Abstract
APOBEC3G is an endogenous host restriction factor that inhibits human immunodeficiency virus (HIV) replication. The antiviral activity of APOBEC3G is dependent upon its incorporation into the virus particle. The mechanisms governing incorporation of APOBEC3G into virus particles are not completely understood. In particular, some investigators have reported that APOBEC3G interacts directly with the nucleocapsid (NC) subunit of Gag, while others have found that an RNA intermediate is required for Gag-APOBEC3G interactions. In this study, we confirmed the RNA dependence of APOBEC3G packaging and performed detailed mapping of the determinants within NC that are required for virion incorporation. Surprisingly, APOBEC3G packaging did not correlate well with the presence of the N-terminal “I,” or interaction, domain within NC. Specifically, Gag constructs containing only the N-terminal region of NC packaged minimal amounts of APOBEC3G, while significant levels of APOBEC3G packaging were achieved with Gag constructs containing the basic linker region of NC. Furthermore, membrane-binding experiments revealed that the basic linker region was essential for the membrane association of APOBEC3G in a Gag-APOBEC3G complex. Fluorescence resonance energy transfer was detected between labeled APOBEC3G in cells and in particles, indicating that APOBEC3G is packaged as a multimer that is bound to packaged RNA. Regions of APOBEC3G-Gag colocalization at the plasma membrane were detected that were distinct from the punctate cytoplasmic bodies where APOBEC3G accumulates within the cell. Together, our results indicate that APOBEC3G multimerizes in an RNA-dependent fashion and that RNA-APOBEC3G multimers are recruited to the plasma membrane and subsequently into virion particles by Gag.
Human immunodeficiency virus type 1 (HIV-1) infection is a major cause of morbidity and mortality worldwide. The ability of HIV to subvert natural human immune defenses has contributed highly to its success in infecting more than 40 million people worldwide. The cytidine deaminase APOBEC3G restricts the replication of retroviruses, including HIV, macaque simian immunodeficiency virus, murine leukemia virus (12, 20, 27, 36), retrotransposons (29), and other viruses containing a reverse transcription step, such as hepatitis B virus (32). APOBEC3G is incorporated into virions exiting the infected cell. Upon infection of the target cell, APOBEC3G causes deamination of cytidines on single-stranded DNA intermediates created during the reverse transcription process. This results in extensive G-to-A nucleotide substitutions in the proviral DNA (12, 18, 20, 39). HIV-1 counteracts the action of APOBEC3G through the action of the viral infectivity factor (Vif), which binds to APOBEC3G and links it to an E3 ligase for polyubiquitination and subsequent degradation by the ubiquitin proteasome pathway (6, 11, 37). A critical aspect of the antiviral function of APOBEC3G is, thus, its ability to be packaged into assembling virions (15, 21, 28).
The HIV-1 Gag protein is responsible for facilitating APOBEC3G incorporation into virions as evidenced by the fact that Gag virus-like particles (VLPs) efficiently incorporate APOBEC3G (10, 31, 38). Furthermore, in vitro binding experiments have shown that APOBEC3G binds efficiently to the nucleocapsid (NC) region of the Gag polyprotein (1, 4, 19, 26, 31, 38). In fact, Gag constructs in which a leucine zipper protein-protein interaction domain replaced the nucleocapsid region produced particles devoid of APOBEC3G (19, 38). The mechanisms by which NC recruits APOBEC3G are not entirely clear. In particular, the role played by RNA and distinct RNA binding regions within NC has been controversial. In vitro binding experiments between NC and APOBEC3G that utilize RNA depletion by RNase A treatment have been reported which support (19, 26, 31, 38) or refute (1, 4) the role that an RNA bridge may play in this interaction.
An essential retroviral assembly function is contained within the N-terminal region of nucleocapsid (2, 3, 33). This assembly function, termed the interaction (“I”) domain, is necessary for Gag-Gag multimerization, formation of detergent-resistant Gag complexes, plasma membrane localization of Gag, and formation of normal density retroviral particles (7, 24, 25). The “I” domain has been implicated in mediating RNA binding to NC through electrostatic interactions of basic residues with cellular or viral RNA. Furthermore, in experiments using a C-terminal NC truncation model in a Gag context, the “I” domain phenotype correlated with the number of basic residues expressed (24). Therefore, it seemed reasonable to hypothesize that the underlying mechanisms governing APOBEC3G incorporation might be linked to the underlying mechanisms governing the “I” domain assembly functions.
In this report, we examine the requirements for APOBEC3G incorporation into HIV VLPs. Surprisingly, we found that APOBEC3G incorporation did not correlate closely with the presence of the I domain. Rather, mapping of APOBEC3G incorporation and RNA revealed a key role for the basic linker region between the two zinc finger domains of NC. The amount of RNA incorporated into particles did correlate with APOBEC3G recruitment to the plasma membrane and packaging into particles. Finally, we developed a fluorescence resonance energy transfer (FRET) assay to detect APOBEC3G-RNA-APOBEC3G complexes and showed that these complexes were packaged into Gag virus-like particles. Together, these data suggest that APOBEC3G is packaged into virions as an RNA-dependent multimer that is recruited to the plasma membrane and into budding virions by the HIV Gag protein.
MATERIALS AND METHODS
Plasmid construction.
This study employed Gag and APOBEC3G expression constructs fused to variants of the codon-optimized version of cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) (Clontech, Palo Alto, CA). Some of the CFP and YFP expression vectors and several of the Gag expression constructs were previously described (7). Briefly, the C-terminal CFP and YFP expression constructs were created by first replacing the enhanced green fluorescent protein (EGFP) gene in pEGFP-N3 (Clontech) with either the CFP gene from pECFP-N1 or the YFP gene from pEYFP-N1. These new constructs were designated pECFP-N3 and pEYFP-N3, respectively (7). The N-terminal YFP expression construct was produced by replacing the EGFP gene in pEGFP-C1 (Clontech) with the YFP gene from pEYFP, using an AgeI site located 5′ to the ATG start site in both plasmids and a BsrGI site located in the 3′ end of both genes prior to the stop codon. This new construct was designated pEYFP-C1. The Gag coding sequences for all constructs were derived from the codon-optimized version of HIV-1 strain HXB2 Gag in expression plasmid pVRC3900 (13). PCR cloning was then used to amplify the gag gene from the pVRC3900 vector, with a HindIII site at the 5′ ATG and a BamHI site at the 3′ end. The amplified gag gene was then ligated into both pEYFP-N3 and pECFP-N3 by digestion of the BamHI and HindIII sites located within the multiple cloning regions. A schematic diagram of the Gag expression constructs employed in this study is shown in Fig. 2B. The APOBEC3G coding sequences for all constructs were derived from plasmid CEM15, which was obtained from Michael Malim, along with a C-terminal hemagglutinin (HA)-tagged version; both have been described previously (27). These constructs are referred to hereafter as APOBEC3G and APOBEC3G-HA. PCR cloning was used to amplify the APOBEC3G gene with an EcoRI site prior to the 5′ ATG codon and an SmaI site at the 3′ end. The amplified APOBEC3G gene was then ligated into pEYFP-N3 by digestion of the EcoRI and SmaI sites located within the multiple cloning region. This new construct was designated A3G-YFP. PCR cloning was used to amplify the APOBEC3G gene with an EcoRI site prior to the 5′ ATG codon and an XmaI site at the 3′ end. The amplified APOBEC3G gene was then ligated into pEYFP-C1 by digestion of the EcoRI and XmaI sites located within the multiple cloning region. This new construct was designated YFP-A3G. PCR cloning was used to amplify the CFP gene from pECFP-N3, placing a SmaI site prior to the 5′ ATG codon and a NotI site at the 3′ end. The YFP gene in A3G-YFP was then removed by digestion with SmaI and NotI, followed by gel purification. The amplified CFP gene was then ligated into the resulting plasmid to yield a new construct designated A3G-CFP.
