Abstract
Since the mid-1990s, lethal infections of koi herpesvirus (KHV) have been spreading, threatening the worldwide production of common carp and koi (both Cyprinus carpio). The complete genome sequences of three KHV strains from Japan, the United States, and Israel revealed a 295-kbp genome containing a 22-kbp terminal direct repeat. The finding that 15 KHV genes have clear homologs in the distantly related channel catfish virus (ictalurid herpesvirus 1) confirms the proposed place of KHV in the family Herpesviridae, specifically in the branch with fish and amphibian hosts. KHV thus has the largest genome reported to date for this family. The three strains were interpreted as having arisen from a wild-type parent encoding 156 unique protein-coding genes, 8 of which are duplicated in the terminal repeat. In each strain, four to seven genes from among a set of nine are fragmented by frameshifts likely to render the encoded proteins nonfunctional. Six of the affected genes encode predicted membrane glycoproteins. Frameshifts or other mutations close to the 3′ ends of coding sequences were identified in a further six genes. The conclusion that at least some of these mutations occurred in vivo prompts the hypothesis that loss of gene functions might be associated with emergence of the disease and provides a basis for further investigations into the molecular epidemiology of the virus.
Fish are the most numerous and species-rich vertebrate group, colonizing diverse aquatic habitats throughout the world (31). Prominent among the freshwater fishes are members of the family Cyprinidae whose most well-known species, the common carp (Cyprinus carpio), has been domesticated and widely distributed from a suspected origin in Europe to freshwater habitats throughout the world (3). Culture of common carp provides a key source of protein for human consumption in Asia, Europe, and the Middle East (3, 35). The practice of selectively breeding common carp for striking body colors, leading to a type called koi, has created a worldwide market for the ornamental fish hobby and competitive showing, where individual fish may exceed $100,000 in value (3). Intensive culture of common carp and large-scale movements of live koi, often in the absence of health certifications, have unfortunately contributed to the rapid distribution of certain diseases. Among the most serious has been the epidemic losses among koi caused by a herpesvirus (22).
Koi herpesvirus (KHV) was first identified in 1998 as the cause of mass mortality among juvenile and adult koi and among common carp cultured in Israel, the United States, and Germany (5, 24, 25). Considerably more information has since been obtained on the properties of this virus, including its host range, the effects of water temperature on disease outcome, development of detection methods, and novel attempts at control (1, 4, 15-17, 20, 26, 27, 34, 37-39). The virus has now been found associated with mass mortality events on most continents, including countries throughout Europe, the United States, Japan, Indonesia, South Africa, Thailand, Taiwan, China, and Malaysia (22, 29, 42, 45). Outbreaks continue to occur among cultured populations of koi in most of these areas and, alarmingly, epidemics have been observed among wild common carp populations in the United Kingdom, Japan and, most recently, several states in the United States (42; P. Bowser and J. Grizzle, unpublished data; K. Way, unpublished data; R. Hedrick, unpublished data).
Taxonomically, members of the family Herpesviridae consist of a linear, double-stranded DNA genome packaged within an icosahedral capsid of characteristic architecture that is surrounded by a proteinaceous tegument layer and finally by a host-derived envelope containing virus glycoproteins (11). Phylogenetically, herpesviruses (HVs) fall into three major clades that are barely related at the sequence level: those that infect mammals, birds, and reptiles (the mammalian HV clade), those that infect fish and amphibians (the fish HV clade), and a single virus that infects an invertebrate, the oyster (the bivalve HV clade). A substantial number of viruses in the mammalian HV clade have been sequenced, but only three in the fish HV clade (ictalurid HV 1, IcHV-1, also known as channel catfish virus, and two ranid HVs [7, 10]) and one in the bivalve HV clade (ostreid HV 1, OsHV-1, also known as oyster HV [12]). It has been proposed that these clades should be classified as three families encompassed by an order, the Herpesvirales (8, 12). The families would be the revised Herpesviridae, the Alloherpesviridae, and the Malacoherpesviridae, respectively.
Early observations demonstrated the clear developmental and morphological affinities of KHV to the HVs (24), but subsequent findings prompted others to suggest that the virus should not be considered a member of the Herpesviridae (26, 38, 39). Key factors in this argument included the estimated genome size of KHV (277 kbp), which is greater than that observed previously among HVs (125 to 245 kbp), and the failure to demonstrate convincing genetic relationships between KHV and recognized HVs from limited sequence analyses. However, comparisons conducted on a more appropriate basis made it clear that KHV is related to fish HVs (46). KHV is related closely to cyprinid HVs 1 and 2 (CyHV-1 and CyHV-2), the agents associated with carp pox and hematopoietic necrosis in goldfish, respectively (28, 41), and distantly to IcHV-1 and ranid HV 1 (RaHV-1, also known as Lucké tumor herpesvirus of frog) (46). Therefore, KHV has been proposed formally as a member of the Alloherpesviridae under the species name Cyprinid herpesvirus 3.
In the present study, we describe the complete genome sequences of three KHV strains isolated from Japan (strain J), the United States (strain U), and Israel (strain I). We confirm that KHV shares significant similarities to fish HVs and belongs as a new species in the family Herpesviridae. The comparative information gained from the three strains also provides insights into epidemiologic features of an emerging disease threatening koi and carp worldwide.
MATERIALS AND METHODS
Growth of viruses.
