Abstract
Huh7 cells constitute a permissive cell line for cell culture of hepatitis C virus (HCV) particles. However, our Huh7 line shows limited permissiveness for HCV. Thus, in this study we set out to determine which host factors are important for conferring permissiveness. To analyze the limited permissiveness of our Huh7 cells, 70 clones were obtained after single-cell cloning of parental Huh7 cells. The cloned Huh7 cells exhibited various levels of HCV pseudoparticles and JFH-1 virus infection efficiency, and some clones were not permissive. A subgenomic replicon was then transfected into the cloned Huh7 cells. While the replication efficiencies differed among the cloned Huh7 cells, these efficiencies did not correlate with infectious permissibility. Flow cytometry showed that CD81, scavenger receptor class B type I, and low-density-lipoprotein receptor expression on the cell surfaces of the Huh7 clones differed among the clones. Interestingly, we found that all of the permissive cell clones expressed CD81 while the nonpermissive cell clones did not. To confirm the importance of CD81 expression for HCV permissiveness, CD81 was then transiently and stably expressed on a nonpermissive Huh7 cell clone, which was consequently restored to HCV infection permissiveness. Furthermore, permissiveness was down-regulated upon transfection of CD81 silencing RNA into a CD81-positive cell clone. In conclusion, CD81 expression is an important determinant of HCV permissiveness of Huh7 cell clones harboring different characteristics.
Hepatitis C virus (HCV) is a worldwide human pathogen, and most infected patients progress to chronic liver disease. The primary therapy for HCV is treatment with pegylated interferon and ribavirin; however, these agents do not cause a marked decline in the virus titers of all treated patients. Thus, the elucidation of native virus-host interactions is necessary to develop new, more effective therapies. However, the lack of a robust cell culture system to produce infectious virions has hampered research. That said, in a great boon to HCV research, a cell culture system that allows the whole life cycle of HCV to be investigated was recently developed (22, 34, 41).
HCV is an enveloped virus that belongs to the Hepacivirus genus in the Flaviviridae family (23). Cell attachment of flaviviruses generally leads to endocytosis of bound virions. Several molecules have been proposed as cell entry receptors of HCV; most of these have been identified based on binding with soluble recombinant E2 protein or HCV-like particles (2, 3, 12, 25, 30, 31). Putative HCV receptors include CD81 (30), low-density-lipoprotein (LDL) receptor (1), scavenger receptor class B type I (SR-BI) (31), and several molecules that induce concentration of viral particles at the cell surface. Infectious HCV pseudoparticles (HCVpp) harboring E1E2 glycoproteins (5, 11) have substantiated the functional roles of the candidate receptors CD81 and SR-BI in HCV entry (5, 6, 15). The importance of CD81 for HCV entry was recently confirmed using cell-cultured HCV particles (22, 34). Furthermore, CD81 is important for postattachment of HCV particles on Huh7 cells (19, 28).
Huh7 and its interferon-cured cells are considered permissive cell lines for HCV particles (22, 34, 41), but our Huh7 cell line shows limited permissiveness. In the present study, we performed single-cell cloning of Huh7 cells and then analyzed heterogeneity. To investigate the host factors important for HCV infection, the Huh7 cell clones were then transiently infected with JFH-1 virus and comparisons of efficiency of replication and expression of candidate receptors were performed.
MATERIALS AND METHODS
Cell culture and single-cell cloning.
Parental Huh7 cells, Huh7.5.1 cells (41) (a generous gift from Francis V. Chisari), and Huh7 cell clones were cultured at 37°C in 5% CO2. Cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, as described previously (18). Parental Huh7 cells were diluted with medium and seeded into 96-well plates at 0.3 cells per well. Seventy single-cell-derived clones were then selected, and after 3 weeks their cells were passaged. The resultant Huh7 cell clones were stored at −80°C until use.
Plasmids.
pJFH-1 (34), pSGR-JFH1/Luc, pSGR-JFH1/Luc-GND (17), and pFGR-JFH1 (10) were generated as previously reported. pFGR-J6/N2X-JFH1 was generated by replacement of the JFH-1 structural region (a core coding region to the BclI site) with pJ6CF (35) (a generous gift from Jens Bukh). pFGR-JFH1/EGFP and pFGR-J6/N2X-JFH1/EGFP were generated by replacement of the neomycin-resistant gene of pFGR-JFH1 and that of pFGR-J6/N2X-JFH1 with the enhanced green fluorescent protein (EGFP) gene from pEGFP-N3 (Clontech, Mountain View, CA). pcDNA3.1-CD81 and the vesicular stomatitis virus (VSV) G protein-expressing construct pCAG-VSVG (27) were kind gifts from Yoshiharu Matsuura (Osaka University, Suita, Japan). The JFH-1 E1E2 expression construct pcDNAdeltaC-E1-E2(JFH1) was a kind gift from Thomas Pietschmann (University of Heidelberg, Heidelberg, Germany), while the murine leukemia virus packaging construct and the luciferase-based transfer vector construct have been described previously (5).
RNA synthesis.
RNA synthesis was performed as described previously (17, 34). Briefly, the plasmids pJFH-1, pSGR-JFH1/Luc, pSGR-JFH1/Luc-GND, pFGR-JFH1/EGFP, and pFGR-J6/N2X-JFH1/EGFP were digested with XbaI and treated with mung bean nuclease (New England Biolabs, Beverly, MA). Digested plasmid DNA fragments were then purified and used as templates for RNA synthesis. HCV RNA was synthesized in vitro by use of a MEGAscript T7 kit (Ambion, Austin, TX). Synthesized RNA was treated with DNase I, followed by acid phenol extraction to remove any remaining template DNA.
