 195 | 71.0 | ORFD13 | OrNV | 118.0 | 85/294 (28) | 3e-28 | |
| 24 | − | 23902 | 21986 | 1,917 | 638 | 83 | 73.2 | LEF-9 | HzNV-1 | 75.9 | 100/386 (25) | 1e-11 | SP |
| 25 | + | 23897 | 24067 | 171 | 56 | −6 | 61.4 | ||||||
| 26 | + | 24208 | 24579 | 372 | 123 | 140 | 62.1 | ||||||
| 27 | − | 24905 | 24708 | 198 | 65 | 128 | 63.6 | ||||||
| 28 | + | 25248 | 28379 | 3,132 | 1,043 | 342 | 69.7 | Immunoglobulin/major histocompatibility complex; serine-rich region | |||||
| 29 | + | 28585 | 28758 | 174 | 57 | 205 | 72.4 | SP; TM | |||||
| 30 | + | 28736 | 29047 | 312 | 103 | −23 | 73.4 | ||||||
| 31 | + | 29010 | 29405 | 396 | 131 | −38 | 69.4 | Bipartite nuclear targeting sequence; histidine-rich region | |||||
| 32 | + | 29407 | 29670 | 264 | 87 | 1 | 73.5 | SP; TM | |||||
| 33 | − | 30922 | 29696 | 1,227 | 408 | 25 | 74.4 | ||||||
| 34 | − | 31976 | 30975 | 1002 | 333 | 52 | 72.0 | Hz1V111 | HzNV-1 | 55.1 | 47/197 (23) | 8e-06 | ATP/GTP-binding site motif A (P-loop) |
| 35 | + | 31975 | 32913 | 939 | 312 | −2 | 67.8 | Bipartite nuclear targeting sequence | |||||
| 36 | + | 32939 | 34171 | 1,233 | 410 | 25 | 74.7 | ||||||
| 37 | − | 36116 | 34281 | 1,836 | 611 | 109 | 74.7 | ||||||
| 38 | − | 37035 | 36136 | 900 | 299 | 19 | 71.3 | DNA ligase (PxORF101) | Plutella xylostella granulovirus | 48.1 | 39/175 (22) | 8e-04 | ATP-dependent DNA ligase, central domain |
| 39 | + | 37025 | 37414 | 390 | 129 | −11 | 73.1 | ||||||
| 40 | − | 38418 | 37471 | 948 | 315 | 56 | 73.5 | ||||||
| 41 | − | 38922 | 38575 | 348 | 115 | 156 | 72.4 | ||||||
| 42 | − | 40184 | 38919 | 1,266 | 421 | −4 | 73.8 | ||||||
| 43 | + | 40246 | 41148 | 903 | 300 | 61 | 77.1 | ||||||
| 44 | − | 42127 | 41297 | 831 | 276 | 148 | 72.0 | Hz1V115 | HzNV-1 | 65.1 | 53/209 (25) | 6e-09 | |
| 45 | + | 42154 | 44145 | 1,992 | 663 | 26 | 64.6 | P74 protein | HzNV-1 | 237.0 | 184/673 (27) | 3e-60 | Baculoviridae p74; bipartite nuclear targeting sequence; leucine zipper pattern; TM |
| 46 | + | 44153 | 46258 | 2,106 | 701 | 7 | 74.4 | Putative helicase 2 (Hz1V060) | HzNV-1 | 72.8 | 111/494 (22) | 1e-10 | ATP/GTP-binding site motif A (P-loop) |
| 47 | + | 46290 | 47771 | 1,482 | 493 | 31 | 71.9 | Leucine-rich repeat; asparagine-rich region profile; SP; TM | |||||
| 48 | − | 49489 | 47822 | 1,668 | 555 | 50 | 72.1 | WD40-like domain | |||||
| 49 | + | 49483 | 52254 | 2,772 | 923 | −7 | 69.4 | LEF-8 | HzNV-1 | 163.0 | 220/997 (22) | 6e-38 | RpoB; TM |
| 50 | + | 52287 | 52886 | 600 | 199 | 32 | 71.7 | ||||||
| 51 | − | 53411 | 52989 | 423 | 140 | 102 | 64.3 | TM | |||||
| 52 | + | 53435 | 54913 | 1,497 | 492 | 23 | 64.9 | Per os infectivity factor 1 (Hz1V055) | HzNV-1 | 122.0 | 73/218 (33) | 9e-26 | SP; TM |
| 53 | + | 54950 | 55294 | 345 | 114 | 36 | 80.0 | Microbody C-terminal targeting signal | |||||
| 54 | − | 55651 | 55289 | 363 | 120 | −6 | 81.8 | Isoleucine-rich region profile; tyrosine-rich region profile; TM | |||||
| 55 | − | 56409 | 55984 | 426 | 141 | 332 | 68.8 | TM | |||||
| 56 | + | 56417 | 56956 | 540 | 179 | 7 | 77.6 | ||||||
| 57 | − | 57991 | 56990 | 1,002 | 333 | 33 | 77.2 | Integrase (Hz1V144) | HzNV-1 | 84.0 | 72/291 (24) | 2e-14 | INT_REC_C (DNA breaking-rejoining enzymes, integrase/recombinases, C-terminal catalytic domain); phage integrase |
| 58 | + | 57993 | 58187 | 195 | 64 | 1 | 75.4 | Hz1V143 | HzNV-1 | 28.8 | 13/49 (26) | 2.7 | SP; TM |
| 59 | + | 58162 | 58371 | 210 | 69 | −26 | 72.9 | TM | |||||
| 60 | − | 59146 | 58442 | 705 | 234 | 70 | 73.9 | Bipartite nuclear targeting sequence; serine-rich region profile | |||||
| 61 | − | 59776 | 59168 | 609 | 202 | 21 | 73.9 | Microbody C-terminal targeting signal | |||||
| 62 | + | 59775 | 60065 | 291 | 96 | −2 | 79.0 | ||||||
| 63 | − | 61244 | 60129 | 1,116 | 371 | 63 | 73.8 | RR2 | Equine herpesvirus 4 | 63.2 | 60/284 (21) | 3e-08 | RR |