FIG. 2.
Nucleocapsid determinants of APOBEC3G incorporation. (A) Schematic illustration of Pr55Gag and position of the N-terminal I domain and selected amino acids. (B) Schematic representation of Gag-CFP constructs subdividing HIV-1 NC. Asterisks indicate the sites of Gag truncation and CFP fusion. The number represents the C-terminal amino acid residue expressed, with the Gag initiator methionine considered residue 1. Arrows represent HIV protease cleavage sites. (C) Gag-CFP fusion constructs illustrated above were cotransfected with A3G-YFP. Supernatants were concentrated through a 20% sucrose cushion. A3G-YFP released in microvesicle contamination is estimated by the YFP fluorescence released when A3G-YFP is cotransfected with pcDNA control and represented less than 5% of the signal seen upon expression of full-length Gag. This value was subtracted, and the resulting numbers of Gag-induced relative fluorescence units (RFU) of A3G-YFP released, normalized to the amount of Gag protein present, are shown. (D) Gag-CFP fusion constructs illustrated above were cotransfected with APOBEC3G. Supernatants were concentrated through a 20% sucrose cushion. CFP fluorescence was used to normalize virus-like particle concentration. Proteins were resolved by SDS-polyacrylamide gel electrophoresis, followed by immunoblot analysis with APOBEC3G antisera and pooled HIV patient sera for Gag-CFP.
The following oligonucleotides were used for PCR amplifications of Gag-CFP constructs. Forward oligonucleotides for all Gag-CFP constructs were GTCAAGCTTGTCGACATGG GCGCCCGCGCCAGC-(F); oligonucleotides used for Gag391-CFP were CGGGATCCCTTCACGATCTT GCGCTG-(R); for Gag405-CFP were CGGGATCCGCAGTTGCGGGCGGTGTG-(R); for Gag411-CFP were CGGGATCCCTTC TTGCGGGGGGCGCG-(R); for Gag426-CFP were CGGGATCCGCAGTCCTTCATCTGGTG-(R); and for Gag432-CFP were CGG GATCCATTAGCCTGTCGCTCGGTG-(R). Oligonucleotides used for PCR amplifications of the A3G-YFP construct were GGAATTCGCCACCATGAAGCCTCACTTCAGAAAC-(F) and TCGCCCGGGTTTTCCTGATTCTG GAG-(R); for the YFP-A3G construct were CGGAATTCGATGAAGCCTCACTTCAGA-(F) and TCCCCCGGGTCAGTTTT CCTGATT-(R); and for the A3G-CFP construct were GGCCCGGGATCCATCGCCACCATGGTGAGCAAG-(F) and GAGTCGC GGCCGCTTTACTTGTACAGCTCGTCCATGCCG-(R).
Cells and transfections.
293T cells were used for all studies, maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum and antibiotics at 37°C in 5% CO2, and grown in 100-cm2 tissue culture dishes. Transfections were performed either with the calcium phosphate-bis-buffered saline transfection method or with Lipofectamine 2000 (Invitrogen, Carlsbad, CA), with 10 μg of total plasmid DNA, unless otherwise stated.
Production and purification of immature Gag VLPs.
Cell culture supernatants from transfected 293T cells were harvested 48 to 72 h posttransfection, clarified by centrifugation, filtered through a 0.45-μm-pore-sized filter, and pelleted through a 20% sucrose cushion (100,000 × g for 2 h at 4°C). The pellets were resuspended in 1.0 ml of phosphate-buffered saline (PBS) and analyzed by scanning cuvette fluorometry using a tunable PTI cuvette fluorometer (Photon Technology International, Lawrenceville, NJ).
Antibodies and immunoblotting.
Particle and lysate preparations were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), followed by transfer to nitrocellulose membranes and immunoblotting. Gag was blotted with sera pooled from HIV-seropositive patients. APOBEC3G antisera was provided by Warner C. Greene through the NIH AIDS Research and Reference Reagent Program (30). Secondary horseradish peroxidase-conjugated antibodies for enhanced chemiluminescence film analysis were obtained from Promega (Madison, WI). Secondary fluorescence-conjugated antibodies for detection with a LI-COR Odyssey imaging system were obtained from LI-COR, Inc. (Lincoln, NE).
Gradient analysis of Gag VLPs.
Gag VLPs, purified as described above, were analyzed by centrifugation on linear 20 to 60% sucrose gradients. Particles resuspended in PBS were overlaid on linear 20 to 60% sucrose gradients. Ultracentrifugation was performed at 100,000 × g overnight at 4°C in a Beckman SW41 rotor. Equal fractions were collected, and the density of each fraction was determined with a refractometer. Samples were subsequently diluted in PBS and pelleted at 100,000 × g for 2 h at 4°C in a Sorvall S45A rotor. Samples were resuspended in 100 μl of PBS. Protein content was analyzed by scanning cuvette fluorometry, and RNA content was analyzed by RiboGreen dye incorporation after treatment with RQ1 DNase according to the manufacturer's instructions. Briefly, 10 μl of each fraction was incubated with 0.1 unit/μl of RQ1 DNase and half of the supplied reaction buffer (5 mM MgSO4, 20 mM Tris-HCl, 0.5 mM CaCl2) in PBS for 2 h at 37°C. Samples were then incubated with 0.1% SDS at 65°C for 20 min to lyse particles. An equivalent volume of RiboGreen (Invitrogen, Carlsbad, CA), diluted 200 times in Tris-EDTA buffer, was then added and allowed to sit in the dark for 10 min. Samples were read on a VersaFluor (Bio-Rad Laboratories, Hercules, CA) cuvette fluorometer along with RNA standards. The original concentration of RNA was then calculated.
Isolation of membrane fractions and analysis by scanning cuvette fluorometry.