Replication assay of JFH-1 subgenomic replicon.
Replication of a JFH-1 subgenomic replicon (SGR-JFH1) in Huh7 cells was detected as described previously (17). Briefly, 2 μg of reporter replicon RNA transcribed from pSGR-JFH1/Luc and pSGR-JFH1/Luc-GND was transfected into 2 × 106 Huh7 cells by electroporation. Transfected cells were immediately transferred to culture medium and seeded into six-well culture plates.
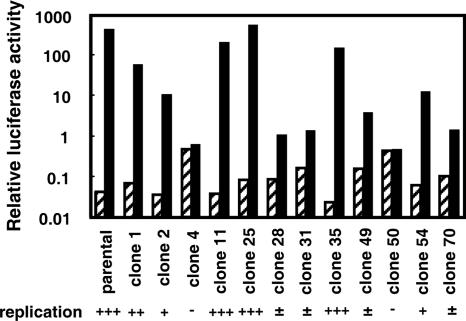
Cells were harvested serially at 4 h (day 0), 24 h (day 1), and 48 h (day 2) after transfection and lysed with 200 μl of cell culture lysis reagent (Promega, Madison, WI). Debris was then removed by centrifugation. Luciferase activities were quantified by use of LUMAT LB9507 (EG & G Berthold, Bad Wildbad, Germany) and a luciferase assay system (Promega). Assays were performed in duplicate, and the results at 24 and 48 h after transfection were normalized and expressed as the relative luciferase activities (RLA) compared to the luciferase activity at 4 h after transfection. The replication efficiency of each cell is indicated in Fig. 3 and Table 1 as follows: −, RLA below 1; ±, RLA between 1 and 10; +, RLA between 10 and 50; ++, RLA between 50 and 100; +++, RLA over 100.
FIG. 3.
Replication of JFH-1 subgenomic replicon in Huh7 cell clones. Reporter replicon RNA was transfected into Huh7 cell clones, and luciferase activities at 4 and 48 h after RNA transfection were then detected. The SGR-JFH1/Luc-GND RNA was used as the negative control. All data indicate the RLA compared to the luciferase activities at 4 h after transfection. The RLA of SGR-JFH1/Luc (solid bars) and SGR-JFH1/Luc-GND (hatched bars) are also shown. All experiments were performed in duplicate, and the data are shown as the means.
TABLE 1.
Permissiveness to infection and expression of candidate receptorsa
| Cell type | Replication efficiency (RLA)b | CD81 expression (MFI)c | SR-BI expression (MFI)d | LDLr expression (MFI)e | HCVpp infectivityf | JFH-1 infectivityg |
|---|---|---|---|---|---|---|
| Huh7 (parental) | +++ | ++ | ++ | + | + | + |
| Clone 1 | ++ | + | ++ | + | + | +++ |
| Clone 2 | + | − | + | + | ± | − |
| Clone 4 | − | − | + | + | ± | − |
| Clone 11 | +++ | − | ++ | + | − | − |
| Clone 25 | +++ | − | ++ | + | ± | − |
| Clone 28 | ± | +++ | ++ | ++ | + | ± |
| Clone 31 | ± | + | ++ | + | + | ++ |
| Clone 35 | +++ | − | ++ | ++ | ± | − |
| Clone 49 | ± | ++ | + | ++ | + | ++ |
| Clone 50 | − | ++ | + | + | + | ± |
| Clone 54 | + | + | ++ | ++ | ++ | ++ |
| Clone 70 | ± | ++ | ++ | + | ++ | ++ |
| Huh7-25-CD81 | +++ | +++ | ++ | + | +++ | +++ |
See Materials and Methods for definitions of symbols.
Detection of replication of the JFH-1 subgenomic replicon on Huh7 clones at 48 h after transfection of SGR-JFH1/Luc RNA.
Detection of CD81 by flow cytometry using JS-81 antibodies on the surfaces of the indicated cells.
Detection of SR-BI by flow cytometry using rat anti-SR-BI antiserum on the surfaces of the indicated cells.
Detection of LDL receptor (LDLr) by flow cytometry using rabbit anti-LDLr antibody (BP5014) on the surfaces of the indicated cells.
Detection of luciferase activities by subtraction from the activity of the nonenveloped control.
Infected foci were detected by immunofluorescence using HCV anticore antibody (2H9).
Production of infectious HCV particles.
HCV particles derived from JFH-1 were prepared as described previously (34). Briefly, in vitro-synthesized RNA was transfected into Huh7 cells by electroporation. Cell culture supernatants were collected 72 h after transfection and passed through a 0.45-μm filter. Filtrated culture medium was then pooled and concentrated using Amicon Ultra-15 (100,000 molecular weight cutoff; Millipore, Bedford, MA). The infectious titer was 1.6 × 104 focus-forming units (ffu) per ml and was determined by immunofluorescence detection of infected foci following infection of naïve parental Huh7 cells. RNA quantification was performed by real-time detection reverse transcription-PCR analysis, as described previously (32), using an ABI Prism 7700 sequence detector system (Applied Biosystems Japan, Tokyo, Japan). The titer was determined to be 4.3 × 108 RNA copies/ml. Concentrated culture medium samples were stored at −80°C until use.
HCV particle infection and immunofluorescence.
Parental Huh7 cells and Huh7 cell clones were seeded at 1 × 104 cells/well in poly-d-lysine-coated 96-well plates (Corning, Inc., Corning, NY), cultured overnight, and then inoculated with serially diluted culture medium containing infectious HCV particles. At 48 h after inoculation, the cells were fixed in methanol for 15 min at −20°C, and the infected foci were visualized by immunofluorescence as described below.