| 64 | − | 61980 | 61216 | 765 | 254 | −29 | 64.3 | ORFC4 | OrNV | 79.0 | 61/236 (25) | 2e-16 | |
| 65 | + | 62197 | 63636 | 1,440 | 479 | 216 | 73.4 | ORFC3 | OrNV | 155.0 | 111/372 (29) | 6e-39 | |
| 66 | − | 64867 | 63731 | 1,137 | 378 | 94 | 70.0 | Per os infectivity factor 2 | OrNV | 323.0 | 164/355 (46) | 1e-89 | SP; TM |
| 67 | + | 64908 | 66170 | 1,263 | 420 | 40 | 71.4 | ||||||
| 68 | + | 66197 | 66460 | 264 | 87 | 26 | 74.2 | ||||||
| 69 | + | 66474 | 67514 | 1,041 | 346 | 13 | 71.9 | ||||||
| 70 | − | 68644 | 68042 | 603 | 200 | 527 | 71.8 | TM | |||||
| 71 | − | 69348 | 68737 | 612 | 203 | 92 | 79.4 | Bipartite nuclear targeting sequence; TM | |||||
| 72 | + | 69422 | 69991 | 570 | 189 | 73 | 76.7 | ||||||
| 73 | + | 69966 | 70226 | 261 | 86 | −26 | 53.3 | Serine-rich region profile; arginine-rich region profile | |||||
| 74 | − | 71298 | 70591 | 708 | 235 | 364 | 69.8 | Putative guanylate kinase | Campylobacter jejuni subsp. jejuni | 36.6 | 33/143 (23) | 0.94 | NK (nucleoside/nucleotide kinase); ATP/GTP-binding site motif A (P-loop) |
| 75 | − | 71931 | 71338 | 594 | 197 | 39 | 69.9 | ||||||
| 76 | + | 71984 | 73846 | 1,863 | 620 | 52 | 72.8 | Bipartite nuclear targeting sequence | |||||
| 77 | + | 73928 | 77275 | 3,348 | 1,115 | 81 | 73.1 | Desmoplakin | Hyphantria cunea nucleopolyhedrovirus | 38.1 | 64/250 (25) | 0.14 | (Smc) chromosome segregation ATPases; endoplasmic reticulum targeting sequence; tRNA-binding arm; leucine zipper pattern; bipartite nuclear targeting sequence |
| 78 | − | 78534 | 77341 | 1,194 | 397 | 65 | 71.7 | ||||||
| 79 | + | 78558 | 79376 | 819 | 272 | 23 | 74.2 | ||||||
| 80 | − | 80517 | 79414 | 1,104 | 367 | 37 | 70.0 | Very late factor 1 | HzNV-1 | 32.7 | 64/286 (22) | 1.8 | INT_REC_C |
| 81 | + | 80516 | 81208 | 693 | 230 | −2 | 75.5 | ||||||
| 82 | + | 81233 | 83284 | 2,052 | 683 | 24 | 71.0 | RR1 | OrNV | 286.0 | 194/614 (31) | 2e-78 | RNR_1; RR1 signature |
| 83 | − | 84675 | 83380 | 1,296 | 431 | 95 | 73.8 | ORFD5 | OrNV | 57.0 | 58/296 (19) | 2e-09 | Bipartite nuclear targeting sequence |
| 84 | + | 84797 | 85438 | 642 | 213 | 121 | 76.3 | ORFD6 | OrNV | 67.0 | 47/174 (27) | 6e-13 | |
| 85 | + | 85561 | 85758 | 198 | 65 | 122 | 71.2 | LEF-5 | OrNV | 37.7 | 15/46 (32) | 1e-04 | Zinc beta-ribbon; cell attachment sequence |
| 86 | − | 86359 | 85850 | 510 | 169 | 91 | 76.9 | ||||||
| 87 | − | 87219 | 86356 | 864 | 287 | −4 | 65.9 | 19 kDa | HzNV-1 | 73.6 | 47/168 (27) | 2e-11 | TM |
| 88 | + | 87173 | 91162 | 3,990 | 1,329 | −47 | 74.0 | Hz1V104 | HzNV-1 | 32.7 | 29/142 (20) | 8.1 | ATP/GTP-binding site motif A (P-loop) |
| 89 | − | 91552 | 91250 | 303 | 100 | 87 | 72.3 | ||||||
| 90 | − | 91842 | 91528 | 315 | 104 | −25 | 74.6 | SP; TM | |||||
| 91 | − | 92296 | 91847 | 450 | 149 | 4 | 67.3 | ||||||
| 92 | − | 92640 | 92464 | 177 | 58 | 167 | 79.1 | SP; TM | |||||
| 93 | − | 93284 | 92697 | 588 | 195 | 56 | 70.8 | ORFD19 | OrNV | 73.2 | 53/187 (28) | 9e-15 | |
| 94 | + | 93286 | 93645 | 360 | 119 | 1 | 78.1 | ORFD18 | OrNV | 35.0 | 27/113 (23) | 1e-03 | |
| 95 | + | 93672 | 94052 | 381 | 126 | 26 | 60.6 | ORFD17 | OrNV | 43.1 | 26/112 (23) | 5e-06 | SP; TM |
| 96 | + | 94085 | 95296 | 1,212 | 403 | 32 | 75.3 | LEF-4 | OrNV | 207.0 | 128/393 (32) | 8e-55 | DNA clamp |
| 97 | + | 95299 | 95844 | 546 | 181 | 2 | 75.6 | ORFD14 | OrNV | 79.0 | 54/168 (32) | 1e-16 | |
| 98 | + | 95834 | 96529 | 696 | 231 | −11 | 70.6 | IAP 3 | Cryptophlebia leucotreta granulovirus | 85.5 | 68/264 (25) | 3e-15 | Baculoviral inhibition of apoptosis protein repeat domain; zinc finger RING-type profile; cysteine-rich region profile; TM |
+, clockwise direction of coding; −, counterclockwise direction of coding.
−, overlap between adjacent ORFs.
SP, signal peptides; TM, transmembrane domains; FAD, flovin adenine dinucleotide.
TABLE 2.