Cells were harvested for analysis 48 h following transfection. Three 10-cm2 dishes of nearly confluent 293T cells were included for each experimental sample. Cells were washed in phosphate-buffered saline, allowed to swell in hypotonic buffer (10 mM Tris-Cl [pH 8.0], plus protease inhibitors) for 20 min on ice, and broken by Dounce homogenization. For nuclease-treated FRET experiments, the lysis buffer was supplemented with 60 μg/ml of RNase A (QIAGEN Inc., Valencia, CA) and 10 units/ml of RQ1 DNase with buffer (Promega Corp., Madison, WI), while the control lysis buffer was treated with 200 units/ml of RNase inhibitor (New England Biolabs Inc., Beverly, MA). The lysates were then adjusted to 0.1 M NaCl, and the nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 10 min. Postnuclear supernatants containing cytosolic and membrane components were then adjusted to 50% iodixanol from a stock solution of 60% iodixanol (Nycomed Pharma, Oslo, Norway). Forty percent and 10% solutions of iodixanol were layered on top of the 50% iodixanol layer. The preparation was centrifuged in a Beckman SW41 rotor at 41,000 rpm for 2 h at 4°C. The membrane fraction was taken from the 10% to 40% iodixanol interface as a 1-ml sample. Each sample was kept at 4°C and analyzed by fluorometry in a PTI T-format scanning cuvette spectrofluorometer (Photon Technology international, Lawrenceville, NJ). For analysis of CFP emission, to compare the relative amounts of CFP present, samples were excited at 433 nm, and an emission scan ranging from 460 to 550 nm was performed. For analysis of YFP emission, to compare the relative amounts of YFP present, samples were excited at 513 nm, with an emission scan ranging from 524 to 534 nm. For FRET analysis, whole cells or particles resuspended in PBS were excited at 433 nm, and an emission scan ranging from 460 to 550 nm was obtained. It should be noted that a constant amount of untagged Gag expression plasmid was cotransfected in each arm of this experiment. Data were collected from at least three different independent experiments for each expression construct.
Fluorescence microscopy analysis of subcellular localization of A3G-YFP and Gag-CFP.
Live HeLa cells expressing A3G-YFP alone or together with Gag-CFP were examined using a Nikon TE2000 microscope equipped with automated filter wheels, a stage, a z-axis motor, and a digital camera. All images were obtained using the 63× objective; serial sectioning was performed using MetaMorph software (Molecular Devices), and deconvolution was performed using constrained iterative algorithms with Autoquant software. For the images shown in Fig. 8F to H, a digital mirror device (Mosaic Digital Diaphragm, Photonic Instruments, St. Charles, IL) and a 488-nm laser were used to specifically photobleach the internal regions of the cell surrounding bright cytoplasmic bodies of A3G-YFP.
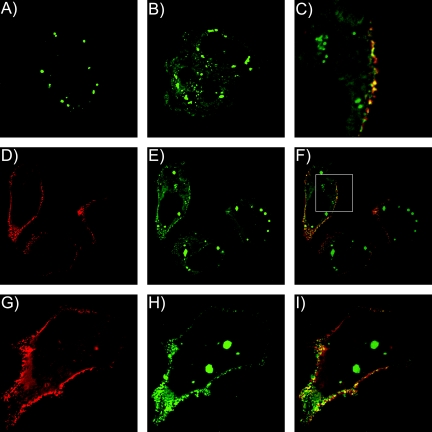
FIG. 8.
Subcellular localization of Gag and A3G-YFP. A3G-YFP and Gag-CFP were expressed in HeLa cells and visualized by optical sectioning with a Nikon TE2000 microscope equipped with an automated stage and z-axis motor, followed by deconvolution using constrained-iterative algorithms. (A) Bright cytoplasmic collections of A3G-YFP are consistent with P body localization. (B) A longer exposure time allowing saturation of P body signal reveals a diffuse cytoplasmic A3G-YFP signal. (C) Magnified section (panel 8F, inset) showing plasma membrane colocalization of Gag (red) and A3G-YFP (green) signals. (D to F) Single-channel images and image overlay indicating Gag signal (red), and A3G-YFP signal (green). (G to I) Similar technique as in panels D to F, but bright collections of A3G-YFP in the cell interiorwere bleached with a 488-nm laser to diminish brightness and facilitate visualization of plasma membrane fluorescence.
Fluorescence microscopy analysis of the subcellular localization of A3G-YFP and A3G-CFP FRET.
Live HeLa cells expressing A3G-YFP alone or together with A3G-CFP were examined using the 63× objective on a Nikon TE2000 microscope. FRET images were obtained by the sensitized emission method, followed by deconvolution with a constrained iterative algorithm (MetaMorph Autoquant software package; Molecular Devices). Briefly, donor and acceptor constants were first generated by the following method. A CFP image and a raw FRET image (CFP excitation/YFP emission) were obtained from cells expressing Gag-CFP alone, and a YFP image and a raw FRET image were obtained from cells expressing Gag-YFP alone. The pixel intensity of the raw FRET image was divided by the pixel intensity of the corresponding CFP image to yield the donor constant, A, of 0.78. This was repeated for YFP to yield the acceptor constant, B, of 0.11. These constants were then used to generate true FRET images from raw FRET images in the following way. Each cell was serially sectioned using MetaMorph software (Molecular Devices) for all three wavelengths (CFP, YFP, and raw FRET). A FRET image was then generated for each z plane using the following formula applied to each pixel: FRET = raw FRET − A × CFP − B × YFP. Finally, the same deconvoluted z plane was selected for the CFP, YFP, and FRET images, and all were treated as follows. MetaMorph imaging software was used to set identical scaling for the YFP and CFP images; scaling of FRET images was set at 50% of the YFP image display value, so that the dimmer FRET images are represented in Fig. 10 with intensities similar to those of the YFP and CFP images. Each set of three images in this figure (CFP/YFP/FRET) was treated in the same manner. Images were colorized as cyan or yellow, exported as red-green-blue tagged image files (TIF), and converted to cyan-magenta-yellow-black (CMYK) images using Adobe Illustrator.
FIG. 10.
Subcellular localization of A3G-CFP and A3G-YFP FRET. (A to C) CFP, YFP, and FRET images obtained from HeLa cells expressing Gag-CFP (negative control). (D to F) CFP, YFP, and FRET images from cells expressing Gag-YFP (negative control). (G to I) CFP, YFP, and FRET images from cells expressing both Gag-CFP and Gag-YFP. (J to L) CFP, YFP, and FRET images from cells expressing A3G-CFP and A3G-YFP (without Gag). Structures consistent with the subcellular localization of P bodies exhibit FRET. (M to O) CFP, YFP, and FRET images from a cell expressing Gag, A3G-CFP, and A3G-YFP. In addition to P body FRET, plasma membrane FRET and a low level of cytoplasmic FRET are shown. Scale bar represents 10 micrometers.
FRET acceptor photobleaching and spectral analysis using laser confocal fluorescence microscopy.
HeLa cells were grown and transfected in cell culture-treated chamber slides and imaged following fixation in 4% paraformaldehyde for 10 min. Images were obtained with a Zeiss LSM 510 laser scanning confocal microscope (Carl Zeiss Inc., Thornwood, NY) equipped with a Meta multichannel detector, making pixel intensity quantitation possible over a range of emission wavelengths. Emission scans were obtained by using Zeiss LSM software following stimulation with a 405-nm laser. A 514-nm laser was used to provide high-intensity pulses of light for photobleaching of YFP to specified regions of interest in the cell. A standard YFP excitation-emission picture was taken before and after the photobleaching.