Cells were permeabilized and blocked for 1 h with BlockAce (Dainippon Sumitomo Pharma, Osaka, Japan) containing 0.3% (vol/vol) Triton X-100. The cells were then washed five times with phosphate-buffered saline (PBS), and anticore monoclonal antibody 2H9 (34) was added at 50 μg/ml in BlockAce. After incubation for 1 h at room temperature, the cells were washed and incubated with a 1:400 dilution of AlexaFluor 488-conjugated or AlexaFluor 546-conjugated anti-mouse immunoglobulin G (IgG) (Molecular Probes, Eugene, OR) with BlockAce. The cells were then washed and examined by fluorescence microscopy (Olympus, Tokyo, Japan). Quantification of infectivity was performed by counting the infected foci, and the assay was performed in triplicate. The infectivity of each clone is indicated in Fig. 3 and Table 1 as follows: −, no infected foci; ±, between 1 and 5 foci; +, between 5 and 10 foci; ++, between 10 and 50 foci; +++, over 50 foci.
Stable and transient expression of CD81.
A trypsinized CD81-negative clone (clone 25) was washed with Opti-MEM I (Invitrogen, Carlsbad, CA) and resuspended at 5 × 106 cells/ml in Cytomix buffer (18). pcDNA3.1-CD81 plasmid DNA (75 μg) was mixed with 400 μl of cell suspension and the mix then transferred to an electroporation cuvette (Precision universal cuvette; Thermo Hybrid, Middlesex, United Kingdom). The cells were then pulsed at 220 V and 950 μF with a Gene Pulser II apparatus (Bio-Rad, Hercules, CA). Transfected cells were immediately transferred to a 75-cm2 flask (Corning) and incubated at 37°C/5% CO2. After 3 days, the cells were passaged and seeded into 10-cm dishes, and G418 (0.8 mg/ml) (Nacalai Tesque, Kyoto, Japan) was then added to the culture medium. Culture medium supplemented with G418 was replaced twice a week. Three weeks later, the colonies were observed and then the cells were trypsinized. CD81-positive cells were obtained and confirmed by the same method as for single-cell cloning of parental Huh7 cells. We obtained a clone, Huh7-25-CD81, in which CD81 was stably expressed.
For transient CD81 expression, 6 μg of pcDNA3.1-CD81 plasmid was transfected into Huh7 clone 25 (Huh7-25) cells (2.5 × 106) by using FuGENE6 transfection reagent (Roche Diagnostics, Indianapolis, IN). After 24 h, cells were passaged and an aliquot used for flow cytometric analysis and virus infection. As controls, Huh7-25 and Huh7-25-CD81 cells were similarly treated by only FuGENE6 and used for flow cytometric analysis and virus infection.
Pseudotype production and infection.
Murine leukemia virus pseudotypes were generated according to methods described previously (5). Briefly, the Gag-Pol packaging construct (3.1 μg), the transfer vector construct (3.1 μg), and the JFH-1 glycoprotein or the VSV-G protein-expressing construct (1 μg) DNAs were transfected into 2.5 × 106 293T cells seeded the day before in 10-cm dishes by use of FuGENE6 transfection reagent (Roche Diagnostics). For the negative control, the constructs (except for the glycoprotein-expressing construct) were similarly transfected. The medium (8 ml/dish) was replaced 6 h after transfection. Supernatants containing the pseudotypes were collected 48 h later and passed through a 0.45-μm filter. The supernatants were stored at −80°C until use.
Target cells were seeded into 48-well plates at a density of 2 × 104 cells/well and incubated overnight at 37°C. A 100-μl aliquot of each diluted supernatant containing pseudotypes was added to each well and incubated for 3 h. The supernatants were removed, and the cells were incubated in regular medium for 72 h at 37°C. Cells were washed once with PBS and lysed with 40 μl/well of cell culture lysis reagent. Luciferase activities were quantified using a luciferase assay system (Promega) as described above. Assays were performed in triplicate. All Huh7 cell clones showed infectivity by the VSV-G pseudoparticle, and infectivity by HCVpp was indicated by the luciferase activity (relative luciferase units [RLU]), determined by subtraction from the activity of the nonenveloped control. The infectivity of each clone is indicated in Table 1 as follows: −, luciferase activity below 1 RLU; ±, activity between 1 and 5,000 RLU; +, activity between 5,000 and 30,000 RLU; ++, activity between 30,000 and 100,000 RLU; +++, activity over 100,000 RLU.
RNA interference.
A 40-pmol amount of silencing RNA (siRNA) duplex for CD81 (Santa Cruz, Inc., Santa Cruz, CA) was electroporated into 2.5 × 106 Huh7 clone 54 (Huh7-54) cells (260 V, 950 μF). Control irrelevant siRNA (siIRR) was designed as described previously (37) and transfected, as was the siRNA of CD81. Cells were then propagated and tested for CD81 expression and JFH-1 virus infection.
Flow cytometric analysis.
