Abstract
Genetic screens have been performed to identify mutants with altered auxin homeostasis in Arabidopsis. A tagged allele of the auxin-overproducing mutant sur2 was identified within a transposon mutagenized population. The SUR2 gene was cloned and shown to encode the CYP83B1 protein, which belongs to the large family of the P450-dependent monooxygenases. SUR2 expression is up-regulated in sur1 mutants and induced by exogenous auxin in the wild type. Analysis of indole-3-acetic acid (IAA) synthesis and metabolism in sur2 plants indicates that the mutation causes a conditional increase in the pool size of IAA through up-regulation of IAA synthesis.
Keywords: metabolism
Indole-3-acetic acid (IAA) represents one of the most important plant hormones, regulating many aspects of plant growth and development, from seed germination to fruit ripening (1). Auxin interacts with other endogenous regulators and external environmental factors such as light or temperature. Strict control of the endogenous IAA concentration is therefore of great importance. The balance between auxin biosynthesis and metabolism, including conjugation, deconjugation, and catabolism, determines the level of the hormone (2, 3). The molecular mechanisms involved in the control of IAA homeostasis are far from fully elucidated, but the recent development of genetic and molecular approaches opens up possibilities for addressing this problem.
In the last decade, the model plant Arabidopsis thaliana has been used to identify several mutants altered in auxin signal transduction, transport, and conjugation (reviewed in ref. 4). Although no auxin auxotrophic mutant has been isolated so far, it has been possible to isolate mutants of Arabidopsis such as superroot1 (5) and superroot2 (6) that overproduce auxin. The phenotype of these mutants can be mimicked by growth of wild-type seedlings on medium containing auxin. sur1 and sur2 seedlings have an elongated hypocotyl, epinastic cotyledons, an increased number of lateral roots, and many adventitious roots originating from the hypocotyl. Both mutants contain an increased amount of free IAA. rooty, a tagged allele of sur1, was identified (7). The ROOTY gene encodes a protein similar to a tyrosine aminotransferase, the role of which in the control of auxin homeostasis has yet to be elucidated.
A tagged allele of sur2 was identified in Arabidopsis lines carrying independent En-1 elements (8, 9). The SUR2 gene was cloned and shown to encode the CYP83B1 protein, which belongs to the large family of plant cytochrome P450-dependent monooxygenases (10). Cytochrome P450 (cyt P450) enzymes are involved in a wide range of secondary metabolic pathways, in the biosynthesis of macromolecules such as cuticule and lignin, and in the biosynthesis of plant growth regulators such as jasmonic acid, gibberellins (reviewed in ref. 11), and brassinosteroids (12–14). Two cyt P450 genes have recently been identified in Arabidopsis that encode proteins catalyzing reactions that potentially can be linked to the indole-acetonitrile-dependent IAA biosynthesis (15). In this article we show that a mutation in the gene encoding the CYP83B1 protein conditionally induces auxin overproduction, suggesting that this cyt P450 is involved in the modulation of IAA biosynthesis in Arabidopsis.
Materials and Methods
Adventitious Root Screen.
Seeds were surface sterilized, sown on MS agar plates [4.3 g/liter MS salts (Sigma)/1% sucrose (pH 6.0) with 1 M KOH/1% agar] and left at 4°C for 48 h. The plates were then placed vertically for 1 day in the light, 3 days in the dark, and an additional 8 days in the light before scoring for adventitious root formation.
Plant Culture for Studying Gene Expression.
Wild-type and mutant seedlings were grown in vitro (as described in ref. 16). For auxin response analysis, seedlings were grown on media supplied with increasing concentrations of IAA or 2,4 dichlorophenoxyacetic acid and harvested 12 days after germination. For short-term responses, 12-day-old wild-type seedlings were transferred to liquid medium with or without 5 μM IAA. Seedlings were then harvested after different incubation times.
DNA Preparation, Amplification, and Sequencing.
Genomic DNA was prepared by using the cetyltrimethylammonium bromide method (17).
Based on the expressed sequence tag (D78598) sequence, specific primers were designed for gene amplification on genomic DNA. Obtained fragments were purified on 1% agarose gel, cloned, or directly sequenced (Genome Express, Montreuil, France). Sequences were obtained from at least three independent amplifications for both alleles and wild type. The data were analyzed by using computer software DNA Strider for Macintosh (18).
Northern Blot Analysis.
Total RNA was prepared as described (19). Twelve micrograms of total RNA was used for Northern blots that were probed with a 32P-labeled SUR2 cDNA fragment. Quantifications were performed with a PhosphoImager BAS2000 image analyzer (Fuji).
Thermal Asymetric Interlaced-PCR (TAIL-PCR).
TAIL-PCR was performed following the published protocol by using the degenerate primers AD1, AD2, and AD3 (20) as well as a further primer, AD4 (NWGSTTMGAACNCGCT). Specific nested primers to the En-1 element were designed.
Digoxigenin-Labeled Probing.
Probes were digoxigenin-labeled by using PCR incorporating 11-digoxigenin-dUTP (Roche Molecular Biochemicals). The probes to the 3′ end of the En-1 element and to the 5′ end of the SUR2 gene were made by using specific primers. Hybridization and detection procedures were carried out as described in the manufacturer's nonradioactive in situ hybridization application manual (Roche Molecular Biochemicals).
