Abstract
The 5′ nontranslated region (NTR) and the X tail in the 3′ NTR are the least variable parts of the hepatitis C virus (HCV) genome and play an important role in the initiation of RNA synthesis. By using subgenomic replicons of the HCV isolates Con1 (genotype 1) and JFH1 (genotype 2), we characterized the genotype specificities of the replication signals contained in the NTRs. The replacement of the JFH1 5′ NTR and X tail with the corresponding Con1 sequence resulted in a significant decrease in replication efficiency. Exchange of the X tail specifically reduced negative-strand synthesis, whereas substitution of the 5′ NTR impaired the generation of progeny positive strands. In search for the proteins involved in the recognition of genotype-specific initiation signals, we analyzed recombinant nonstructural protein 5B (NS5B) RNA polymerases of both isolates and found some genotype-specific template preference for the 3′ end of positive-strand RNA in vitro. To further address genotype specificity, we constructed a series of intergenotypic replicon chimeras. When combining NS3 to NS5A of Con1 with NS5B of JFH1, we observed more-efficient replication with the genotype 2a X tail, indicating that NS5B recognizes genotype-specific signals in this region. In contrast, a combination of the NS3 helicase with NS5A and NS5B was required to confer genotype specificity to the 5′ NTR. These results present the first genetic evidence for an interaction between helicase, NS5A, and NS5B required for the initiation of RNA synthesis and provide a system for the specific analysis of HCV positive- and negative-strand syntheses.
The hepatitis C virus (HCV) is an enveloped positive-strand RNA virus belonging to the genus Hepacivirus in the family Flaviviridae. The genome of HCV encompasses a single ∼9,600-nucleotide-long RNA molecule carrying primarily one large open reading frame, flanked by nontranslated regions (NTRs), and being translated into one polyprotein. The polyprotein precursor is cleaved by cellular and viral proteases into at least 10 different products (for a review, see reference 4). The structural proteins core, E1, and E2 are located in the amino terminus of the polyprotein, followed by p7 and the nonstructural proteins (NS) NS2, NS3, NS4A, NS4B, NS5A, and NS5B. NS2 and the amino-terminal domain of NS3 comprise the NS2-3 protease responsible for cleavage between NS2 and NS3. NS3 is a multifunctional protein, consisting of an amino-terminal serine-protease domain required for the processing of the NS3-to-NS5B region and a carboxy-terminal helicase/nucleoside triphosphatase domain. NS4A is a cofactor that activates the NS3 protease by forming a heterodimer. The hydrophobic protein NS4B induces the formation of a cytoplasmic vesicular structure, designated membranous web, which appears to contain the replication complex of HCV (16). NS5A is a phosphoprotein comprising basally phosphorylated (p56) and hyperphosphorylated (p58) forms (22). An important role for NS5A in viral replication is suggested by the finding that most of the cell culture-adaptive mutations described so far are located within a central region of NS5A (6, 36) and that the loss of hyperphosphorylation by the inhibition of casein kinase I activates RNA replication (43, 48). Furthermore, NS5A binds to RNA (21) and the recently obtained crystal structure of domain I suggests that this region forms a dimer that could exert this function (62). NS5B is the RNA-dependent RNA polymerase (RdRp) of HCV, the key enzyme of viral RNA replication.
The NTRs are among the most conserved parts of the viral genome due to their multiple functions in viral translation and replication. The 5′ NTR is a highly structured region of 340 to 341 nucleotides in length and contains an internal ribosome entry site (IRES) (64) that directs cap-independent translation of the open reading frame. In addition, it has been shown that the first 125 nucleotides are essential for RNA replication but that the entire 5′ NTR is required for maximal replication efficiency (15, 27). It is generally believed that the function of the 5′ NTR in RNA replication, namely, the initiation of positive-strand synthesis, is exerted by the complementary sequence, corresponding to the 3′ end of the negative strand, which adopts a secondary structure that is different from the mirror image of the 5′ NTR (54, 59). The 3′ NTR of the positive-strand RNA has a tripartite structure: (i) a variable region that is in part dispensable for replication in vivo (71) and in vitro but seems to be important for efficient RNA replication (13, 72); (ii) a poly(U/UC) tract of variable length, which is essential in vivo and in vitro and which requires a minimal length of 26 to 50 nucleotides (13, 72); and (iii) the very 3′ end of the HCV genome, designated X tail. It comprises an almost invariant 98-nucleotide sequence (28, 60) containing three highly conserved stem-loop structures (8), which are all critical for RNA replication in vitro and in vivo (13, 71, 72). Further important cis-acting RNA elements are located in the coding region of NS5B (74), one of which undergoes a kissing-loop interaction with stem-loop II (SLII) in the X region that is essential for RNA replication (14).
Although the secondary structures of the termini of positive- and negative-strand RNAs have been defined (8, 20, 54, 59) and the regions involved in RNA replication have been analyzed in great detail using subgenomic replicons (13, 15, 72, 73), little is known about the critical protein RNA interactions governing the initiation and regulation of RNA synthesis. This is due mainly to the lack of appropriate model systems, which is notoriously difficult for many positive-strand RNA viruses, since most of the factors involved in this process function only in cis and many of the proteins are active only as part of a polyprotein. In the case of alphaviruses, it was possible to study RNA replication by separate expression of RNAs containing the cis-acting noncoding elements and the nonstructural proteins to dissect the requirements for the initiation of negative- and positive-strand syntheses (33). For flaviviruses, some of the nonstructural proteins could be supplied in trans (26, 34, 35), but for HCV, trans-complementation was limited to particular NS5A mutants (1). Some limited insight into basic mechanisms of RNA synthesis was obtained by in vitro analysis of purified enzymes. For instance, it has been shown that NS5B can initiate RNA synthesis de novo (41, 44, 76), which is also thought to occur in vivo, but although de novo initiation does not work on all templates with equal efficiencies (51), there is no restriction to HCV sequences (55). In the case of the NS3 helicase, specific binding to the 3′ NTR of positive and negative strands was shown (3), but although a direct involvement of this enzyme in RNA replication is obvious, the precise role has yet to be defined. Therefore, it is still not clear which of the nonstructural proteins are actively involved in HCV RNA synthesis.
Chimeras between related viruses have been a valuable tool for the analysis of many aspects of the viral life cycle, and the genetic diversity among HCV isolates already provides a starting point to create chimeric virus genomes. HCV sequences are grouped into six genotypes, differing up to 30 percent in nucleotide sequence, and several subtypes (58). Most of the HCV isolates that have been tested in cell culture belong to genotype 1, replicate very poorly, and have to gain adaptive mutations for efficient RNA amplification, as exemplified by the genotype 1b isolate Con1 (6, 36, 37, 39). HCV isolates of other genotypes have not yet been established to replicate in vitro, with the exception of a genotype 2a isolate, designated JFH1, which amplifies to high levels without the need for cell culture adaptation (24). The availability of HCV isolates of different genotypes efficiently replicating in cell culture allows us now for the first time to establish a system to analyze the determinants involved in RNA replication based on intergenotypic chimeras.
By using subgenomic replicons of HCV Con1 and JFH1, we developed a system to dissect the initiation of negative- and positive-strand RNA syntheses and identified genotype-specific replication signals in the conserved parts of the NTRs. Moreover, we identified a specific NS5B interaction site in the 3′ X region and show that the NS3 helicase, NS5A, and NS5B are part of the initiation complex involved in progeny positive-strand synthesis.
MATERIALS AND METHODS
Cell cultures.
Cell monolayers of the human hepatoma cell line Huh7 (42) were grown in Dulbecco modified Eagle medium (DMEM; Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U per ml of penicillin, 100 μg per ml streptomycin, and 10% fetal calf serum. Huh7-Lunet cells refer to a Huh7 cell clone that was generated with a selectable replicon and cured from HCV by treatment with a specific inhibitor. Huh7-Lunet cells are more permissive for HCV replication than naïve Huh7 cells (14).
Plasmid constructs.
All sequence numbering refers to the position of the corresponding amino acid or nucleotide of a complete HCV genome cloned by our group (HCV Con1; GenBank accession number AJ238799) or of the isolate JFH1, cloned by T. Wakita (GenBank accession no. AB047639) (24, 25, 66). Note that all JFH1-based in vitro transcripts contain one additional guanosine at the 5′ end to allow for efficient transcription by T7 RNA polymerase, which is not included in accession no. AB047639 and is disregarded in nucleotide numbering. All constructs were made using standard PCR and cloning procedures (52).
