Abstract
We demonstrate that a genetically engineered human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) composed mainly of p66 or p51 subunits can be incorporated into virus-like particles (VLPs) when coexpressed with HIV-1 Pr55gag. VLP-associated RT exhibited a detergent-resistant association with immature cores during sucrose gradient equilibrium centrifugation, suggesting that RT is incorporated into VLPs. However, RT that retains downstream integrase (IN) is severely inhibited in terms of incorporation into VLPs. Results from immunofluorescence tests reveal that RT-IN is primarily localized at the perinuclear area and exhibits poor colocalization with Gag. IN removal leads to a redistribution of RT throughout the cytoplasm and improved RT incorporation into VLPs. Similar results were observed for RT-IN in which alanine was substituted for 186-Lys-Arg-Lys-188 residues of the IN putative nuclear localization signal, suggesting that IN karyophilic properties may partly account for the inhibitory effect of IN on RT incorporation. Although the membrane-binding capacity of RT was markedly reduced compared to that of wild-type Gag or Gag-Pol, the correlation of membrane-binding ability with particle incorporation efficiency was incomplete. Furthermore, we observed that membrane-binding-defective myristylation-minus RT can be packaged into VLPs at the same level as its normal myristylated counterpart. This suggests that the incorporation of RT into VLPs is independent of membrane affinity but very dependent on RT-Gag interaction. Results from a genetic analysis suggest that the Gag-interacting regions of RT mainly reside in the thumb subdomain and that the RT-binding domains of Gag are located in the matrix (MA) and p6 regions.
In a late stage of human immunodeficiency virus (HIV) replication, molecules of the viral gag-encoded structural protein precursor Pr55gag self-assemble into virus-like particles (VLPs) and bud outward from the cell membrane (15, 25, 54). Coincidental with virus budding, Pr55gag is cleaved by viral protease (PR) into four major products: matrix (MA; p17), capsid (CA; p24), nucleocapsid (NC; p7), and the C-terminal p6 domain (23, 27, 30, 36, 38, 48). PR-mediated virus particle processing is essential for viral infectivity but is not required for virus assembly or budding (19, 29, 42, 43). HIV type 1 (HIV-1) PR is encoded by the pol gene. In HIV, Pol and Gag are translated from the same mRNA template, with the 5′ end of the pol reading frame overlapping with gag's reading frame. During Gag translation, −1 ribosomal frameshift events occur at a 5 to 10% frequency, resulting in Pol being synthesized as the polyprotein precursor Pr160gag-pol (26). The proteolytic cleavage of Pr160gag-pol by PR yields protease, reverse transcriptase (RT), and integrase (IN) in addition to Gag products.
HIV-1 Pr160gag-pol does not assemble into VLPs per se but is capable of interacting with Pr55gag and being incorporated into VLPs (21, 46, 47). Although a myristic acid moiety that is cotranslationally attached to the Gag N terminus is essential for Pr55gag plasma membrane targeting and virus particle production (5, 40), the Pr160gag-pol N-terminal myristylation signal appears to be dispensable for incorporating Pr160gag-pol into VLPs (13, 41). This suggests that such incorporation is largely dependent on the Pr160gag-pol interaction with assembling Pr55gag molecules.
Results from several genetic analysis studies indicate that the C-terminal third of the gag coding sequence in Gag-Pol is responsible for incorporating Pr160gag-pol into VLPs (13, 14, 24, 47). This region corresponds with the area identified as crucial to Gag oligomerization and assembly (1-4, 18, 31, 32, 34, 35, 44, 52). Accordingly, domains involved in Gag-Gag interactions may also play a major role in directing Gag-Pol incorporation into VLPs via interactions between Pr160gag-pol and Pr55gag. However, HIV-1 Pr160gag-pol (which carries some of the CA mutations that sharply diminish virus particle production) can still be incorporated into VLPs when coexpressed with Pr55gag (12). This agrees with the idea that Gag particle assembly may involve multiple domains along the Gag precursor and suggests that Pr160gag-pol mutants can be rescued into VLPs provided they retain domains capable of interacting with Pr55gag. As expected, HIV-1 Pr160gag-pol lacking this sequence upstream of the Pol or PR domains is severely defective in assembly into VLPs (13, 14), suggesting that the N-terminal Gag domain of HIV-1 Pr160gag-pol is the major determinant of Gag-Pol incorporation into VLPs.
One research team has recently demonstrated that HIV-1 Pol can be efficiently incorporated into VLPs at approximately 70% of the level of wild-type Gag-Pol, suggesting that HIV-1 Gag-Pol may be incorporated into virus particles independent of the upstream gag coding sequence (11). Another group reports that murine leukemia virus Pol can be incorporated into virions in the absence of an upstream Gag domain (6). These results suggest that retroviral Pol may contain undefined signals capable of directing Pol incorporation into virus particles. In a like manner, human foamy virus Pol expressed from separate spliced mRNA is naturally incorporated into virus particles without Gag-Pol formation (57).
Although HIV-1 Pol appears to possess domains capable of interacting with Pr55gag, evidence of Pol-Gag interaction remains to be substantiated. To address this issue and to define the domains responsible for putative interactions between HIV-1 Pol and Gag, we constructed several HIV-1 RT-IN expression vectors with progressive truncations at the C terminus. After coexpressing each construct with Pr55gag, we used Western blotting to determine the ability of each mutant to be packaged into VLPs. Results from our analyses indicate that HIV-1 RT composed mainly of p66 or p51 RT subunits was efficiently incorporated into VLPs and that such incorporation occurred independent of their membrane-binding capacities. IN-retaining RT recombinants were severely inhibited in terms of incorporation into VLPs, suggesting that IN plays an interfering role in this process. Results from genetic analyses suggest that the C-terminal region of p51RT is required for the efficient incorporation of RT into VLPs and that the MA and p6 regions of Pr55gag are both required for efficient packaging of RT into VLPs.
MATERIALS AND METHODS
Plasmid construction.
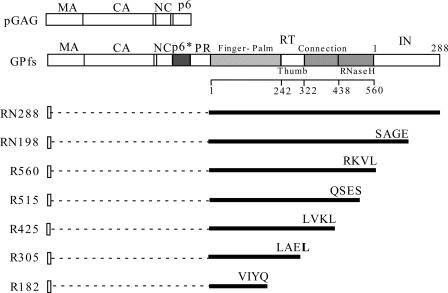
The parental HIV-1 proviral plasmid used in this study was derived from HXB2. To construct HIV-1 RT-IN recombinants, a series of HIV-1 Gag-Pol truncated mutants containing stop codons in HIV proviral positions at nucleotide (nt) 4823, 4243, 4094, 3824, 3467, or 3095 (33) were introduced into an HIV-1 Gag-Pol deletion mutant, Δ(Gag+ PR), in which the sequence upstream of RT (HIV-1 proviral nt 836 to 2546) was deleted (13). The resultant constructs were designated RN198, R560, R515, R425, R305, and R182, respectively (Fig. 1). In this report the Δ(Gag+ PR) construct expressing HIV-1 RT-IN is referred to as RN288.