Sequencing of Footprint Alleles.
DNA was prepared from 10-day-old mutant F3 seedlings. The region encompassing the En-1 insertion site within the SUR2 gene was PCR-amplified and sequenced on an Applied Biosystems Prism 310 fluorescent DNA analyzer.
Quantitative Analysis of IAA and Indole-3-Acetonitrile (IAN).
Between 10 and 50 mg of tissue was frozen with liquid nitrogen and homogenized in Eppendorf tubes. One milliliter of 50 mM phosphate buffer containing 0.02% diethyldithiocarbamic acid and internal standards (250 pg of [13C6]IAA and 20 ng of [13C1]IAN per sample) were added. Samples were extracted for 6 h at 4°C and centrifuged. Supernatants were acidified with 1 M HCl to pH 2.7 and purified by using Amberlite XAD-7 resin. Absorbed compounds were eluted with dichloromethane, dried under vacuum, and methylated with diazomethane. Subsequently, samples were dissolved in heptane, sialylated with N,o-bis(trimethylsilyl)-trifluoroacetamide/0.1% trimethylchlorosilane, and 1 μl of each was splitless injected into a GC/MS system (21). The mass spectrometer (JEOL JMS SX102/102) was operated in selected reaction monitoring (SRM) mode; for IAA analysis, reactions monitored were m/z 261.118 to m/z 202.105 (endogenous IAA) and m/z 267.137 to m/z 208.125 (internal standard). IAN measurement was based on reactions: m/z 228.108 to m/z 213.085 for endogenous and m/z 229.112 to m/z 214.088 for internal standard IAN.
Quantitative Analysis of IAA Conjugates and Catabolites.
For conjugate analysis, 250–500 mg of plant tissue was used. After homogenization, samples were extracted with the same buffer as for IAA measurements, but additionally containing 30% methanol. The internal standards mixture, composed of D4oxoIAA, D5-N-(indole-3-acetyl)-Ala (D5IAAla), D5-N-(indole-3-acetyl)-Leu (D5IALeu), [13C6]IAAsp, and [13C6]IAGlu, was added at a concentration of 1 ng per sample. Samples were extracted for 24 h at 4°C, centrifuged, and purified on C18 solid-phase extraction columns. Eluate from that column was methylated with diazomethane and passed through Amberlite XAD-7 resin. Compounds absorbed on the resin were eluted with a mixture of acetone and ethyl acetate, dried under vacuum, and analyzed by GC/MS (M.K., unpublished results).
Metabolic Profiles.
Metabolic profiles were obtained as described (22) with the following modifications. Wild-type and sur2 plants were grown in the liquid culture system for 5–6 days, after which time the growth medium was replaced with fresh medium containing 1 μM [14C]indole or 5 μM [1′-14C]IAA (American Radiolabeled Chemicals, St. Louis) or a mixture of 5 μM l-tryptophan (l-Trp) and 5 μM l-D5-Trp. For all experiments the incubation time was 12 h in the dark. Plants were homogenized and extracted with either 50 mM phosphate buffer (pH 7) containing 30% methanol (for IAA metabolites) or 80% methanol (for indole metabolites). Samples purified on Isolute C18 solid phase extraction columns were analyzed by HPLC (Waters, Millipore) with a radioactivity detector (Reeve Analytical Ltd., Glasgow, U.K.) as described (22), except for indole metabolites, for which the methanol in the mobile phase was increased to 20% for the initial conditions. Compounds from D5 labeling experiment were separated on HPLC, by using the same conditions as used for indole metabolites; 5-min (4-ml) fractions were collected and analyzed by GC/MS.
Results
Identification of an Adventitious Rooting Mutant.
A population of 3,000 lines of Arabidopsis (Col0) carrying approximately 15,000 independent En-1 elements (8, 9) were screened for mutants that produced increased numbers of adventitious roots originating from the hypocotyl. For each independent line, 25 seedlings were screened, resulting in the identification of four seedlings from line I-33, which formed an abnormally high number of adventitious roots and which also had epinastic cotyledons. The mutant I-33 was crossed to the previously described adventitious root mutants sur1 and sur2 and thereby shown to be allelic to sur2.
Identification and Isolation of the SUR2 Gene.
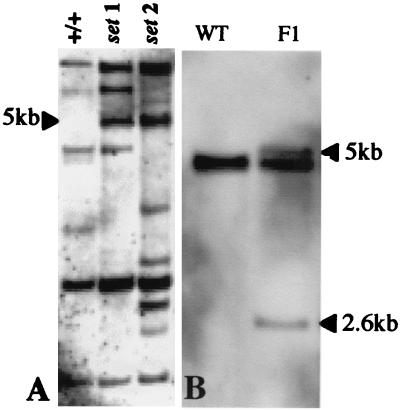
Southern hybridization revealed up to 10 En-1 elements to be present within the I-33 line (data not shown). A F2 population segregating for the sur2 mutant phenotype was generated by outcrossing line I-33 with Col0. F2 plants showing a wild-type phenotype were genotyped in the F3 generation. DNA prepared from 30 F2 plants and probed with the 3′ En probe failed to reveal a single En-1 element that cosegregated with the mutant and hemizygous F2 plants. The En-1 transposable element used in generating the AMAZE population is autonomous, leading to the possibility that the En-1 element had been excised, leaving a footprint mutation. To circumvent the difficulties caused by this event, the analysis was performed by using pools of hemizygous or wild-type F2 plants. DNA was prepared from pools of five +/+ F2 lines and pools of five sur2/+ F2 lines, restricted with EcoRV, then Southern hybridized with a 3′ En-1 probe. A 5-kb band was present in the sur2/+ samples and absent from the +/+ pool, providing a candidate for the En-1 element causing the sur2 phenotype (Fig. 1A).