GbNV homologues present in baculoviruses HzNV-1 and OrNVa
| Baculovirus genes | Homologous ORF
|
||
|---|---|---|---|
| GbNV | HzNV-1 | OrNVb | |
| 38K | 1 | Hz1V010 | ? |
| vp91 | 2 | Hz1V046 | ? |
| pif-3 | 3 | Hz1V088 | ? |
| odv-e56 | 5 | Hz1V076 | ? |
| dnapol | 12 | Hz1V131 | C17 |
| ac81 | 14 | Hz1V033 | C14 |
| lef-9 | 24 | Hz1V075 | ? |
| p74 | 45 | Hz1V011 | ? |
| lef-8 | 49 | Hz1V090 | ? |
| pif-1 | 52 | Hz1V055 | ? |
| pif-2 | 66 | Hz1V123 | C2 |
| vlf-1 | 80 | Hz1V121 | ? |
| lef-5 | 85 | Hz1V101 | D7 |
| 19kda | 87 | Hz1V103 | ? |
| lef-4 | 96 | Hz1V098 | D16 |
| dnaligase | 38 | Hz1V036 | ? |
| helicase 2 | 46 | Hz1V060 | ? |
| rr2 | 63 | Hz1V073 | ? |
| desmoplakin | 77 | ? | |
| rr1 | 82 | Hz1V095 | D8 |
| iap-3 | 98 | Hz1V135 | ? |
| Hz1V138 | |||
| 7 | Hz1V013 | ? | |
| 13 | C15 | ||
| 17 | Hz1V051 | C20 | |
| 19 | D11 | ||
| 22 | D12 | ||
| 23 | D13 | ||
| 34 | Hz1V111 | ? | |
| 44 | Hz1V115 | ? | |
| 57 (integrase) | Hz1V144 | ? | |
| 58 | Hz1V143 | ? | |
| 64 | C4 | ||
| 65 | Hz1V068 | C3 | |
| 83 | D5 | ||
| 84 | D6 | ||
| 88 | Hz1V104 | ? | |
| 93 | D19 | ||
| 94 | D18 | ||
| 95 | Hz1V124 | D17 | |
| 97 | D14 | ||
The fifteen baculovirus core genes and their homologues are in bold.
?, in the portion of the OrNV genome which is not yet determined.
There are six baculovirus core genes (p47, lef-8, lef-9, lef-4, vlf-1, and lef-5) required for the transcription and expression of late and very late viral genes. These transcription-specific lef genes are essential for regulating virus propagation during infection. Strikingly, five of them, ORF49 (lef-8), ORF24 (lef-9), ORF96 (lef-4), ORF80 (vlf-1), and ORF85 (lef-5), are also present in the GbNV genome; a p47 homologue was the only one which could not be detected. LEF-8, LEF-9, LEF-4, and P47 are the subunits of the baculovirus DNA-dependent RNA polymerase. LEF-8 contains a conserved C-terminal motif of DNA-directed RNA polymerase which is critical for late gene promoter activation (49). LEF-4 has RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities and plays the role of capping mRNA (28). LEF-9 contains a motif which is part of the catalytic center of DNA-dependent RNA polymerase, but its function in LEF-9 is not clear (34). LEF-5 functions as a transcription initiation factor (13). The putative LEF-5 of GbNV comprises 65 aa and is similar in size to its homologue in OrNV (78 aa) (52). It seems to be N-terminally truncated, since it is much shorter than the LEF-5 proteins of baculoviruses (230 to 315 aa) and HzNV-1 (241 aa). However, a C-terminal zinc ribbon-encoding domain required for the maximal late transcription activity (13, 15) is highly conserved (Fig. 3), suggesting that GbNV and OrNV encode an active LEF-5. VLF-1 is required for normal capsid assembly and possesses an essential function in the viral DNA packaging process (50). GbNV and HzNV-1 lack a p47 homologue. It might not exist in their genomes, or it was not detectable due to low sequence conservation.
FIG. 3.
Alignments of the putative LEF-5 zinc ribbon domains from AcMNPV, HzNV-1, OrNV, and GbNV. Identical and biochemically similar amino acid residues are shaded in black and gray, respectively. Potential Zn-coordinating amino acids are marked with asterisks.
Six out of the 12 baculovirus core genes encoding structural proteins were identified in GbNV. These include all of the homologues, ORF45 (p74), ORF52 (pif-1), ORF66 (pif-2), and ORF3 (pif-3), required for per os infection of insect midgut cells by the occlusion-derived virions (ODV) of baculoviruses. In addition, ORF5 and ORF2 are homologues of baculovirus ODV envelope protein-encoding gene odv-e56 (5) and of vp91, encoding a baculovirus protein of 91 kDa present in both the capsid and the envelope surrounding the capsid of ODVs (45). Baculovirus P74 and PIF-1 are present only in the ODV envelope (9, 14, 31). Even though PIF-2 and PIF-3 have not yet been localized, it is conceivable that these functionally conserved proteins are also present in the ODV envelope. Baculovirus ODVs enter the midgut epithelial cells by attaching and fusing their viral envelopes with cell membranes (10, 20). These highly conserved envelope proteins involved in oral infectivity suggest a similar entry mechanism shared by GbNV, HzNV-1, and baculovirus ODVs. Thus, it is most likely that the basic infection mechanism of theses viruses is evolutionarily conserved. In contrast, homologues of the envelope fusion protein GP64 or F-protein in baculovirus budded viruses (BVs) were not found either in GbNV or in HzNV-1.