RESULTS
Specificity of APOBEC3G packaging into Gag VLPs.
We first confirmed the findings of other groups that Gag alone was sufficient to package APOBEC3G. Gag VLPs were produced with 293T cells in the presence and absence of APOBEC3G-HA overexpression. When APOBEC3G-HA was expressed alone, it was not released into the cell culture medium. However, Gag expression and VLP production were sufficient and necessary to release APOBEC3G into the cell culture medium (Fig. 1A). In order to facilitate quantitative studies of the specificity of APOBEC3G packaging into Gag VLPs, a fluorescence system for tracking the relative amounts of APOBEC3G and Gag was developed using an APOBEC3G-YFP fusion protein (hereafter referred to as A3G-YFP). Packaging of A3G-YFP into Gag VLPs was readily demonstrated (Fig. 1B), indicating that APOBEC3G-fluorescent protein fusions could be useful tools to further characterize the determinants of APOBEC3G packaging.
FIG. 1.
APOBEC3G incorporation into Gag VLPs. (A) APOBEC3G bearing an HA tag was expressed in 293T cells in the presence or absence of Gag. Shown are immunoblots probed with anti-HA antibody (top) and pooled HIV patients’ sera (bottom). (B) A3G-YFP or YFP alone was coexpressed with Gag-CFP, and the amounts of YFP incorporated into Gag-CFP particles and corresponding cell lysates were quantified using a scanning cuvette fluorometer. Results are shown as a percentage of total relative fluorescence units (%RFU) released (supernatant/supernatant + cell).
Efficient APOBEC3G incorporation requires the NC basic linker region.
The packaging of APOBEC3G into virions or VLPs has been shown by a number of groups to be dependent on NC (1, 4, 19, 26, 31, 38). We sought to better define the region of NC required for APOBEC3G packaging and to determine if APOBEC3G packaging sequences correlate directly with the “I” domain that localizes to NC (7, 24, 25). In order to do this, we utilized a panel of Gag protein constructs fused to CFP that spans the NC region (Fig. 2B). These constructs have been extensively characterized for the production of particles of normal density, for membrane-binding properties, and for their ability to interact with detergent-resistant membrane fractions (8, 24, 25). We reasoned, therefore, that we could correlate these properties with APOBEC3G packaging.
VLPs were collected and purified from 293T cells cotransfected with A3G-YFP and each of the Gag-CFP fusion constructs. Relative levels of Gag were determined by CFP fluorescence, and relative APOBEC3G levels were determined by YFP fluorescence. Approximately equivalent expression levels of APOBEC3G in each of the cell lysates examined were verified by YFP fluorescence (data not shown). Very small amounts of APOBEC3G were released in the absence of Gag (Fig. 1B), presumably in microvesicles that copurify with the VLPs. The small amount of A3G-YFP released in the absence of Gag (always less than 5% of values in the presence of Gag) was subtracted from the fluorescence values in the presence of each of the Gag-CFP constructs, and the Gag-dependent packaging of A3G-YFP was plotted (Fig. 2C). Values were expressed as YFP relative fluorescence units (counts/s) divided by CFP relative fluorescence units (counts/s).
Gag377-CFP expresses MA, CA, and SP1 of Gag and failed to package APOBEC3G (Fig. 2C). Gag384-CFP includes the minimal “I” domain (24) yet unexpectedly also failed to package APOBEC3G. Additional constructs that did not contain the basic linker region (between amino acids 405 and 411) packaged very little APOBEC3G, despite being released into the medium in the form of VLPs. This was true for Gag405-CFP, a Gag-CFP fusion construct that includes the intact N-terminal zinc finger domain, which failed to package APOBEC3G above background levels (Fig. 2C). However, addition of the six-amino-acid basic linker region represented in Gag411-CFP produced a sharp increase in packaging efficiency. Interestingly, constructs that contain the basic linker plus the second zinc finger region but lack p6 incorporated an enhanced amount of APOBEC3G compared to that of the wild-type Gag, using this assay.
To ensure that this packaging phenotype was not an artifact of the YFP fusion construct, these experiments were repeated with an untagged APOBEC3G construct. In this experiment, the VLP pellets were normalized for loading by measuring the amount of Gag as indicated by CFP fluorescence, and the incorporated APOBEC3G protein was detected by Western blotting on a LI-COR fluorescence imaging system. Gag411-CFP, a construct containing the basic linker, demonstrated APOBEC3G packaging, as did those constructs that included the second zinc finger and the entire NC region (Fig. 2D). Together, these results confirm the dependence of APOBEC3G packaging on the nucleocapsid region and suggest that the basic linker is required for incorporation. Strikingly, the packaging requirements of APOBEC3G were distinct from those of the “I” domain, which have been mapped more proximally to the N-terminal domain of NC (24).
RNA content of particles correlates with APOBEC3G content.
Both APOBEC3G and the NC subunit of Gag are known to bind RNA. The role of RNA binding in packaging has not been completely resolved. While some investigators report RNA-dependent packaging (26, 31, 38), others have reported a direct APOBEC3G-NC interaction (1, 4). If the ability of NC to bind to cellular RNA is the prime determinant of APOBEC3G packaging into HIV VLPs, then we would expect that the amount of RNA packaged in VLPs would correlate with the quantity of APOBEC3G packaged. Furthermore, truncated Gag constructs that lack subdomains of NC may package less cellular RNA. In order to test these hypotheses, VLPs from several of the constructs shown in Fig. 2B were purified by equilibrium density sedimentation on a linear sucrose gradient. Equal fractions were collected, treated with DNase, and analyzed for protein content by CFP fluorescence and for RNA content by incorporation of the fluorescent dye RiboGreen (Molecular Probes). The RNA content of each construct, expressed as an RNA/Gag fluorescence ratio, present in the 1.16 g/ml particle peak is shown in Fig. 3A, and a representative curve demonstrating cofractionation of total RNA and Gag is shown in Fig. 3B. We found measurable RNA incorporation in constructs representing the “minimal” I domain (Fig. 3A, Gag384), which did not increase substantially when the entire N-terminal domain was included (Fig. 3A, Gag405). Inclusion of the NC basic linker substantially increased the RNA/Gag ratio (Fig. 3A, Gag411), and inclusion of full-length NC enhanced this ratio further (Fig. 3A, Gag432). The pattern of RNA incorporation that we observed closely mimicked the relative levels of incorporation of A3G-YFP (compare Fig. 3A and Fig. 2C). These data support a model in which APOBEC3G binds to the cellular RNA that is incorporated into the developing virion and indicate that the quantity of APOBEC3G incorporated is greatest in those particles with the highest RNA content, which in this experiment was the result of nonspecific interactions between NC and cellular RNA. Interestingly, the very low level of APOBEC3G packaged in the absence of the basic linker region (Fig. 2C), despite packaging of some cellular RNA by Gag384 and Gag405, argues that there may be either a specific contribution of the basic linker region of NC for packaging or a threshold level of RNA incorporation that dictates packaging.
FIG. 3.