Figure 1.
(A) A single En-1 element cosegregates with pooled sur2/+ F2 plants and is absent from +/+ F2s. The DNA from five pooled +/+ F2 lines and from two sets of five pooled F2 sur2/+ lines (sets 1 and 2) was Southern blotted and probed with the 3′ end of the En-1 element. A single hybridizing band of 5 kb is indicated with an arrow; this is present in both sur2/+ pooled samples but is absent from the +/+ sample. (B) An En-1 insertion disrupts the cyt P450 gene CYP83B1. DNA was Southern blotted and probed with the 5′ end of CYP83B1. Arrows indicate the bands migrating at 5.0 kb and 2.6 kb, which result from disruption of the cyt P450 gene.
Isolation of Flanking DNA by TAIL-PCR.
To determine whether the 5-kb band represented the insertion causing the mutation, DNA that had been restricted with EcoRV was size selected between 4.5 and 5.5 kb and then subjected to TAIL-PCR (20), by using degenerate primer AD1, AD2, AD3, or AD4 together with three nested En-1 specific primers. The AD4 reaction amplified a product of approximately 600 bp in the tertiary PCR that was absent in the Col0 control (data not shown). DNA sequence analysis demonstrated almost complete homology to the 5′ end of a cDNA encoding the cytochrome P450 CYP83B1 (10).
DNA prepared from +/sur2 and +/+ plants was probed with the region of the cytochrome P450 gene spanning the putative insertion position to verify that the CYP83B1 gene identified by homology to the TAIL-PCR product contains an En-1 insertion. The +/+ sample produced a single band, whereas the +/sur2 produced three bands (Fig. 1B), one corresponding to the wild-type gene copy and the other two resulting from the En-1 insertion disrupting the gene.
Cosegregation of the Mutant Phenotype with an Insertion in the CYP83B1 Gene.
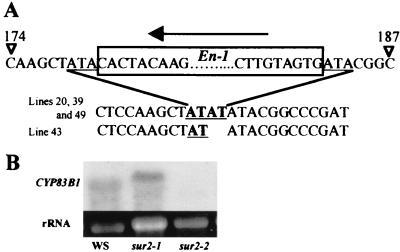
The putative En-1 insertion within the CYP83B1 gene causing the sur2 mutant phenotype was verified by examining the cosegregation of the insert with the mutant phenotype. A PCR diagnostic was established by using En-1 and CYP83B1 specific primers. All 11 F2 mutants produced a diagnostic PCR product, demonstrating the presence of the En-1 element within the CYP83B1 gene (data not shown). The En-1 element was inserted 180 bp 3′ to the putative start codon and had resulted in a characteristic A/T-rich target site duplication (Fig. 2A) (23, 24). Of the 39 phenotypically wild-type F2 plants, 19 produced the diagnostic PCR product and were all genotyped as sur2/+ in the F3 generation (data not shown). Of the 20 F2 plants that failed to produce a diagnostic PCR product, 16 were shown to be +/+, but the remaining four were demonstrated to be sur2/+ and thus did not show cosegregation of the En-1 element with the mutant phenotype. The region of the SUR2 gene containing the insertion site of the En-1 element was amplified and sequenced for the four F2 lines that did not demonstrate cosegregation. Three of the sequences showed an identical 4-base insertion (TATA) at the site of the En-1 insert, whereas the fourth showed a 2-base insertion (TA), also at the En-1 insertion site (Fig. 2A).
Figure 2.
(A) Footprint mutations in the four lines that did not show the cosegregation between the sur2 phenotype and the En-1 insertion. The 3-base target site duplication (ATA) is underlined. Bases in bold and underlined represent the insertion footprints. An arrow shows the 5′ to 3′ orientation of the En-1 element. (B) Expression of SUR2 in sur2–1 and sur2–2 alleles. A Northern blot with total RNA from wild-type (WS) and sur2–1 and sur2–2 homozygous seedlings was hybridized with the full-length cDNA as a probe.
Sequencing and CYP83B1 Expression Analysis in the sur2–1 and sur2–2 Alleles.