Cells were seeded in 10-cm dishes (Corning) and cultured overnight. Then, subconfluent cells were harvested either by trypsinization or by treatment with 0.05% EDTA in PBS. Parental Huh7 cells and Huh7 cell clones (1 × 106) were incubated with or without 1 μg mouse anti-CD81 antibody (JS-81; Pharmingen, Franklin Lakes, NJ) for 30 min at 4°C and washed with PBS. The cells were then incubated with fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG secondary antibody (Cappel, Durham, NC) at 1:100 for 30 min at 4°C, washed repeatedly, and resuspended in PBS containing 1% (vol/vol) formaldehyde. SR-BI expression of each cell was tested using rat anti-human SR-BI antiserum (1:50) and FITC-conjugated anti-rat IgG secondary antibody (Cappel) by the same method as for CD81 (2). Rat preimmune serum was used as the negative-control primary antibody. LDL receptor expression was tested using rabbit anti-LDL receptor antibody (BP5014 at 1:10; Acris Antibodies GmbH, Hiddenhausen, Germany) and FITC-conjugated anti-rabbit IgG secondary antibody (Cappel). Analyses were performed using an EPICS ALTRA MultiCOMP unit (Beckman Coulter, Fullerton, CA) and FACSCalibur (Becton Dickinson, Franklin Lakes, NJ). The expression levels are indicated in Table 1 as follows: −, mean fluorescence intensity (MFI) in relative units was below 1; +, MFI in relative units was between 1 and 3; ++, MFI in relative units was between 3 and 6; +++, MFI in relative units was over 6.
Analysis of cell surface CD81 expression levels and HCV infection.
For EGFP virus production, in vitro-synthesized RNA was transfected into Huh7.5.1 cells by electroporation. Cell culture supernatants of transfected cells were collected and concentrated as described above. The infectious titer was also determined as described above. Huh7-70 and Huh7-25-CD81 cells were seeded in six-well plates at 1 × 105 cells/well 24 h before virus inoculation. Cells were inoculated with EGFP virus (multiplicity of infection, 2) for 4 h, followed by a PBS wash. The inoculated cells were cultured in complete medium and then harvested at 24, 48, 72, and 96 h after inoculation. CD81 expression and GFP-positive cells were analyzed by FACSCalibur as described above, using allophycocyanin-conjugated anti-mouse IgG (R&D Systems, Minneapolis, MN) as a secondary antibody at 1:10. All experiments were performed in triplicate, and analysis was performed using CellQuest Pro software (Becton Dickinson). CD81 expression on uninfected and infected cells was calculated from the geometric MFI of the each quadrant plot. The results are given as MFIs ± standard deviations.
RESULTS
Parental Huh7 cell shows limited permissiveness for JFH-1 virus.
A parental Huh7 cell was infected with JFH-1 virus and infectivity assessed by counting the infected foci in anticore immunofluorescence. The number of infected foci increased linearly with lower doses of virus infection (<50 ffu/well). However, the number of infected foci did not increase with higher doses of infection (>200 ffu/well) (Fig. 1). Furthermore, when the parental Huh7 cell infected with JFH-1 was passaged, the infected cells did not expand (data not shown). Thus, the original parental Huh7 cell had limited permissiveness for JFH-1 infection.
FIG. 1.
Infectivity of JFH-1 virus in parental Huh7 cell. Parental Huh7 cells were seeded at 1 × 104 cells/well and infected with JFH-1 virus at 3 to 800 ffu/well for 48 h in 96-well plates. Infected cells were visualized with immunofluorescence using anticore antibody (2H9), and the foci were counted. All experiments were performed in triplicate, and the data are shown as means ± standard deviations.
Huh7 cell clones have different levels of permissiveness for HCVpp and JFH-1 virus infections.
The parental Huh7 cell did not show dose-dependent permissiveness for JFH-1 infection. Thus, single-cell cloning of this Huh7 cell was performed by limiting dilution, and the efficiency of virus infection for each clone was then investigated. Seventy clones were obtained. Among the isolated cloned cells, 65 clones were first screened with JFH-1 virus infection, and the other 5 clones were not tested, because of their slower growth. We found different numbers of focus formation in the clones (46 positive and 19 negative). Among them, we selected seven positive (clones 1, 28, 31, 49, 50, 54, and 70) and five negative (clones 2, 4, 11, 25, and 35) clones for further analysis. Next, these 12 cell clones were infected with HCVpp and JFH-1 virus. Interestingly, the efficiencies of both virus infections of the Huh7 cell clones differed among the clones (Fig. 2; Table 1). Furthermore, some differences were observed between HCVpp and JFH-1 virus permissiveness. Four clones that were not permissive for JFH-1 virus showed slight permissiveness for HCVpp (clones 2, 4, 25, and 35). In addition, three clones were more permissive for both HCVpp and JFH-1 virus than for the parental Huh7 cell (clones 1, 54, and 70). However, clone 11 was not permissive at all for either virus infection. Thus, the parental Huh7 cell population was most likely heterogeneous and included cells with different characteristics.
FIG. 2.
Infectivity of HCVpp and JFH-1 virus in Huh7 cell clones. (A) Target cells (2 × 104 cells) were inoculated with supernatant containing HCVpp for 3 h in 48-well plates. After 72 h, cells were harvested and the luciferase activities quantified. All experiments were performed in triplicate, and infectivity is indicated as the RLU minus the activity of the nonenveloped negative control. (B) Target cells (1 × 104 cells) were infected with the same titer of JFH-1 virus for 48 h in 96-well plates. Infected cells were visualized with immunofluorescence using anticore antibody (2H9), and the foci were counted. All experiments were performed in triplicate, and the data are shown as means ± standard deviations.
Replication of JFH-1 subgenomic replicon in Huh7 cell clones does not correlate with the efficiency of virus infection.
In a previous study, a subgenomic replicon of JFH-1 was shown to efficiently replicate in Huh7 cells (18). Thus, to investigate whether replication efficiency correlated with infectious efficiency, we transiently transfected SGR-JFH1/Luc RNA into each Huh7 cell clone and measured the resultant RLA. At 48 h posttransfection, levels of replication of the replicon in the Huh7 cell clones differed among the clones (Fig. 3), but the efficiency did not correlate with JFH-1 virus infectivity (Table 1).