Six GbNV ORFs are homologous to nonconserved baculovirus genes. They are ORF38 (dnaligase), ORF46 (helicase 2), ORF82 (rr1), ORF63 (rr2), ORF98 (iap-3), and ORF77 (desmoplakin) (Table 2). Their homologues are also present in HzNV-1, except for desmoplakin (Table 2). DNA ligase belongs to the DNA ligase III family and catalyzes the formation of a phosphodiester bond at the site of a single-strand break in duplex DNA (54). In baculoviruses, it is conserved only in GVs. Presently, two types of DNA helicase genes, helicase and helicase 2, have been found in baculoviruses. helicase is a conserved core gene in the baculoviruses, is essential for the initiation of viral DNA replication, and may contribute to other functions. It reveals low similarity to helicase genes of other viruses and organisms. It is part of the helicase superfamily I, which takes part in multiple aspects of cellular and viral DNA and RNA metabolism, e.g., replication, recombination, repair, transcription, or RNA processing. A homologue of the baculovirus core helicase gene was not detected in the GbNV genome. helicase 2 accounts for circa 40% of helicase in size and is so far unique to all sequenced GVs, two lepidopteran-specific NPVs, HzNV-1, and GbNV (ORF46). Baculovirus helicase 2 is not a functional analogue of the core helicase and is likely involved in DNA recombination or repair (43). However, given that some helicases function as complexes with other proteins and in multiple processes in vivo and that GbNV helicase 2 is distantly homologous to those of baculoviruses, it might have a role in viral DNA replication. Ribonucleotide reductase (RR) functions in nucleotide metabolism by reducing ribonucleotides to deoxyribonucleotides for DNA synthesis and DNA repair (32). It normally consists of two subunits encoded by two different genes, rr1 (RR large subunit) and rr2 (RR small subunit) (37). RR1 and/or RR2 is widely dispersed among many baculoviruses and other DNA viruses, e.g., in herpesviruses, asfarviruses, poxviruses, iridoviruses, phycodnaviruses, and nimavirus (36). Even though dnaligase, helicase 2, rr1, and rr2, involving viral DNA metabolism, are present both in many baculoviruses and in nudiviruses (HzNV-1 and GbNV), phylogenetic analyses indicated that they are not conserved ancient genes (data not shown). ORF98 encodes an IAP homologue. IAPs are known as inhibitors of apoptosis, preventing programmed cell death by blocking activation of the host caspase, which is required for apoptotic death (35). They are frequently found as single gene or multigene families in baculoviruses, herpesviruses, iridoviruses, ascoviruses, and asfarviruses. Interestingly, another multigene family, the baculovirus repeat ORF genes, representing a class of viral DNA-binding proteins and distributed among many baculoviruses, poxviruses, iridoviruses, ascoviruses, and bacteriophages, were not identified in GbNV (3, 23, 52). The predicted product of ORF77 showed similarity to an internal region of a human desmoplakin, an essential constituent of intracellular junctions, which can also be found in several DNA viruses (11, 16).
A C-terminal catalytic domain of a phage integrase was found to be encoded by putative ORF57. Integrases are a large group of site-specific DNA recombinases, which function in DNA rearrangement and are identified in several organisms and viruses, where they play a role in the integration and excision of viral genomes and decatenation of newly replicated DNA (39). Thus, the ORF57-encoded protein may be involved in the processing or packaging of virus genomic DNA. ORF7 contains a flavin adenine dinucleotide-dependent thiol oxidase signature and might encode a putative thiol oxidoreductase protein. Thiol oxidoreductase (sulfiredoxin) is essential for the antioxidant function of peroxiredoxins by reducing peroxiredoxin-(S-hydroxy-S-oxocysteine) to the active form of peroxiredoxin-(S-hydroxycysteine) and is likely to be involved in the repair of proteins containing cysteine-sulfinic acid modifications and in signaling pathways involving protein oxidation (4, 7). The ORF74-encoded protein showed the highest amino acid sequence similarity (14.9%) to putative guanylate kinase (GK) in the bacterial enteric pathogen Campylobacter jejuni subsp. jejuni (40). It contains a nucleoside kinase domain and an ATP/GTP-binding site motif, suggesting that ORF74 encodes a GK. GK is an essential enzyme in the nucleotide biosynthetic pathway, catalyzing the reversible transfer of the terminal phosphoryl group of ATP to (d)GMP (38).
Gene order.
The arrangement of orthologous genes in the GbNV genome was compared to those of the OrNV, HzNV-1, and baculovirus genomes. There were no regions of organizational similarity between baculoviruses and the GbNV, OrNV, and HzNV-1 genomes, respectively. However, two gene clusters, each containing two ORFs homologous to ORFs in GbNV, have a colinear arrangement in the genome of HzNV-1 (Fig. 2). Compared to the partial genome of OrNV, there are five regions of colinearly arranged ORFs (Fig. 2). These regions will likely increase when the entire genome of OrNV is sequenced. The observed patterns of conserved gene arrangements also foster the conclusion that GbNV and OrNV are more closely related to each other than either is to HzNV-1, as was suggested by gene content analyses.
Repeat regions.
Homologous repeat regions (hr's) are characterized by direct repeats (dr's) containing an imperfect palindromic core and located at different positions in the genome and are a common feature of large, circular, invertebrate dsDNA viruses. Baculovirus hr's function as both origins of DNA replication and enhancers of gene transcription (29, 42). Instead of hr's, fourteen short dr regions were detected in GbNV (Table 3), which account for 0.6% of the GbNV genome and are distributed throughout the genome. GbNV dr's were up to 96% AT rich and contained two or three copies of tandemly arranged repeat sequences, ranging from 11 to 42 bp in size (Table 3). The dr's within each region were not homologous to those in other regions or to those in baculoviruses HzNV-1 and OrNV. dr1 -4, -5, -7, -8, -10, -11, and -12 overlapped with predicted ORFs. Perfect and imperfect palindromes ranging in size from 4 to 9 bp were found in dr1 -2, -3, -6, -10, -12, and -14 (Table 3). The palindromes showed limited similarity to each other. In addition, a 36-bp AAC-repeat region was located inside GbNV ORF17 and a 51-bp TTA-repeat region was located between ORF20 and -21. Most strikingly, an unusual AT-repeat sequence of 176 bp was located inside ORF54 (Fig. 2). AT-rich repeats were identified in HzNV-1 and OrNV, as well as in hymenopteran NPVs and in some lepidopteran NPVs and GVs. The function of these repeat regions has yet to be elucidated.
TABLE 3.