Analysis of CYP83B1 expression in wild-type and sur2–1 and sur2–2 mutants was performed on total RNA, by using the full-length cDNA as a probe. A transcript of 1.6 kb was detected in the wild type. In the sur2–1 allele a transcript with a higher molecular weight could be detected, but no transcript was detectable in sur2–2 mutants, even after a long exposure time (Fig. 2C). Genomic DNA was sequenced for both alleles and wild type. sur2–1 shows a 61-bp insertion (data not shown), 441 bp downstream of the transcription initiation codon ATG, leading to the formation of a stop codon, 142 amino acids after the first Met. This insertion corresponds to a duplication of part of the CYP83B1 gene itself and to an unknown sequence that may be due to a DNA repair mechanism associated with the failure of the insertion of the T-DNA. The sur2–2 allele does not show any mutation within the ORF region of the gene sequence or of the 180 bp upstream of the ATG. Failure to amplify the 850 bp upstream of the ATG by PCR suggested that a rearrangement probably had occurred in this region. Collectively, these results confirm that SUR2 encodes CYP83B1.
SUR2 Gene Expression Is Induced by Auxin.
SUR2 (as CYP83B1) has already been shown to be expressed in all organs and preferentially in the roots (10). It was demonstrated that wounding enhanced the expression of the gene (10).
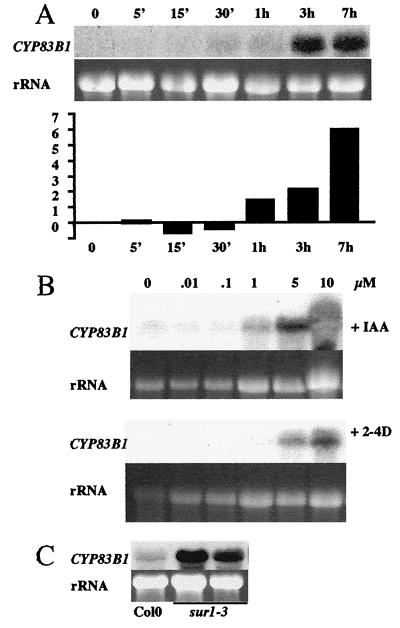
Sequence analysis of the promoter region identified an auxin-responsive element, TGTCTC. Auxin-responsive elements are found in promoters of primary/early auxin-response genes and specifically interact with auxin-responsive factors that mediate auxin response (25). We have examined the expression of SUR2 after short-term IAA induction (Fig. 3A). Twelve days after germination wild-type seedlings were transferred to liquid medium without or with auxin. In the control experiment without auxin, SUR2 was induced after the seedlings had been incubated for 30 min in liquid medium, which probably reflects a stress response (data not shown). When 5 μM IAA was added to the medium, a clear induction could be observed after 1 h. The expression was induced 2-fold after 1 h and could reach 5–6 times the wild-type level after 7 h in the presence of IAA (Fig. 3A).
Figure 3.
SUR2 is induced by auxin. Northern blots with total RNA from 10-day-old wild-type or mutant seedlings were hybridized with the full-length cDNA as a probe. (A) RNAs from wild-type seedlings, after transfer to liquid medium containing 5 μM IAA for 0–7 h. In the histogram, the level of expression of the SUR2 gene in the presence of auxin was normalized against the level of expression of the gene in absence of auxin. (B) RNAs from wild-type seedlings grown in media containing a range of concentration of IAA or 2,4 dichlorophenoxyacetic acid between 0 and 10 μM. (C) RNAs from wild-type Col0 seedlings and two independent batches of homozygous sur1–3 seedlings.
When seedlings were grown for 12 days on media containing increasing concentrations of IAA or 2,4 dichlorophenoxyacetic acid, a clear induction of the SUR2 gene expression could be observed. SUR2 was induced 2-fold by 1 μM IAA (Fig. 3B). The level of expression was between 6- and 9-fold higher when seedlings were grown in the presence of 5 μM IAA or 2,4 dichlorophenoxyacetic acid, respectively. Five micromolar piclorame, which can phenocopy sur2 phenotype in wild type (6), also strongly induced SUR2 expression in wild type (data not shown). Auxin could not induce SUR2 in the auxin-resistant mutant axr1 (data not shown), but the gene was strongly expressed in auxin-overproducing sur1–3 mutants (Fig. 3C). The cytokinin benzylaminopurine or the ethylene precursor 1-aminocyclopropane-1-carboxylique acid had no effect on the expression of SUR2 (data not shown).
sur2 Mutants Show Increased Endogenous Levels of IAA Throughout the Seedling.
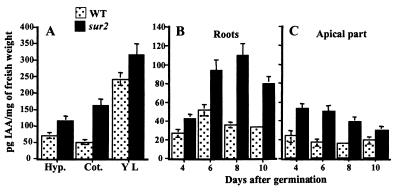
It was previously shown that sur2 mutants contained higher levels of free IAA (6). The endogenous content of free IAA is higher compared with wild type in all organs analyzed, demonstrating that the mutation causes up-regulation of the IAA pool in organs not normally believed to synthesize significant amounts of IAA (Fig. 4 A and B). Previously it has been shown that the sur2 phenotype is transient because it reverts to a wild-type phenotype 12–15 days after germination (6). This transient phenotype correlates with a decrease in the IAA content observed in the aerial part of the seedling and in the leaves when they start to expand (Fig. 4C). A similar trend is observed in the roots from days 8 to 10 (Fig. 4B). This similarity could be a result of the seedlings gaining the capacity, after the initial stages of growth, to modulate the endogenous auxin level through induction of additional homeostatic mechanisms, as observed in other plants during early seedling growth (K.L. et al., unpublished data).