Expression of CD81 on Huh7 cell clones is correlated with HCV permissiveness.
The Huh7 cell clones displayed differing levels of permissiveness for HCVpp and JFH-1 virus infections, and replication of the replicon did not correlate with infectivity. Thus, the difference in infectivity was most likely due to host factors related to the initial phase of infection. Previous studies using pseudotype particles bearing envelope proteins of HCV have shown that CD81 is a candidate receptor of HCV (5, 6, 9, 15, 38). Therefore, we investigated CD81 expression on our Huh7 cell clones by flow cytometry using an anti-CD81 antibody. We found that the levels of CD81 expression on the Huh7 cells differed among the clones (Fig. 4). All of the CD81-negative clones (clones 2, 4, 11, 25, and 35) were also negative for JFH-1 virus infectivity. However, CD81-positive clones (clones 1, 28, 31, 49, 50, 54, and 70) showed HCV permissiveness at different levels (Table 1). Interestingly, clones 28 and 50 expressed relatively high levels of CD81 but low levels of permissiveness. This may have been due to a lower replication efficiency of these clones, although clones 49 and 70 also had low replication capacities but were permissive of HCV infection. Thus, the low permissiveness of clones 28 and 50 is most likely due to other, as yet unknown mechanisms (Table 1). To confirm that the different expression levels of CD81 among the clones were not due to the cell-harvesting conditions, we harvested using two different techniques, namely, trypsinization and EDTA treatment. Neither method affected the results. This finding suggests that CD81 expression is highly correlated with HCV infectivity, although the level of CD81 expression did not necessarily correlate with JFH-1 virus infectivity among these Huh7 cell clones (Table 1).
FIG. 4.
CD81 expression on the surfaces of Huh7 cell clones. Huh7 cell clones were seeded in 10-cm dishes and cultured overnight. Then, subconfluent cells were harvested either by trypsinization or by treatment with 0.05% EDTA in PBS. Cells (1 × 106) were incubated with 1 μg of mouse anti-CD81 monoclonal antibody (JS-81) and subsequently stained with FITC-conjugated goat anti-mouse IgG. The negative control represents cells incubated with only secondary antibody. The analysis was performed by EPICS ALTRA MultiCOMP (Beckman Coulter). The x and y axes show fluorescence intensity and relative number of stained cells, respectively.
Expression levels of SR-BI and the LDL receptor on Huh7 cell clones do not correlate with permissiveness.
CD81 expression correlated highly with infectivity of Huh7 cell clones. In a previous report, CD81 expression level determined permissiveness, as shown by a transient-transfection experiment (40). However, the levels of CD81 expression among the clones from this study did not correlate with virus infectivity (Table 1), suggesting that multiple factors determine the level of infectivity. As previous studies have suggested that SR-BI and the LDL receptor play important roles in HCV infection (4, 20, 33), we next investigated the expression of these molecules on the surfaces of our Huh7 cell clones. The expression of these molecules was also detected by flow cytometry. Small differences in the expression levels of SR-BI and the LDL receptor compared to that of CD81 were observed (Fig. 5; Table 1). In terms of virus permissiveness, SR-BI and LDL receptor expression did not display the high correlation seen with CD81 (Fig. 5; Table 1).
FIG. 5.
SR-BI and LDL receptor on the surfaces of Huh7 cell clones. Huh7 cell clones (1 × 106 cells) were incubated with rat anti-SR-BI antiserum or rabbit anti-LDL receptor antibody and subsequently stained with FITC-conjugated secondary antibody. The negative control represents cells incubated with rat preimmune serum (SR-BI) or only secondary antibody (LDL receptor [LDLr]). The analysis was performed by FACSCalibur (Becton Dickinson). The x and y axes show fluorescence intensity and relative number of stained cells, respectively. (A) Expression of SR-BI on parental Huh7 cells, clone 2 (the cell clone with the lowest expression level), and clone 11 (the cell clone with the highest expression level). (B) Expression of LDL receptor on parental Huh7 cells, clone 4 (the cell clone with the lowest expression level), and clone 28 (the cell clone with the highest expression level).
CD81 expression restores HCV infection permissiveness in a nonpermissive Huh7 cell clone.
To confirm the importance of CD81 expression for HCV permissiveness, CD81 was expressed on a nonpermissive, non-CD81-expressing Huh7 cell clone (Huh7-25) (Fig. 4; Table 1) and the cells were then infected with JFH-1 virus. When CD81 was transiently and stably expressed on Huh7-25 cells, these cells were restored to permissiveness. The CD81-positive cells of transiently and stably transfected cells were 45.1 and 80.6%, respectively, and infectivities (infected foci) were 58.0 ± 7.9 and 257.7 ± 14.6, respectively, with inoculation of the same titer of JFH-1 virus (Fig. 6A). A clone in which CD81 was stably expressed (Huh7-25-CD81) had high infectivity for JFH-1 virus (Fig. 6A), although the replication efficiency was equal to that of Huh7-25 cells (Fig. 6B). In addition, the infected foci indicated a linear, dose-dependent increase in virus dose (Fig. 7). Thus, ectopic expression of CD81 in a nonpermissive Huh7 cell clone restored HCV infection permissiveness. Furthermore, the Huh7-25-CD81 cell clone supported the highest permissiveness for HCV infection.
FIG. 6.