The two basic replicons Luc Con and Luc JFH used in this study refer to plasmid constructs pFK-I341PI-Luc/NS3-3′/Con1/ET/δg and pFK-I341PI-Luc/NS3-3′/JFH1 (68), respectively. Both plasmids contain the T7 promoter sequence fused to the 5′ NTR, followed by the poliovirus IRES (PI), the firefly luciferase (Luc) gene, the encephalomyocarditis virus IRES, the NS3-to-NS5B coding sequence, the 3′ NTRs of the respective isolates, the hepatitis delta virus genomic ribozyme, and the T7 terminator sequence, followed by an SpeI restriction site in the case of Con1 and SpeI and MluI restriction sites in the case of JFH1. pFK-I341PI-Luc/NS3-3′/Con1/ET/δg is identical to pFK-I341PI-Luc/NS3-3′/Con1/ET (36), harboring cell culture-adaptive mutations in NS3 (E1202G and T1280I) and NS4B (K1846T), with the exception of the 3′ X sequence, which was reverted to the genotype 1 consensus sequence and fused to the hepatitis delta virus genomic ribozyme (cloned from peu3aHDV [50]) and the T7 terminator sequence (obtained from pX8dT [53]) to generate the appropriate 3′ ends of replicon in vitro transcripts. ΔGDD constructs were described previously and have an in-frame deletion of 10 amino acids (MLVCGDDLVV) encompassing the GDD motif of NS5B (36, 68). Replicons with chimeric 5′ NTRs were constructed by using unique SbfI and PmeI restriction sites, present in all constructs, 5′ of the T7 promoter and 3′ adjacent to the 5′ NTR, respectively. 5′ J6 and 5′ H77 were amplified from plasmids pJ6CF and pCV-H77 (accession no. AF177036 and NC_004102, respectively; both generous gifts of J. Bukh, NIH, Bethesda, MD) and flanked with SbfI and PmeI sites by PCR. Chimeric X regions were created using a conserved NheI restriction site within SLII of the X tail and SpeI sites following the T7 terminator sequence. Constructs 1-156Con and 157-341Con were cloned using the same SbfI and PmeI restriction sites and an internal AgeI restriction site which is conserved between JFH1 and Con1. Individual parts of the JFH1 5′ NTR were mutated to correspond to the Con1 sequence by standard PCR mutagenesis techniques with constructs 1-4Con, 1-4/I′Con, IIz′Con, and IIy′Con, referring to the stem-loop structures in the 3′ end of the negative strand (see Fig. 4B) (59).
FIG. 4.
Mapping of genotype-specific signals in the 3′ X tail and 5′ NTR. (A) Secondary structure of SLI of the X tail (8). The consensus sequences of genotypes 1 and 3 to 6 are given, and deviations found in the consensus sequence of genotype 2 are indicated by arrows. (B) Schematic representation of the secondary structures determined for the 3′-terminal 156 nucleotides of HCV negative-strand RNA (59), complementary to the 5′ NTR of the positive strand. Note that numbering starts at the 3′ end. Differences between the Con1 and JFH1 sequences are indicated by filled circles. The open circle indicates an isolate-specific deviation of the JFH1 sequence at position 78 which was excluded from the analysis. (C) Impact of single-nucleotide replacements in the 3′ X-tail sequence on HCV replication efficiency. Luc/ubiquitin JFH1 replicon RNAs with a homologous 3′ X tail (X JFH), with individual nucleotide exchanges in the 3′ X tail (C72U, UA74/75AU, and U81C, as indicated in panel A), or with the entire 3′ X-tail sequence derived from Con1 (X Con) were transfected into Huh7 cells, and 4, 24, 48, and 72 h after transfection, cell lysates were analyzed for luciferase activity. Luciferase activity is given as the change in RLU (n-fold) relative to the value obtained 4 h after transfection. The structure of the replicon Luc/ubi JFH used in this experiment is given at the top. luc, firefly luciferase; ubi, ubiquitin; VR, variable region and poly(U/UC) tract of the HCV 3′ NTR; X, X tail (for a detailed explanation, refer to the legend to Fig. 1A). (D) Analysis of genotype-specific signals in the 5′ NTR. Luc JFH replicons with authentic 5′ NTRs (JFH1), 5′ NTRs derived from Con1 (5′ Con), or chimeric JFH 5′ NTRs containing parts of Con1 clustered according to the secondary structure prediction of the 3′ end of the negative strand (B) were electroporated into Huh7 cells. Transfected cells were analyzed for luciferase activity at 4 h and 24 h after transfection. The ratio of luciferase activity at 24 h to that at 4 h after transfection is shown. For further explanation, refer to the text.
The monocistronic replicon Luc/ubi JFH refers to plasmid pFK I389Luc_ubi_NS-3′/JFH and contains the 5′ NTR, the first 16 codons of the core open reading frame, in frame fused with the genes encoding firefly luciferase, human ubiquitin, and the NS3-to-NS5B coding region, followed by the 3′ NTR, the genomic ribozyme of hepatitis delta virus, and the T7 terminator sequence. All HCV parts in this plasmid were derived from HCV isolate JFH1. Point mutations (C72U, UA74/75AU, and U81C) in the 3′ X region of this construct were introduced by standard PCR techniques and numbered according to their positions in the X region, starting with 1 at position 9549 of the reference H77 strain (accession no. AF009606) (58).
The plasmid encoding NS5B of Con1 lacking the 21 C-terminal amino acids (ΔC21) and C-terminally fused to the His6 tag in a pET21b vector was a generous gift of Zhi Hong (Valeant Pharmaceuticals) (55). The NS5BΔC21 coding region of JFH1 was first subcloned into pQE60 (QIAGEN, Hilden, Germany) by engineering NcoI and BglII restriction sites into forward and reverse primers, respectively. To obtain pET21 NS5BΔC21 JFH, the N-terminal coding sequence of the gene was again amplified by PCR to add an XbaI restriction site by using primer S_pET21Xba (CCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGGCTTCCATGTCATACTCCTGGACC) and an appropriate reverse primer, cut with XbaI and BsrGI, and fused to a BsrGI-BlpI fragment from pQE60 NS5BΔC21 JFH by using XbaI and BlpI restriction sites in pET21b.
Plasmid pFK I341PiLucNS3-3′JFH/5B,3′Con was used to generate replicon JFH/5BCon. Its construction was based on pFK-I341PI-Luc/NS3-3′/JFH1 by the replacement of the NS5B coding region and the 3′ NTR with the corresponding Con1 sequence. The NS5A/NS5B junction was set after amino acid 2442 of the JFH1 isolate and generated by PCR.
pFK I341PiLucNS3-3′Con/NS5B,5′,3′ JFH was generated by replacing the NS5B coding region following amino acid 2419, the 5′ NTR, and the 3′ NTR with the corresponding sequence of isolate JFH1 in pFK-I341PI-Luc/NS3-3′/Con1/ET/δg. This plasmid was used to transcribe replicon Con/5B,5′,X JFH and was the starting point for other plasmids encoding intergenotypic replicase chimeras. In the case of NS3 helicase chimeras, the region encoding amino acids 1209 to 1647 of Con1 was exchanged for that encoding amino acids 1213 to 1651 of JFH1 by PCR and by using a conserved NsiI restriction site. The junction between NS4B and NS5A was set at position 1124/1125 of the Con1 polyprotein.
The sequences of all PCR-derived DNA fragments were verified by DNA sequencing using an ABI 310 sequencer (Applied Biosystems) and BigDye version 1.1 (Applied Biosystems) according to the manufacturer's protocol.
A detailed description of particular cloning strategies and primer sequences is available upon request.
In vitro transcription.
In vitro transcripts of HCV replicons or PCR products were generated using a protocol described previously (36). All replicon constructs used in this study contained the genomic ribozyme of hepatitis delta virus, followed by the T7 terminator fused to the HCV 3′ NTR, and were transcribed without linearization. Prior to transcription, DNA was extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. In vitro transcription reaction mixtures contained 80 mM HEPES, pH 7.5, 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM of each nucleoside triphosphate, 1 U/μl RNasin (Promega, Mannheim, Germany), 0.05 μg/μl plasmid DNA, and 0.6 U/μl T7 RNA polymerase (Promega). After the reaction mixture was incubated for 2 h at 37°C, an additional 0.3 U/μl T7 RNA polymerase was added and the reaction mixture was incubated for another 2 h. Transcription was terminated by the addition of 1 U RNase-free DNase (Promega) per μg plasmid DNA and a 30-min incubation at 37°C. After extraction with acidic phenol and chloroform, RNA was precipitated with isopropanol and dissolved in RNase-free water. The concentration was determined by measurement of the optical density at 260 nm, and RNA integrity was checked by agarose gel electrophoresis.
Electroporation of HCV replicons.
For electroporation, single-cell suspensions of Huh7 or Huh7-Lunet cells were prepared by trypsinizing monolayers, detaching the cells from the culture dish by rinsing them with complete DMEM, washing them once with phosphate-buffered saline, counting the cells, and resuspending them at 107 per ml in cytomix (65) containing 2 mM ATP and 5 mM glutathione. In vitro-transcribed RNA (2 to 10 μg) was mixed with 400 μl of the cell suspension by pipetting and, after electroporation, immediately transferred to 12 ml complete DMEM. Cells were seeded in aliquots and harvested for a luciferase assay or RNA preparation at the time points specified for each experiment. The electroporation conditions were 975 μF and 275 V, using a Gene Pulser II system (Bio-Rad, Munich, Germany) and a cuvette with a gap width of 0.4 cm (Bio-Rad). For Northern hybridization analysis 7 μg (see Fig. 3B) or 10 μg (see Fig. 3A) of in vitro-transcribed replicon RNA was used for each electroporation; cells of several electroporations were combined and then seeded in aliquots. For time points up to 24 h, cells corresponding to an entire electroporation were seeded on a 10-cm dish, and for later time points, half the amount of cells was plated.
FIG. 3.