KHV strain J was isolated from a dead koi on a farm in Japan. Strain U was isolated in 2003 from moribund koi from a producer in the eastern United States who had attended a koi show in New York. Strain I was isolated in 1998 from adult koi during large mortalities in a producer plant on the coastal plain of Israel. The viruses were isolated and passaged in the KF-1 koi fin cell line (24). Bulk virus preparations were made (passages 3, 12, and 30 for J, U, and I, respectively) and purified by sucrose gradient ultracentrifugation (16). Genome DNA was purified by phenol extraction and ethanol precipitation.
DNA sequencing.
The complete genome sequences were determined commercially (Hitachi Instruments Service Co., Ltd., Tokyo, Japan) by standard shotgun sequencing. Briefly, fragments in the 2- to 5-kbp size range generated from purified viral DNA by passage through a syringe needle were cloned into pUC118 to construct a plasmid library. The KHV DNA inserts were sequenced from both ends of the plasmids by using the M13 forward and reverse primers with a BigDye Terminator v3.1 cycle sequencing kit and ABI 3700 and 3730xl DNA analyzers.
DNA sequence analysis.
The DNA sequences were assembled by using Phrap (14), employing the quality files and default settings to produce a consensus sequence that was edited by using Consed (18). The final consensus sequence in each case represented an average eightfold redundancy at each base position. Gap closure was achieved by primer walking of gap-spanning clones and sequencing of PCR products. The sequence traces were checked carefully at positions where coding regions were fragmented in order to rule out errors. The genome termini in U were located precisely by PCR amplification of virion DNA that had been flush ended and ligated to a partially double-stranded adaptor oligonucleotide as described previously (13), using a primer matching part of the adaptor plus 5′-CGCAGTAGGCCTTGACCAGCA-3′ for the left terminus or 5′-AGGGTCTGAACATGGCTTAGG-3′ for the right terminus. The locations of the termini determined from sequencing of 12 plasmid clones of each product were assumed to be identical in J and I.
The GCG suite (Accelrys) and other programs (19, 33) were used for standard analyses of the sequences. ATG-initiated open reading frames (ORFs) of >50 codons were evaluated initially for protein coding potential by using GAMBLER, which semiautomates analysis of the output from genome assembler software, assigns ORFs automatically, and carries out homology searching (40). Additional homology searches were carried out by using the FastA (36) and BLAST (2) tools, and alignments were made by using CLUSTAL W (44). The preliminary gene map was refined by using GCG Codonpreference, which measures codon preference and third position codon G+C bias on the basis of statistics generated from standard genes. Since KHV happens to be strongly biased in these measures, the analysis proved useful in revising the preliminary map, as a result of which most ORFs were confirmed as likely to be protein coding and several ambiguities were clarified. Four large KHV ORFs with a high probability of encoding proteins were used as the standards in Codonpreference: ORF79, ORF92, ORF107, and ORF62. Additional criteria routinely used in HV genome analysis were used, in particular the lack of extensive overlap between coding regions and the presence of polyadenylation signals downstream from ORFs or families of similarly oriented ORFs. Splicing is relatively rare in HVs, and predictions for KHV based on the presence of appropriately located splice donor and acceptor signals were kept to a minimum.
The availability of an incomplete sequence database for the majority of the related CyHV-1 genome (T. Waltzek and R. Hedrick, unpublished data) enabled most predicted protein-coding regions in KHV (including splice sites) to be confirmed by homology and helped clarify many remaining difficulties. A few aspects of the interpretation were also strengthened by comparisons with homologous ORFs in the more distantly related IcHV-1 genome (7).
Analysis of mutated regions.
Regions containing frameshift mutations in one or another strain were amplified by standard PCR from U at passages 2 and 12 (the former from infected cell DNA and latter from infected cell DNA and purified virion DNA). In addition, data were obtained for a fourth KHV strain (CO5-53-3) at passage 0 (i.e., directly from infected fish tissue) and 2. The CO5-53-3 strain was obtained in 2005 from a moribund wild common carp during a mortality episode in the lower San Joacquin River, California. Standard PCRs were carried out, and the amplicons were sequenced directly using the PCR primers.
RESULTS AND DISCUSSION
Genome characteristics.
The size of the KHV genome as determined from the sequences is 295 kbp (295,271, 295,146, and 295,138 bp for J, U, and I, respectively). Restriction endonuclease digestion of the DNA preparations with NotI or XbaI yielded identical profiles for the three strains, with fragment sizes as predicted from the sequences (data not shown). The genome has a 22-kbp direct repeat at each terminus (22,437, 22,469, and 22,485 bp for J, U, and I, respectively). The overall nucleotide composition for each is 59.2% G+C, and the frequency of the CG dinucleotide is as expected from this value.
The genomes are highly similar to each other at the sequence level, with U and I more closely related to each other than either is to J. For example, in respect of single nucleotide substitutions (not counting duplicates in the terminal repeat), J differs from U and I at 181 of the 217 loci that are not conserved in all three strains; that is, there is a nucleotide difference every 1.5 kbp on average. Of the remaining 36 nonconserved loci, I differs from J and U at 32 loci, and U differs from J and I at 4 loci. These relationships imply a history in which an ancestral KHV strain gave rise initially to two branches: the J lineage that led eventually to J and the U/I lineage, which subsequently split into the branches leading to U and I. This conclusion is also consistent with the pattern of differences due to insertions and deletions.
Genetic content.