RNA content of particles correlates closely with APOBEC3G content. Gag-CFP fusion constructs were transfected into 293T cells. Supernatants were purified on linear 20 to 60% sucrose gradients. Fractions were collected from the top of the gradient, treated with RQ1 DNase, stained with RiboGreen, and subjected to analysis by fluorometry. (A) RNA content of particles in peak fractions is shown normalized to Gag-CFP content (RNA:Gag ratio). RFU, relative fluorescence units. (B) Sedimentation pattern of Gag-CFP VLPs (black squares) and associated RNA (gray circles).
The NC basic linker is required for membrane recruitment of APOBEC3G.
Particle budding is known to occur at the plasma membrane in both primary T cells and 293T cells. Therefore, we determined whether Gag constructs that packaged APOBEC3G also recruited it to membrane fractions. 293T cells were cotransfected with A3G-YFP and the panel of Gag-CFP truncation mutants, and membranes were isolated via flotation on iodixanol gradients. The membrane-enriched fraction was collected and assayed along with the cell lysates for APOBEC3G and Gag protein content by YFP and CFP fluorescence intensity, respectively. Results are expressed as the percentage of membrane fluorescence compared to total cell lysate fluorescence (Fig. 4). Marked recruitment of cellular APOBEC3G to cellular membranes was observed upon expression of Gag (Fig. 4A). The recruitment of APOBEC3G to the membrane fraction largely mirrored the results described above for particle incorporation: those constructs that lacked the basic linker failed to recruit APOBEC3G into membrane fractions. In parallel, we measured the membrane flotation of Gag (Fig. 4B). Remarkably, Gag membrane association did not demonstrate a requirement for the basic linker (Fig. 4B). The association of Gag constructs bearing only the N-terminal subdomain of NC with membranes is consistent with our previous description of the I domain (24) and indicates that the determinants mapped as the I domain are distinct from the packaging requirements for APOBEC3G. These data support a more specific role for the basic linker region in APOBEC3G interaction and recruitment to cellular membranes.
FIG. 4.
Nucleocapsid determinants of APOBEC3G membrane recruitment. Gag-CFP fusion constructs illustrated in Fig. 2B were cotransfected with A3G-YFP. The protein content of membrane-enriched fractions generated by flotation on iodixanol step gradients was determined by fluorescence spectrophotometry. The percentage of protein present in the membrane fraction was calculated by dividing by the amount of protein present in the total cell lysate. (A) YFP signal was used to determine the amount of APOBEC3G present in membrane fractions for each indicated construct. RFU, relative fluorescence units. (B) CFP signal was used to determine the amount of Gag protein present in membrane fractions for each indicated construct.
RNase disrupts APOBEC3G binding to NC.
Indirect evidence has indicated the importance of RNA in the recruitment of APOBEC3G into Gag VLPs. To directly assess the question of RNA involvement, a glutathione S-transferase (GST) sedimentation assay was employed. Cell lysates expressing APOBEC3G-HA were added to each Gag subunit, fused to GST, and conjugated to glutathione Sepharose beads. After extensive washing, it was found that APOBEC3G-HA bound only to the NC region and not to other Gag subunits (Fig. 5A). When cell lysates were treated with nucleases, this binding was abolished (Fig. 5B). These results are consistent with those already published by several groups, further supporting an important role for RNA in particle incorporation of APOBEC3G (26, 31, 38).
FIG. 5.
RNase disrupts APOBEC3G binding to NC. (A) Cell lysates from 293T cells expressing APOBEC3G-HA were added to glutathione agarose beads containing the indicated bacterially purified Gag subunits. After extensive washing, the glutathione beads were analyzed by SDS-PAGE, followed by both Coomassie blue staining and immunoblot analysis with anti-HA for APOBEC3G-HA. In, 5% of input. (B) As described above, following the addition of RNase and DNase to the cell lysates prior to performing GST pulldown.
APOBEC3G multimers bind to RNA in cells and particles.
FRET between CFP and YFP moieties has been used to detect interactions between Gag molecules in living cells. Briefly, stimulation of a CFP molecule at 433 nm normally leads to the emission of a strong peak at 475 nm, a shoulder from 490 to 510 nm, and a broad trail from 510 to 550 nm (Fig. 6A, A3G-CFP emission spectra). YFP molecules are maximally stimulated at 514 nm; therefore, if a YFP molecule is in contact (less than 100 angstroms) from an excited CFP molecule, the overlap in CFP emission and YFP excitation will allow energy transfer from the CFP to the YFP molecule. This will lead to the YFP molecule emitting at 527 nm.
FIG. 6.
APOBEC3G is packaged as multimers that interact with RNA. (A) The indicated constructs were cotransfected into 293T cells. Cells were resuspended in PBS and read directly with a scanning cuvette fluorometer. FRET was measured by stimulating the CFP fluorophore at 433 nm, and the FRET peak was observed at 527 nm. Gray squares, FRET curve for A3G-YFP/A3G-CFP and GagCFP/GagYFP (open squares). Open circles, pEYFP coexpressed with A3G-CFP. Closed triangles, GagCFP VLPs. (B) Relative levels of cellular YFP expression are shown for the experiment depicted in panel A, as determined by peak YFP output following excitation of cell lysates at 514 nm. (C) Cell lysates prepared by treatment with hypotonic buffer and Dounce homogenization. Half of the cell lysates were treated with RNase A and DNase RQ1 prior to FRET analysis, while RNA in the control lysates were preserved with RNase inhibitor. Gray squares, FRET curve for A3G-YFP/A3G-CFP; open circles, loss of FRET in nuclease-treated A3G-YFP/A3G-CFP lysates. Closed triangles, A3G-CFP curve and treated (closed diamonds). (D) Relative levels of cellular YFP expression are shown for the experiment depicted in panel A, as determined by peak YFP output following excitation of cell lysates at 514 nm. (E) VLPs created by coexpressing pVRC3900Gag with the indicated constructs in 293T cells. Supernatants were concentrated through a 20% sucrose cushion, resuspended in PBS, and analyzed by scanning cuvette fluorometry. Open circles, FRET curve for A3G-CFP/A3G-YFP and for A3G-CFP/YFP-A3G (gray boxes). Closed triangles, A3G-CFP coexpressed with pEYFP. (F) Relative levels of VLP YFP content are shown for the experiment depicted in panel C, as determined by peak YFP output following excitation of VLPs at 514 nm. Note that transfection of an untagged Gag expression construct was included in each of the transfections at a constant amount.
APOBEC molecules are known to dimerize when binding to RNA (14, 35). Therefore, in an effort to develop an assay to detect A3G-RNA-A3G complexes in live cells, A3G-YFP and A3G-CFP were coexpressed in 293T cells, and the emission spectra of whole cells, resuspended in PBS, were obtained with a spectrofluorometer using an excitation wavelength of 433 nm. Cells expressing A3G-YFP and A3G-CFP exhibited a curve representative of efficient fluorescence energy transfer (Fig. 6A, gray squares), as did Gag-CFP and Gag-YFP (Fig. 6A, open squares), while cells expressing A3G-CFP and a control YFP resulted in a CFP emission peak with no YFP emission peak (Fig. 6A, open circles). Identical results were obtained with YFP-A3G (results not shown). Similar levels of YFP emission were obtained upon YFP excitation for each experimental condition, indicating that the protein levels were similar (Fig. 6B).