Impact of 3′ X-tail and 5′ NTR chimeras on negative- and positive-strand syntheses of replicon Luc JFH. Analysis of transient replication of Luc JFH replicons with different NTR sequences by Northern hybridization. (A) Huh7 cells were transfected by electroporation with Luc JFH replicons harboring homologous NTR (5′ X JFH), 5′ NTR, or 3′ X-tail sequences derived from Con1 (5′ Con/X JFH, 5′ JFH/X Con, or 5′ XCon, respectively) or with a replication-deficient control replicon (ΔGDD) and analyzed for positive- and negative-strand RNA syntheses at 0 to 72 h after transfection. β-Actin RNA levels were used to normalize for loading in each lane. (B) Huh7-Lunet cells were transfected with the same replicons as those used for panel A and analyzed 0 to 12 h after electroporation. Replicon Luc JFH 5′ X Con was excluded from this analysis due to inefficient replication. Cells were harvested at the time points postelectroporation indicated above lanes 2 to 21. Ten micrograms of total RNA was analyzed by Northern hybridization with a 32P-labeled negative- or positive-sense riboprobe to detect HCV positive-strand RNA [(+)RNA] or negative-strand RNA [(−)RNA], respectively, or with a negative-sense riboprobe specific for β-actin as given on the right. Four micrograms of total RNA from naïve Huh7 cells was used as a negative control (Huh7). Luc JFH in vitro transcripts (107 or 108) of positive-sense [(+)RNA] or negative-sense [(−)RNA] orientation were loaded to quantify HCV RNA, as shown above the two outer left or right lanes, respectively.
Luciferase assay.
For the luciferase activity assay, cells were washed twice with phosphate-buffered saline and scraped off the plate into 350 μl ice-cold lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT). One hundred microliters of cleared lysate was mixed with 360 μl assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 15 mM K2PO4, pH 7.8) and, after the addition of 200 μl of a 200 μM luciferin solution, measured in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany) for 20 s. All luciferase assays were done in duplicate measurements. Luciferase activity was expressed as the increase in relative light units (RLU) (n-fold) relative to the luciferase activity measured 4 h after transfection unless stated otherwise.
Preparation of total RNA and quantification of HCV RNA by Northern hybridization.
The preparation of total RNA and the quantification of HCV RNA by Northern hybridization have been described previously (36). In brief, total RNA from cells was prepared by a single-step isolation method (9), denatured by treatment with 5.9% glyoxal in 50% dimethyl sulfoxide and 10 mM sodium phosphate buffer, pH 7.0, and analyzed after denaturing agarose gel electrophoresis by Northern hybridization. Prior to hybridization, the membrane was stained with methylene blue and cut ∼1 cm below the 28S rRNA band. The upper strip containing the HCV replicon RNA was hybridized with a 32P-labeled negative-sense riboprobe complementary to the NS4B-to-NS5A region (nucleotides 5979 to 6699) for the detection of viral positive-strand RNA or with a positive-sense riboprobe encompassing the same region for the detection of HCV negative-strand RNA. The lower strip that was hybridized with a β-actin-specific antisense riboprobe was used to correct for total RNA amounts loaded in each lane of the gel. Specific bands were quantified by phosphorimaging with a Molecular Imager FX scanner (Bio-Rad, Munich, Germany), and the number of HCV molecules was determined by comparison with a serial dilution of in vitro transcripts corresponding to a known number of positive or negative strands of subgenomic replicons mixed with 2 μg of total RNA from naïve Huh7 cells loaded in parallel onto the gel. In vitro transcripts were checked by agarose gel electrophoresis to ensure that almost all RNAs were full length and quantified by measurement of the optical density at 260 nm.
Expression and purification of JFH1 and Con1 RdRp.
The NS5BΔC21 coding regions of HCV JFH1 and Con1, C-terminally fused to a hexahistidine tag, were expressed in Escherichia coli BL21(DE3) cells. Bacterial cells were grown to an optical density at 600 nm of 0.8, induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside, incubated for 4 h at room temperature with shaking, and sedimented for 10 min at 6,000 × g. The pellet of a 100-ml culture was lysed in 4 ml lysis buffer I (LBI; 100 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM MgCl2, 2% Triton X-100, 2 mg/ml lysozyme [Roche Biochemicals, Mannheim, Germany]) and 1 U/ml Benzonase (Merck, Darmstadt, Germany) for 30 min on ice and centrifuged for 10 min at 20,000 × g at 4°C. The supernatant (S1) was removed, the pellet was resuspended in 5 ml of LBII (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 2% Triton X-100, 10 mM imidazole, 30% glycerol, 10 mM 2-mercaptoethanol), and the suspension was sonicated in 1-ml aliquots five times for 20 s at an output control setting of 6 at 4°C using a Branson 450 sonifier and a cup horn with a cooling device. After a 10-min centrifugation at 20,000 × g, the supernatant (S2) was mixed with 500 μl of a 1:1 Ni-nitrilotriacetic acid (NTA) agarose slurry (QIAGEN, Hilden, Germany), incubated for 30 min at 4°C with mild shaking, and centrifuged for 5 min at 500 × g. The supernatant (S3) was discarded, the Ni-NTA agarose was washed four times with 4 ml of LBII containing 50 mM imidazole, and bound protein was eluted with 1.5 ml of LBII containing 250 mM imidazole. Purified NS5B was quantified by a modification of the method of Lowry and stored in small aliquots at −70°C.
RdRp assay.
RdRp assays were performed essentially as described previously (40). In brief, 200 ng purified NS5B from JFH1 or 1 μg NS5B from Con1, corresponding to a final concentration of 150 nM or 750 nM, respectively, was incubated for 1 h at 22°C with 0.5 μg of template RNA in a total volume of 25 μl containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM DTT, 25 mM KCl, 1 mM EDTA, 20 U of RNasin (Promega), 10 μCi of [α-32P]CTP (3,000 Ci/mmol; GE Healthcare), 10 μM cold CTP, 1 mM ATP and UTP, and 5 mM GTP. After the addition of 10 μl 3 M Na-acetate and 20 μg glycogen, samples were extracted with phenol-chloroform and nucleic acids were precipitated with 0.7 volumes of isopropanol. RNAs were analyzed by denaturing glyoxal-agarose gel electrophoresis. To generate in vitro-transcribed RNAs corresponding to the 3′ portion of the HCV positive- or negative-strand genome, the corresponding region of appropriate plasmids was amplified by PCR. Primers S/T7/9191 (TTGTAATACGACTCACTATAGGGACCAAGCTCAAACTCACTCCA) and A9605Con (ACATGATCTGCAGAGAGGCC) or primers S/T7/9191 and A9678JFH (ACATGATCTGCAGAGAGACC) were used on plasmid Luc JFH or Luc JFH 5′ X Con, respectively, to generate template DNA for the transcription of 3′ (+)X JFH or 3′ (+)X Con. Oligonucleotide S_5′ Con (GCCAGCCCCCGATTGGGGGC) or S_5′ JFH (ACCTGCCCCTAATAGGGGCG) or S_5′ G_JFH (GACCTGCCCCTAATAGGGGCG) with A_T7_341 (TTGTAATACGACTCACTATAGGGTGCACGGTCTACGAGACC) was applied to amplify the template 3′ (−)Con, 3′ (−)JFH, or 3′ (−)C JFH, respectively, from plasmid Luc JFH 5′ X Con or Luc JFH. All PCR products were treated with Klenow enzyme to generate blunt ends following the instructions of the manufacturer (New England Biolabs) and purified by phenol-chloroform extraction. In vitro transcription and purification of transcripts were performed as described above. In vitro-transcribed RNAs were denatured for 3 min at 95°C and cooled on ice prior to their use in RdRp assays.
RESULTS
Replication kinetics of genotype 1b and 2a replicons.
To gain more insight into the differences between cell culture-adapted Con1 and the wild-type JFH1 isolate, we compared the replication kinetics of subgenomic luciferase reporter replicons derived from these two isolates (Fig. 1A) by using luciferase activity as a correlate for RNA replication (31, 36). In vitro transcripts corresponding to each replicon were transfected into Huh7-Lunet cells, a cured replicon cell clone highly permissive for HCV RNA replication (30, 68), and compared to replication-deficient Con1- and JFH1-based RNAs lacking the GDD motif of NS5B polymerase (ConΔGDD and JFHΔGDD). As shown in Fig. 1B, within the first 8 h after transfection, luciferase activity was due mainly to the translation of the input RNA, since no significant differences were observed with that of the negative control ConΔGDD, which seemed to be somewhat more stable than JFHΔGDD. For Con1, luciferase activity slightly and constantly increased over time up to 30-fold of the input values at 72 h after transfection. In contrast, reporter gene activity increased rapidly for the JFH1-derived replicon, reaching 1,000-fold of the input levels by 24 h after transfection and staying at the same high level at later time points, indicating that host cell determinants might limit further amplification. Similar results were obtained with monocistronic replicons, in which the luciferase gene was fused to NS3 via ubiquitin, resembling the translational properties of authentic genomes, and with bicistronic replicons expressing luciferase under the translational control of the HCV IRES (data not shown).
FIG. 1.