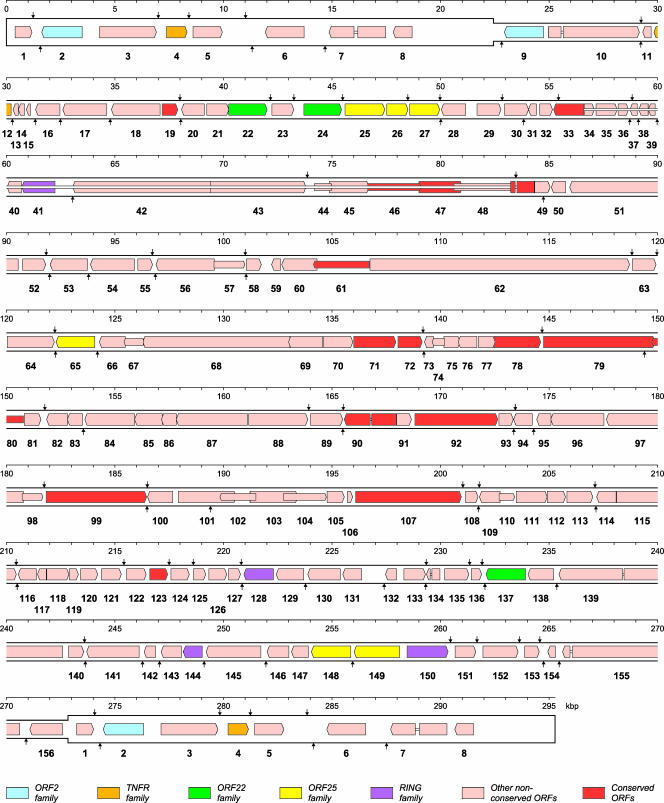
The close relationship among the three strains resulted in the same predicted gene map for each, with the exception of a number of fragmented ORFs discussed below. Figure 1 shows the predicted layout of protein-coding genes in an ancestral (wild-type) KHV as envisaged prior to gene fragmentation events. The genetic complement is 156, with 8 duplicated in the terminal repeat, yielding a total of 164 in the genome. Table 1 lists the features of KHV genes for which information was derived from bioinformatic analysis.
FIG. 1.
Gene layout in wild-type KHV. The locations of predicted protein-coding ORFs (based on the strain J coordinates) are shown as defined in the key at the foot, with conserved genes (those with clear IcHV-1 homologs) differentiated from nonconserved genes, which include five gene families. Introns are depicted as narrow bars. Nomenclature is given with the ORF prefix omitted, and the terminal repeat is shown in a thicker format than the rest of the genome. Predicted polyadenylation sites are shown above and below the genome for rightward- and leftward-oriented ORFs, respectively.
TABLE 1.
KHV genes for which predictions were made regarding functional properties or mutations
| Gene | Properties or putative functiona | Identityb | Coding region mutationc |
|---|---|---|---|
| ORF2 | ORF2 family | ||
| ORF4 | TNFR family; soluble TNFR | 30%, 185 | |
| ORF9 | ORF2 family | ||
| ORF12 | TNFR family; soluble TNFR | 34%, 110 | |
| ORF16 | G protein-coupled receptor | 21%, 240 | Frameshifted once in J |
| ORF19 | Related to IcHV-1 ORF5; deoxynucleoside (deoxyguanosine) kinase | 30%, 179 | |
| ORF22 | ORF22 family | ||
| ORF23 | Small subunit of ribonucleotide reductase | 64%, 321 | |
| ORF24 | ORF22 family | ||
| ORF25 | ORF25 family; membrane glycoprotein | ||
| ORF26 | ORF25 family; membrane glycoprotein | Broken at least thrice in the same places in J, U, and I | |
| ORF27 | ORF25 family; membrane glycoprotein | Broken once in different places in U and I | |
| ORF28 | Similar to bacterial NAD-dependent epimerase/dehydratase | 24%, 249 | |
| ORF29 | Multiple transmembrane protein | ||
| ORF30 | Membrane glycoprotein | Broken once in the same place in U and I | |
| ORF31 | Similar to eukaryotic DUF614 proteins | 34%, 116 | |
| ORF32 | Similar to a family of Singapore grouper iridovirus proteins | 32%, 95 | |
| ORF33 | Related to IcHV-1 ORF62; ATPase subunit of terminase | 29%, 664 | |
| ORF39 | Multiple transmembrane protein | ||
| ORF40 | Membrane glycoprotein | Broken at least twice in the same places in J, U and I, and once more in J; frameshifted once more in U | |
| ORF41 | RING family; RING finger protein | Motif | |
| ORF46 | Related to IcHV-1 ORF63; primase | Motif | |
| ORF47 | Related to IcHV-1 ORF64 | 21%, 428 | |
| ORF48 | Similar to protein kinases | 27%, 123 | |
| ORF54 | Zinc-binding protein | Motif | |
| ORF55 | Thymidine kinase | 41%, 171 | Frameshifted once in J |
| ORF61 | Related to IcHV-1 ORF54 | 23%, 259 | |
| ORF62 | OTU-like cysteine protease domain; also present in IcHV-1 ORF65 | Motif | |
| ORF64 | Multiple transmembrane glycoprotein; nucleoside transporter | 23%, 433 | Broken twice in J |
| ORF65 | ORF25 family; membrane glycoprotein | ||
| ORF68 | Similar to myosin and related proteins and possibly to IcHV-1 ORF22 | 21%, 867 | |
| ORF71 | Related to IcHV-1 ORF25; DNA helicase | 25%, 523 | |
| ORF72 | Related to IcHV-1 ORF27; capsid triplex protein | 22%, 333 | |
| ORF78 | Related to IcHV-1 ORF28; capsid protease and scaffolding protein | 30%, 104 | |
| ORF79 | Related to IcHV-1 ORF57; DNA polymerase | 25%, 1076 | |
| ORF80 | Related to IcHV-1 ORF60 | 29%, 199 | |
| ORF81 | Multiple transmembrane protein | ||
| ORF82 | Multiple transmembrane protein | ||