FIG. 1.
Schematic representatives of HIV-1 Gag, Gag-Pol, and RT-IN expression constructs. pGAG and GPfs express Pr55gag and Pr160gag-pol, respectively. Indicated are the HIV Gag protein domains MA (matrix), CA (capsid), NC (nucleocapsid), and p6 and the pol-encoded p6*, PR, RT, and IN; numbers refer to amino acid residue positions. p66RT subdomain boundaries are also indicated. Dashed lines represent deleted sequences. Designated construct numbers indicate IN or RT terminal amino acid residue positions. The final three or four amino acid residues in each truncated construct are shown; inserted or altered amino acids are in boldface.
To introduce a hemagglutinin (HA) tag at the RN288 N terminus, two primers were used to amplify the DNA fragment of nt 2548 to 3002 using HIVgpt (39) as a template: 5′-TCAGATCGATATCCATACGATGTGCCAGATTACGCCCCCATTAGCCCTATTGAGACTG-3′ as the forward primer (containing a flanking ClaI restriction site and HA tag-coding nucleotides) and 5′-CCTGTGGAAGCACATTGTACT-3′ as the reverse primer. The purified PCR product was digested with ClaI and EcoRV and ligated into the RN288 construct, producing a new construct designated as hRN288 with the inserted HA tag located between the RT N terminus and the remaining N-terminal 14 gag codons. Each Gag-Pol truncated mutation was introduced into hRN288, yielding the hRN198, hR560, hR515, hR425, hR305, and hR182 constructs. To construct hRN(NLS−) containing alanine substitutions in the IN putative nuclear localization signal (NLS) motif residues 186-KRK-188, two primers were used to amplify DNA fragments using HIVgpt as a template: 5′-CCACAATTTTGCAGCTGCAGGGGGGATT-3′ as the forward primer and 5′-TTTCGTCGACCTAATCCTCATCCTG-3′ as the reverse primer. The resultant amplicons served as a reverse primer for a second PCR round using 5′-GGATTAGATATCAGTACAATG-3′ as a forward primer. Amplified DNA fragments were digested with EcoRV and SalI and ligated into hRN288. Myristylation-minus (Myr−) versions of the RT-IN constructs were generated by recombining Myr-Δ(Gag+ PR) (13) with each Gag-Pol truncation construct. Mutant constructs were analyzed using restriction enzyme digestion or confirmed by DNA sequencing. The HIV-1 Pr55gag expression plasmid pGAG and the Pr160gag-pol expression plasmid GPfs have been described previously (13).
Cell culture and transfection.
293T and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. Confluent 293T cells were trypsinized, split 1:10, and seeded into 10-cm dish plates 24 h before transfection. For each construct, 293T cells were transfected with 20 μg plasmid DNA using the calcium phosphate precipitation method (20), with 50 μM chloroquine added to enhance transfection efficiency. When the wild-type (wt) or mutant GPfs was cotransfected with pGAG, 10 μg of the construct and 10 μg of pGAG were used. For HeLa cell transfection, plasmid DNA was mixed with Lipofectamine 2000 (Invitrogen) at a ratio of 1 μg to 2.5 μl; the transfection procedure was performed according to the manufacturer's protocols.
Western immunoblot analysis.
Culture medium from transfected 293T cells was filtered (0.45-μm pore size) and centrifuged through 2 ml of 20% sucrose in TSE (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) plus 0.1 mM phenylmethylsulfonyl fluoride (PMSF) at 4°C for 40 min at 274,000 × g (SW41 rotor at 40,000 rpm). Viral pellets were suspended in IPB (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 1% Triton X-100, 0.02% sodium azide) plus 0.1 mM PMSF. Cells were rinsed with ice-cold phosphate-buffered saline (PBS), scraped from each plate, collected in 1 ml PBS, and pelleted at 2,500 rpm for 5 min. Cell pellets were resuspended in 250 μl IPB plus 0.1 mM PMSF prior to microcentrifugation at 4°C for 15 min at 13,700 × g (14,000 rpm) to remove cell debris. Either supernatant or cell samples were mixed with equal volumes of 2× sample buffer (12.5 mM Tris-HCl [pH 6.8], 2% SDS, 20% glycerol, 0.25% bromophenol blue) and 5% β-mercaptoethanol and boiled for 5 min.
Samples were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto nitrocellulose membranes that were subsequently blocked with 3% gelatin in Tris-buffered saline containing 0.05% Tween 20 (TBST). This mixture was incubated with the primary antibody in 1% gelatin-TBST on a rocking platform for 1 h at room temperature. Membranes were washed three times for 10 min each with TBST and rocked for 30 min with the secondary antibody in 1% gelatin-TBST. Blots were rewashed three times in TBST for 10 min each prior to detecting membrane-bound antibody-conjugated enzyme activity using enhanced chemiluminescence. To detect HIV Gag proteins we used an anti-p24gag monoclonal antibody (mouse hybridoma clone 183-H12-5C) at a 1:5,000 dilution. To detect HIV RT or Gag-Pol deletion mutants, we used a mouse anti-HIV-1 RT monoclonal antibody, mouse anti-HA monoclonal antibody, or HIV-positive human serum at a 1:10,000 dilution as the primary antibody. Our secondary antibody was either a goat anti-human or rabbit anti-mouse (horseradish peroxidase)-conjugated antibody at a 1:15,000 dilution. Procedures for horseradish peroxidase activity detection followed the manufacturer's protocols (Pierce).
Isolation of immature HIV-1 cores.
The procedure for HIV-1 core isolation has been described by Wyma et al. (56). Briefly, a 30 to 70% discontinuous sucrose gradient in TSE buffer was kept at 4°C for at least 4 h prior to overlaying with 0.2 ml TSE buffer containing 15% sucrose and 1% Triton X-100. This layer was covered with 0.2 ml TSE buffer containing 7.5% sucrose to prevent the premature mixing of detergent with the concentrated HIV-1 VLPs. VLPs concentrated via centrifugation through 20% sucrose cushions were suspended in 0.1 ml PBS and applied to the top of the barrier layer prior to centrifugation at 100,000 × g (Beckman SW-55 rotor) at 4°C for 16 h. Ten 0.5-ml fractions were collected from top to bottom. In addition to measuring the sucrose density of each fraction, the sample was assayed for the HIV-V1 CA and RT proteins using the Western blotting procedure described above.
Laser scanning immunofluorescence microscopy.