Structure and replication kinetics of genotype 1b and 2a replicons. (A) Structures of the replicons Luc Con (genotype 1b) and Luc JFH (genotype 2a) used for transient replication assays. luc, firefly luciferase; EI, encephalomyocarditis virus IRES; VR, variable region and poly(U/UC) tract of the HCV 3′ NTR; X, X tail. Coding sequences are indicated by rectangles, and noncoding regions are indicated by oval forms. Con1 sequences are indicated in white with black letters, and JFH1-derived sequences are indicated by black, filled forms with white lettering. This code was kept throughout the whole study. Adaptive mutations E1202G, T1280I, and K1846T in Con1 are indicated by black dots. (B) Transient replication of Luc Con and Luc JFH. Huh7-Lunet cells were transfected with 5 μg of in vitro transcripts corresponding to Luc Con, Luc JFH, ConΔGDD, or JFHΔGDD. Luciferase activity was determined in cell lysates prepared at 2, 4, 8, 12, 18, 24, 48, and 72 h posttransfection. Data were normalized for transfection efficiency among identical replicons as determined by measurement of the luciferase activity 2 h after transfection. Electroporations and luciferase assays were performed in duplicate. Note the logarithmic scale of the ordinate and the abscissa.
The dramatic boost of the JFH1 signal within 12 to 24 h after transfection, which was preceded by an 8-h lag phase similar to that observed with Con1, argued that the efficiency of RNA synthesis was the main clue to the different replication kinetics of the two isolates. Therefore, we focused on a possible role for the NTRs containing the signals in the initiation and regulation of negative- and positive-strand syntheses.
The 5′ NTR and 3′ X tail contain genotype-specific signals.
The NTRs are among the most conserved parts of the HCV genome. The 5′ NTR differs in 25 positions between Con1 and JFH1. The last 98 nucleotides of the 3′ NTR are almost invariant (28, 60). However, while almost all HCV genotypes share the same X consensus sequence, the consensus sequence of genotype 2 differs in four positions of the 3′-terminal SLI (see Fig. 4A); in addition, some isolate-specific differences have been reported (28, 73). To evaluate whether genotype-specific signals are important for efficient JFH1 replication, we exchanged the original 5′ NTR and the X tail of the 3′ NTR either individually or simultaneously in the Luc JFH replicon (Fig. 1A) to obtain replicons with heterologous NTRs and analyzed them for replication capability. Since the luciferase expression of these replicons was directed by the PI, any possible influence of translation by the HCV IRES should have been avoided (15, 36). Figure 2A shows that heterologous NTRs had a strong impact on JFH1 replication efficiency. The exchange of the 5′ NTR or X tail in replicon 5′ Con/X JFH or 5′ JFH/X Con, respectively, resulted in a 1.5-log reduction of luciferase counts 24 h after transfection compared to what was observed with a replicon with wild-type NTRs (5′ X JFH). However, chimeric replicons 5′ Con/X JFH and 5′ JFH/X Con met wild-type luciferase values at later time points because the wild-type replicon reached a plateau of luciferase activity at 48 h after transfection. The luciferase expression of replicon Luc JFH 5′ X Con was more severely impaired and never reached input levels, indicating that the individual negative effects of the heterologous 5′ NTR and X tail were additive.
FIG. 2.
Impact of heterologous NTR sequences on replication efficiency of Con1 and JFH1 replicons. (A) Effect of the heterologous 5′ NTR and 3′ X tail on replication efficiency of a Luc JFH replicon. Luc JFH replicon RNAs with homologous NTRs (5′ X JFH), 5′ NTR, or 3′ X-tail sequences derived from Con1 (5′ Con/X JFH, 5′ JFH/X Con, or 5′ X Con, respectively) or a replication-deficient control replicon (ΔGDD) were transfected into Huh7 cells; transfected cells were seeded and harvested at 4, 24, 48, and 72 h after transfection, and cell lysates were analyzed for luciferase activity. Luciferase activity is given as the change in RLU (n-fold) relative to the value obtained 4 h after transfection. VR, variable region and poly(U/UC) tract of the HCV 3′ NTR; X, X tail. (B) Genotype specificity of replication signals in the 5′ NTR. Luc JFH replicons with 5′ NTRs derived from isolates JFH1 and J6 (both genotype 2a), H77 (genotype 1a), and Con1 (genotype 1b) were transfected into Huh7 cells; luciferase activity was determined as described above. JFH1 and J6 5′ NTRs differed at positions 4, 78, and 302 and Con1 and H77 at positions 11, 12, 13, 34, 35, 204, and 243. (C) Heterologous NTR sequences affect the replication efficiency of Luc Con replicons. Luc Con replicon RNAs with homologous NTRs (5′ X Con), 5′ NTR, or 3′ X-tail sequences derived from JFH1 (5′ JFH/X Con, 5′ Con/X JFH, or 5′ X JFH, respectively) were transfected into Huh7 cells and analyzed for replication efficiency as described above. Schematic drawings of replicons are shown at the top of panels A and C. JFH1- or Con1-derived sequences are given in black or white, respectively; portions given in gray are variable within one panel. For a detailed description of the luciferase replicons, refer to the legend to Fig. 1A.
To exclude isolate-specific effects, we analyzed the impact of 5′ NTRs from another genotype 2a (J6 [70]), and a genotype 1a isolate (H77 [69]) on JFH1 replication (Fig. 2B). The 5′ NTR of J6 had no significant effect on JFH1 replication, as expected from the small degree of divergence between the JFH1 and J6 sequences, whereas the 5′ NTR of H77 reduced JFH1 replication efficiency similarly to the Con1 sequence, consistent with the idea that the 5′ NTR contains genotype- but not isolate-specific signals. To further confirm the genotype specificity of the NTRs, we used the Luc Con replicon (Fig. 1A) and replaced the 5′ NTR and X tail with the respective JFH1 counterparts (Fig. 2C). Again, homologous NTRs were the most efficient (5′ X Con), the exchange of the 5′ NTR or X tail resulted in a mild reduction of replication efficiency for 5′ Con/X JFH and 5′ JFH/X Con, and the effects were additive when both heterologous NTR sequences were combined (5′ X JFH1). The reduction in replication efficiency relative to that of the wild-type sequence was less severe than for JFH1, most likely due to the generally much lower replication efficiency and slower kinetics of Con1.
Taken together, these results indicate that the 5′ NTR and the X tail contain genotype-specific signals, which are important for efficient HCV RNA replication.
Impact of chimeric 3′ X-tail and 5′ NTR sequences on negative- and positive-strand syntheses.
The NTRs of positive-strand RNA viruses contain signals important for the initiation of RNA synthesis. The 3′ X region of the HCV genome has been shown to be essential for RNA replication (13, 29, 71, 72) and should contain signals for the initiation of negative-strand synthesis. Progeny positive-strand synthesis is thought to be regulated by the 3′ end of the viral negative strand. Therefore, the reduction of replication efficiency by heterologous NTRs should be due to a specific impairment of the initiation of negative-strand synthesis in the case of the 3′ X tail and of positive-strand synthesis in the case of the 5′ NTR. To prove this hypothesis, we transfected Luc JFH replicons with authentic or Con1-derived 5′ NTRs and/or 3′ X tails into Huh7 cells and analyzed positive- and negative-strand synthesis at different time points after transfection (Fig. 3A). The amounts of positive-strand RNA obtained for constructs with authentic and chimeric NTRs resembled closely the results of luciferase expression shown in Fig. 2A. Replication efficiency was highest for a replicon harboring authentic NTRs (5′ X JFH) (Fig. 3A, upper panel, lanes 2 to 5); replicons containing either a Con1-derived 5′ NTR (5′ Con/X JFH) (lanes 6 to 9) or a 3′ X tail (5′ JFH/X Con) (lanes 10 to 13) were similarly impaired in positive-strand RNA levels at 24 to 72 h after transfection compared to input levels (0 h), whereas RNA replication was barely detectable for the chimeric replicon containing the 5′ NTR and 3′ X tail from Con1 (5′ X Con) (lanes 14 to 17), demonstrating that the RNA replication capability of this construct was severely disturbed. Likewise, the ratios of positive- to negative-strand HCV RNA at 24, 48, and 72 h after transfection were also altered with the different chimeras (Fig. 3A, upper and middle panels). In the case of the wild-type replicon (5′ X JFH), the average positive- to negative-strand RNA ratio (±standard deviation) was about 8.7 (±2.5). The exchange of the 3′ X region resulted in an increased positive- to negative-strand RNA ratio of 16.1 (±3.8), due mainly to a decreased level of negative strands (5′ JFH/X Con) (Fig. 3A, middle panel, lanes 10 to 13). In contrast, the chimera with the heterologous 5′ NTR (5′ Con/X JFH), which contains the signals for progeny positive-strand synthesis, had a significantly reduced positive- to negative-strand RNA ratio (4.7 ± 1.2), indicating that indeed positive-strand RNA synthesis was primarily impaired. These results, which are representative of two independent experiments, were corroborated by short-term kinetic analyses (Fig. 3B). The kinetics of negative-strand synthesis were identical for 5′ X JFH and 5′ Con/X JFH, with the first detectable signal at 4 h posttransfection and steadily increasing up to 12 h after transfection (Fig. 3B, middle panel, compare lanes 2 to 6 and 7 to 11), demonstrating that a heterologous 5′ NTR had no effect on negative-strand synthesis. In contrast, the heterologous 3′ X tail decreased the negative-strand synthesis of the 5′ JFH/X Con replicon to undetectable levels within the time span of this experiment (Fig. 3B, middle panel, lanes 12 to 16).
In summary, although the overall replication efficiencies of the replicons 5′ Con/X JFH and 5′ JFH/X Con were reduced to similar levels, the underlying mechanisms were very different. The heterologous 3′ X tail specifically affected negative-strand synthesis, whereas alteration of the 5′ NTR decreased the positive- to negative-strand RNA ratio, suggesting that progeny positive-strand synthesis was impaired. Therefore, subgenomic replicons with chimeric NTRs provide an excellent model to study the factors involved in the initiation of negative- and positive-strand synthesis independently.