| ORF83 | Multiple transmembrane protein | ||
| ORF87 | Frameshifted once in J | ||
| ORF90 | Related to IcHV-1 ORF37 | 21%, 524 | |
| ORF92 | Related to IcHV-1 ORF39; major capsid protein | 22%, 555 | |
| ORF94 | Trypsin-like serine protease | 30%, 245 | Frameshifted once in U |
| ORF98 | Uracil-DNA glycosylase | 26%, 226 | |
| ORF99 | Related to IcHV-1 ORF46; membrane glycoprotein | 21%, 373 | |
| ORF104 | Serine-threonine protein kinase | 28%, 189 | |
| ORF105 | Among the least convincing protein-coding regions | Broken twice in J | |
| ORF107 | Related to IcHV-1 ORF56 | 29%, 186 | |
| ORF108 | Different C terminus in J due to 3-bp insertion | ||
| ORF112 | Contains double-stranded nucleic acid-binding domain (helix-turn-helix) | Motif | |
| ORF114 | Multiple transmembrane protein; similar to cellular protein of unknown function (Danio rerio XP_684064) | 24%, 286 | |
| ORF115 | Membrane glycoprotein | ||
| ORF116 | Membrane glycoprotein | Frameshifted once in I | |
| ORF123 | Related to IcHV-1 ORF49; deoxyuridine triphosphatase | 32%, 131 | |
| ORF124 | Membrane glycoprotein | ||
| ORF125 | Broken once in I | ||
| ORF126 | Membrane glycoprotein | ||
| ORF128 | RING family; RING finger protein; SPRY protein; TRIM-like protein | 30%, 192 | Broken once in I |
| ORF131 | Membrane glycoprotein | ||
| ORF132 | Membrane protein | ||
| ORF134 | Interleukin-10 | 23%, 182 | |
| ORF136 | Membrane glycoprotein | ||
| ORF137 | ORF22 family | ||
| ORF138 | Membrane glycoprotein | ||
| ORF139 | Membrane glycoprotein related to poxvirus B22R proteins | 23%, 985 | Broken twice in I |
| ORF140 | Thymidylate kinase | 62%, 194 | |
| ORF141 | Large subunit of ribonucleotide reductase | 56%, 804 | |
| ORF144 | RING family; RING finger protein | Motif | |
| ORF146 | Membrane glycoprotein | ||
| ORF148 | ORF25 family; membrane glycoprotein | ||
| ORF149 | ORF25 family; membrane glycoprotein | ||
| ORF150 | RING family; RING finger protein | Motif | |
| ORF153 | Multiple transmembrane protein |
Genes encoding proteins with minor predicted features only, such as signal peptides, hydrophobic regions, or immunoglobulin domains, are not included. These details are available in the database accessions.
The percent sequence identity and length of alignment over which it features were derived using BLASTP for proteins that have counterparts in other organisms (all E-values <e−5 except for ORF61, where it was 0.072). Data in italics refer to homologs in IcHV-1, and data in regular type relate to homologs in other organisms. In some cases, only a recognized motif is conserved; that in ORF46 is present in primases from a wide range of organisms.
Fragmented genes are described as “broken” where mutations are located well within the coding region and likely to ablate function and as “frameshifted” where they are close to the 3′ end and unlikely to ablate function. Identification of strains considered to have the wild-type gene in the latter case was much less certain than in the former, as also noted in the footnote to Table 2.
We view the gene layout as substantially accurate, but expect improvements to be made in the future. Alterations are most likely to come in the form of additional small ORFs, further splicing, and removal of present ORFs (e.g., perhaps ORF58 and ORF105, which we consider as the least likely ORFs to encode proteins). Counting the terminal repeat once, KHV has an ORF density of 0.57 per kbp, somewhat lower than IcHV-1 (0.68 per kbp), even though the great majority of the genome is predicted to be protein coding. Indeed, despite a substantially greater genome size, KHV is predicted to have fewer genes than the largest human HV, human cytomegalovirus (165 genes in a genome of 236 kbp, or 0.70 per kbp; 13).