Infectivity in CD81-transfected Huh7 cells. (A) CD81 was transiently and stably expressed on a Huh7 nonpermissive clone (Huh7-25). The expression of CD81 on the cell surface was examined by flow cytometry. Bars indicate the populations of the CD81-positive cells. Cells were inoculated with JFH-1 virus (1 × 106 copies of HCV RNA) for 3 h. Virus was then removed and cells incubated at 37°C for 48 h. Infected cells were detected by immunofluorescence using anticore antibody. (B) Replication of SGR-JFH1 in Huh7-25 cells and Huh7-25 cells in which CD81 was stably expressed (Huh7-25-CD81 cells).
FIG. 7.

Dependency of permissiveness on virus concentration in Huh7-25-CD81 cells. Huh7-25-CD81 cells (1 × 104) were infected with JFH-1 virus at 10 to 160 ffu/well in 96-well plates. Infected cells were visualized and assayed as described in the legend for Fig. 1. All experiments were performed in triplicate, and the data are shown as means ± standard deviations. The upper-left panel shows a magnified version of the area in the square at the lower left.
Knockdown of CD81 expression reduced HCV infection permissiveness.
A permissive and CD81-expressing Huh7 cell clone (Huh7-54) was transfected with siRNA for CD81. At 48 h after transfection, the expression of CD81 on the cell surface declined by about 60% compared to that for the cell transfected with siIRR (Fig. 8A). The CD81 knockdown cell was then infected with JFH-1 virus, and infectivity was determined. Infectivity of the CD81 knockdown cell declined by about 80% compared to that for the siIRR-transfected cell or mock-transfected cells (Fig. 8B). Thus, CD81 expression was shown to be an important factor in HCV infection permissiveness. Taken together, the level of CD81 expression is also a determinant of the level of HCV permissiveness in a cell clone.
FIG. 8.
siRNA silencing of CD81 expression on Huh7-54 cells and JFH-1 virus infection. (A) CD81-positive Huh7-54 cells transfected with either siIRR or siRNA of CD81 (siCD81) were stained with anti-CD81 antibody (JS-81) at 48 h posttransfection and analyzed by flow cytometry. (B) siRNA-transfected Huh-54 cells were inoculated with the same titer of JFH-1 virus (1 × 106 copies HCV RNA) for 3 h. Virus was then removed, and cells were incubated at 37°C for 48 h. Infected cells were visualized and assayed as described in the legend for Fig. 1. All experiments were performed in triplicate, and the data are shown as means ± standard deviations.
CD81 expression levels followed by HCV infection.
CD81 expression on the cell surface may alter with culture conditions or HCV infections. To test this, we produced infectious HCV particles harboring the EGFP gene. J6/JFH1 EGFP virus was derived from pFGR-J6/N2X-JFH1/EGFP, and JFH-1 EGFP virus was from pFGR-JFH1/EGFP. J6/JFH1 EGFP virus was produced more efficiently from synthetic RNA-transfected cells than wild-type JFH-1 EGFP virus (data not shown). We thus used J6/JFH1 EGFP virus to inoculate Huh7-25-CD81 and Huh7-70 cells. Cell surface CD81 expression and HCV infection were detected simultaneously by flow cytometry. In Fig. 9A, results of the fluorescence-activated cell sorting analysis are given as quadrant plots of Huh7-25-CD81 cells inoculated with J6/JFH1 EGFP virus at 96 h after inoculation. The infected cells were observed by a shift to positive GFP fluorescence, and CD81 expression was detected by an anti-CD81 antibody. Under this experimental condition, HCV infections were detected in both cell types at 24 h after inoculation (Fig. 9B) (0.5% and 0.6% for Huh7-25-CD81 and Huh7-70 cells, respectively). The ratio of infected cells increased substantially in Huh7-25-CD81 cells (Fig. 9B) (27.7% at 96 h postinoculation); however, the ratio of infected cells was not increased in Huh7-70 cells (Fig. 9B) (0.9% at 96 h). On the other hand, the CD81 expression level of Huh7 cells increased until 2 days after cell passages and then declined gradually (data not shown). This kinetic property was also confirmed with mock-infected Huh7-25-CD81 and Huh7-70 cells (Fig. 9C) (incubation was started 24 h after passage). After J6/JFH1 EGFP virus inoculations, CD81 expression levels on the cell surfaces of uninfected and infected cells were also analyzed (Fig. 9C). CD81 expression levels of uninfected cells were not different from those of mock-infected cells. Interestingly, CD81 expression levels of infected Huh7-25-CD81 cells were higher than those of uninfected cells at 48 and 72 h after virus infection (Fig. 9C, left panel). However, CD81 expression of infected cells also decreased gradually, as observed for uninfected cells. The CD81 expression level of Huh7-70 cells was lower than that of Huh7-25-CD81 cells, and the difference in CD81 expression level between uninfected and infected cells was not clear for Huh7-70 cells (Fig. 9C, right panel).
FIG. 9.
Kinetics of cell surface CD81 expression and HCV infection. Huh7-25-CD81 and Huh7-70 cells were seeded into six-well plates at a density of 1 × 105 cells/well. Twenty-four hours later, J6/JFH1 EGFP virus (multiplicity of infection, 2) was inoculated. Cells were harvested at 24, 48, 72, and 96 h after inoculation and analyzed for infection and cell surface CD81 expression by FACSCalibur. The experiments were performed as described in the legend for Fig. 4, using anti (α)-mouse IgG-allophycocyanin (APC) as a secondary antibody. (A) Huh7-25-CD81 cells were harvested 96 h after inoculation and analyzed by fluorescence-activated cell sorting. Relative numbers of cells in the respective quadrants are given. (B) The proportion of infected cells was determined at each time point postinoculation and plotted for Huh7-25-CD81 and Huh7-70 cells. Mean values from triplicate experiments are given. (C) The cell surface CD81 expression (MFI) of infected cells, uninfected cells, and mock-infected cells is plotted at each time point after inoculation for Huh7-25-CD81 and Huh7-70 cells. Mean values and standard deviations are given.