Mapping of genotype-specific signals in the 5′ NTR and 3′ X tail.
To gain more insight into the nature of the genotype-specific signals in the NTRs, we mapped the responsible nucleotide sequences. In the case of the X region of the 3′ NTR, we chose the monocistronic replicon Luc/ubi JFH1 (Fig. 4C), resembling an authentic HCV genome more closely than the bicistronic Luc JFH replicons. The overall replication efficiency of this replicon was very similar to that of the Luc JFH used in the previous experiments (data not shown), and the exchange of the X region with the heterologous Con1 sequence resulted in a 1-log decrease in luciferase counts at 24 to 72 h after transfection (Fig. 4C), consistent with our previous results. All four differences in the X regions of JFH1 and Con1 were located in SLI (Fig. 4A) and mutated individually in the monocistronic replicon. As shown in Fig. 4C, the flipping of UA to AU at position 74/75 had no effect on replication efficiency; the C72U mutant was only slightly impaired, but the mutation of U at position 81 to C affected the replication of the corresponding replicon almost as much as conversion of the entire X region to the Con1 counterpart, indicating that this position was most critical in conferring genotype specificity to the X region.
Mapping studies of the 5′ NTR were done by using Luc JFH replicons (JFH1) (Fig. 4D). The replacement of the first half of the 5′ NTR with the Con1 counterpart (1-156Con) resulted in significantly reduced replication efficiency, whereas a replicon containing the second half of the Con1 5′ NTR (157-341Con) was barely affected. The fact that replicon 1-156Con was almost as impaired as the replicon chimera with the entire Con1 5′ NTR (5′ Con) indicated that the important genotype-specific signals were located primarily in the 5′ half of the NTR. Since the complementary sequence of this region was required for progeny positive-strand synthesis, we clustered 12 of the 13 differences between Con1 and JFH1 on the basis of previously published secondary structure predictions for the 3′ end of the negative strand (54, 59) (Fig. 4B): a 1-nucleotide insertion and six alterations at the 3′ end, encompassing a nonpaired region and the first stem-loop (1-4/I′Con), three substitutions in the second stem-loop (IIz′Con), and two in the third stem-loop (IIy′Con). The alterations in the nonpaired region at the very 3′ end of the negative strand were also analyzed separately (1-4Con), whereas one additional difference at position 78 of the JFH1 sequence was not analyzed because it was restricted to JFH1 and not found in other genotype 2a isolates. Surprisingly, the replication of none of these constructs was significantly impaired in comparison to that of the JFH1 wild-type replicon (Fig. 4D).
Taken together, genotype-specific signals are contained primarily at positions 81 and 72 of the X region in the 3′ NTR as well as in the first half of the 5′ NTR.
Analysis of recombinant NS5B polymerase for genotype-specific template recognition in vitro.
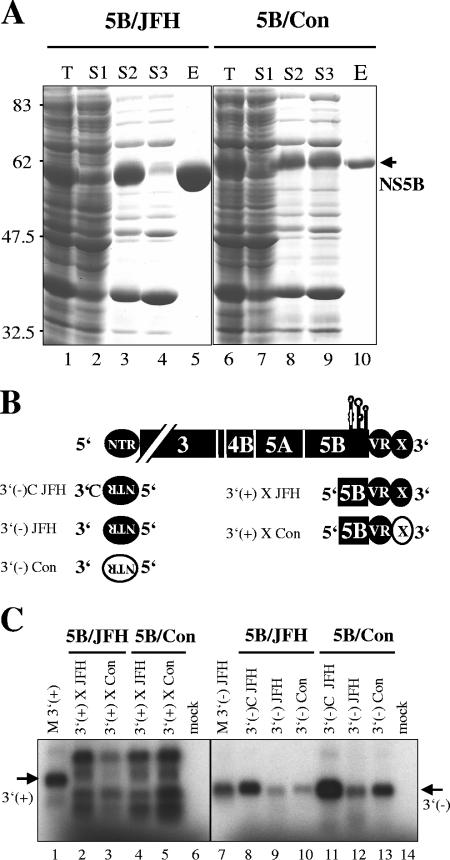
Helicase, NS5A, and NS5B have an intrinsic ability to bind to RNA (19, 21, 38) and could in principle recognize genotype-specific replication signals in the NTRs. However, the most obvious candidate was the NS5B polymerase, which is capable of de novo initiation of RNA synthesis on templates derived from the 3′ termini of the viral genome (41, 44, 51, 76). To address whether NS5B was responsible for genotype specificity, we expressed C-terminally truncated NS5B of JFH1 and Con1 in E. coli and purified both enzymes to near homogeneity (Fig. 5A). We then performed in vitro polymerization assays by using templates corresponding to the 3′ ends of viral positive- and negative-strand RNA to determine genotype specificity (Fig. 5B). To compensate for the lower specific activity of Con1 RdRp (V. Lohmann, unpublished data), we used five times the amount of 5B/Con compared to that of 5B/JFH in these assays.
FIG. 5.
Purification of JFH1 NS5BΔC21 and analysis of genotype-specific template recognition in vitro. (A) Expression of NS5B from isolates JFH1 (lanes 1 to 5) and Con1 (lanes 6 to 10) in E. coli and purification by differential solubilization and affinity chromatography. Both proteins lack 21 C-terminal amino acids and are fused to a His6 tag. T, total bacterial lysate after induction; S1, supernatant 1 after treatment of bacterial cells with LBI and centrifugation (note that NS5B was not soluble under these conditions); S2, supernatant 2 obtained from solubilization of the pellet remaining from S1; S3, supernatant 3 after incubation of S2 with Ni-NTA agarose; E, eluted protein. Numbers on the left refer to the sizes (in kDa) of reference proteins run on the same gel. For a detailed explanation, refer to Materials and Methods. (B) Schematic representation of different template RNAs used for in vitro RdRp assays. Portions corresponding to Con1 sequences are given in white with black letters, and JFH1-derived sequences are indicated by black, filled forms with white lettering. The positions of cis-acting RNA elements in the coding sequence of NS5B are indicated by schematic stem-loop drawings. Sequences corresponding to the 3′ terminus of the negative strand are given upside down and in 3′-to-5′ orientation. VR, variable region and poly(U/UC) tract of the HCV 3′ NTR; X, X tail. (C) Analysis of genotype-specific template recognition in vitro by NS5B of isolates JFH1 and Con1. Different in vitro-transcribed RNAs as indicated above each lane and as shown schematically in panel B were incubated with 200 ng purified NS5BΔC21 from isolate JFH1 (5B/JFH) or 1 μg Con1 (5B/Con) in the presence of [α-32P]CTP. Reaction products were separated by denaturing agarose-glyoxal gel electrophoresis; the gel was dried and subjected to autoradiography. mock, elution buffer instead of purified polymerase was added to the reaction; M 3′ (+), in vitro-transcribed, radiolabeled RNA corresponding to nucleotides 9259 to 9678 of the HCV JFH1 positive strand, identical in size to template 3′ (+) X JFH; M 3′ (−) JFH, in vitro-transcribed, radiolabeled RNA corresponding to the 3′-terminal 340 nucleotides of the negative strand of HCV JFH1, identical in size to template 3′ (−) JFH. Note that 3′ (−)C JFH and 3′ (−) Con encompass 341 nucleotides in length. The sizes of the template RNAs are indicated by arrows on the left [3′ (+)] and right [3′ (−)]. For a detailed explanation of the different template RNAs, refer to the text.
In the case of the positive-strand RNA, we used a JFH1 template that included SL3.1-3.3 in the NS5B coding sequence (Fig. 5B) (74), because it has been shown that an SL3.2 kissing-loop interaction with SLII in the X region is essential for HCV replication (14) and might contribute to the initiation of HCV RNA synthesis by NS5B in vitro. We used either the authentic 3′ end of JFH1 [3′ (+)X JFH] or the same sequence with the X region of Con1 [3′ (+)X Con]. The patterns of reaction products were complex but very similar for both templates and polymerases (Fig. 5C, lanes 2 to 5). A fraction of the newly synthesized RNAs was larger than the template (Fig. 5C, lane 1) and was most likely generated by elongation of the 3′ end of the template, resembling so-called “copy-back” synthesis (5). Another fraction was heterogeneous in size but smaller than the template and therefore probably generated by de novo initiation. Although the mode and site of initiation in vitro did not resemble the mechanism expected in vivo, there was a moderate but clear increase in the amount of all types of reaction products, when the X region of the template was homologous to the polymerase (Fig. 5C, compare lanes 2 to 3 and 4 to 5). Using several preparations of template RNA, this difference was 1.6- to 2.3-fold for JFH1 RdRp and 1.1- to 1.8-fold for Con1 RdRp. Since these templates differed at only four nucleotides in SLI of the X tail (Fig. 4A), these data indicated that NS5B interacted specifically with this region.