Figure 4.
Endogenous content of total IAA in different organs of the seedlings. Free IAA was analyzed by mass spectrometry (SRM) in different apical organs (A). (B and C) In the root system (B) and in the apical part (C) of both wild-type and mutant seedlings. Quantification was performed in three independent experiments. Hyp., hypocotyl; Cot., cotyledons; Y.L., young leaves.
The sur2 Phenotype Can Be Restored to a Wild-Type Phenotype by Exogenous IAA or Low pH Medium.
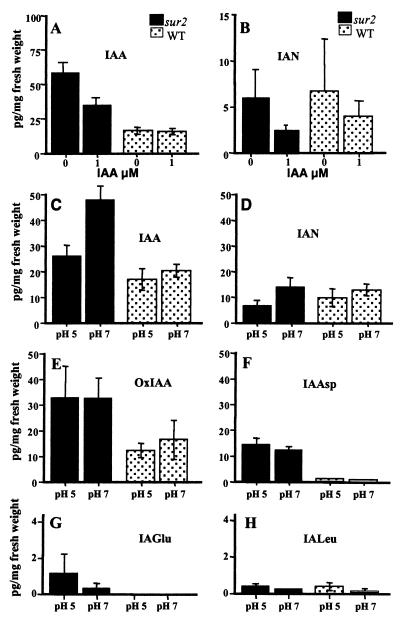
In vitro, the phenotype was heterogeneous, with most of the seedlings blocked in development at the cotyledon stage, whereas a minority developed like the wild type (6). In an attempt to understand this heterogeneity, the mutants were subjected to a range of climatic conditions and media compositions. Two conditions were able to rapidly restore the wild-type phenotype. Surprisingly, when sur2 seedlings were grown in the presence of 1 μM IAA the majority of the seedlings were able to develop, in contrast to those grown in the absence of auxin (Fig. 5 A and B). Similarly, the sur2 mutants showed nearly normal cotyledons as well as expanded leaves when grown on a pH 5 medium (Fig. 5C). In contrast, the sur2 phenotype was even stronger with increasing pH, with seedlings blocked at the cotyledon stage at pH 7 (Fig. 5D). Both exogenous IAA and low pH were able to significantly decrease the endogenous level of IAA in the mutants (Fig. 6 A and C) but had no significant effect on wild type. Because IAN has been postulated to be an auxin precursor in Arabidopsis (reviewed in ref. 26), we also measured the endogenous level of IAN in the wild-type and sur2 seedlings treated under the different pH and IAA conditions. Interestingly, we observed that both low pH and 1 μM IAA significantly decrease endogenous IAN levels in the mutant (Fig. 6 B and D).
Figure 5.
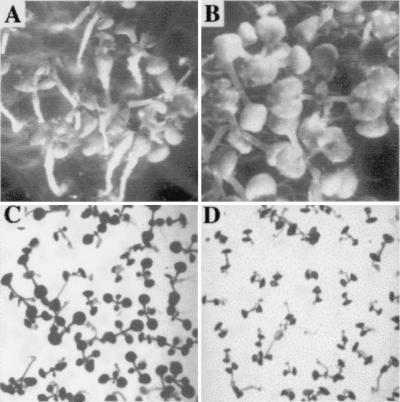
Homozygous sur2 seedlings, 3 weeks after germination, grown on media without auxin (A) or containing 1 μM IAA (B). Homozygous sur2 seedlings, 12 days after germination, grown on medium at pH 5 (C) or pH 7 (D).
Figure 6.
Quantification of metabolites. Endogenous IAA (A) and IAN (B) contents in wild-type and sur2 seedlings, 3 weeks after germination, on a medium without auxin or containing 1 μM IAA. IAA (C), IAN (D), oxindole-3-acetic acid (E), IAAsp (F), IAGlu (G), and IAleu (H) were analyzed by mass spectrometry (SRM) in the wild-type and mutant seedlings grown for 12 days on medium at pH 5 or pH 7. Quantification was performed in three independent experiments.
Metabolic Profile of Exogenous IAA: Quantitative Analysis of the IAA Conjugates and Catabolites.