Confluent HeLa cells were split 1:80 onto coverslips 24 h before transfection. Two days posttransfection, cells were fixed at 4°C for 20 min with ice-cold PBS containing 3.7% formaldehyde, washed once with PBS and once with DMEM plus 10% heat-inactivated calf serum (DMEM-calf serum), and permeabilized at room temperature for 10 min in PBS plus 0.2% Triton X-100. Samples were incubated with the primary antibody for 1 h and secondary antibody for 30 min. Following each incubation, samples were subjected to three washes (5 to 10 min each) with DMEM-calf serum. Primary antibody concentrations were as follows: anti-HIV-1 p24CA, 1:500; anti-HA (Sigma), 1:200; fluorescein isothiocyanate-conjugated anti-HA, 1:40; or anti-HIV-1 RT monoclonal antibody, 1:500. A rabbit anti-mouse rhodamine-conjugated antibody at a 1:100 dilution served as the secondary antibody (Cappel, ICN Pharmaceuticals, Aurora, Ohio). After the final DMEM-calf serum wash, the coverslips were washed three times with PBS and mounted in 50% glycerol in PBS for viewing. Images were analyzed and photographs taken using an epifluorescence microscope (Olympus AX-80) or laser scanning confocal microscope (Olympus FV300). For RT/Gag colocalization quantification (see Fig. 5L, O, and R, below), we used the colocalization finder plug-in of the ImageJ image analysis software (http://rsb.info.nih.gov/ij/).
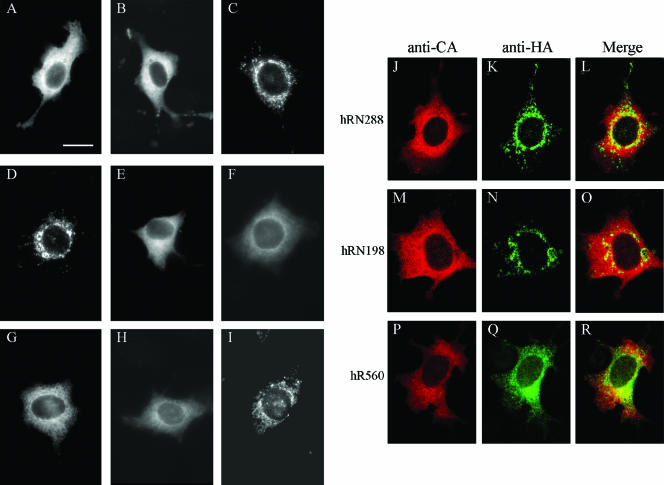
FIG. 5.
Subcellular localization of HIV-1 RT. HeLa cells grown on coverslips were transfected with pGAG (A), PR-defective GPfs (B), hRN288 (C), hRN198 (D), hR560 (E), hR515 (F), hR425 (G) hR305 (H), or hR182 (I) or transfected with pGAG plus hRN288 (J), hRN198 (M), or hR560 (P). At 48 h posttransfection, cells were fixed and permeabilized for immunofluorescent staining as described in Materials and Methods. Primary antibodies were either mouse anti-HIV-1 p24CA (A and B) or anti-HA (C to I); the secondary antibody was rhodamine-conjugated rabbit anti-mouse. For double staining (J to R), HIV-1 Gag was detected with the anti-HIV-1 p24CA monoclonal antibody followed by the rhodamine-conjugated rabbit anti-mouse antibody (J, M, and P); the HA-tagged proteins were probed with a fluorescein isothiocyanate-conjugated anti-HA antibody (K, N, and Q). Merged green and red fluorescence images are shown in panels L, O, and R. Mock-transfected HeLa cells and cells not exposed to the primary antibody yielded no signal (data not shown). Bar, 10 μm.
Equilibrium flotation centrifugation.
At 48 h posttransfection, 293T cells were rinsed twice, pelleted in PBS, and resuspended in TE buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA) containing 10% sucrose and complete protease inhibitor cocktail (Roche). Cell suspensions were subjected to sonication followed by low-speed centrifugation. Postnuclear supernatant (200 μl) was mixed with 1.3 ml of 85.5% sucrose in TE buffer, placed at the bottom of a centrifuge tube, and covered with a layer of a sucrose mixture (7 ml 65% sucrose and 3 ml 10% sucrose in TE buffer) prior to centrifugation at 100,000 × g for 16 to 18 h at 4°C. Ten fractions were collected from the top of the centrifuge tube, and proteins in each fraction were precipitated with ice-cold 10% trichloroacetic acid prior to Western immunoblotting.
Electron microscopy.
Virus-containing supernatant was centrifuged through 20% sucrose cushions. Concentrated viral samples were placed onto carbon-coated, UV-treated 200 mesh copper grids for 2 min as described elsewhere (45). Sample-containing grids were rinsed for 15 s in water, dried with filter paper, and stained for 1 min in filtered 1.3% uranyl acetate. Excess staining solution was removed by applying filter paper to the edge of each grid. Grids were allowed to dry before viewing with a JOEL JEM-2000 EXII transmission electron microscope. Images were collected at ×30,000 and ×60,000.
RESULTS
HIV-1 RT incorporation into virus particles is mainly determined by the p51RT C-terminal sequence.
We previously demonstrated that when the HIV-1 Pr160gag-pol deletion mutant Δ(Gag+ PR) lacks a sequence upstream of RT, it is excluded from VLPs (13). Following Kleiman et al.'s results suggesting that the domain responsible for incorporating HIV-1 Pol into VLPs resides within the RT region (11), we tested the possibility of HIV-1 RT containing undefined domains capable of interacting with Pr55gag. Specifically, we constructed a series of HIV-1 RT-IN mutants with progressively larger truncations at the C terminus of a Δ(Gag+PR) construct referred to as RN288. As shown in Fig. 1, the RN288 construct primarily expressed HIV-1 RT-IN, RN198 was truncated within IN, the R560 and R425 constructs were terminated at the C terminus of p66RT and p51RT, respectively, and the R515, R305, and R182 constructs were truncated at specific RT residues.