To determine whether RNA was a necessary component of these APOBEC3G complexes, cell lysates from cells expressing A3G-YFP and A3G-CFP were prepared and treated with RNase A and DNase RQ1, while RNA in control lysates was preserved by the addition of RNase inhibitor. Control lysates expressing A3G-YFP and A3G-CFP exhibited a curve representative of efficient fluorescence energy transfer (Fig. 6C, shaded squares), while nuclease-treated lysates resulted in a CFP emission peak with no YFP emission peak (Fig. 6C, open circles). The A3G-CFP emission curve remained unchanged by nuclease treatment (Fig. 6C, closed triangles, closed diamonds). The lack of FRET in the nuclease-treated lysates was not a result of lower protein concentration or degradation since similar levels of YFP emission were obtained upon YFP excitation for each experimental condition (Fig. 6D). This establishes that FRET is an effective tool with which to measure the presence of APOBEC3G multimers bound to RNA and that RNA is required for APOBEC3G multimerization.
We next employed A3G-CFP/A3G-YFP FRET to determine whether APOBEC3G is incorporated into particles as a multimeric complex bound to RNA. Gag VLPs were generated in cells expressing Gag, A3G-YFP, and A3G-CFP. The emission spectra of these particles were obtained with a spectrofluorometer using an excitation wavelength of 433 nm. Particles containing A3G-CFP and A3G-YFP (Fig. 6E, open circles) or A3G-CFP and YFP-A3G (Fig. 6E, gray squares) exhibited a curve representative of efficient fluorescence energy transfer. However, particles containing A3G-CFP and a control YFP resulted in a CFP emission peak with no YFP emission peak (Fig. 6E, closed triangles). Increased YFP overexpression was used in this experiment to increase the free YFP content of VLPs. We conclude that the FRET detected in Gag VLPs represents APOBEC3G multimers bound to RNA that have become incorporated in VLPs.
Lack of support for a direct Gag-APOBEC3G interaction by FRET.
To further analyze the Gag-APOBEC3G interaction in living cells and particles, we asked whether we could detect a direct Gag-APOBEC3G interaction by a FRET assay. Cells expressing Gag-YFP and Gag-CFP exhibited a curve representative of efficient fluorescence energy transfer (Fig. 7A, gray circles) as a positive control. In contrast, cells expressing Gag-YFP and A3G-CFP revealed a CFP emission peak with no YFP emission peak, indicating that no FRET occurred (Fig. 7A, open squares). Results for Gag-YFP and A3G-CFP were identical to those of the negative control of Gag-YFP coexpressed with CFP (Fig. 7A, closed triangles). Similar levels of YFP emission were obtained upon YFP excitation for each experimental condition, indicating that differential protein levels did not account for the differences in positive controls and the test molecules (Fig. 7B).
FIG. 7.
Lack of FRET between APOBEC3G and Gag. (A) The indicated constructs were cotransfected into 293T cells. Cells were resuspended in PBS and read directly with a scanning cuvette fluorometer. FRET was measured by stimulating the CFP fluorophore at 433 nm, and the FRET peak was observed at 527 nm. Gray circles, FRET curve for Gag-YFP/Gag-CFP. Open squares, Gag-YFP coexpressed with A3G-CFP and pECFP (closed triangles). (B) Relative levels of cellular YFP expression are shown for the experiment whose results are depicted in panel A, as determined by peak YFP output following excitation of cell lysates at 514 nm. (C) VLPs created by coexpressing the indicated constructs in 293T cells. Supernatants were concentrated through a 20% sucrose cushion, resuspended in PBS, and analyzed by scanning cuvette fluorometry. Gray circles, FRET curve for Gag-CFP/Gag-YFP. Open squares, Gag-CFP coexpressed with A3G-YFP and pEYFP (closed triangles). (D) Relative levels of VLP YFP content are shown for the experiment whose results are depicted in panel C, as determined by peak YFP output following excitation of VLPs at 514 nm.
Gag VLPs were then generated in cells expressing Gag, Gag-CFP, and A3G-YFP. In the same system, A3G-YFP was replaced by Gag-YFP as a FRET-positive control and YFP-Mem (Clontech) as a FRET-negative control. The emission spectra of these particles were obtained with a spectrofluorometer using an excitation wavelength of 433 nm. Particles containing Gag-CFP and Gag-YFP exhibited a curve representative of efficient fluorescence energy transfer (Fig. 7C, gray circles), while particles containing Gag-CFP and A3G-YFP resulted in a CFP emission peak with no YFP FRET peak (Fig. 7C, open squares). The lack of FRET in particles containing Gag-CFP and A3G-YFP was not due to lower concentrations of A3G-YFP in this experiment, as similar levels of YFP emission were obtained for each experimental condition (Fig. 7D). These results indicate that we were unable to detect direct interactions between Gag and APOBEC3G in cells or VLPs using this technique. Interpretation of these results must include the recognition that a negative result in a CFP-YFP FRET assay does not rule out a direct interaction, as FRET can be dependent on both the position and the orientation of the CFP and YFP molecules (9, 23).
APOBEC3G is recruited to the plasma membrane by Gag.
APOBEC3G has been described as diffuse in the cytoplasm of a cell but has recently been shown to collect in dense cytoplasmic bodies identified as mRNA processing (P) bodies by Wichroski and colleagues (35). We assessed the subcellular localization of APOBEC-YFP using wide-field deconvolution fluorescence microscopy. Very bright, punctate collections of A3G-YFP were noted in the cytoplasm of most transfected cells, consistent with the report of Wichroski et al. (Fig. 8A). Under these conditions, diffusely cytoplasmic APOBEC3G could be visualized only under conditions allowing saturation of pixels in P bodies (Fig. 8B). We observed little evidence of APOBEC3G on the plasma membrane, even in these overexposed images. When Gag-CFP was coexpressed with A3G-YFP, a mixed pattern was observed. The most intense A3G-YFP signal was consistently present in P bodies, but a subset of cells demonstrated plasma membrane A3G-YFP signal (Fig. 8D to F, leftmost cell). Note that plasma membrane colocalization was not apparent in the majority of cells (Fig. 8E). When it was observed, however, the colocalization appeared in bright punctate patterns along the plasma membrane, suggestive of sites of particle assembly, as indicated in Fig. 8C (a higher magnification view of the cell shown in Fig. 8E). In many cells in which A3G-YFP was expressed, the extreme brightness of the P body did not allow assessment of potential colocalization with Gag at the plasma membrane. To better examine these cells, we performed laser photobleaching of the interior region surrounding the P bodies, followed by a longer exposure of the remaining fluorescence signal in the cell. Figure 8G to I shows one such cell in which Gag-CFP is in red, A3G-YFP is shown in green, and plasma membrane colocalization is demonstrated as yellow pixels (Fig. 8H). Little apparent colocalization of Gag was noted in the P bodies, but there appeared to be recruitment of A3G-YFP from the cytoplasm to punctate spots on the plasma membrane. We conclude from these data that, although overexpressed A3G-YFP appears most intensely in P bodies, a subset of A3G-YFP is recruited to the plasma membrane, where it colocalizes with Gag in the developing particle.