Structures of GbNV DRs
| dr no. or type of sequence | Length of repeat sequence unit (bp) | No. of copies | Copy position in genome and sequencea | % AT | Total length of repetitive sequence (bp) |
|---|---|---|---|---|---|
| 1 | 18 | 2 | 18431 TTAAATTTTACGATTAAA 18448 | 89 | 36 |
| 18451 TTAAAATTTACGTTTAAA 18468 | |||||
| 2 | 19 | 2 | 20931 TGATTTATATATAATAAAA 20949 | 95 | 38 |
| 20951 TGATTTATATAAAATAAAA 20969 | |||||
| 3 | 11 | 3 | 28545 TATATTTTATA 28555 | 94 | 33 |
| 28556 TATATTTTATA 28566 | |||||
| 28567 TGCATTTTATA 28577 | |||||
| 4 | 19 | 2 | 35239 TCATTTA-CTGATTT-TATT 35256 | 84 | 37 |
| 35257 TCATTTATC-GATTTATATT 35275 | |||||
| 5 | 26 | 2 | 41250 TATATAATACATTTTTAATTATTTTA 41275 | 96 | 50 |
| 41276 TATATAATACA-TTTT-ATTATATTA 41299 | |||||
| 6 | 25 | 2 | 52895 TTGTATTTAATAAAAATTATTAAAT 52919 | 92 | 50 |
| 52920 TTGTATTTAATAAAGATTATTACAT 52944 | |||||
| 7 | 19 | 2 | 57325 TTTTGTATAAATTTTTTAA 57343 | 92 | 38 |
| 57345 TTTTCTATAAATATGTTAA 57363 | |||||
| 8 | 15 | 2 | 58686 GATGAAGACGAAGAC 58700 | 57 | 30 |
| 58701 GATGAAGAAGATGAC 58715 | |||||
| 9 | 19 | 2 | 67596 TAATATT-GTAAAA-TAAA 67612 | 92 | 36 |
| 67614 TAATATATGTAAAAGTAAA 67632 | |||||
| 10 | 15 | 2 | 69891 TAAAAAATAATTATA 69905 | 97 | 30 |
| 69906 TAAAAGATAATTATA 69920 | |||||
| 11 | 42 | 2 | 70004 TTCTCGCTCTAGATCACGATCAAAATCTCGATCTCGTTCTAA 70045 | 58 | 84 |
| 70046 TTCTCGTTCGAGATCGCGATCTAAATCTCGATCTCGTTCTAA 70087 | |||||
| 12 | 29 | 2 | 72339 AAAAAAATGATATTAT-TACTATTT-CTCGA 72367 | 83 | 58 |
| 72368 AAAAAAATTATATTTACTACT-TTTCCTC-A 72396 | |||||
| 13 | 21 | 2 | 85774 TATGTAATGTAAAA-AAAAAA 85793 | 93 | 41 |
| 85795 TATGTAATATAAAATAAAAAA 85815 | |||||
| 14 | 31 | 2 | 91175 TTATGTAAGTAAAA-GAAATTATATA-A-TTA 91203 | 90 | 60 |
| 91204 TTTTGTAAATAAAAAGAAA-TATATTTAGTTA 91234 | |||||
| AAC repeat | 3 | 12 | 15448 (AAC)12 15483 | 66 | 36 |
| TTA repeat | 3 | 17 | 18927 (TTA)17 18977 | 100 | 51 |
| AT repeat | 2 | 88 | 55300 (AT)88 55475 | 100 | 176 |
The palindromic sequences are marked in bold. Dashes indicate sequence gaps.
Phylogeny and evolution.
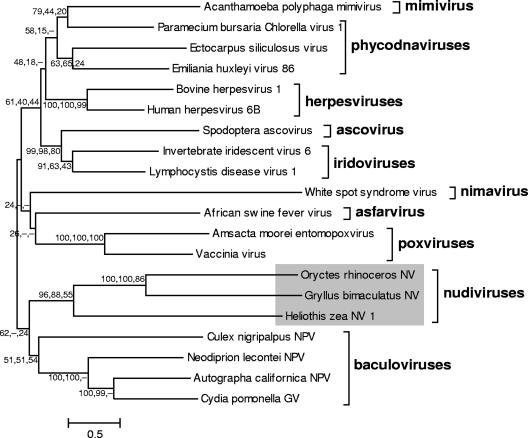
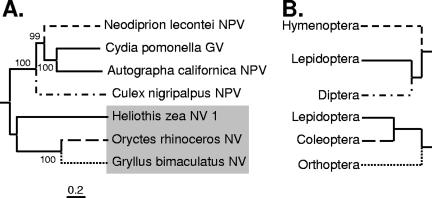
To reveal the phylogenetic relationship of GbNV, its putative DNA polymerase sequence (ORF12) was aligned and analyzed with 19 DNA polymerase sequences from different families of large eukaryotic dsDNA viruses. As shown in Fig. 4, there is evidence, which is >55% bootstrap-supported, that GbNV, HzNV-1, and OrNV form a monophyletic clade. The closest neighbor of GbNV is OrNV, which is not unexpected considering the similarities of gene sequences, gene content, and gene order (see above) between these two viruses. We therefore concluded that GbNV belongs to the recently suggested Nudivirus genus, which appears to share a common ancestor with the Baculoviridae. The long branches suggest that the nudiviruses diverged early, evolved separately, and existed before the split between dipteran baculoviruses and the ancestors of hymenopteran and lepidopteran baculoviruses. This concept is further supported by the midpoint-rooted tree based on the concatenated deduced amino acid sequences of five conserved baculovirus core gene homologues (lef-4, lef-5, dnapol, pif-2, and ac81) present in GbNV, OrNV, and HzNV-1 (Fig. 5). These analyses indicate that baculoviruses and nudiviruses form two monophyletic groups, and that GbNV, OrNV, and HzNV-1 have similar but larger genetic distances between each other than those between members of the recently proposed reclassified Baculovirus genera Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus (24). The extensive divergence of nudiviruses is also illustrated by the large differences in their genome sizes: 96.9 kb (GbNV), 228.1 kb (HzNV-1) (8), and 127 kb (OrNV) (51). Furthermore, our phylogenetic analyses allowed for the first time rooting the baculovirus tree based on conserved baculovirus core genes (Fig. 5). It demonstrates that Culex nigripalpus nucleopolyhedrovirus is indeed the earliest branch of baculoviruses, followed by hymenopteran and lepidopteran baculoviruses (19, 24).
FIG. 4.