DISCUSSION
CD81 is a candidate receptor for HCV and plays an important, as yet unknown role in HCV infection. Pileri et al. were the first to demonstrate a relationship between CD81 and HCV when they found that the envelope protein E2 of HCV bound to the large extracellular loop of CD81 (30). However, experiments that use recombinant proteins are limited in that they can only provide information regarding molecular interactions. The development of pseudoparticles with HCV envelope proteins (HCVpp) made possible the investigation of HCV infection and cell entry, and some candidate receptors for HCV have been proposed previously (2, 3, 12, 25, 30, 31). The involvement of CD81 in HCV entry was also ascertained by the HCVpp system. The HCVpp system has shed some light on the role of cell surface molecules involved in the early steps of the HCV life cycle. On the other hand, a cell culture system to produce HCV bearing the genotype 2a genome has recently been established (22, 34, 41). This system enables investigation of the whole HCV life cycle.
The Huh7 cell line is a human hepatoma cell line, established in 1982 (29), that is now recognized as being permissive for HCV particles (34). Since our Huh7 cell showed limited permissiveness, in this study we performed single-cell cloning of a Huh7 cell and found that the parental Huh7 cell produced a heterogeneous population upon culture (Table 1). The heterogeneity of subsequent infectivity may have been due to the limited permissiveness of the parental Huh7 cells. Further analysis of these Huh7 cell clones revealed that CD81 expression determined permissiveness with high correlation (Fig. 4; Table 1). Moreover, given that HCV replication efficiency was not changed by CD81 expression (Fig. 6B), this molecule must be important in the early steps of HCV infection. However, the level of CD81 expression on the Huh7 cell clones did not necessarily correlate with HCVpp and JFH-1 virus infectivity, with some clones displaying high permissiveness but relatively low CD81 expression and vice versa (Table 1). It is thus likely that replication efficiency is related to the appearance of infected foci, since the translation of HCV core protein is affected by HCV RNA replication. For example, Huh7 clone 1 indicated relatively low permissiveness for HCVpp but high permissiveness for JFH-1 virus, while clone 28 indicated the opposite. These differences may have arisen from the postinfection steps of virus infection, as the JFH-1 system depends on HCV infection and replication and the HCVpp system depends on HCV infection and pseudotype gene expression. Nonetheless, multiple factors are predicted to play a role in HCV infection, in addition to CD81.
When the siRNA for CD81 was transfected into a CD81-positive cell clone and expression subsequently down-regulated, the permissiveness for HCVpp (38) and JFH-1 virus was also down-regulated (Fig. 8). On the other hand, when a Huh7 cell line was transfected with serial doses of CD81 expression vector, the transfected cells indicated permissiveness according to the level of CD81 expression (40). Although the MFIs differed between transiently and stably transfected cells, more clonal CD81 expression was observed for stably transfected cells than for transiently transfected cells (Fig. 6A). This may account for the difference in infectivity between transiently and stably transfected cells.
SR-BI and the LDL receptor are other putative molecules thought to be involved in HCV infection (1, 31), and their relationships to HCV have been investigated using recombinant proteins and HCVpp (5, 6, 15). The expression levels of SR-BI and the LDL receptor on our Huh7 cell clones differed slightly among the clones (Fig. 4); however, their expression did not appear to determine HCV permissiveness, unlike with CD81. On the other hand, it was recently reported that CD81 and SR-BI function cooperatively and cholesterol dependently to initiate HCV entry (16). In the present study, JFH-1 virus infection levels varied among the Huh7 cell clones, and thus each molecule may have a threshold expression level that determines HCV permissiveness.
CD81 expression level on the cell surface may be changed with cell culture condition and HCV infection. Therefore, it is important to analyze a dynamic expression of CD81. In fact, higher CD81 expression was observed for infected Huh7-25-CD81 cells than for uninfected cells (Fig. 9C, left panel). However, this difference was not clear with Huh7-70 cells (Fig. 9C, right panel). This discrepancy may be due to the different CD81 expression levels between the cells. Huh7-25-CD81 cells express higher levels of CD81 on the cell surface (Fig. 6 and 9; Table 1), and when such cells are infected with HCV, CD81 molecules might be stabilized to keep higher expression levels on the cell surface. Alternatively, CD81 expression in Huh7-25-CD81 cells may be controlled differently than in other Huh7 cell clones because CD81 is expressed from the transfected vector in Huh7-25-CD81 cells. Taken together, a more detailed analysis will be necessary for a dynamic expression of CD81 and HCV infection.
CD81 is a member of the tetraspanin family, and its functions are unclear. It is known that CD81 is a component of the tetraspanin web on the plasma membrane (21) and that the homologous region shared with CD9 is involved in egg-sperm fusion (42). Thus, CD81 may play an important role in cell-virus fusion through the tetraspanin web. Having said that, it is unclear what kinds of molecules associate with CD81 on human hepatic cells. Since CD81 and other tetraspanins are thought to interact with various molecules (13), including integrins (36), GPR56 (24), 14-3-3 (8), and signaling enzymes (7, 37, 39), various signal transductions through CD81 and multiprotein complexes may be involved in the level of HCV permissiveness. Thus, the tetraspanin-enriched microdomain on permissive cell lines may be necessary for virus-host interaction (14, 26).