The KHV genome contains 15 genes (colored red in Fig. 1) that have clear homologs in IcHV-1. The data for IcHV-1 and other HVs (7-9) indicate that these genes encode proteins involved in capsid morphogenesis (ORF92, ORF72, and ORF78, encoding two structural components of the capsid shell and a candidate protease involved in capsid maturation), nucleotide metabolism (ORF19 and ORF123 encoding deoxynucleoside kinase and deoxyuridine triphosphatase, respectively), DNA replication (ORF79, ORF71, and ORF46, encoding DNA polymerase, helicase, and a candidate primase), and DNA packaging (ORF33, which encodes the putative ATPase subunit of terminase; the three exons that comprise this coding region are shown in Fig. 1 as connected by introns). In addition, the conserved genes include one encoding a large membrane glycoprotein (ORF99) and five encoding proteins with unknown functions (ORF47, ORF61, ORF80, ORF90, and ORF56). This modest number of convincingly conserved genes highlights the substantial evolutionary distance between KHV and IcHV-1 (26, 46). When the ranid HV sequences (10) are taken into account, only 13 genes are convincingly conserved: the 15 genes conserved between KHV and IcHV-1 as described above, less ORF19 and ORF123. These findings indicate that the fish HV clade is considerably more divergent overall than the mammalian HV clade, in which 43 genes have been inherited from a common ancestor (8). Nonetheless, KHV clearly represents a new HV species, as concluded previously (23, 46). It is likely that the number of conserved genes in the fish HV clade will increase as data for other species facilitate more sensitive comparisons, but not to the number observed in the mammalian HV clade. As is the case with distantly related viruses in the mammalian HV clade (8), conserved genes are located centrally in the KHV and IcHV-1 genomes, and their arrangement is not conserved.
The remaining 141 KHV genes are in the nonconserved set. These are colored pink in Fig. 1, except for five families of related genes. The RING family encodes four proteins containing a zinc ion-coordinating motif known as the RING finger. This motif is ubiquitous among the HVs and is encoded by representatives of the three clades. The tumor necrosis factor receptor (TNFR) family encodes two versions of a secreted form of TNFR, which presumably have roles in immune evasion, as demonstrated with the poxviruses (43). The ORF2 and ORF22 families encode two and three proteins, respectively. The ORF25 family encodes six membrane glycoproteins, of which two are fragmented, one (ORF27) at different locations in U and I and one (ORF26) in all three strains.
The putative products of several KHV genes are convincingly related to enzymes in addition to those described above. These include the large and small subunits of ribonucleotide reductase (ORF141 and ORF23, respectively), thymidine kinase (ORF55), thymidylate kinase (ORF140), uracil-DNA glycosylase (ORF98), serine protease (ORF94), and serine-threonine protein kinase (ORF104). KHV also encodes proteins related to G protein-coupled receptors (ORF16), eukaryotic DUF614 proteins (ORF31), a family of iridovirus proteins (ORF32), nucleoside transporters (ORF64), a cellular protein of unknown function (ORF114), and a poxvirus B22R protein (ORF139) that is likely to be involved in immune evasion (30). Like some members of the mammalian HV clade (6), KHV encodes a protein that is clearly related to interleukin-10 (ORF134), which may modulate host immune responses. Other KHV proteins bear limited similarities to bacterial NAD-dependent epimerase/dehydratases (ORF28), protein kinases (ORF48), zinc-binding proteins (ORF54), cysteine proteases (ORF62), myosin (ORF68), and nucleic acid-binding proteins (ORF112). It is apparent from the conserved and nonconserved gene sets that KHV evolution has been characterized by gene capture from the cell or other viruses.
Strain evolution.
As mentioned above, the three KHV strains differ from each other at only a small number of loci. A few of the insertions and deletions represent mutations (usually frameshifts) in one or more strains that disrupt coding regions. It should be noted that the identification of affected ORFs depends upon the accuracy of the predicted gene set and, moreover, that genes mutated in all strains may only be identified in certain circumstances. This is possible when such genes are members of a family or are related to genes in other organisms or where the encoded proteins contain characteristic features, such as those of a membrane protein with a signal sequence and transmembrane region.
Despite these analytical limitations, 15 ORFs (10% of the complement) appear to be mutated in one or more strains (Table 1). Of these, the unmutated forms of 11 ORFs encode proteins with features implying that their assignment as protein-coding regions at least is correct. It is notable that the majority of mutated ORFs encode membrane glycoproteins and that none of the conserved genes is affected. Nine genes (listed as “broken” in Table 1) are probably rendered nonfunctional by frameshifts located centrally in the coding regions, and the wild-type gene is identified readily. Six genes (listed as “frameshifted”) might retain function since the mutations are close (sometimes very close) to the 3′ ends of the ORFs, making it more difficult to identify the wild-type gene (see the assumptions explained in the footnotes to Tables 1 and 2). Nonetheless, even though identification of the precise number of mutated genes in each genome is problematic, all three sequenced strains are evidently multiple mutants derived from a wild-type ancestor.
TABLE 2.