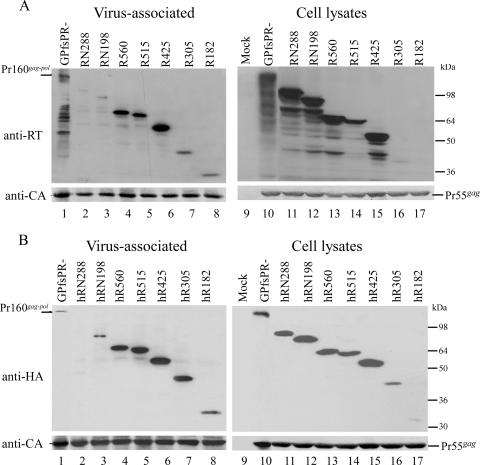
Each construct was coexpressed with a Pr55gag expression plasmid (pGAG). The ability of individual encoded proteins to be incorporated into VLPs was determined by Western blotting. As shown in Fig. 2A, the particle incorporation of RN288 was severely inhibited, with only trace amounts of virus-associated RN288 detected (lane 2); this is consistent with our previous results (13, 14). The RT-associated products observed with GPfsPR− cotransfections (Fig. 2A, lane 1) may have been derived from cellular RT-associated degradation products (Fig. 2A, lane 10) packaged into VLPs via interaction with Pr55gag; these bands did not appear when probed with an anti-p24CA monoclonal antibody (Fig. 2B, lanes 1 and 10). The amount of virus-associated RN198 was slightly higher than that of RN288, suggesting that partial IN removal improves RT-IN incorporation into VLPs (Fig. 2A, lane 3).
FIG. 2.
HIV-1 RT expression and incorporation into VLPs. 293T cells were cotransfected with 10 μg pGAG and 10 μg of PR-defective GPfs, GPfsPR−, or the indicated construct. Cells and supernatant were collected at 48 h posttransfection for protein analysis as described in Materials and Methods. Cell samples corresponding to 4% of total cell lysates and supernatant samples corresponding to 50% of total recovered viral pellets were fractionated by 10% SDS-PAGE. (A) HIV-1 RT and Gag-Pol were probed with an anti-RT polyclonal antibody. Since R182 is smaller in size, we must consider the possibility that it was electrophoresed off the gel (lane 17). However, even when we repeated this experiment and carefully monitored the gel, we still failed to detect R182. (B) HA-tagged RT proteins were detected with an anti-HA monoclonal antibody. Membranes from top panels A and B were stripped and reprobed using a monoclonal antibody directed against HIV-1 p24CA. Note that the Pr160gag-pol bands (B, lanes 1 and 10) did not appear when probed with the anti-HA antibody but were detected by the anti-p24CA antibody. Molecular size marker positions are shown on the right.
We noted that complete IN deletion resulted in the strongly enhanced incorporation of RT into VLPs, as demonstrated by R560, R515, and R425 (Fig. 2A, lanes 4 to 6). This suggests that p66RT or p51RT may interact with Pr55gag prior to being packaged into VLPs and that the presence of the downstream IN sequence may prevent RT incorporation into VLPs, possibly by interfering with the RT-Gag interaction. In contrast, VLP-associated R305 and R182 levels were relatively lower, presumably due (at least in part) to low expression levels of R305 and R182 (Fig. 2A, lanes 7 and 8). However, detected virus-associated RT levels may not serve as an accurate reflection of particle incorporation efficiency, since anti-RT antibody binding capacity may be significantly reduced in RT with large sequence deletions. To test this idea, we added one HA tag to each construct and probed the virus-associated RT-IN derivatives with an anti-HA monoclonal antibody. With one exception, the results indicate that the addition of an HA tag resulted in only a small difference in the level of virus-associated proteins (Fig. 2A and B). The exception was R305, in which the level of detected virus-associated HA-tagged protein was significantly increased compared to its non-HA-tagged counterpart. This suggests that the low level of virus-associated R305 may be due to a reduced binding capacity for the anti-RT antibody. In contrast, the relatively low level of virus-associated R182 was consistent whether it contained an HA tag or not (Fig. 2A and B, lane 8). It is likely that the inefficient incorporation of R182 into VLPs is the result of a combination of low steady-state expression and defective interaction with Gag.
Nonionic detergent-resistant association between HIV-1 RT and immature viral cores.
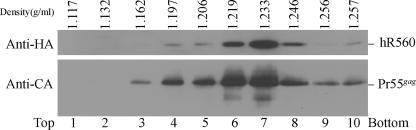
Since supernatant cultures containing VLP were centrifuged through 20% sucrose cushions for 40 min, we believe that recovered RT in the pelleted medium was either virus associated or VLP enveloped. We adapted the method described in reference 56 to isolate viral cores and to test whether the released RT was indeed virus associated and embedded in viral cores. Specifically, we concentrated virus-containing medium from cells cotransfected with pGAG and hR560 by centrifugation through 2-ml 20% sucrose cushions. Resuspended pellets were layered over 30 to 70% sucrose gradients containing a layer of 1% Triton X-100 prior to centrifugation at 100,000 × g for 16 h, after which fractions were collected and analyzed for sucrose density and viral proteins. As expected, hR560 and Pr55gag were found in the same sediment and with the same peak fraction at a density of 1.233 g/ml (Fig. 3), similar to the density of immature HIV cores (56). Consistent results were observed in the non-HA-tagged counterpart (R560). These results strongly suggest that HIV-1 RT is not released in vesicle form but is incorporated into HIV-1 cores, which are resistant to nonionic detergent treatment.
FIG. 3.
Sucrose density gradient fractionation of virus-associated HIV-1 RT. 293T cells were cotransfected with HA-tagged R560 (hR560) and pGAG. At 48 h posttransfection, culture supernatant was collected, filtered, and pelleted through 20% sucrose cushions. Viral pellets were resuspended and centrifuged through 30 to 70% sucrose gradients containing a layer of 1% Triton X-100 as described in Materials and Methods. Ten fractions of equal size were collected from the top of each gradient after 16 h of centrifugation. In addition to measuring the density of each fraction, we used Western immunoblotting to analyze Gag and RT protein levels. Fraction densities (in g/ml) are indicated at the top.
The membrane-binding capacity of the RT-IN derivatives is not completely compatible with their particle incorporation efficiency.
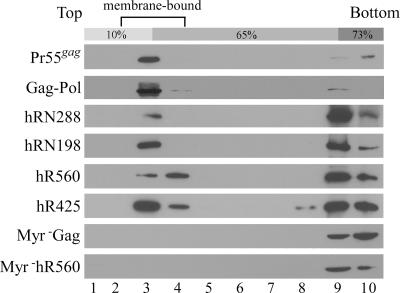
Since all of the RT-IN derivatives retained N-terminal 14 MA amino acid residues, each encoded protein was presumably myristylated. N-terminal myristylation signal is required for both HIV-1 Gag membrane binding and virus particle production (5, 40). We therefore tested for correlations between the membrane-binding capacities of the RT-IN recombinants and particle incorporation efficiency by subjecting postnuclear transfection lysates to equilibrium membrane flotation centrifugation. Our results show that most of the wt Gag was membrane associated (Fig. 4). In contrast, hRN288 was severely defective in terms of cell membrane binding (i.e., membrane-associated hRN288 represented less than 10% of the total hRN288). The membrane-binding capacities of hRN198, hR560, and hR425 were all significantly reduced, with over 50% of their total encoded proteins not observed floating with the membrane fraction (data not shown). In the absence of an HA tag, the membrane flotation profiles of the R425, R560, and RN288 constructs were all similar to those of their HA-tagged counterparts (data not shown), suggesting that the HA tag exerted no major effect on RT binding to cell membranes. As expected, the membrane-binding capacity of myr− Gag and myr− hR560 (negative controls) was abolished by the myr− mutation.