APOBEC3G-APOBEC3G interactions were observed in structures consistent with P bodies by confocal spectral analysis.
The laser confocal microscope allows the spectral separation of light emitted from selected pixels when it is equipped with a multichannel detector. To further establish the presence of A3G-A3G FRET and to determine the localization of this FRET, we analyzed the emission spectra from different regions of the cell before and after photobleaching the A3G-YFP acceptor molecule. If a transfer of energy were occurring between the CFP and YFP molecules, the obliteration of YFP molecules by photobleaching would lead to retention of energy by the CFP molecules and an increase in CFP fluorescence. We had previously performed spectral analysis with a Zeiss LSM510 Meta confocal microscope to demonstrate the interaction of Gag-CFP and Gag-YFP on the plasma membrane (7). We repeated this analysis using Gag-CFP and Gag-YFP as positive controls in parallel with our APOBEC test constructs. Briefly, HeLa cells expressing Gag-CFP and Gag-YFP were imaged showing YFP localization before and after bleaching of the YFP at 514 nm (Fig. 9A and B). An emission scan of the cells shown in Fig. 9A and B was performed using excitation of CFP at 405 nm. The spectrum from a bleached section of the plasma membrane (region of interest 1 [ROI1]) revealed that after bleaching, the YFP peak at 530 nm was reduced while the CFP peak at 475 nm increased in fluorescent intensity (Fig. 9C). The emission spectra from a control region of the plasma membrane (region of interest 2) remained unchanged (Fig. 9D). This confirms the existence of FRET between Gag-CFP and Gag-YFP molecules on the plasma membrane. Next, we employed this technique to determine the presence of a FRET interaction between A3G-CFP and A3G-YFP in structures consistent with P body subcellular localization. Figure 9E and F shows the localization of A3G-YFP in these cells before and after bleaching the ROI1. The emission spectra from pixels contained within the P body at ROI1 reveal that the decrease in the YFP peak following bleaching is accompanied by a significant increase in the CFP peak (Fig. 9G). P body pixels in the nearby but unbleached region of interest 2 show unchanged spectra (Fig. 9H). Bleached cytoplasmic locations close to ROI1 exhibit neither a FRET peak nor an increase in CFP fluorescence following bleaching (data not shown). This confirms that A3G-A3G interactions can be observed by FRET and occur primarily in P bodies.
FIG. 9.
Analysis of A3G-A3G interactions by confocal microscopy and fluorescence acceptor photobleaching. (A) Gag-CFP and Gag-YFP were cotransfected in HeLa cells, and images were obtained with a Zeiss LSM 510-Meta confocal microscope. The image represents YFP excitation-emission before photobleaching. The arrows indicate the selected plasma membrane region of interest to be bleached (ROI1) and the control region (ROI2). (B) The same cell as that shown in panel A is depicted following photobleaching at 514 nm in the indicated square. (C) Emission scans were obtained from region of interest 1, with excitation at 405 nm (CFP excitation), before (dashed line) and after (solid line) photobleaching of cells shown in panels A and B. (D) Emission scans were obtained from the control region of interest 2, with excitation at 405 nm, before (dashed line) and after (solid line) photobleaching of cells shown in panels A and B. (E) A3G-CFP and A3G-YFP were cotransfected in HeLa cells, and the image shows the distribution of A3G-YFP prior to bleaching. The regions of interest selected are consistent with P body localization. (F) The same cell as that shown in panel E is shown after photobleaching of the indicated square. (G) Spectra obtained from ROI1 before (dashed line) and after (solid line) photobleaching from cells shown in panels E and F. (H) Spectra obtained from ROI2 before (dashed line) and after (solid line) photobleaching from cells shown in panels E and F. Scale bar represents 10 micrometers.
APOBEC3G multimers are observed in P bodies and at the plasma membrane following Gag expression.
In order to image the subcellular location of A3G-A3G interactions during Gag expression, we next employed FRET microscopy with living cells. Using distinct filter sets, we obtained individual CFP, YFP, and CFP-YFP (FRET) images and corrected each FRET image for donor bleedthrough and acceptor cross-stimulation using the algorithm described in Materials and Methods. Obtaining true FRET images using filter sets is complicated, even with the best filters and software algorithms, so we included known positive controls that were treated identically to the APOBEC3G expression experiments. Controls included HeLa cells expressing Gag-CFP alone (Fig. 10A to C) and cells expressing Gag-YFP alone (Fig. 10D to F). These two negative controls were critical to establish that bleedthrough and cross-stimulation from CFP and YFP, respectively, can effectively be measured and subtracted from the final FRET image. We next employed this technique to examine the location of FRET in cells expressing Gag-CFP and Gag-YFP. Consistent with the findings shown in Fig. 9A to D and our previously published findings (7) Gag-Gag FRET was detected at the plasma membrane and at distinct intracellular puncta (Fig. 10I). Finally, we employed this assay to observe the locations of A3G-A3G FRET with and without Gag expression. Consistent with the results shown in Fig. 9E to H, cells expressing A3G-CFP and A3G-YFP exhibit FRET only in the P bodies (Fig. 10J to L). However, the expression of Gag together with A3G-CFP and A3G-YFP resulted in detectable FRET signal at the plasma membrane in a subset of cells examined (Fig. 10M to O). This is consistent with a model in which Gag recruits A3G-RNA-A3G multimers to the plasma membrane.
DISCUSSION
A number of published reports have established that the NC region of Gag is necessary for APOBEC3G packaging into virions or VLPs. The majority of reports support an essential role for RNA in mediating the APOBEC-NC interaction (26, 31, 38), while some investigators have found evidence for a direct protein-protein interaction (1, 4). Our findings support an RNA-dependent incorporation of APOBEC3G into particles, using several lines of evidence. First, in vitro interactions of APOBEC3G and NC were shown to be RNase sensitive, as reported by others. Second, we found that the incorporation of APOBEC3G into a series of serially truncated Gag protein constructs correlated well with the RNA-to-Gag ratio of the released particles. Finally, data from our FRET studies provide indirect evidence for the requirement of RNA in packaging by revealing both that RNA is required for APOBEC3G-APOBEC3G interactions and that these APOBEC3G multimers are subsequently packaged into VLPs. Under the same experimental conditions in which APOBEC3G-APOBEC3G FRET was detected, and in which Gag-Gag FRET is readily detected, we failed to detect Gag-APOBEC3G FRET. Although this may be due to the fact that there is not a direct protein-protein interaction between Gag and APOBEC3G in cells or virions, we recognize that the position and orientation of the CFP and YFP tags on Gag and APOBEC3G may simply not support FRET. The presence of a flexible linker region between both proteins and the fluorescent moieties should have minimized these potential orientation effects (9, 23), but a negative result still is not definitive. Taken together, our findings support a model in which APOBEC3G multimers form on cellular RNA as part of an RNA-protein complex that is subsequently recruited into the developing particle through interactions with NC.