For analysis of the 3′ end of the negative strand [3′ (−)], we used the sequences complementary to the entire 5′ NTR in three different variants (Fig. 5B): the authentic Con1 sequence [3′ (−)Con], the authentic published JFH1 sequence [3′ (−) JFH] terminating with uridine, and the JFH1 sequence with an additional cytidine at the 3′ end [3′ (−)C JFH], resembling the 5′ end of the in vitro-transcribed replicon RNAs used in this and several other studies (24, 66). Both RdRps produced only one dominant reaction product corresponding in size to the template (Fig. 5C, compare lanes 8 to 13 with lane 7), indicating that de novo initiation was the preferred reaction when these templates were used. Although the templates were used with very different efficiencies, both RdRps exhibited the same preferences. The authentic JFH1 negative-strand 3′ end [3′ (−)JFH] yielded the smallest amount of reaction products (Fig. 5C, lanes 9 and 12); 3′ (−)Con was 1.2- to 1.4-fold and 1.7- to 1.9-fold more efficient for JFH1 and Con1 RdRp (lanes 10 and 13), respectively, whereas 3′ (−)C JFH resulted in more than 10-fold increased levels of RNA synthesis for both RdRps compared to the same template lacking the terminal cytidine (compare lanes 8 and 11 to lanes 9 and 12, respectively). These results indicated that guanosine was the preferred nucleotide for de novo initiation in vitro and that this parameter was more important for efficient RNA synthesis than the genotype of the template. Although the Con1 template also allowed de novo initiation starting with guanosine, it was used with much lower efficiencies by both RdRps than 3′ (−)C JFH, implying that NS5B had no clear genotype-specific template preference for the 3′ end of the negative strand in vitro.
In summary, recombinant NS5B proteins from both Con1 and JFH1 showed some genotype-specific preference for templates with a homologous X region. In contrast, templates comprising the 3′ end of the negative strand were utilized by both RdRps without evident genotype specificity.
Construction of intergenotypic replicon chimeras.
To further substantiate the findings obtained with purified RdRp and to identify further HCV nonstructural proteins conferring genotype specificity to the NTRs, we constructed intergenotypic replicase chimeras in which we exchanged RNA binding proteins. Since we expected from the in vitro assays that the polymerase would be an important determinant to confer genotype specificity to the NTRs, we chose the 5′ NTR and 3′ NTR according to the genotype of the NS5B sequence in the chimeric replicons (Fig. 6A). We started with replicon Luc JFH (Fig. 1A) and replaced the NS5B coding region, the 3′ NTR, and the 5′ NTR with the corresponding Con1 sequences (JFH/5B Con) (Fig. 6A), but unfortunately, the replication of this RNA was too low and indistinguishable from that of the negative control (Fig. 6B and Table 1). Likewise, when we exchanged only the NS5B coding region and the variable parts of the 3′ NTR, the construct was still replication deficient in the transient replication assay and the more sensitive colony formation assay using a selectable replicon harboring neomycin phosphotransferase instead of luciferase (data not shown). Therefore, we used the Luc Con replicon as a starting point (Fig. 1A) and replaced NS5B, the 5′ NTR, and the 3′ NTR with the equivalent JFH1 counterparts (Con/5B JFH) (Fig. 6A). This chimeric replicon was clearly replication competent, with a slightly reduced efficiency compared to that of its parental Luc Con (Fig. 6B, open diamonds compared to open squares) and an efficiency similar to that of Luc Con/5′ X JFH (Table 1). We then used Con/5B JFH and replaced the Con1 coding sequences of other HCV nonstructural proteins with known RNA binding capability with those of JFH1 (NS3 helicase and NS5A), either independently or in combination (Con/5AB JFH, Con/Hel,5B JFH, and Con/Hel,5AB JFH) (Fig. 6A). Replicons Con/5AB JFH and Con/Hel,5B JFH were still replication competent but with a clearly reduced efficiency compared to that of the chimera containing only NS5B of JFH1 (Fig. 6B, filled diamonds and open circles compared to open diamonds). In contrast, the combination of helicase, NS5A, and NS5B of JFH1 in the Con1 backbone was even more efficient than the Luc Con replicon, at least at time points later than 24 h after transfection (Fig. 6B, filled circles versus open squares), indicating that a helicase-NS5A interaction might be critical for efficient RNA replication. However, the fact that this replicon was far less efficient than Luc JFH (Fig. 6B, filled squares, and Table 1) pointed to the direction that additional interactions within the nonstructural proteins were still disturbed in this chimera.
FIG. 6.
Structure and replication competence of intergenotypic replicase chimeras. (A) Schematic structures of replicons harboring a chimeric replicase. White areas with black lettering represent Con1-derived sequences, and black areas with white lettering refer to JFH1. Adaptive mutations E1202G, T1280I, and K1846T in the Con1 sequence are indicated by black dots. luc, firefly luciferase; EI, encephalomyocarditis virus IRES; VR, variable region and poly(U/UC) tract of the HCV 3′ NTR; X, X tail. For further details, refer to the legend to Fig. 1A. (B) Replication competence of intergenotypic replicase chimeras. Luciferase replicon RNAs derived from either JFH1 (Luc JFH) or Con1 (Luc Con) or replicon RNAs harboring chimeric NS3-to-NS5B coding regions or a replication-deficient control replicon (ΔGDD) were transfected into Huh7-Lunet cells by electroporation; transfected cells were seeded and harvested at 4, 24, 48, and 72 h after transfection, and cell lysates were analyzed for luciferase activity. Luciferase activity is given as the change in RLU (n-fold) relative to the value obtained 4 h after transfection. Electroporations and luciferase assays were performed in duplicate; data represent the averages and standard deviations of at least four single values. Note the logarithmic scale of the ordinate.
TABLE 1.
Replication efficiencies of intergenotypic replicase chimeras compared to those of parental replicons
| Replicon | Genotype | Replication efficiency (fold RLU)a |
|---|---|---|
| Luc JFH | 5′ X JFH | (2.3 ± 0.05) × 102 |
| Luc Con | 5′ X Con | (2.9 ± 0.1) × 101 |
| 5′ X JFH | (6.7 ± 0.8) × 10° | |
| JFH/5B Con | 5′ X JFH | (7.8 ± 5.8) × 10−3 |
| 5′ X Con | (2.5 ± 0.2) × 10−3 | |
| Con/5B JFH | 5′ X JFH | (5.8 ± 0.9) × 10° |
| 5′ Con/X JFH | (8.5 ± 0.1) × 10° | |
| 5′ JFH/X Con | (1.1 ± 0.04) × 10−1 | |
| 5′ X Con | (2.6 ± 0.08) × 10−1 | |
| Con/5AB JFH | 5′ X JFH | (1.3 ± 0.2) × 10° |
| 5′ Con/X JFH | (4.1 ± 0.6) × 10° | |
| 5′ JFH/X Con | (7.5 ± 0.9) × 10−2 | |
| 5′ X Con | (2.4 ± 0.4) × 10−1 | |
| Con/Hel,5B JFH | 5′ X JFH | (6.6 ± 0.3) × 10−1 |
| 5′ Con/X JFH | (7.6 ± 0.8) × 10−1 | |
| 5′ JFH/X Con | (1.1 ± 0.1) × 10−1 | |
| 5′ X Con | (9.5 ± 0.6) × 10−2 | |
| Con/Hel,5AB JFH | 5′ X JFH | (4.5 ± 0.2) × 101 |
| 5′ Con/X JFH | (9.7 ± 1.3) × 10° | |
| 5′ JFH/X Con | (6.1 ± 0.8) × 10° | |
| 5′ X Con | (1.3 ± 0.2) × 10° | |
| ΔGDD | 5′ X Con | (3.5 ± 0.1) × 10−3 |
Since Con1-based replicons containing polymerase, helicase, and/or NS5A from JFH1 all replicated, albeit with various efficiencies, these intergenotypic replicase chimeras were the appropriate tool to study determinants conferring genotype specificity to the NTRs.
Role of NS5B, helicase, and NS5A in the initiation of RNA synthesis.
The next step in identifying the determinants for the recognition of replication signals in the NTRs consisted of analyzing the intergenotypic replicase chimeras in the context of NTRs from different genotypes. We tested all Con1-based intergenotypic replicase chimeras (Fig. 6A) in every possible combination of the 5′ NTR and X tail derived from JFH1 and Con1, respectively, to analyze the genotype preferences of the mixed-replicase complexes. In the case of Con/5B JFH, harboring only NS5B of JFH1, both replicons containing the 3′ X tail of JFH1 behaved similarly and were more efficient than those harboring the X tail of Con1 (Fig. 7A), irrespective of the origin of the 5′ NTR. The preference of NS5B for its own X region, but not for the 5′ NTR (or rather the 3′ end of the negative strand), resembled and confirmed the results obtained with purified polymerase in vitro (Fig. 5C), again indicating that NS5B was the critical determinant for the recognition of genotype specificity in SLI of the X region. However, the fact that the 5′ NTR of Con1 yielded a slight, reproducible increase in replication efficiency in both settings (Fig. 7A, compare open and filled symbols) suggested that additional factors besides NS5B were important in conferring genotype specificity to the 5′ NTR. The addition of JFH1 NS5A (Fig. 7B) or the JFH1 NS3 helicase (Fig. 7C) led in principle to outcomes similar to those for isolated NS5B of JFH1 in the context of NS3-5A of Con1: a strong preference for the 3′ X tail of JFH1 and either a weak bias for the Con1 5′ NTR for NS5AB (Fig. 7B) or no preference at all for this region in the case of Con/Hel,5B JFH (Fig. 7C). Only when we combined all three factors, helicase, NS5A, and NS5B, in one replicon did genotype-dependent recognition of the NTRs resemble the picture of the entire JFH1 replicase (compare Fig. 7D and 2A). In this case, both NTRs derived from JFH1 were most efficient in RNA synthesis (5′ X JFH), the exchange of either the 5′ NTR or the 3′ X tail resulted in a significant decrease in replication efficiency (5′ Con/X JFH or 5′ JFH/X Con, respectively), and the replacement of the 5′ NTR and 3′ X tail at once led to an additive reduction in the luciferase expression achieved with the respective replicon (5′ X Con). Therefore, helicase, NS5A, and NS5B together are necessary and sufficient to recognize genotype-specific signals in the 5′ NTR. A second conclusion that could be drawn from these experiments relied on the fact that a heterologous X region selectively impaired negative-strand synthesis, whereas a heterologous 5′ NTR affected the generation of progeny positive strands (Fig. 3). Based on this observation, it seems plausible that those nonstructural proteins conferring genotype specificity to the X region or the 5′ NTR are directly involved in the initiation of negative- or positive-strand RNA synthesis, respectively. Therefore, interactions between the NS3 helicase, NS5A, and NS5B seem to be important for the initiation of positive-strand synthesis. Since the differences in the X region were limited to one particular area in the tip of SLI, probably leaving space for the binding of only one protein, it was not clear from our results whether helicase and NS5A were also involved in the initiation of negative-strand synthesis.