FIG. 4.
Membrane flotation analysis of RT proteins. 293T cells were transfected with pGAG, GPfsPR−, or the designated expression constructs. Two days posttransfection, cells were harvested, homogenized, and subjected to equilibrium flotation centrifugation analysis as described in Materials and Methods. Ten fractions were collected from the top downward. Fraction aliquots were resolved by SDS-PAGE (10%) and probed with a monoclonal antibody directed against HIV-1 p24CA or the HA tag. During ultracentrifugation, membrane-bound Gag proteins floated to the 10 to 65% sucrose interface and became enriched in fraction 3.
Although hRN198 showed a slightly higher membrane-binding capacity than hR560, the efficiency of its incorporation into VLPs was much lower than that of hR560. This suggests that membrane-binding capacity is not a primary determinant of RT packaging into VLPs. In addition, we observed that the nonmyristylated versions of R560 or R425 were efficiently packaged into VLPs and capable of floating with Pr55gag into membrane fractions during equilibrium flotation centrifugation (data not shown). This supports our previous finding (13) that N-terminal myristylation is not required for Gag-Pol incorporation into VLPs.
Improper subcellular localization of expressed RT-IN proteins may partly account for their reduced incorporation into VLPs.
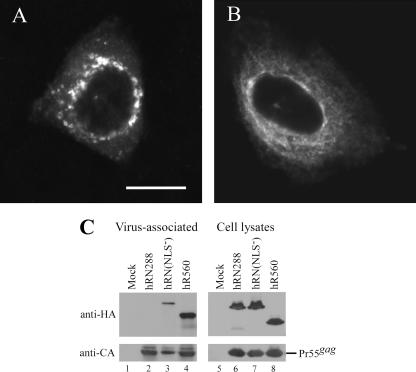
Improper transport or subcellular localization may contribute to reduced RT-IN incorporation into VLPs. To test this possibility we performed immunofluorescence experiments to reveal the subcellular distribution of each HIV-1 RT-IN derivative. As shown in Fig. 5, wt Gag (Fig. 5A) and Gag-Pol (Fig. 5B) were detected throughout the cellular cytoplasm with a heterogeneous punctate staining pattern and a clear perinuclear ring. In contrast, hRN288 (Fig. 5C) and hRN198 (Fig. 5D) were both enriched in the perinuclear area, which fits with their inefficient incorporation into VLPs. We also found that IN removal leads to RT redistribution throughout the cytoplasm, as observed in hR560 (Fig. 5E), hR515 (Fig. 5F), hR425 (Fig. 5G), and hR305 (Fig. 5H). The R560 and R425 cellular localization patterns are similar to their HA-tagged counterparts (data not shown), suggesting that HA tagging exerts no major effect on the subcellular distribution of RT-IN derivatives. The majority of hR182 transfectants (Fig. 5I) showed sparse punctate spots throughout the observed cells, which is consistent with its low level of steady-state expression. Combined, these results indicate a strong correlation between subcellular distribution patterns and particle incorporation efficiency. In addition, hRN288 and hRN198 colocalized with the coexpressed Pr55gag at very low levels: 2.11% and 1.12%, respectively. The percentage of hR560 that colocalized with the coexpressed Pr55gag was 4.76%—in other words, at two and four times the respective efficiencies of hRN288 and hRN198 (Fig. 5L, O, and R). The inability of mislocalized hRN288 and hRN198 to interact with Pr55gag at substantial levels at least partly accounts for the lack of particle incorporation.
Substitution mutations in the IN putative nuclear localization signal lead to RT-IN redistribution with concomitant improved viral incorporation.
Since amino acid residue signaling for nuclear localization has been found within IN (8, 17), RT-IN localization in the perinuclear area may be attributed to IN karyophilic properties. Accordingly, disruption of the putative NLS may alleviate RT-IN perinuclear trapping. To test this possibility, we replaced IN putative NLS motif 186-KRK-188 in hRN288 residues with alanine; the resulting hRN(NLS−) construct was expressed in HeLa cells. Similar to the IN deletion effect illustrated in Fig. 5E to H, the NLS− mutation caused a significant redistribution of RT-IN throughout the cytoplasm (Fig. 6A versus B), although approximately 30% of the hRN(NLS−) transfectants still showed substantial perinuclear fluorescence. This suggests that IN karyophilic properties at least partly account for RT-IN localization in the perinuclear area, which may in turn inhibit the packaging of RT-IN into VLPs. When correlated with the redistributed subcellular RT-IN following an NLS motif mutation, hRN(NLS−) particle incorporation efficiency was markedly enhanced after taking into account the Gag VLP level (Fig. 6C, lane 3).
FIG. 6.
Effect of IN putative nuclear localization mutation on RT-IN subcellular localization and particle incorporation. (A and B) Subcellular localization of HIV-1 RT-IN. HeLa cells were transfected with hRN288 (A) or hRN(NLS−) (B); the two are identical except for alanine substitutions for the 186-Lys-Arg-Lys-188 residues in the IN nuclear localization motif. HA-tagged RT-IN proteins were detected as described in the legend for Fig. 5. Bar, 10 μm. (C) Incorporation of HIV-1 RT-IN into VLPs. 293T cells were cotransfected with pGAG and each indicated construct. At 48 h posttransfection, cells and culture supernatant were collected, prepared, and subjected to Western immunoblotting. HA-tagged RT proteins and HIV-1 Gag were probed with anti-HA and anti-p24CA antibodies, respectively.
Gag MA and p6 regions are both required for incorporating RT into VLPs.