Unrooted ME phylogenetic tree based on 2,518 sites of the DNA polymerases of 20 viruses from various families. Distances were calculated using Poisson correction. The homogeneous substitution pattern among lineages with the gamma-distributed rate among sites (Gamma parameter 2.02) is employed for reconstruction of the tree. Gaps and missing data are included for calculation of informative sites using pairwise deletion. The robustness of the tree was tested using bootstrap analyses (1,000 replicates), and the percent values (ME, NJ, MP) are given next to the nodes. The names of the selected virus families are indicated on the tree. The GbNV, OrNV, and HzNV-1 clade is indicated on a gray background. The GenBank accession numbers of the viral DNA polymerase (from top to bottom) are YP_142676, NP_048532, NP_077578, YP_293784, NP_045328, NP_050219, AAC54632, NP_149500, NP_078724, NP_478036, NP_042783, NP_064832, NP_063712, ABF93350, ABO45345, NP_690550, NP_203396, YP_025217, NP_054095, and NP_148895. The scale bar represents a distance of 50%.
FIG. 5.
(A) Midpoint-rooted NJ phylogenetic tree based on the concatenated amino acid sequences (1,805 sites) of the lef-4, lef-5, dnapol, pif-2, and ac81 genes from GbNV, OrNV, HzNV-1, and four selected baculoviruses. Distances were calculated using Poisson correction. Homogeneous substitution patterns among lineages with uniform rates among sites are employed for reconstruction of the tree. Gaps and missing data are excluded from analyses. The robustness of the tree was tested using bootstrap analyses (1,000 replicates), and the percent values (NJ) are given next to the nodes. ME and MP analyses revealed similar topologies for the tree. The group of GbNV, OrNV, and HzNV-1 is indicated on a gray background. (B) Evolutionary relationships of the orders of insects that are infected by the baculoviruses and nudiviruses shown in panel A (12). The branching line style indicates the group affiliations of the insect hosts. The scale bar represents a distance of 20%.
Although phylogenetic analyses of their DNA polymerases provide evidence that nudiviruses and baculoviruses may have shared a common ancestor, the tree of a single gene would never be sufficient alone for such a significant conclusion. Large dsDNA virus genomes especially resemble a variegated blend of genes of different origins. Horizontal gene transfer from other organisms, recombination with related viruses, and domain and gene duplication and deletion are the major evolutionary forces which shape the genetic composition of virus genomes (47). For instance, a number of baculovirus gene homologues are present in a wide range of dsDNA viruses of insects, e.g., entomopoxviruses, ascoviruses, iridoviruses, and others. However, none of these genes belong to the set of baculovirus core genes. The presence of 15 baculovirus core gene homologues in GbNV and HzNV-1, which are supposed to be involved in fundamental mechanisms of the infection process, such as oral infection of insect hosts, viral DNA replication, and transcription, strongly suggests that these genes are essential and functionally conserved during the evolution of these viruses. In bacteria, it is suggested that essential genes are evolutionarily more conserved than nonessential genes (26) and the same holds true for viruses. Considering that the proteins encoded by these genes are involved in complex protein-protein interactions, e.g., in virus attachment to host cells or as subunits of a putative RNA polymerase, it is very unlikely that the genes were integrated into nudivirus genomes by independent horizontal gene transfer events. Hypothesizing a recent horizontal gene transfer of these baculovirus core homologues to GbNV and HzNV-1 is further unjustifiable since (i) these homologues are spread throughout the genomes of GbNV and HzNV-1, (ii) the identities between amino acids encoded by GbNV and HzNV-1 ORFs and baculovirus ORFs are low, ranging from 19 to 46%, and (iii) none of the single core gene trees would support this assumption (data not shown). Notably, compared to the 16 baculovirus core gene homologues identified in HzNV-1 (52), the number of shared core genes of baculoviruses and nudiviruses dropped by only one after the GbNV genome was completed. No additional baculovirus core gene was identified in GbNV. Consequently, it is most reasonable to conclude that these highly conserved core genes are the genomic footprints of a common ancestor of baculoviruses and nudiviruses rather than the result of horizontal gene transfer. Comparing the distantly related genomes of HzNV-1 and GbNV further suggested that the nudiviruses GbNV, HzNV-1, and OrNV do not harbor more than 29 presently detectable conserved core genes with each other, of which 15 are shared with baculoviruses (Table 2).
Previous studies suggested an ancient coevolutionary relationship between baculoviruses and their insect hosts, which did not result in a cocladogenesis with their insect host orders (19). The proposed phylogeny of the baculoviruses and nudiviruses does not mirror that of insect hosts, where Coleoptera are more closely related to Lepidoptera than to Orthoptera, and Diptera are more closely related to Lepidoptera than to Hymenoptera (Fig. 5) (12). Such phylogenetic incongruence of viruses and their insect hosts suggests that host ecology rather than host phylogeny was the main driving force of virus evolution. As was previously pointed out for baculoviruses (19), dipteran larvae develop in aquatic habitats, whereas lepidopteran and hymenopteran sawfly larvae often coexist in the same habitat and show similar feeding behaviors. Thus, Lepidoptera and Hymenoptera might be exposed to each other's baculoviruses rather than to dipteran-specific baculoviruses, explaining the pattern of relationship among baculoviruses (19). As for nudiviruses, their orthopteran and coleopteran insect hosts lay eggs in soil; the larvae live either in soil (Coleoptera) or on the soil surface (Orthoptera); the adults live in similar terrestrial habitats. Again, the shared ecological niches among evolutionarily distantly related insect hosts may explain why GbNV and OrNV are more closely related to each other than to HzNV-1. However, it has to be noted that nothing is known about the natural distribution and host range of HzNV-1, since this virus was detected only in infected lepidopteran cell lines (6).