FIG. 7.
NTR preferences of intergenotypic replicase chimeras. Genotype Con1-based luciferase replicons harboring the sequences of different nonstructural proteins from isolate JFH1 were fused to the 5′ NTR and 3′ X tail from JFH1 (5′ X JFH), Con1 (5′ XCon), or mixed NTRs (5′ Con/X JFH and 5′ JFH/X Con) and tested for replication efficiency as described in the legend to Fig. 6B. Schematic drawings of replicons are shown at the top of panels A to D. JFH1- or Con1-derived sequences are given in black or white, respectively; portions given in gray are variable within one panel. luc, firefly luciferase; EI, encephalomyocarditis virus IRES; VR, variable region and poly(U/UC) tract of the HCV 3′ NTR; X, X tail. For a detailed description of the luciferase replicons, refer to the legend to Fig. 1A.
In summary, we were able to show that NS5B RdRp was the critical component for the recognition of genotype-specific signals in the X region of the 3′ NTR, whereas the NS3 helicase, NS5A, and NS5B together were required to confer genotype specificity to the 5′ NTR, indicating that the interaction between these three proteins plays a critical role in the initiation of HCV positive-strand synthesis.
DISCUSSION
The present study provides evidence that the conserved regions in the NTRs of the HCV genome contain genotype-specific signals regulating the initiation of RNA synthesis. This observation combined with the development of efficient intergenotypic replicon chimeras allowed us for the first time to analyze which proteins are involved in the initiation of HCV negative- and positive-strand syntheses. Our results indicate that helicase, NS5A, and NS5B participate in the initiation of progeny positive-strand RNA synthesis and provide a starting point to characterize the interaction between these proteins in more detail.
Analysis of HCV replication in cell culture has long been focused on genotype 1 isolates due to the lack of isolates from other genotypes efficiently replicating in cell culture. A common feature of genotype 1 genomes is the need for adaptive mutations enhancing RNA replication to detectable levels (6, 7, 31, 36, 37, 39). In striking contrast, the genotype 2a isolate JFH1 supports HCV replication in cell culture to even higher levels than adapted genotype 1 isolates without requiring prior adaptation (24, 61). The Con1 replicon which was used in the present study contained three cell culture-adaptive mutations (E1202G, T1280I, and K1846T), which stimulated replication in cell culture cooperatively (36). The mechanism underlying cell culture adaptation is still unknown for NS3 mutations (E1202G and T1280I), while the mutation in NS4B (K1846T) reduces the level of NS5A hyperphosphorylation (2, 12), which might be responsible for the adaptive phenotype (12). The effects of adaptive mutations on RNA replication in the context of intergenotypic chimeras between Con1 and JFH1 were hardly predictable, especially since the design of some chimeras required the swapping of the helicase (including mutation T1280I) and NS5A. Therefore, the overall replication efficiencies of particular intergenotypic chimeras as shown in Fig. 6 might be influenced by the presence or absence of the Con1-adaptive mutations. However, given the multiple interactions between all HCV NS proteins, the efficiency of chimeric replication complexes will be determined by a variety of parameters and thus must be interpreted with caution.
Our results indicate that one major difference between JFH1 and Con1 lies in the efficiency of the initiation of RNA synthesis and that the RdRp of JFH1 might have properties that are critical for this performance. This theory is based on several observations. (i) The time required for a complete RNA replication cycle of a subgenomic replicon seems to be 8 to 12 h for JFH1 and Con1, as indicated by the first increase of luciferase activity over background (Fig. 1B). However, within the next 12 h, JFH1 reaches up to 1,000-fold of input luciferase levels, whereas Con1 gains less than 10-fold. Therefore, JFH1 either generates a higher number of active replication complexes within the initial 8 to 12 h that will, however, not yet initiate RNA synthesis or produces more progeny RNA per replication complex. (ii) The replacement of the 5′ NTR and X region of JFH1 with their Con1 counterparts dramatically reduces JFH1 replication efficiency by more than 2 orders of magnitude (Fig. 2A), whereas the equivalent manipulation had less severe effects on Con1, resulting in replicons with almost comparable replication efficiencies (Fig. 2C). We have shown that these changes directly impair the initiation of RNA synthesis; therefore, it seems likely that a higher initiation rate of RNA synthesis is a critical component of the extraordinary replication efficiency of JFH1. (iii) Purified RdRp of JFH1 has a 5- to 10-fold-higher specific activity than Con1 RdRp, although elongation rates were comparable for both enzymes (V. Lohmann, unpublished observations), arguing that JFH1 NS5B is more efficient at the initiation of RNA synthesis. (iv) An intergenotypic replicase chimera based on NS3 to NS5A of JFH1 and NS5B of Con1 was not viable (Fig. 6). This is well in line with previous observations demonstrating that even the intragenotypic exchange of NS5B resulted in a reduced replication efficiency (63), whereas in another study, NS5B of a genotype 2b isolate in the background of genotype 1b NS3 to NS5A was barely replication competent and required compensatory mutations for efficient replication (17). In contrast, our intergenotypic replicon chimeras harboring NS3 to NS5A from Con1 and NS5B of JFH1 replicated with high efficiencies (Fig. 6), again indicating that NS5B of JFH1 might have special properties that account to some extent for the unique properties of this HCV isolate. Further biochemical and structural analysis of NS5B will be required to better understand the contribution of the RdRp to the efficient replication of JFH1.
Genetic studies of HCV RNA-protein interactions have been complicated by the fact that all RNA and protein functions in HCV RNA replication in cell culture have to be provided in cis, with the exception of certain NS5A functions (1). Therefore, it has not been possible to dissect the interaction of individual proteins and RNA elements in a cell-based replication assay like it has been for, e.g., alphaviruses (33). The identification of genotype-specific signals in the NTR in combination with intergenotypic replicase chimeras as described in this study allows us for the first time to study the determinants for the initiation and regulation of negative- and positive-strand HCV RNA syntheses in a genetic system. Chimeras between closely related enteroviruses have already been successfully used to define determinants in the polymerase important for strand-specific RNA synthesis (10). Similar strategies might also be applicable to other pairs of closely related positive-strand RNA viruses, such as West Nile virus/Kunjin virus, different dengue virus subtypes, and bovine viral diarrhea virus/classical swine fever virus.
Little is known about the actual mechanism of the initiation of HCV negative-strand RNA synthesis. Our results indicate that one step in this process involves an interaction between NS5B and the apical part of the stem of SLI in the X region (Fig. 4A). This is in line with a previous study showing that the sequence and structure of this part of SLI is very critical for HCV RNA replication (73). The slight preference of purified NS5B for an RNA template of the same genotype (Fig. 5C) suggests that the genotype specificity in the case of SLI is mediated by a protein-RNA interaction. However, we cannot formally exclude that an RNA structure within NS5B or in the variable region of the 3′ NTR directs genotype specificity to SLI, analogous to the kissing-loop interaction that was found between SL3.2 in the NS5B coding sequence and SLII in the X region (14). This possibility also holds true for other, yet unknown, cis-acting RNA elements in the NS protein-coding region that could be involved in long-term RNA-RNA interactions with the 3′ end of the negative strand and thereby mediate genotype specificity. One obvious limitation of our system is the dependence on genotype diversity in a particular region. Therefore, only SLI is accessible to our analysis within the X region, because the consensus sequences of SLII and SLIII are invariant among all genotypes. Previous in vitro studies have shown specific binding of NS5B to stem-loop structures in its own coding region (32) and to SLII in the X tail (45). This might indicate either that NS5B binds sequentially to different regions of the 3′ NTR or that multiple copies of NS5B simultaneously interact with this region to form a higher-ordered complex. The latter hypothesis is in line with in vitro studies demonstrating that NS5B oligomerizes and displays cooperativity (18, 67). It is also possible that other viral proteins such as NS3 and NS5A are involved in the initiation of negative-strand RNA synthesis but were not identified in our analysis due to their binding to the invariant or the more variable parts of the 3′ NTR. For example, it has been shown that the NS3 helicase, NS5A, and NS5B all efficiently bind to poly(U) (19, 21, 38), suggesting a functional role for the poly(U/UC) tract in the initiation of RNA synthesis. In addition, specific binding to the 3′ NTR was shown for the NS3 helicase (3) but not for NS5A (21). Potential interactions of helicase and NS5A with the variable region of the 3′ NTR could be identified with intergenotypic replicase chimeras but were not in the scope of the present study.