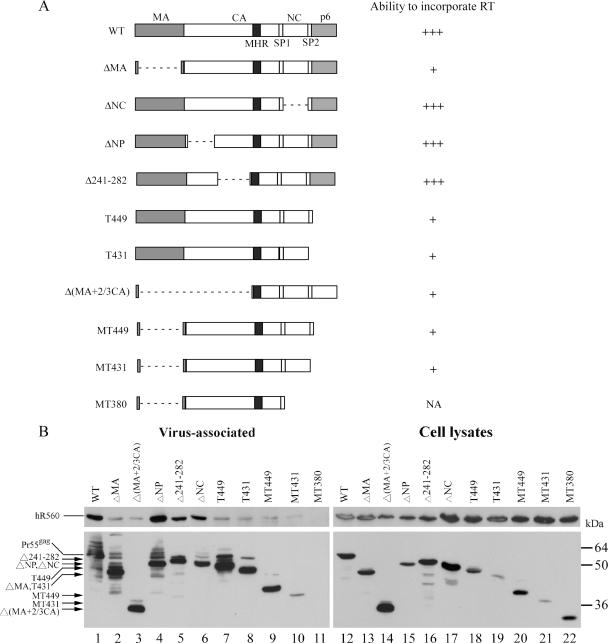
The same evidence showing that HIV-1 Gag particles can efficiently incorporate coexpressed RT suggests that HIV-1 Pr55gag can interact with RT and recruit it into VLPs. To map the boundary of the Gag sequence responsible for RT incorporation, we coexpressed hR560 with a series of HIV-1 gag mutants containing various gag coding sequence deletions or truncations (Fig. 7A). When the mutations involved either the CA (ΔNP, Δ241-282, lanes 4 and 5) or NC (ΔNC, lane 6) domains, the Gag mutants incorporated RT with an efficiency comparable to that of wt (lane 1). When they involved either the MA [ΔMA, Δ(MA+2/3CA)] and/or p6 (T449, T431, and MT449) domains, the Gag mutants were strongly defective in incorporating RT into VLPs (lanes 2, 3, and 7 to 9). These results suggest that the MA and p6 domains of Pr55gag are both required for the efficient incorporation of HIV-1 RT into VLPs.
FIG. 7.
Mapping of RT-binding domains on Gag. (A) Schematic representatives of HIV-1 gag mutants. Four major domains are indicated: MA, CA, NC, and p6. Also indicated are the space peptides SP1 and SP2 and a major homology region (MHR) in CA. Each Gag mutant contains an internal deletion (dashed line) and/or a C-terminal truncation mutation in the gag coding sequence. The ability of each mutant to incorporate HIV-1 RT is summarized on the right as follows: +++, RT incorporation efficiency comparable to that of wt (≥80% of wt); +, efficiency between 2 and 10% of wt; NA, not applicable. Note that MT380 is severely defective in VLP production. (B) Incorporation of HIV-1 RT into virus-like particles. 293T cells were cotransfected with hR560 and the designated constructs. Two days posttransfection, culture supernatant and cells were collected, prepared, and subjected to 10% SDS-PAGE. hR560 and Gag proteins were probed with anti-HA and anti-p24CA monoclonal antibodies, respectively. Levels of virus-associated Gag and RT in each sample were quantified by scanning immunoblot band densities. RT/Gag protein level ratios were calculated for each sample and normalized to that of wt in parallel experiments. Percentages of virus-associated RT denote the ability of each mutant to package RT. Molecular mass marker positions are shown on the right. Predicted molecular masses corresponding to Gag deletion mutant sizes were as follows: Δ241-282, 51 kDa; ΔNP and ΔNC, 50 kDa; T449, 49 kDa; T431, 47 kDa; ΔMA, 44 kDa; MT449, 39 kDa; MT431, 37 kDa; Δ(MA+2/3CA) and MT380, 27 to 28 kDa. Note that the immunoblot shown in panel B (left) was intentionally overexposed to allow the assembly-defective gag mutant to be viewed. p24-associated degraded bands usually appeared as exposure was increased.
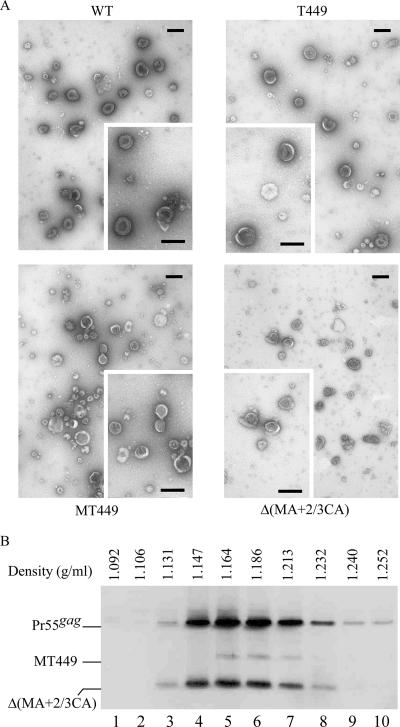
The results shown in Fig. 7B indicate that with the exceptions of MT431 and MT380, all of the gag mutants were capable of producing substantial amounts of VLPs. This is consistent with previous results (53) and supports the idea that coexpressed hR560 has no noticeable effect on VLP production. That both T449 and MT449 can efficiently produce VLPs suggests that the p6 domain is dispensable for VLP budding in 293T cells—an idea that is consistent with evidence showing that in certain circumstances the p6 domain is not required for virus budding in 293T cells (16). Since cultured supernatant was centrifuged through 20% sucrose cushions, we assumed that the recovered Gag would be present in pelleted particles. To confirm that the recovered Gag in large deletion mutants and/or p6-deleted mutants was indeed from VLPs, we prepared and viewed viral supernatant samples of T449, MT449, and Δ(MA+2/3CA) with a transmission electron microscope and observed spherical immature wt and mutant Gag particles with electron-dense cores (Fig. 8A).
FIG. 8.
Analysis of HIV-1 Gag VLPs. (A) Transmission electron microscopy of concentrated culture supernatant from 293T cells expressing the wt or the indicated gag deletion mutant. Bars, 200 nm. (B) Sucrose density gradient fractionation of HIV-1 Gag VLPs. 293T cells were transfected with wt, MT449, or Δ(MA+2/3CA). At 48 to 72 h posttransfection, culture supernatants were collected and pelleted through 20% sucrose cushions. Viral pellets were suspended in TSE buffer. To make direct comparisons with wt HIV-1 particle densities, wt viral pellets were spun through the same sucrose density gradient (20 to 60%) with pooled pellets containing MT449 and Δ(MA+2/3CA) at 274,000 × g for 16 h. Fractions were collected, measured for sucrose density, and analyzed for Gag protein levels by immunoblotting. Fraction densities are indicated at the top. The relatively lower signal of MT449 was due to loss of the viral pellets prior to pooling them together.
Some virion-size vesicles lacking cores were noted, but particles containing electron-dense cores were not detected in mock-transfected samples (data not shown). Significant amounts of T449 particles had electron-dense cores localized laterally in comparison to wt particles with more centralized electron-dense cores (Fig. 8A). Particles formed by the large deletion mutants MT449 and Δ(MA+2/3CA) were more heterogeneous in size.