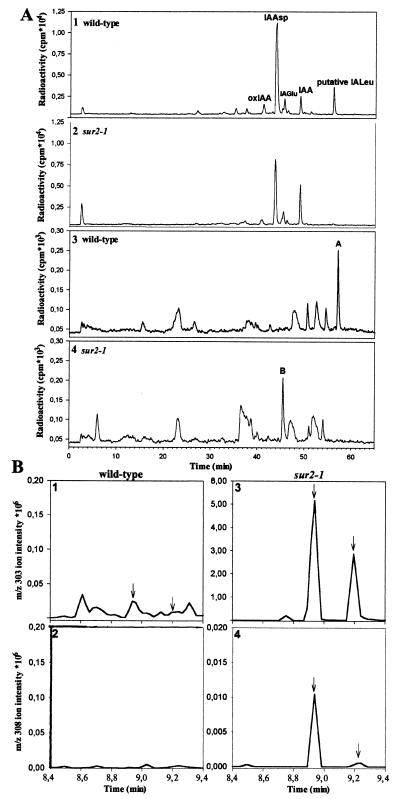
Exogenous IAA metabolism is shown in Fig. 7A (1 and 2). The sur2 profile is very similar to that of wild type. Furthermore, sur2 plants can synthesize at least a limited set of the IAA conjugates. The peaks at retention time (RT) 43.8 and 45.6 min representing IAAsp and IAGlu amide-type conjugates were previously identified in Arabidopsis (22). A low abundance peak observed in both profiles at RT 37.5 min is the oxindole-3-acetic acid sugar conjugate, followed by free 2-oxoIAA at RT 40.9 min, and the peak at 49.1 min represents unmetabolized IAA. The peak at RT 56.1 min in the wild-type profile is missing in the sur2 plants. GC/MS analysis of extracts prepared from larger amounts of wild-type material allowed us to identify this compound as N-(indole-3-acetyl)-leucine (IALeu) (M.K., unpublished results). Assays of exogenous IAA metabolism in liquid cultures are prone to artifacts and are difficult to control quantitatively (22). To overcome this problem, we used a GC/MS-based method to directly quantify levels of the endogenous IAA conjugates and catabolites in sur2 and wild type. IALeu (Fig. 6H) and IAAla (data not shown) were indeed detectable in sur2 at a level similar to those in the wild type. IALeu, together with IAAla and IAPhe, is believed to be hydrolyzable and likely to play an important role in IAA homeostasis. Apparently sur2 can synthesize these conjugates; however, once IAA starts to accumulate, this ability is probably lost and the metabolism switched to form catabolites rather then hydrolyzable conjugates to cope with the increasing levels of the free hormone. Therefore, 6 days after germination, wild-type plants still produce IALeu (Fig. 7A1) from exogenous IAA, whereas sur2 has lost this capacity (Fig. 7A2). The concentration of the endogenous 2-oxindole-3-acetic acid, a major catabolite in Arabidopsis, is 2-fold higher in sur2 compared with wild type (Fig. 6E). Endogenous levels of IAAsp and IAGlu are also increased by at least 12- to 15-fold in the sur2 mutant (Fig. 6 F and G). Both of these conjugates should be considered as catabolites rather then storage forms of IAA (22, 27). These results are in agreement with the IAA-overproducing phenotype, causing accumulation of the auxin catabolites and conjugates (28, 29). We observe that the pH of the medium has no effect on IAA conjugation or metabolism.
Figure 7.
Metabolic profiles. (A) Metabolic profiles of [1′-14C]IAA in the wild-type (1) and sur2 (2) seedlings and of [14C]indole in the wild-type (3) and sur2 (4) seedlings. (B) SRM analysis of the D5-labeling products. Endogenous IAAld peaks (arrows) in the wild type (1) and sur2 (3) correspond to D5-labeled IAAld peaks (arrows) (2 and 4).
Metabolic Profiles of Exogenous Indole.
Metabolic profiles for exogenous [14C]indole are presented in Fig. 7A 3 and 4. Exogenous indole is rapidly incorporated into tryptophan, so the observed labeled products can originate from either indole or tryptophan. Indole metabolism in sur2 plants (Fig. 7A4) differs significantly from wild type (Fig. 7A3) with unidentified metabolite A (RT 57.2 min) totally missing in the mutant. Conversely, the peak at RT 45.5 min (Fig. 7B) appears as a major product of the sur2 indole metabolism. It has RT properties expected for indole-3-acetaldehyde (IAAld) and indole-3-acetaldoxime (IAOx). Full-scan GC/MS and GC-SRM-MS analysis of the corresponding fractions from in vivo l-D5-Trp labeling experiments showed the presence of IAAld (Fig. 7B). In the mutant, characteristic double peaks of cis and trans trimethylsilyl derivatives of IAAld (in enol form) were easily observed, with intensities several times higher than the peaks of heavy labeled IAAld (Fig. 7B 3 and 4). In wild type, only a small endogenous IAAld peak and no peaks from heavy labeled IAAld are observed (Fig. 7B 1 and 2). On the incubation with a mixture of unlabeled and labeled Trp, we expected the isotope ratio to be 1:1, which was not the case. Therefore unlabeled IAAld must have accumulated in sur2 plants before the experiment. It can be concluded that the sur2 mutation leads to a change in the tryptophan metabolism, causing accumulation of IAAld. Nevertheless, the IAA peak (RT 49.1 min) was not present in the indole profile (Fig. 7A4), and direct measurements of the IAA pool showed a 2- to 3-fold increase in sur2 (Figs. 4 and 6). In the labeling experiments only minute amounts of labeling were going through from IAAld to IAA, indicating that the conversion of IAAld into IAA was very slow. Therefore it is unlikely that the high amount of IAA will be linked to IAAld accumulation.
Discussion
In this article we have described the isolation of the SUR2 gene. The pooled analysis approach adopted in this study has allowed the difficulties linked to cloning genes tagged with autonomous transposable elements to be overcome. This approach may possibly be used to facilitate the cloning of genes in plants containing autonomous transposons.