In this study, we have shown that replicons harboring a 5′ NTR of a heterologous HCV genotype are directly impaired in progeny positive-strand synthesis (Fig. 3), thereby providing the first experimental system to study this process in cell culture. Using this approach, we were able to define those viral proteins that are involved in the recognition of the signals regulating positive-strand RNA synthesis, namely, the NS3 helicase, NS5A, and NS5B. These signals mapped primarily to the first half of the 5′ NTR (Fig. 4D), corresponding to the three 3′-terminal stem-loops of the HCV negative strand, which were recently defined in two independent studies (54, 59). Surprisingly, we were not able to further map genotype-specific signals in this region (Fig. 4B), probably because the effects of the individual mutations were below the detection limit of our assays. Alternatively, the binding of helicase, NS5A, and NS5B or several copies of one of these proteins to different sites might be stabilized by protein-protein interactions; in this case, the presence of one specific binding region might be strong enough for the docking of the whole protein complex. In line with this hypothesis, it has been shown that purified helicase binds to the 3′-terminal stem-loop of the negative-strand RNA (3), whereas NS5B seems to bind further upstream (23), and in addition, several potential binding sites could exist in this region for NS5A (21). Additional mapping analyses implying different intergenotypic replicase chimeras might help to identify distinct binding regions for the individual nonstructural proteins in the 3′ NTR of the HCV negative strand and to define the protein-protein interaction sites between helicase and NS5A.
Our data imply that helicase, NS5A, and NS5B are part of an initiation complex for RNA synthesis. Previous studies already suggested interactions of NS3 and NS5B (46, 75) and N5A and NS5B (56, 57) and several other interactions among the nonstructural proteins (11). These hints, together with results obtained with purified replication complexes showing equal stoichiometries of the NS proteins in these structures (47), indicate that the nonstructural proteins altogether act as a complex supporting different functions in the viral life cycle: regulation of translation and replication, induction of membrane alterations, positive- and negative-strand RNA syntheses, and probably retrieval of RNA for packaging into viral particles. It is getting clearer that NS5A plays a central role in the regulation of several of these processes. NS5A is a hotspot for cell culture-adaptive mutations, resulting in an altered phosphorylation pattern (2, 6) and increased RNA replication efficiency. In turn, these adaptive mutations seem to interfere with HCV virion formation (66), suggesting that the phosphorylation state of NS5A might regulate RNA replication and packaging, probably via host cell proteins such as human VAP-A (12) and the cellular kinase CKI (43, 48, 49). The finding that NS5A indeed binds to RNA (21) and the crystal structure of domain I of NS5A (62), showing that an NS5A dimer creates a deep basic groove that could bind single- and eventually also double-stranded RNA, already suggested a direct involvement of NS5A in RNA synthesis, which is further confirmed by our results. The actual role of NS5A and the precise composition of the initiation complex for HCV RNA synthesis have yet to be defined.
In conclusion, intergenotypic replicase chimeras will be a useful tool for the mapping of protein-RNA and protein-protein interactions among viral nonstructural proteins, in particular the newly identified interplay between helicase, NS5A, and NS5B, and will provide deeper insight into the mechanisms governing the initiation and regulation of HCV RNA synthesis.
Acknowledgments
We are grateful to Irina Anossova, Perdita Backes, and Michael Rohr for generating some of the replicon constructs, T. Wakita, J. Bukh, Z. Hong, M. A. Billeter, and K.-K. Conzelmann for generously supplying plasmid DNAs, and Peter Friebe for critically reading the manuscript.
This work was funded within the National Genome Research Network, NGFN, by the German Ministry for Research and Education.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appel, N., T. Pietschmann, and R. Bartenschlager. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, R., and A. Dasgupta. 2001. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 75:1708-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 5.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Cornell, C. T., R. Perera, J. E. Brunner, and B. L. Semler. 2004. Strand-specific RNA synthesis determinants in the RNA-dependent RNA polymerase of poliovirus. J. Virol. 78:4397-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77:5401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 101:13038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friebe, P., J. Boudet, J.-P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, D. J., M. Stahlhut, O. Flores, D. B. Olsen, D. J. Hazuda, R. L. Lafemina, and S. W. Ludmerer. 2006. A genotype 2b NS5B polymerase with novel substitutions supports replication of a chimeric HCV 1b:2b replicon containing a genotype 1b NS3-5A background. Antivir. Res. 69:24-30. [DOI] [PubMed] [Google Scholar]
- 18.Gu, B., L. L. Gutshall, D. Maley, C. M. Pruss, T. T. Nguyen, C. L. Silverman, J. Lin-Goerke, S. Khandekar, C. Liu, A. E. Baker, D. J. Casper, and R. T. Sarisky. 2004. Mapping cooperative activity of the hepatitis C virus RNA-dependent RNA polymerase using genotype 1a-1b chimeras. Biochem. Biophys. Res. Commun. 313:343-350. [DOI] [PubMed] [Google Scholar]
- 19.Gwack, Y., D. W. Kim, J. H. Han, and J. Choe. 1996. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 225:654-659. [DOI] [PubMed] [Google Scholar]
- 20.Honda, M., M. R. Beard, L. H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417-36428. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko, T., Y. Tanji, S. Satoh, M. Hijikata, S. Asabe, K. Kimura, and K. Shimotohno. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320-326. [DOI] [PubMed] [Google Scholar]
- 23.Kashiwagi, T., K. Hara, M. Kohara, J. Iwahashi, N. Hamada, H. Honda-Yoshino, and T. Toyoda. 2002. Promoter/origin structure of the complementary strand of hepatitis C virus genome. J. Biol. Chem. 277:28700-28705. [DOI] [PubMed] [Google Scholar]
- 24.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 25.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64:334-339. [DOI] [PubMed] [Google Scholar]
- 26.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, Y. K., C. S. Kim, S. H. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290:105-112. [DOI] [PubMed] [Google Scholar]
- 28.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, H., H. Shin, E. Wimmer, and A. V. Paul. 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 78:10865-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 40.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 41.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 43.Neddermann, P., M. Quintavalle, C. Di Pietro, A. Clementi, M. Cerretani, S. Altamura, L. Bartholomew, and R. De Francesco. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78:13306-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh, J. W., T. Ito, and M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 46.Piccininni, S., A. Varaklioti, M. Nardelli, B. Dave, K. D. Raney, and J. E. McCarthy. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 277:45670-45679. [DOI] [PubMed] [Google Scholar]
- 47.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quintavalle, M., S. Sambucini, C. Di Pietro, R. De Francesco, and P. Neddermann. 2006. The α isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J. Virol. 80:11305-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintavalle, M., S. Sambucini, E. Summa, L. Orsatti, F. Talamo, R. De Francesco, and P. Neddermann. 2007. Hepatitis C virus NS5A is a direct substrate of CKI-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J. Biol. Chem. 282:5536-5544. [DOI] [PubMed] [Google Scholar]
- 50.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reigadas, S., M. Ventura, L. Sarih-Cottin, M. Castroviejo, S. Litvak, and T. Astier-Gin. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur. J. Biochem. 268:5857-5867. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 53.Schnell, M. J., T. Mebatsion, and K. K. Conzelmann. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 13:4195-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuster, C., C. Isel, I. Imbert, C. Ehresmann, R. Marquet, and M. P. Kieny. 2002. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J. Virol. 76:8058-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shim, J. H., G. Larson, J. Z. Wu, and Z. Hong. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimakami, T., M. Hijikata, H. Luo, Y. Y. Ma, S. Kaneko, K. Shimotohno, and S. Murakami. 2004. Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J. Virol. 78:2738-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 58.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 59.Smith, R. M., C. M. Walton, C. H. Wu, and G. Y. Wu. 2002. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J. Virol. 76:9563-9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka, T., N. Kato, M. J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 61.Targett-Adams, P., and J. McLauchlan. 2005. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J. Gen. Virol. 86:3075-3080. [DOI] [PubMed] [Google Scholar]
- 62.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tripathi, R. L., P. Krishnan, Y. He, T. Middleton, T. Pilot-Matias, C. M. Chen, D. T. Lau, S. M. Lemon, H. Mo, W. Kati, and A. Molla. 2007. Replication efficiency of chimeric replicon containing NS5A-5B genes derived from HCV-infected patient sera. Antivir. Res. 73:40-49. [DOI] [PubMed] [Google Scholar]
- 64.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van den Hoff, M. J., A. F. Moorman, and W. H. Lamers. 1992. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res. 20:2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Windisch, M. P., M. Frese, A. Kaul, M. Trippler, V. Lohmann, and R. Bartenschlager. 2005. Dissecting the interferon-induced inhibition of hepatitis C virus replication by using a novel host cell line. J. Virol. 79:13778-13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 71.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, C., Z. Cai, Y. C. Kim, R. Kumar, F. Yuan, P. Y. Shi, C. Kao, and G. Luo. 2005. Stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3) helicase activity by the NS3 protease domain and by HCV RNA-dependent RNA polymerase. J. Virol. 79:8687-8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]