We performed a sucrose density gradient fractionation analysis as an additional test of whether large gag deletion mutations affect Gag packing or formed particle density. In order to make comparisons with wt HIV-1 particle densities, viral pellets of MT449 and Δ(MA+2/3CA) were spun with wt pellets through the same sucrose density gradient. Results shown in Fig. 8B indicate that wt, MT449, and Δ(MA+2/3CA) cosedimented with the wt Gag proteins and banded in fractions with densities between 1.16 and 1.18 g/ml (Fig. 8B). We also found that VLP-associated wt and mutant Gag proteins were pelletable following treatment of VLP-containing supernatant with trypsin or 0.5% Triton X-100 (data not shown), indicating that the mutant particles were enveloped in the membrane and that viral assembly cores were resistant to nonionic detergent. Combined, these results suggest that the released Gag proteins were VLP associated and that the pelletable medium Gag protein levels reflected VLP levels.
DISCUSSION
We found that the efficiency of RT-IN incorporation into VLPs can be dramatically enhanced by IN removal (Fig. 2). The conclusion that HIV-1 RT can be incorporated into VLPs may be tempered by the use of a high-level expression system, but several lines of evidence suggest that RT is incorporated into VLPs due to an association with Gag. The cosedimentation of VLP-associated hR560 with immature viral cores during sucrose density gradient centrifugation corroborates the hypothesis that HIV-1 RT can be incorporated into Pr55gag particles (Fig. 3). Furthermore, evidence that RT without membrane-binding ability can cofloat with Pr55gag in membrane fractions and be recruited into VLPs strongly supports the proposal that HIV-1 RT is incorporated into VLPs via interaction with Gag.
HIV-1 RT incorporation into VLPs was observed with R560 and R425 independent of the N-terminal myristate moiety and HA tagging. R305 and R182 and their HA-tagged versions all showed low steady-state expression levels, suggesting that removal of the C-terminal region may affect RT stability. In spite of its low expression level, hR305 was nevertheless incorporated into VLPs at a level comparable to those of R560 and R425, suggesting that the sequence from C terminal to residue 305 (covering the connection and RNase H subdomain of RT) is not required for RT incorporation into VLPs. However, as demonstrated by R182, the removal of the thumb subdomain markedly impaired RT incorporation. Thus, it is very likely that the domains responsible for RT incorporation into VLPs exist between RT residues 183 and 305. Some mutations in this region have been shown to block p66/p51 heterodimer formation (49). It remains to be determined if the truncation mutations affect RT dimerization and consequently diminish RT incorporation into VLPs. Since R182 and its HA-tagged versions repeatedly showed relatively low expression levels, the instability or low expression level of R182 may contribute (at least in part) to its inefficient particle incorporation.
Even though myristylation-minus mutations blocked RT binding to cell membranes, Myr−R560 was incorporated into VLPs to the same extent as its myristylated counterpart. Further, the membrane-binding capacity of the RT-IN constructs did not fully correlate with their efficiency of incorporation into VLPs (Fig. 4). These results suggest that (i) the presence of the myristate moiety in the N terminus of RT exerts a negligible effect on the incorporation of RT into VLPs and (ii) membrane-binding capacity is not a major determinant of RT incorporation into VLPs. Although all of the RT-IN constructs retained the N-terminal myristylation signal, it is unknown why their cell membrane-binding capacities were reduced to various levels. One possibility is that the myristate moiety is partially masked to different degrees by the conformations exhibited by individual recombinant proteins, resulting in variations in membrane-binding capacity.
The marked increase in the ability to incorporate HIV-1 RT-IN into VLPs after IN removal implies IN interference with RT-IN particle incorporation. HIV-1 IN contains a required NLS for nuclear transport of the viral preintegration complex (PIC) (8, 17). In addition, results from recent studies show that other cellular factors may interact with IN and facilitate IN enzymatic function during viral integration into cell chromosomes (51). In the Gag-Pol context, these karyophilic properties of IN may be sequestered in a manner that does not affect Gag-Pol transport or incorporation into VLPs. Once the sequence upstream of Pol or RT is removed, the IN functional domains involved in nuclear transport may be exposed and thus interfere with RT-IN transport and VLP incorporation by trapping RT-IN in perinuclear areas via interaction with nuclear transport and/or other cellular factors. This hypothesis is supported by the findings that substituting alanine for the IN putative NLS motif 186-KRK-188 leads to redistribution and enhanced RT-IN incorporation in a manner similar to that of IN-deleted mutations (Fig. 6). Further, the presence of IN may block RT-Gag interaction by covering the Gag-binding site on RT via a cis or trans interaction with RT. Several research teams have proposed interactions between HIV-1 IN and RT (22, 50, 55, 59). Such scenarios may explain why RN288 and RN198 cannot colocalize with Gag and are consequently poorly incorporated. RN288 and RN198 localization in the nucleus has not yet been observed with a confocal microscope. Perhaps the combined presence of an N-terminal myristate moiety and RT prevents RT-IN nuclear transport. Unlike RN288 (which shows defective incorporation into VLPs), HIV-1 Pol with or without a PR deletion can be packaged into VLPs at about 70% of wt Gag-Pol efficiency (11). It may be that the HIV-1 Pol codons used by Kleiman's research group were optimized in order to permit more stable and stronger protein expression, which would counteract the interfering effect of IN on Pol incorporation.
VLPs produced by Gag mutants lacking the MA and/or p6 domains were deficient in R560, suggesting that both domains are required for incorporating RT into VLPs. This finding is supported by the Kleiman group report that p6 removal significantly affects HIV-1 Pol incorporation into VLPs (11). Combined results from our earlier studies suggest that strong RT-Gag interaction is required for RT incorporation and that such interaction requires the MA and p6 domains. It has not yet been determined if MA and p6 can interact with RT individually or if putative MA-RT and p6-RT interactions involve different RT regions. Since assembly-competent Δ(MA+2/3CA) is incapable of incorporating RT, the C-terminal CA assembly domain considered essential for particle formation apparently is not involved in Gag-RT interaction. Furthermore, even though our results suggest that the MA and p6 domains in the Gag precursor context may interact with RT, it is unknown whether mature MA and p6 are capable of interacting with RT. Some researchers have proposed that MA plays a role in the postentry stage of the HIV-1 life cycle (8, 10, 28, 58), and the MA has thus far been the only Gag protein identified as part of the HIV-1 preintegration complex (8, 37). The NC domain has recently been identified as playing a facilitating role in integration (7, 9), implying that it may be part of the PIC but tends to become dissociated during PIC isolation.
Acknowledgments
We thank H.-C. Chiu and H.-H. Chu for reagents and technical assistance and Y.-I. Yu for taking the electron microscopy photos. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: hybridoma clone 183 H12-5C from Bruce Chesebro and anti-HIV-1 RT from Stuart LeGrice.