The differences between the host ecological niches provide a hint for the divergent evolution of baculoviruses and nudiviruses. Members of the Baculoviridae develop an OB, and only larval stages can be perorally infected, whereas apparent infections of adult stages are not recorded. The presence of nonhomologous OB proteins in dipteran and lepidopteran/hymenopteran baculoviruses (44) even suggests that baculovirus OB might have been invented twice during baculovirus evolution, or that the genes were somehow replaced by nonorthologous gene displacement. Horizontal cycling of virus infection through all life stages of baculovirus hosts is most likely hampered by the different habitats often occupied by larvae and adults and the different feeding behaviors of larvae (biting mouth parts) and adults (sucking mouth parts). Baculovirus OBs increase the persistence of infectious viruses in the environment and consequently facilitate horizontal virus transmission. Thus, the formation of an OB can be considered as a key evolutionary transition of viruses with insect hosts, whose developmental stages occupy rather-different ecological niches, providing only limited opportunities for horizontal transmission. The evolutionary constraint of these viruses is to optimize horizontal transmission by using a persistent virus form, e.g., an OB. By contrast, at least for nonoccluded GbNV and OrNV, it is known that they perorally infect not only larvae but also adult beetles and crickets, which colonize habitats similar to those of their larvae and also have biting mouth parts. In these cases, cycling of virus infections through the different developmental stages might be less intricate and OB formation is of a smaller evolutionary advantage. Due to the lack of OB, however, these viruses may have evolved closer host association and more complex transmission modes in the form of latent and persistent infections (6). Taken together, it is suggested that the evolution of baculoviruses and nudiviruses was shaped by the interaction between the viruses and their hosts and was driven by the specific ecological niches the insects colonize. The phylogenetic affiliation of GbNV also suggests that the common ancestor of baculoviruses and nudiviruses did not date back to the ancient origin of arthropods; otherwise GbNV should be the earliest branch in the phylogenetic trees (Fig. 4 and 5).
In summary, based on genetic and phylogenetic analyses, GbNV is most closely related to HzNV-1 and OrNV, and nudiviruses are more distantly related to the baculoviruses. The main similarities between baculoviruses and nudiviruses are covalently closed, dsDNA genomes, rod-shaped nucleocapsids, virus replication in the nucleus, and the presence of 15 core genes suggesting similar mechanisms of midgut infection and late viral RNA production. On the other hand, baculoviruses and nudiviruses differ in gene content and genome organization, cytopathology, and infection of adults and most likely in host range. The proposed relationship of baculoviruses and nudiviruses raises the question of whether nudiviruses should be reconsidered taxonomically as a subfamily within the family Baculoviridae or whether a new family needs to be established which, together with baculoviruses, may form a distinct virus order. This answer cannot be given on the basis of phylogenetic analyses alone, but must be based on the specific definition of demarcation criteria defining these taxa. Formation of OB alone, however, should not be used as a distinguishing criterion of virus families, since OB formation can also be observed in other insect-specific virus clades within Poxviridae and Reoviridae (2, 27). Hence, much more needs to be known about virion properties and infection and replication strategies, as well as the host range and virus ecology of nudiviruses. Nevertheless, the genome sequence of GbNV casts the first light on the genomic characteristics and the evolution of this highly divergent group of viruses.
Acknowledgments
This study was funded by a grant from the Deutsche Forschungsgemeinschaft to J.A.J. (Je245-7).
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belloncik, S., and H. Mori. 1998. Cypoviruses. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Press, New York, NY.
- 3.Bideshi, D. K., S. Renault, K. Stasiak, B. A. Federici, and Y. Bigot. 2003. Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J. Gen. Virol. 84:2531-2544. [DOI] [PubMed] [Google Scholar]
- 4.Biteau, B., J. Labarre, and M. B. Toledano. 2003. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425:980-984. [DOI] [PubMed] [Google Scholar]
- 5.Braunagel, S. C., D. M. Elton, H. Ma, and M. D. Summers. 1996. Identification and analysis of an Autographa californica nuclear polyhedrosis virus structural protein of the occlusion-derived virus envelope: ODV-E56. Virology 217:97-110. [DOI] [PubMed] [Google Scholar]
- 6.Burand, J. P. 1998. Nudiviruses, p. 69-90. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Press, New York, NY.
- 7.Chang, T. S., W. Jeong, H. A. Woo, S. M. Lee, S. Park, and S. G. Rhee. 2004. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem. 279:50994-51001. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, C. H., S. M. Liu, T. Y. Chow, Y. Y. Hsiao, D. P. Wang, J. J. Huang, and H. H. Chen. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024-9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faulkner, P., J. Kuzio, G. V. Williams, and J. A. Wilson. 1997. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 78:3091-3100. [DOI] [PubMed] [Google Scholar]
- 10.Granados, R. R., and K. A. Lawler. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108:297-308. [DOI] [PubMed] [Google Scholar]
- 11.Green, K. J., D. A. Parry, P. M. Steinert, M. L. Virata, R. M. Wagner, B. D. Angst, and L. A. Nilles. 1990. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J. Biol. Chem. 265:2603-2612. [PubMed] [Google Scholar]
- 12.Grimaldi, D., and M. S. Engel. 2005. Evolution of the insects. Cambridge University Press, New York, NY.
- 13.Guarino, L. A., W. Dong, and J. Jin. 2002. In vitro activity of the baculovirus late expression factor LEF-5. J. Virol. 76:12663-12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutiérrez, S., I. Kikhno, and M. López-Ferber. 2004. Transcription and promoter analysis of pif, an essential but low-expressed baculovirus gene. J. Gen. Virol. 85:331-341. [DOI] [PubMed] [Google Scholar]
- 15.Harwood, S. H., L. Li, P. S. Ho, A. K. Preston, and G. F. Rohrmann. 1998. AcMNPV late expression factor-5 interacts with itself and contains a zinc ribbon domain that is required for maximal late transcription activity and is homologous to elongation factor TFIIS. Virology 250:118-134. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto, Y., T. Hayakawa, Y. Ueno, T. Fujita, Y. Sano, and T. Matsumoto. 2000. Sequence analysis of the Plutella xylostella granulovirus genome. Virology 275:358-372. [DOI] [PubMed] [Google Scholar]
- 17.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211-234. [DOI] [PubMed] [Google Scholar]
- 19.Herniou, E. A., J. A. Olszewski, D. R. O'Reilly, and J. S. Cory. 2004. Ancient coevolution of baculoviruses and their insect hosts. J. Virol. 78:3244-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, H. M., and J. P. Burand. 1993. Saturable attachment sites for polyhedron-derived baculovirus on insect cells and evidence for entry via direct membrane fusion. J. Virol. 67:1860-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huger, A. M. 1985. A new virus disease of crickets (Orthoptera: Gryllidae) causing macronucleosis of fatbody. J. Invertebr. Pathol. 45:108-111. [Google Scholar]
- 22.Huger, A. M., and A. Krieg. 1991. Baculoviridae. Nonoccluded baculoviruses, p. 287-319. In J. R. Adams and J. R. Bonami (ed.), Atlas of invertebrate viruses. CRC Press, Inc., Boca Raton, FL.