Presence of selected mutations in coding regions of KHV genes
| Gene | PCR primers (5′-3′) | Mutationa | Presence of wild type (+) or mutant (-) markerb
|
|||||
|---|---|---|---|---|---|---|---|---|
| J | I | U P2 | U P12 | CO5-53-3 P0 | CO5-53-3 P2 | |||
| ORF16 | GTTGTCCTCATAGGACGCCAT | 2-bp insertion | - | + | + | + | ND | + |
| CTGATCTTCTACACGCCGATG | ||||||||
| ORF27 | CTGCTCCTGCTGCCATCACCG | 1-bp deletion | + | + | + | - | ND | + |
| GCCAGGAGGTCCAAGTGTCAC | ||||||||
| ORF30 | CACTCTCCACGAACGAGACCG | 23-bp deletion | + | - | - | - | + | + |
| TGTAGGTGCCGCGGTAGACTC | ||||||||
| ORF40 | TTGAAACGGTGAGGCAGCCAT | 2-bp deletion | + | + | Mixture | - | + | + |
| CAACTGCAACATACCGTCAAG | ||||||||
| ORF55 | GTCTCACGGGGTTCAGGATGG | 2-bp deletion | - | + | + | + | + | + |
| CAAAGTCGCTCAGAGCAAGTG | ||||||||
| ORF87 | GTTCAGCGCCTCTGCCATGAG | 14-bp deletion | - | + | + | + | + | + |
| ATCGACGACAGCCAACTCTTC | ||||||||
| ORF94 | GTCTCTGTGCTCCTGGGAGAC | 1-bp deletion | + | + | - | - | + | + |
| CAGCGTCTACACCGACGTGAG | ||||||||
| ORF108 | CTGCCCTCAACTCTTCTACCT | 3-bp insertion | - | + | + | + | + | + |
| TGTCATCGGACGCACCAAGCA | ||||||||
| ORF116 | ACAAGATGTAAAGGTCCAGAC | 2-bp deletion | + | - | + | + | + | + |
| ACCTTCAACTAACATGGGTCT | ||||||||
The mutations in ORF27 and ORF30 are located well within the coding regions, making it obvious which is the wild-type sequence. Mutations in the other genes listed are near the 3′ ends of coding regions, making identification of the wild type much less certain. For the purposes of this summary, the wild-type gene is assumed to correspond to the sequence most frequently found.
P0, not passaged in cell culture. P2, passage 2. P12, passage 12. ND, not determined.
Since the sequenced DNAs were obtained from KHV strains passaged in cell culture, it is possible that some of the mutations occurred after isolation from infected fish. Unfortunately, the tissues from which the strains had been isolated were no longer available, and it was not possible to examine the unpassaged viruses. However, some of the mutations were examined in an earlier passage of U (passage 2, as opposed to passage 12), and in infected cell DNA from passaged and unpassaged forms of a fourth KHV isolate (CO-530-3). The distribution of mutations in a subset of the loci in Table 1 is summarized in Table 2. The combined data indicate that mutations in three genes arose in vivo, since they are present in more than one of the sequenced viruses: that in ORF30 in U and I and those in ORF26 and ORF40 in J, U, and I. Mutations in six genes (ORF16, ORF55, ORF87, ORF94, ORF108, and ORF116) are present in a single strain each and therefore may have arisen in vivo or in vitro. Mutations in two genes (ORF27 and ORF40) may have arisen in vitro, since the relevant sequences differ between passages 2 and 12 of U (Table 2; Table 1 indicates that ORF40 was already broken in all three genomes in vivo prior to the additional mutation). We conclude that mutations occurred in vivo in ORF26 and ORF40 prior to divergence of the three strains from a wild-type parent, with a mutation then arising in vivo in ORF30 after the U/I lineage had diverged from the J lineage. In the absence of the original infected tissues, it was not possible to determine whether subsequent mutations occurred in vivo or in vitro.
The sequence comparisons indicate that the three sequenced KHV strains arose via the loss of genetic functions, as evidenced by frameshifted coding regions, with at least some of the cognate mutations having occurred in vivo. Elsewhere among large DNA viruses, fragmented genes have been documented extensively in the Poxviridae, particularly the Orthopoxvirus genus, where up to 16% of genes in a single virus may be fragmented (21). These genes are invariably not required for virus growth in cell culture and are therefore presumed to contribute to some aspect of growth in the host. The simplest interpretation is that some orthopoxviruses have become associated with their hosts relatively recently and have diverged from an ancestral virus by losing certain functions either because these functions are not required or because they reduce fitness. Gene fragmentation was also observed in the bivalve HV, OsHV-1, which had not been passaged in cell culture, suggesting that this might have contributed to the striking pathogenicity (and perhaps increased host range) of this emerging virus in farmed shellfish (12). The apparent loss of KHV gene functions, particular among those encoding membrane glycoproteins that may be associated with host specificity and critical to virulence, presents a provocative parallel. Nonetheless, a role for gene loss in the emergence of KHV is currently speculative.
Origins of KHV.
The epidemiologic and pathogenic features of KHV-associated disease are new, and we doubt that they were overlooked previously (23, 46). The earliest known archival evidence indicates the presence of KHV among wild common carp in the United Kingdom as early as 1996 (K. Way, unpublished data), preceding observations of the disease in koi in Germany that were first recorded in 1997 (5). The active and often unregulated movements of large numbers of koi have contributed to a rapid spread of the virus presumably from these origins (22). The first cases of the disease in the eastern United States (strain U) occurred in 1998 following a koi show in New York that involved fish from Israel (24), a finding consistent with the high degree of similarity between U and I. Causal links for the origin of KHV among common carp in Japan are less defined (42).
Despite these observations, the origins of KHV remain obscure. It is possible that it has derived from an innocuous virus of C. carpio or another of the more than 2,000 species of cyprinid fishes, escaping from a long-standing equilibrium with its host and adapting via increased virulence to the conditions of aquaculture, where susceptible hosts are in abundant supply, constantly renewed, and transported worldwide. In this respect, findings with KHV have relevance beyond carp aquaculture, since similar phenomena have been observed with herpesviruses of other intensively cultured animals, including Marek's disease virus in chickens, where the evolution of increasingly virulent strains is exacerbated by vaccination (32), and perhaps OsHV-1 in oysters (12).