The N-terminal subdomain of NC has been shown to be sufficient to allow the formation of particles of normal retroviral density. The “I” domain, which is located within this region, is widely believed to contribute to Gag-Gag multimerization, mediated by NC-RNA interactions. We were surprised in this study to find that the minimal “I” domain of Gag, an essential assembly determinant, did not correspond precisely with the packaging requirements for APOBEC3G. Rather, in this study only those constructs that included the first zinc finger and basic linker packaged APOBEC3G. In contrast to our study, Luo et al. (19) showed that one Gag protein truncated proximal to the first zinc finger packaged APOBEC3G. While the reason for this discrepancy is not certain, we suggest that it reflects the fluorescence quantitation performed in our study versus the semiquantitative Western blot analysis performed in the study by Luo et al. In agreement with their results, however, we observed a minor amount of APOBEC3G in all truncated constructs (Fig. 2D). The quantitative packaging data, combined with evidence that membrane recruitment requires the same critical region, emphasizes the importance of the basic linker region for APOBEC3G interaction and packaging.
A unique aspect of the present study was the finding that Gag recruits APOBEC3G to the plasma membrane. Three lines of evidence support this finding. First, membrane flotation data indicated a redistribution of APOBEC3G to cellular membranes following Gag expression. Second, APOBEC3G colocalized with Gag at the plasma membrane. Third, APOBEC3G-RNA-APOBEC3G complexes, which were found predominantly in P bodies in the absence of Gag expression, were detected at the plasma membrane in cells cotransfected with Gag. Recruitment of APOBEC3G to the plasma membrane is somewhat intuitive, since Gag is responsible for the incorporation of RNA and APOBEC3G, and Gag must interact with membranes for intracellular trafficking and budding from the cell. It is tempting to speculate that Gag recruits APOBEC3G at early stages following translation on cytoplasmic ribosomes, in conjunction with viral genomic RNA. In this model, a Gag-RNA-APOBEC3G complex would then traffic along endosomal pathways to the plasma membrane for budding (or to the multivesicular body in the infected macrophage). We were not able to demonstrate significant colocalization of Gag and APOBEC3G within the cytoplasm of the cell, in particular in the punctate cytoplasmic collections reported by others to be P bodies (34, 35). We note that occasional faint colocalization was seen at these sites (data not shown). However, we were able to detect colocalization of Gag with APOBEC3G at the plasma membrane, especially in cells in which the intensely bright signal from P bodies was reduced by photobleaching. It is possible that the population of APOBEC3G present in the dense P bodies is itself incorporated into particles, but perhaps it is more likely that Gag recruits APOBEC3G from the cytoplasmic pool that then multimerizes on the viral RNA. Kozak et al. recently reported the binding of APOBEC3G to a pool of RNA that reversibly shuttles between polysomes and stress granules (16); it may be this pool that is packaged in the VLP in the absence of HIV genomic RNA. We note the caveat that the intensity of the signal observed in our microscopic study results from overexpression of APOBEC3G fused to YFP rather than endogenous APOBEC3G. However, these results are consistent with those of others who have reported very intense cytoplasmic (P body) collections of APOBEC3G (34, 35).
We found that multimeric APOBEC3G complexes were packaged into VLPs. Opi et al. reported a monomeric APOBEC3G mutant (C97A) that was packaged and remained catalytically active (22). However, the majority of APOBEC3G in cells is present in an RNase-sensitive multimeric form (22), and several reports have suggested that APOBEC3G binds to itself in cells (14, 35) in an RNA-dependent manner (35), producing either the head-to-tail dimers traditionally associated with APOBEC3G family members (14) or the high-molecular-mass complexes observed in activated T cells and transfected 293T cells (5, 17). We used a FRET assay to demonstrate that APOBEC3G multimers were incorporated into virions. These oligomers were also detected in cell lysates and could be disrupted by RNase treatment. These data suggest that APOBEC3G multimerizes on cellular or viral RNA, and it is APOBEC3G multimers complexed with RNA that are recruited into developing particles. It makes sense that these complexes represent the enzymatically active enzyme during the reverse transcription process, as they are recruited directly into the viral core that becomes the nucleus for reverse transcription upon infection of a cell. It is also possible that monomeric APOBEC3G is formed during the reverse transcription process as the RNA template is degraded.
In summary, our results provide strong support for the importance of RNA in mediating the incorporation of APOBEC3G into virions or VLPs. This incorporation is dependent upon regions of NC that allow efficient RNA incorporation, especially the basic linker region. We suggest a model in which APOBEC3G multimerizes on RNA and the APOBEC3G-RNA complex is then recruited by Gag into the developing particle.
Acknowledgments
This work was supported by R01 AI40338 and R01 AI67101.
We thank Michael Malim for providing the APOBEC3G-HA expression construct. We also thank Aaron Derdowski for help with FRET fluorometry and confocal microscopy, Jason Hammonds for help with VLP production and purification, and Kelie Reece for helpful suggestions. We thank the Winship Cancer Institute Cell Imaging and Microscopy Core for their service and assistance.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 67:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 6.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 7.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding, L., A. Derdowski, J. J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.dos Remedios, C. G., and P. D. Moens. 1995. Fluorescence resonance energy transfer spectroscopy is a reliable “ruler” for measuring structural changes in proteins. Dispelling the problem of the unknown orientation factor. J. Struct. Biol. 115:175-185. [DOI] [PubMed] [Google Scholar]
- 10.Douaisi, M., S. Dussart, M. Courcoul, G. Bessou, R. Vigne, and E. Decroly. 2004. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 321:566-573. [DOI] [PubMed] [Google Scholar]
- 11.Feng, F., A. Davis, J. A. Lake, J. Carr, W. Xia, C. Burrell, and P. Li. 2004. Ring finger protein ZIN interacts with human immunodeficiency virus type 1 Vif. J. Virol. 78:10574-10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y., W.-P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 15.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The Anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281:29105-29119. [DOI] [PubMed] [Google Scholar]
- 17.Kreisberg, J. F., W. Yonemoto, and W. C. Greene. 2006. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 203:865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 19.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X.-F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defense by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 21.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 22.Opi, S., H. Takeuchi, S. Kao, M. A. Khan, E. Miyagi, R. Goila-Gaur, Y. Iwatani, J. G. Levin, and K. Strebel. 2006. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 80:4673-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson, G. H., D. W. Piston, and B. G. Barisas. 2000. Forster distances between green fluorescent protein pairs. Anal. Biochem. 284:438-440. [DOI] [PubMed] [Google Scholar]
- 24.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 27.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 28.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 29.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 30.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 12:591-601. [DOI] [PubMed] [Google Scholar]
- 31.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 32.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 33.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wichroski, M. J., K. Ichiyama, and T. M. Rana. 2005. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J. Biol. Chem. 280:8387-8396. [DOI] [PubMed] [Google Scholar]
- 35.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 37.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 38.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]