- 23.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jehle, J. A., G. W. Blissard, B. C. Bonning, J. S. Cory, E. A. Herniou, G. F. Rohrmann, D. A. Theilmann, S. M. Thiem, and J. M. Vlak. 2006. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 151:1257-1266. [DOI] [PubMed] [Google Scholar]
- 25.Jehle, J. A., M. Lange, H. Wang, Z. Hu, Y. Wang, and R. Hauschild. 2006. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 346:180-193. [DOI] [PubMed] [Google Scholar]
- 26.Jordan, I. K., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin. 2002. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12:962-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King, L. A., N. Wilkinson, D. P. Miller, and S. A. Marlow. 1998. Entomopoxviruses. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Press, New York, NY.
- 28.Knebel-Morsdorf, D., I. Quadt, Y. Li, L. Montier, and L. A. Guarino. 2006. Expression of baculovirus late and very late genes depends on LEF-4, a component of the viral RNA polymerase whose guanyltransferase function is essential. J. Virol. 80:4168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kool, M., P. M. van den Berg, J. Tramper, R. W. Goldbach, and J. M. Vlak. 1993. Location of two putative origins of DNA replication of Autographa californica nuclear polyhedrosis virus. Virology 192:94-101. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 31.Kuzio, J., R. Jaques, and P. Faulkner. 1989. Identification of p74, a gene essential for virulence of baculovirus occlusion bodies. Virology 173:759-763. [DOI] [PubMed] [Google Scholar]
- 32.Lammers, M., and H. Follmann. 1983. The ribonucleotide reductases: a unique group of metalloenzymes essential for cell-proliferation. Struct. Bonding 54:27-91. [Google Scholar]
- 33.Lange, M., H. Wang, Z. Hu, and J. A. Jehle. 2004. Towards a molecular identification and classification system of lepidopteran-specific baculoviruses. Virology 325:36-47. [DOI] [PubMed] [Google Scholar]
- 34.Lu, A., and L. K. Miller. 1997. Regulation of baculovirus late and very late gene expression, p. 193-216. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, NY.
- 35.Manji, G. A., R. R. Hozak, D. J. LaCount, and P. D. Friesen. 1997. Baculovirus inhibitor of apoptosis functions at or upstream of the apoptotic suppressor P35 to prevent programmed cell death. J. Virol. 71:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, L. K., and L. A. Ball (ed.). 1998. The insect viruses. Plenum Press, New York, NY.
- 37.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681-706. [DOI] [PubMed] [Google Scholar]
- 38.Oeschger, M. P., and M. J. Bessman. 1966. Purification and properties of guanylate kinase from Escherichia coli. J. Biol. Chem. 241:5452-5460. [PubMed] [Google Scholar]
- 39.Okano, K., A. L. Vanarsdall, V. S. Mikhailov, and G. F. Rohrmann. 2006. Conserved molecular systems of the Baculoviridae. Virology 344:77-87. [DOI] [PubMed] [Google Scholar]
- 40.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 41.Payne, C. C. 1974. The isolation and characterization of a virus from Oryctes rhinoceros. J. Gen. Virol. 25:105-116. [DOI] [PubMed] [Google Scholar]
- 42.Pearson, M., R. Bjornson, G. Pearson, and G. Rohrmann. 1992. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science 257:1382-1384. [DOI] [PubMed] [Google Scholar]
- 43.Pearson, M. N., and G. F. Rohrmann. 1998. Characterization of a baculovirus-encoded ATP-dependent DNA ligase. J. Virol. 72:9142-9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perera, O. P., S. M. Valles, T. B. Green, S. White, C. A. Strong, and J. J. Becnel. 2006. Molecular analysis of an occlusion body protein from Culex nigripalpus nucleopolyhedrovirus (CuniNPV). J. Invertebr. Pathol. 91:35-42. [DOI] [PubMed] [Google Scholar]
- 45.Russell, R. L., and G. F. Rohrmann. 1997. Characterization of P91, a protein associated with virions of an Orgyia pseudotsugata baculovirus. Virology 233:210-223. [DOI] [PubMed] [Google Scholar]
- 46.Schäffer, A. A., L. Aravind, T. L. Madden, S. Shavirin, J. L. Spouge, Y. I. Wolf, E. V. Koonin, and S. F. Altschul. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29:2994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shackelton, L. A., and E. C. Holmes. 2004. The evolution of large DNA viruses: combining genomic information of viruses and their hosts. Trends Microbiol. 12:458-465. [DOI] [PubMed] [Google Scholar]
- 48.Theilmann, D. A., G. W. Blissard, B. Bonning, J. A. Jehle, D. R. O'Reilly, G. F. Rohrmann, S. Thiem, and J. M. Vlak. 2005. Baculoviridae, p. 177-185. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: the eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, New York, NY.
- 49.Titterington, J. S., T. K. Nun, and A. L. Passarelli. 2003. Functional dissection of the baculovirus late expression factor-8 gene: sequence requirements for late gene promoter activation. J. Gen. Virol. 84:1817-1826. [DOI] [PubMed] [Google Scholar]
- 50.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2006. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J. Virol. 80:1724-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlak, J. M., A. M. Huger, J. A. Jehle, and R. G. Kleespies. The Oryctes rhinoceros virus (OrV) (unassigned). In B. W. J. Mahy and M. H. V. van Regenmortel (ed.), Encyclopedia of virology, 3rd ed., in press. Academic Press, New York, NY.
- 52.Wang, Y., M. M. van Oers, A. M. Crawford, J. M. Vlak, and J. A. Jehle. 2007. Genomic analysis of Oryctes rhinoceros virus reveals genetic relatedness to Heliothis zea virus 1. Arch. Virol. 152:519-531. [DOI] [PubMed] [Google Scholar]
- 53.Wolf, Y. I., I. B. Rogozin, N. V. Grishin, and E. V. Koonin. 2002. Genome trees and the tree of life. Trends Genet. 18:472-479. [DOI] [PubMed] [Google Scholar]
- 54.Zimmerman, S. B., J. W. Little, C. K. Oshinsky, and M. Gellert. 1967. Enzymatic joining of DNA strands: a novel reaction of diphosphopyridine nucleotide. Proc. Natl. Acad. Sci. USA 57:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]