The SUR2 gene encodes the cyt P450 CYP83B1. Mutations in the SUR2 gene clearly induced an increase in the IAA pool size in all tissues analyzed, including those not normally thought to produce auxin. We analyzed the metabolic pathways thought to control the IAA pool size, namely catabolism, conjugation, and biosynthesis. As expected, on increased endogenous IAA concentrations, formation of oxindole-3-acetic acid and the catabolically related conjugates IAAsp and IAGlu was increased, thereby clearly demonstrating that the sur2 mutation was not affecting this part of the homeostatic control. The IAA conjugation process was also not affected by the mutations as the contents of IALeu and IAAla were not significantly different from that of the wild type.
The SUR2 gene was induced by stress, by in planta overproduction of auxin as in the sur1 mutant, and by exogenous auxin. This result suggests that SUR2 might act as a negative feedback regulator in one of the IAA biosynthesis pathways under different environmental conditions.
The sur2 mutant showed a conditional phenotype that was strongest during early seedling development, reverting to a more wild-type phenotype after 12–15 days. The restoration of sur2 to a wild-type phenotype was induced by growing the seedlings in the presence of exogenous IAA or on a low pH medium. This restoration coincided with a clear reduction in the content of free IAA, which was correlated with a decrease in IAN content. IAN has been described as the IAA precursor in the Trp–IAOx–IAN–IAA pathway in Arabidopsis (reviewed in ref. 26). Our results suggest that a mutation in the SUR2 gene leads to an up-regulation of the Trp–IAOx–IAN–IAA pathway.
To verify this hypothesis we analyzed IAA biosynthesis in a feeding experiment in which conversion of indole and tryptophan, two of the most likely precursors in IAA biosynthesis, was monitored. Interestingly, we did not observe significant modifications in the Trp–IAOx–IAN–IAA pathway as expected, but a dramatic accumulation of IAAld in sur2 instead. IAAld is the precursor of IAA in the Trp–indole-3-pyruvic acid–IAAld–IAA pathway, which may indicate a deregulation of this pathway in the sur2 mutant. However, IAAld has been also postulated to be an intermediate in the Trp–IAOx–IAN–IAA pathway (30, 31). Labeling experiments showed that only minute amounts of label were transferred from IAAld to IAA, indicating that no real up-regulation of the Trp–indole-3-pyruvic acid–IAAld–IAA pathway occurred. The lack of IAN accumulation in the mutant can probably be explained by the rapid interconversion of this intermediate into IAA. Then why do the mutants accumulate IAAld? It is known from animal systems that oximes can be metabolized in two ways: reduction to imine followed by hydrolysis or conversion to aldehyde, or P450-mediated dehydration to a nitrile (32). A similar model can explain the sur2 phenotype. Hull et al. (15) have recently proposed IAOx as an intermediate not only for glucosinolate synthesis, but also as the first intermediate in the IAN-dependent pathway for IAA biosynthesis. A mutation downstream of IAOx will thus potentially lead to an increase in the flux of IAN to IAA. Alternatively, IAOx can be reduced to the corresponding imine, which can then undergo pH-dependent hydrolysis to IAAld. This model is favored by our observation that IAAld accumulates in sur2 independently of an up-regulation of the Trp–indole-3-pyruvic acid–IAAld–IAA pathway. It is also supported by the fact that the conversion from IAOx is highly pH dependent, with higher activity at low pH than at high pH. Our observation that the sur2 phenotype is restored to a wild-type phenotype at low pH can be interpreted as follows. At low pH significant amounts of IAOx are converted to IAAld, which accumulates and is only converted to IAA to a limited extent. Conversely, at high pH the conversion of IAOx to IAAld is blocked, leading to increased pool sizes of IAOx that feed into the IAN pathway. This conclusion is also supported by the effect of pH on the IAN content observed in sur2. One of the most surprising results in this study was that 1 μM exogenous IAA could also restore the phenotype of sur2 to a wild-type phenotype by significantly reducing the endogenous IAN level. One explanation could be that exogenous IAA modifies endogenous pH and in this way modulates the conversion of IAOx to IAAld.
It should be noted, however, that a direct conversion pathway from IAOx to IAA or indole-3 ethanol has been postulated before (31), and, based on our data, the existence of such a pathway cannot be precluded. Interestingly, this pathway uses IAAld and not IAN as an intermediate from which IAA or indole-3 ethanol is formed by oxidation or reduction.
From our results we can conclude that the sur2 mutation causes increased synthesis of IAA, which is not mediated via the Trp–indole-3-pyruvic acid–IAAld–IAA pathway. The exact mechanism resulting in higher synthesis remains to be fully elucidated, but our observations indicate that the SUR2 gene is most probably involved in the regulation of the Trp–IAOx–IAN–IAA pathway. These results are strongly supported by the fact that the CYP83B1 (SUR2) enzyme has recently been shown to catalyze the N-oxidation of IAOx, which is the first step in indole glucosinolate biosynthesis (Bak et al., unpublished data). These results also suggest that IAOx is at the branch point of two biosynthetic pathways. Therefore a mutation in SUR2 would lead to an up-regulation of IAA synthesis through the disruption of the indole glucosinolate pathway.
Acknowledgments
This project was funded by the Commission of the European Communities (Contract ERBBIOC4 CT 960487) and the Biotechnology and Biological Sciences Research Council (M.B.), the University of Nottingham Research Fund (A.M.), the Swedish Natural Science Research Council (G.S.), and the Institut National de la Recherche Agronomique Research Fund (C.B.).