On the other hand, the level of CD81 expression on the Huh7 cell clones did not correlate with the level of permissiveness, indicating that CD81-independent molecules were also involved in permissiveness. Recently, our laboratory and others have indicated that heparan sulfate proteoglycan (HSPG) may play an important role in the initial cell surface binding of HCV particles (19, 28). Since HCV particles are thought to be concentrated by HSPG on the surfaces of cells, the differences in infectivity among Huh7 cell clones may be due to differences in the expression levels or types of HSPG. Furthermore, other unknown molecules that harbor affinity with HCV particles may also be important. A more detailed analysis is clearly required.
In conclusion, we investigated HCV permissiveness and host factors by use of cell-cultured infectious particles and a heterogeneous population of Huh7 cells derived from a single cell. We discovered that HCV particle permissiveness is determined by CD81 expression with high correlation. However, the level of permissiveness of each Huh7 cell clone is not explained by only CD81 expression levels, suggesting that another host factor(s) is involved.
Acknowledgments
This work was partially supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science and from the Ministry of Health, Labor, and Welfare of Japan; by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO); and by Research on Health Sciences Focusing on Drug Innovation from the Japan Health Sciences Foundation.
Huh7.5.1 cells were a kind gift from Francis V. Chisari. pJ6CF plasmid DNA was a kind gift from Jens Bukh. pcDNA3.1-CD81 plasmid DNA and pCAG-VSVG plasmid DNA were kind gifts from Yoshiharu Matsuura. pcDNAdeltaC-E1-E2(JFH1) was a kind gift from Thomas Pietschmann.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Agnello, V., G. Ábel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, H., R. Cerino, M. Arcuri, M. Hoffmann, P. Schürmann, M. I. Adah, B. Gissler, X. Zhao, V. Ghisetti, B. Lavezzo, H. E. Blum, F. von Weizsäcker, A. Vitelli, E. Scarselli, and T. F. Baumert. 2005. Scavenger receptor class B type I and hepatitis C virus infection of primary Tupaia hepatocytes. J. Virol. 79:5774-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth, H., C. Schäfer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 4.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudoparticles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 7.Berditchevski, F., K. F. Tolias, K. Wong, C. L. Carpenter, and M. E. Hemler. 1997. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272:2595-2598. [DOI] [PubMed] [Google Scholar]
- 8.Clark, K. L., A. Oelke, M. E. Johnson, K. D. Eilert, P. C. Simpson, and S. C. Todd. 2004. CD81 associates with 14-3-3 in a redox-regulated palmitoylation-dependent manner. J. Biol. Chem. 279:19401-19406. [DOI] [PubMed] [Google Scholar]
- 9.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Date, T., M. Miyamoto, T. Kato, K. Morikawa, A. Murayama, D. Akazawa, J. Tanabe, S. Sone, M. Mizokami, and T. Wakita. An infectious and selectable full-length replicon system with hepatitis C virus JFH-1 strain. Hepatol. Res., in press. [DOI] [PubMed]
- 11.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 12.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemler, M. E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6:801-811. [DOI] [PubMed] [Google Scholar]
- 14.Hemler, M. E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397-422. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapadia, S. B., H. Barth, T. Baumert, J. A. McKeating, and F. V. Chisari. 2007. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 81:374-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, T., T. Date, M. Miyamoto, M. Sugiyama, Y. Tanaka, E. Orito, T. Ohno, K. Sugihara, I. Hasegawa, K. Fujiwara, K. Ito, A. Ozasa, M. Mizokami, and T. Wakita. 2005. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 43:5679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 19.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 21.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wölk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 24.Little, K. D., M. E. Hemler, and C. S. Stipp. 2004. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-G_q/11 association. Mol. Biol. Cell 15:2375-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houlès, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 26.Martin, F., D. M. Roth, D. A. Jans, C. W. Pouton, L. J. Partridge, P. N. Monk, and G. W. Moseley. 2005. Tetraspanins in viral infections: a fundamental role in viral biology? J. Virol. 79:10839-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuura, Y., H. Tani, K. Suzuki, T. Kimura-Someya, R. Suzuki, H. Aizaki, K. Ishii, K. Moriishi, C. S. Robison, M. A. Whitt, and T. Miyamura. 2001. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology 286:263-275. [DOI] [PubMed] [Google Scholar]
- 28.Morikawa, K., Z. Zhao, T. Date, M. Miyamoto, A. Murayama, D. Akazawa, J. Tanabe, S. Sone, and T. Wakita. Infectious hepatitis C virus particle binding to the Huh7 cell surface is mediated by glycosaminoglycans and its internalization by CD81. J. Med. Virol., in press.
- 29.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 30.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 31.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 33.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 34.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 36.Yang, X., O. V. Kovalenko, W. Tang, C. Claas, C. S. Stipp, and M. E. Hemler. 2004. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J. Cell Biol. 167:1231-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yauch, R. L., F. Berditchevski, M. B. Harler, J. Reichner, and M. E. Hemler. 1998. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell 9:2751-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X. A., A. L. Bontrager, and M. E. Hemler. 2001. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta(1) integrins. J. Biol. Chem. 276:25005-25013. [DOI] [PubMed] [Google Scholar]
- 40.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, G. Z., B. J. Miller, C. Boucheix, E. Rubinstein, C. C. Liu, R. O. Hynes, D. G. Myles, and P. Primakoff. 2002. Residues SFQ (173-175) in the large extracellular loop of CD9 are required for gamete fusion. Development 129:1995-2002. [DOI] [PubMed] [Google Scholar]