This work was supported by grants VGH93-309 from the Taipei Veterans General Hospital and NSC94-2320-B-010-035 from the National Science Council, Taiwan, Republic of China.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Abdurahman, S., S. Hoglund, L. Goobar-Larsson, and A. Vahlne. 2004. Selected amino acid substitutions in the C-terminal region of human immunodeficiency virus type 1 capsid protein affect virus assembly and release. J. Gen. Virol. 85:2903-2913. [DOI] [PubMed] [Google Scholar]
- 2.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is critical for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchschacher G. L., Jr., L. Yu, F. Murai, T. Friedmann, and A. Miyanohara. 1999. Association of murine leukemia virus Pol with virions, independent of Gag-Pol expression. J. Virol. 73:9632-9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckman, J. S., W. J. Bosche, and R. J. Gorelick. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carteau, S., S. C. Batson, L. Poljak, J. F. Mouscadet, H. de Rocquigny, J. L. Darlix, B. P. Roques, E. Kas, and C. Auclair. 1997. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J. Virol. 71:6225-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casella, C. R., L. J. Raffini, and A. T. Panganiban. 1997. Pleiotropic mutations in the HIV-1 matrix protein that affect diverse steps in replication. Virology 228:294-306. [DOI] [PubMed] [Google Scholar]
- 11.Cen, S., M. Niu, J. Saadatmand, F. Guo, Y. Huang, G. J. Nabel, and L. Kleiman. 2004. Incorporation of Pol into human immunodeficiency virus type 1 Gag virus-like particles occurs independently of the upstream Gag domain in Gag-Pol. J. Virol. 78:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien, A.-I., W.-H. Liao, D.-M. Yang, and C.-T. Wang. 2006. A domain directly C-terminal to the major homology region of human immunodeficiency type 1 capsid protein plays a crucial role in directing both virus assembly and incorporation of Gag-Pol. Virology 348:84-95. [DOI] [PubMed] [Google Scholar]
- 13.Chiu, H. C., S. Y. Yao, and C.-T. Wang. 2002. Coding sequence upstream of the human immunodeficiency virus type 1 reverse transcriptase domain in Gag-Pol are not essential for incorporation of the Pr160gag-pol into virus particles. J. Virol. 76:3221-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu, H. C., W.-H. Liao, S.-W. Chen, and C.-T. Wang. 2004. The human immunodeficiency virus type 1 carboxyl-terminal third of capsid sequence in Gag-Pol is essential but not sufficient for efficient incorporation of Pr160gag-pol into virus particles. J. Biomed. Sci. 11:398-407. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 1998. HIV Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganser-Pornillos, B. K., U. K. von Schwedler, K. M. Stray, C. Aiken, and W. I. Sundquist. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristylation in morphogenesis and infectivity of human immunodeficiency virus type1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham, R., and A. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 21.Halwani, R., A. Khorchid, S. Cen, and L. Kleiman. 2003. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. J. Virol. 77:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hehl, E. A., P. Hoshi, G. V. Kalpana, and V. R. Prasad. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 78:5056-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. A. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processing, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, M., and M. A. Martin. 1997. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J. Virol. 71:4472-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter, E. 1994. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin. Virol. 5:71-83. [Google Scholar]
- 26.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature (London) 331:280-283. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan, A. H., M. Manchester, and R. Swanstorm. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 68:6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohl, N. E., E. A. Emini, W. E. Schleif, L. J. Davis, J. C. Heimbach, R. A. F. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leis, J., D. Baltimore, J. B. Bishop, J. Coffin, E. Fleissner, S. P. Goff, S. Oroszlan, H. Robinson, A. M. Skalka, H. M. Temin, and V. Vogt. 1988. Standardized and simplified nomenclature for proteins common to all retroviruses. J. Virol. 62:1808-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, C., J. Hu, R. S. Russell, A. Roldan, L. Kleiman, and M. A. Wainberg. 2002. Characterization of a putative α-helix across the capsid-sp1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao, W.-H., and C.-T. Wang. 2004. Characterization of human immunodeficiency virus type 1 Pr160gag-pol mutants with truncations downstream of the protease domain. Virology 329:180-188. [DOI] [PubMed] [Google Scholar]
- 34.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melamed, D., M. Mark-Danieli, M. Kenan-Eichler, O. Kraus, A. Castiel, N. Laham, T. Pupko, F. Glaser, N. Ben-Tal, and E. Bacharach. 2004. The conserved carboxy terminus of the capsid domain of human immunodeficiency virus type 1 Gag protein is important for virion assembly and release. J. Virol. 78:9675-9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. R. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslation modifications, and evidence for alternative gag precursors. J. Virol. 62:3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navia, M. A., and B. M. McKeever. 1990. A role for the aspartyl protease from the human immunodeficiency type 1 (HIV-1) in the orchestration of virus assembly. Ann. N. Y. Acad. Sci. 616:73-85. [DOI] [PubMed] [Google Scholar]
- 39.Page, K. A., N. R. Landau, and D. R. Littman. 1990. Construction and use of a human immunodeficiency virus: vector for analysis of virus infectivity. J. Virol. 64:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pal, R., M. S. Reitz, Jr., E. Tschanchler, R. C. Gallo, M. G. Sarngadharan, and F. D. M. Veronese. 1990. Myristylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res. Hum. Retrovir. 6:721-730. [DOI] [PubMed] [Google Scholar]
- 41.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng, C., B. K. HO, T. W. Chang, and N. T. Chang. 1989. Role of human immunodeficiency virus type 1-specific protease in core particle maturation and viral infectivity. J. Virol. 63:2550-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scarlata, S., and C. Carter. 2003. Role of HIV-1 Gag domains in viral assembly. Biochim. Biophys. Acta 1614:62-72. [DOI] [PubMed] [Google Scholar]
- 45.Scholz, I., B. Arvidson, D. Huseby, and E. Barklis. 2005. Virus particle core defects caused by mutations in the human immunodeficiency virus capsid N-terminal domain. J. Virol. 79:1470-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, A. J., N. Srinivasakumar, M.-L. Hammarskjold, and D. Rekosh. 1993. Requirements for the incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 67:2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srinivasakumar, N., M.-L. Hammarskjold, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 49.Tachedjian, G., H.-E. G. Aronson, and S. P. Goff. 2000. Analysis of mutations and suppressor affecting interactions between the subunits of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 97:6334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tasara, T., G. Maga, M. O. Hottiger, and U. Hübscher. 2000. HIV reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett. 507:39-44. [DOI] [PubMed] [Google Scholar]
- 51.van Maele, B., and Z. Debyser. 2005. HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev. 7:26-43. [PubMed] [Google Scholar]
- 52.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, C.-T., H.-Y. Lai, and J.-J. Li. 1998. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J. Virol. 72:7950-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 55.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55gag in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 58.Yu, X., Q. C. Yu, T. H. Lee, and M. Essex. 1992. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J. Virol. 66:5667-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, K., C. Dobard, and S. A. Chow. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]