The sequence comparisons prompt the hypothesis that intensive culture of common carp and koi, combined with large-scale movements of live koi, may have favored transmission of genetically deficient KHV strains of enhanced virulence. Testing of this hypothesis will include further extensive comparisons of KHV isolates that have not been passaged in cell culture in order to understand the extent and timescale of gene loss and specific mutagenesis studies in order to assess any contribution of gene loss to pathogenicity. Whether or not further adaptation might occur, more active control measures, including listing of the KHV disease by international organizations charged with disease control, are probably needed to reduce future economic and ecologic impacts of this important viral pathogen.
Nucleotide sequence accession numbers.
The genome sequences for KHV strains J, U, and I were submitted to GenBank/DDJB under accession numbers AP008984, DQ657948, and DQ177346, respectively.
Acknowledgments
This research was supported in part by the Tokyo University of Marine Science and Technology, by the United Kingdom Medical Research Council, and by a grant from the Center for Study of Emerging Diseases (Israel) and BARD (United States-Israel Binational Agricultural Research and Development Fund project no. IS-3539-04CR).
We are grateful to Charles Cunningham for technical support in preparing and sequencing PCR products and to Duncan McGeoch for helpful comments on the manuscript.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Adkison, M. A., O. Gilad, and R. P. Hedrick. 2005. An enzyme linked immunosorbent assay (ELISA) for detection of antibodies to the koi herpesvirus (KHV) in the serum of koi, Cyprinus carpio. Fish Pathol. 40:53-62. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Balon, E. K. 1995. Origin and domestication of the wild carp, Cyprinus carpio: from Roman gourmets to the swimming flowers. Aquaculture 129:3-48. [Google Scholar]
- 4.Bercovier, H., Y. Fishman, R. Nahary, S. Sinai, A. Zlotkin, M. Eyngor, O. Gilad, A. Eldar, and R. P. Hedrick. 2005. Cloning of the koi herpesvirus (KHV) gene encoding thymidine kinase and its use for a highly sensitive PCR-based diagnosis. BMC Microbiol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretzinger, A., T. Fischer-Scherl, M. Oumouna, R. Hoffmann, and U. Truyen. 1999. Mass mortalities in koi carp, Cyprinus carpio, associated with gill and skin disease. Bull. Eur. Assoc. Fish. Pathol. 19:182-185. [Google Scholar]
- 6.Chang, W. L., N. Baumgarth, D. Yu, and P. A. Barry. 2004. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J. Virol. 78:8720-8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J., and M. D. Davison. 1995. Identification of structural proteins of channel catfish virus by mass spectrometry. Virology 206:1035-1043. [DOI] [PubMed] [Google Scholar]
- 10.Davison, A. J., C. Cunningham, W. Sauerbier, and R. G. McKinnell. 2006. Genome sequences of two frog herpesviruses. J. Gen. Virol. 87:3509-3514. [DOI] [PubMed] [Google Scholar]
- 11.Davison, A. J., R. Eberle, G. S. Hayward, D. J. McGeoch, A. C. Minson, P. E. Pellett, B. Roizman, M. J. Studdert, and E. Thiry. 2005. Herpesviridae, p. 193-212. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, England.
- 12.Davison, A. J., B. L. Trus, N. Cheng, A. C. Steven, M. S. Watson, C. Cunningham, R.-M. Le Deuff, and T. Renault. 2005. A novel class of herpesvirus with bivalve hosts. J. Gen. Virol. 86:41-53. [DOI] [PubMed] [Google Scholar]
- 13.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. G. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 14.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 15.Gilad, O., S. Yun, M. A. Adkison, K. Way, N. H. Willits, H. Bercovier, and R. P. Hedrick. 2003. Molecular comparison of isolates of an emerging fish pathogen, koi herpesvirus, and the effect of water temperature on mortality of experimentally infected koi. J. Gen. Virol. 84:2661-2667. [DOI] [PubMed] [Google Scholar]
- 16.Gilad, O., S. Yun, K. B. Andree, M. A. Adkison, A. Zlotkin, H. Bercovier, A. Eldar, and R. P. Hedrick. 2002. Initial characteristics of koi herpesvirus and development of a polymerase chain reaction assay to detect the virus in koi, Cyprinus carpio koi. Dis. Aquat. Org. 48:101-108. [DOI] [PubMed] [Google Scholar]
- 17.Gilad, O., S. Yun, F. Zagmutt-Vergara, C. M. Leutenegger, H. Bercovier, and R. P. Hedrick. 2004. Concentrations of a Koi herpesvirus (KHV) in tissues of experimentally infected Cyprinus carpio koi as assessed by real-time TaqMan PCR. Dis. Aquat. Org. 60:179-187. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 19.Goto, N., K. Kurokawa, and T. Yasunaga. 2000. CONSERV: a tool for finding exact matching conserved sequences in biological sequences. Genome Informatics 11:307-308. [Google Scholar]
- 20.Gray, W. L., L. Mullis, S. E. LaPatra, J. M. Groff, and A. Goodwin. 2002. Detection of koi herpesvirus DNA in tissues of infected fish. J. Fish Dis. 25:171-178. [Google Scholar]