Abbreviations
- IAN
indole-3-acetonitrile
- IAOx
indole-3-acetaldoxime
- IAAld
indole-acetaldehyde
- IAAsp
N-(indole-3-acetyl)-Asp
- IAGlu
N-(indole-3-acetyl)-Glu
- IAAla
N-(indole-3-acetyl)-Ala
- IALeu
N-(indole-3-acetyl)-Leu
- Trp
tryptophan
- cyt P450
cytochrome P450
- RT
retention time
- SRM
selected reaction monitoring
- TAIL-PCR
thermal asymetric interlaced-PCR
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260502697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260502697
References
- 1.Davies P J. In: The Plant Hormones: Their Nature, Occurrence and Functions. Davies P J, editor. Dordrecht, The Netherlands: Kluwer; 1995. pp. 1–12. [Google Scholar]
- 2.Bartel B. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
- 3.Normanly J. Physiol Plant. 1997;100:431–442. [Google Scholar]
- 4.Bennett M, Kieber J, Giraudat J, Morris P. In: Arabidopsis: Annual Plant Reviews. Anderson M, Roberts J A, editors. U.K.: Scheffield Academic; 1998. pp. 107–150. [Google Scholar]
- 5.Boerjan W, Cervera M-T, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Montagu M V, Inzé D. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delarue M, Prinsen E, Van Onckelen H, Caboche M, Bellini C. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- 7.Gopalraj M, Tseng T S, Olszweski N. Plant Physiol. 1996;111:S–114. [Google Scholar]
- 8.Baumann E, Lewald J, Saedler H, Schulz B, Wisman E. Theor Appl Genet. 1998;97:729–734. [Google Scholar]
- 9.Wisman E, Cardon G H, Fransz P, Saedler H. Plant Mol Biol. 1998;37:989–999. doi: 10.1023/a:1006082009151. [DOI] [PubMed] [Google Scholar]
- 10.Mizutani M, Ward E, Ohta E. Plant Mol Biol. 1998;37:39–52. doi: 10.1023/a:1005921406884. [DOI] [PubMed] [Google Scholar]
- 11.Durst F, O'Keefe D. Drug Metab Drug Interact. 1995;12:171–187. doi: 10.1515/dmdi.1995.12.3-4.171. [DOI] [PubMed] [Google Scholar]
- 12.Szekeres M, Németh K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Rédei G P, Nagy F, Schell J, Koncz C. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Nagpal P, Vitart V, McMorris T C, Chory J. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 14.Choe S, Dilkes B P, Fujioka S, Takatuso S, Sakurai A, Feldmann K A. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hull A, Vij R, Celenza J L. Proc Natl Acad Sci USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. . (First Published February 18, 2000; 10.1073/pnas.040569997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santoni V, Bellini C, Caboche M. Planta. 1994;192:557–566. [Google Scholar]
- 17.Doyle J J, Doyle J L. Focus (Rochester, NY) 1990;12:13–15. [Google Scholar]
- 18.Marck C. Nucleic Acids Res. 1988;15:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagrimini L M, Burkhart W, Moyer M, Rothstein S. Proc Natl Acad Sci USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y-G, Mitukawa N, Oosumi T, Whittier R F. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 21.Edlund A, Eklöf S, Sundeberg B, Moritz T, Sandberg G. Plant Physiol. 1995;108:1043–1047. doi: 10.1104/pp.108.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Östin A, Kowalyczk M, Bhalerao R P, Sandberg G. Plant Physiol. 1998;118:285–296. doi: 10.1104/pp.118.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aarts M G M, Dirkse W G, Stiekema W J, Pereira A. Nature (London) 1993;263:715–717. doi: 10.1038/363715a0. [DOI] [PubMed] [Google Scholar]
- 24.Speulman E, Metz P L J, van Arkel G, te Lintel Hekkert B, Stiekema W J, Pereira A. Plant Cell. 1999;11:1853–1866. doi: 10.1105/tpc.11.10.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilfoyle T, Hagen G, Ulmasov T, Murfett J. Plant Physiol. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Normanly J, Bartel B. Curr Opin Plant Biol. 1999;2:207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- 27.Tuominen H, Östin A, Sandberg G, Sundberg B. Plant Physiol. 1994;106:1511–1520. doi: 10.1104/pp.106.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sitbon F, Östin A, Sundberg B, Olsson O, Sandberg G. Plant Physiol. 1993;101:313–320. doi: 10.1104/pp.101.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sitbon F, Sundberg B, Olsson O, Sandberg G. Plant Physiol. 1991;95:480–485. doi: 10.1104/pp.95.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helmlinger J, Rausch T, Hilgenberg W. Phytochemistry. 1987;26:615–618. [Google Scholar]
- 31.Rajagopal R, Larsen P. Planta. 1972;103:45–54. doi: 10.1007/BF00394605. [DOI] [PubMed] [Google Scholar]
- 32.Mahtews J M, Black S R, Burka L T. Xenobiotica. 1998;28:767–777. doi: 10.1080/004982598239182. [DOI] [PubMed] [Google Scholar]