- 21.Gubser, C., S. Hué, P. Kellam, and G. L. Smith. 2004. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 85:105-117. [DOI] [PubMed] [Google Scholar]
- 22.Haenen, O. L. M., K. Way, S. M. Bergmann, and E. Areil. 2004. The emergence of koi herpesvirus and its significance to European aquaculture. Bull. Eur. Assoc. Fish Pathol. 24:293-307. [Google Scholar]
- 23.Hedrick, R. P., O. Gilad, S. C. Yun, T. S. McDowell, T. B. Waltzek, G. O. Kelley, and M. A. Adkison. 2005. Initial isolation and characterization of a herpes-like virus (KHV) from koi and common carp. Bull. Fish. Res. Agen. 2:1-7. [Google Scholar]
- 24.Hedrick, R. P., O. Gilad, S. Yun, J. V. Spangenberg, G. D. Marty, R. W. Nordhausen, M. J. Kebus, H. Bercovier, and A. Eldar. 2000. A herpesvirus associated with mass mortality of juvenile and adult koi, a strain of common carp. J. Aquat. Anim. Health. 12:44-57. [DOI] [PubMed] [Google Scholar]
- 25.Hedrick, R. P., G. D. Marty, R. W. Nordhausen, M. Kebus, H. Bercovier, and A. Eldar. 1999. An herpesvirus associated with mass mortality of juvenile and adult koi Cyprinus carpio. Fish Health Newsl. Am. Fish. Soc. 27:7. [DOI] [PubMed] [Google Scholar]
- 26.Hutoran, M., A. Ronen, A. Perelberg, M. Ilouze, A. Dishon, I. Bejerano, N. Chen, and M. Kotler. 2005. Description of an as yet unclassified DNA virus from diseased Cyprinus carpio species. J. Virol. 79:1983-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilouze, M., A. Dishon, and M. Kotler. 2006. Characterization of a novel virus causing a lethal disease in carp and koi. Microbiol. Mol. Biol. Rev. 70:147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung, S. J., and T. Miyazaki. 1995. Herpesviral hematopoietic necrosis of goldfish, Carassius auratus (L.). J. Fish Dis. 18:211-220. [Google Scholar]
- 29.Liu, H., X. Shi, L. Gao, and Y. Jiang. 2002. Study on the etiology of koi epizootic disease using the method of nested-polymerase chain reaction assay (nested-PCR). J. Huazhong Agric. Univ. 21:414-418. [Google Scholar]
- 30.McFadden, G., and S. H. Nazarian. 2006. Immune evasion by poxviruses. Fut. Virol. 1:123-132. [Google Scholar]
- 31.Moyle, P. B., and J. J. Cech. 2004. An introduction to ichthyology. Prentice Hall, London, England.
- 32.Nair, V. 2005. Evolution of Marek's disease: a paradigm for incessant race between the pathogen and the host. Vet. J. 170:175-183. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa, I., K. Kurokawa, M. Nakata, A. Yamashita, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neukirch, M., and U. Kunz. 2001. Isolation and preliminary characterization of several viruses from koi (Cyprinus carpio) suffering gill necrosis and mortality. Bull. Eur. Assoc. Fish Pathol. 21:125-135. [Google Scholar]
- 35.New, M. B. 1997. Aquaculture and the capture fisheries. World Aquaculture 1997(June):11-30. [Google Scholar]
- 36.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perelberg, A., M. Smirnov, M. Hutoran, A. Diamant, Y. Bejerano, and M. Kotler. 2003. Epidemiological description of a new viral disease afflicting cultured Cyprinus carpio in Israel. Is. J. Aquat. Bamidgeh. 55:5-12. [Google Scholar]
- 38.Pikarsky, E., A. Ronen, J. Abramowitz, B. Levavi-Sivan, M. Hutoran, Y. Shapira, M. Steintz, A. Perelberg, D. Soffer, and M. Kotler. 2004. Pathogenesis of acute viral disease induced in fish by carp interstitial nephritis and gill necrosis virus. J. Virol. 78:9544-9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronen, A., A. Perelberg, J. Abramowitz, M. Hutoran, S. Tinman, I. Bejerano, M. Steinitz, and M. Kotler. 2003. Efficient vaccine against the virus causing a lethal disease in cultured Cyprinus carpio. Vaccine 21:4677-4684. [DOI] [PubMed] [Google Scholar]
- 40.Sakiyama, T., H. Takami, N. Ogasawara, S. Kuhara, T. Kozuki, K. Doga, A. Ohyama, and K. Horikoshi. 2000. An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci. Biotechnol. Biochem. 64:670-673. [DOI] [PubMed] [Google Scholar]
- 41.Sano, T., H. Fukuda, M. Furukawa, H. Hosoya, and Y. Moriya. 1985. A herpesvirus isolated from carp papilloma in Japan. Fish Shell Pathol. 32:307-311. [Google Scholar]
- 42.Sano, M., T. Ito, J. Kurita, T. Yanai, N. Watanabe, M. Satoshi, and T. Iida. 2004. First detection of koi herpesvirus in cultured common carp Cyprinus carpio in Japan. Fish Pathol. 39:165-168. [Google Scholar]
- 43.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377-423. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu, C., M. C. Weng, J.-R. Shiau, and S.-Y. Lin. 2004. Detection of koi herpesvirus in koi Cyprinus carpio in Taiwan. Fish Pathol. 39:109-110. [Google Scholar]
- 46.Waltzek, T. B., G. O. Kelley, D. M. Stone, K. Way, L. Hanson, H. Fukuda, I. Hirono, T. Aoki, A. J. Davison, and R. P. Hedrick. 2005. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J. Gen. Virol. 86:1659-1667. [DOI] [PubMed] [Google Scholar]