Abstract
We documented that the NF-κB signaling pathway was rapidly induced following human cytomegalovirus (HCMV) infection of human fibroblasts and that this induced NF-κB activity promoted efficient transactivation of the major immediate-early promoter (MIEP). Previously, we showed that the major HCMV envelope glycoproteins, gB and gH, initiated this NF-κB signaling event. However, we also hypothesized that there were additional mechanisms utilized by the virus to rapidly upregulate NF-κB. In this light, we specifically hypothesized that the HCMV virion contained IκBα kinase activity, allowing for direct phosphorylation of IκBα following virion entry into infected cells. In vitro kinase assays performed on purified HCMV virion extract identified bona fide IκBα kinase activity in the virion. The enzyme responsible for this kinase activity was identified as casein kinase II (CKII), a cellular serine-threonine protein kinase. CKII activity was necessary for efficient transactivation of the MIEP and IE gene expression. CKII is generally considered to be a constitutively active kinase. We suggest that this molecular characteristic of CKII represents the biologic rationale for the viral capture and utilization of this kinase early after infection. The packaging of CKII into the HCMV virion identifies that diverse molecular mechanisms are utilized by HCMV for rapid NF-κB activation. We propose that HCMV possesses multiple pathways to increase NF-κB activity to ensure that the correct temporal regulation of NF-κB occurs following infection and that sufficient threshold levels of NF-κB are reached in the diverse array of cells, including monocytes and endothelial cells, infected in vivo.
Human cytomegalovirus (HCMV) infects a majority of individuals and persists for the life of the host (18, 48). In immunocompetent individuals, HCMV infection is rarely associated with disease, although it can cause mononucleosis (18, 48) and is associated with certain cancers (15, 57, 61) and cardiovascular diseases (reviewed in references 23, 45, 61, and 64). In immunocompromised individuals, HCMV infection causes severe and often fatal diseases (18, 48). Because viral gene expression and the ensuing viral replication are paramount steps in the manifestation of HCMV-mediated disease, understanding of the initiation of the viral life cycle is necessary for uncovering mechanisms of viral pathogenesis.
Following infection, the major immediate-early promoter (MIEP) is transactivated (11, 31, 46, 62, 63, 65), a requirement for generation of the major immediate-early (IE) transcripts, viral replication, and production of infectious virus (20, 21, 28, 31, 32, 44). In the murine CMV system, mutants that lack the murine MIEP enhancer show no observable pathogenesis in vivo (27). Together, these studies suggest a key role for the transactivation of the MIEP in the pathobiology of HCMV infection. Mechanistically, cellular transcription factors induced rapidly following viral infection appear to be responsible for the upregulation of the MIEP. NF-κB is one transcription factor that has been implicated in the transactivation of the MIEP following primary infection or during reactivation of latent virus (14, 20, 21, 30, 38, 50, 51, 53, 73-75). NF-κB activity is upregulated within 5 min postinfection (10, 73, 74), consistent with its role in the rapid upregulation of the MIEP. Early studies using deletion mutant MIEP reporter constructs offered the first clues that NF-κB could transactivate the MIEP (14, 53). Recent results from our laboratory (20, 21) and others (13, 38, 49) using pharmacological or dominant-negative inhibitors present additional evidence for the critical role that this cellular transcription factor plays in the efficient transactivation of the MIEP and, consequently, IE gene expression.
In normal cells, NF-κB is a tightly controlled transcription factor that is regulated by the action of a group of proteins termed the IκBs (reviewed in references 6 and 29). IκBα is the prototype IκB and is responsible for regulating acute and rapid release of NF-κB. The IκB kinase (IKK) complex is required to activate NF-κB. The IKKs directly phosphorylate critical serine residues in the NH2 terminus of IκBα (Ser32 and Ser36), leading to its ubiquitination and degradation and, ultimately, to the release of free NF-κB (29). When this tightly regulated system of NF-κB regulation is perturbed, aberrant changes in cellular function, survival, and development occur (17, 35, 37).
In our studies on the regulation of NF-κB activity following infection, we observed that increased NF-κB activity was seen as early as 5 min after infection and was due to the binding of HCMV glycoproteins to cognate host receptors (73, 74). Additional studies examining glycoprotein-mediated signaling (9, 12, 16, 59), as well as the recent identification of HCMV entry receptors and the rapid signaling that ensues following glycoprotein binding to these receptors is consistent with the results of our earlier studies (24, 69, 70). Initial binding of the virus to the host cell results in the release of preformed stores of NF-κB that is largely dependent on the activation of the IKK complex (20, 74). We hypothesized that changes in NF-κB activity served to temporally regulate viral gene expression. To test that possibility, we initiated studies to address whether virus-induced NF-κB upregulated viral gene expression. We showed that NF-κB activity was required for the efficient transactivation of the MIEP and the expression of IE genes (20) and then showed that NF-κB activity was required for efficient viral replication and the production of infectious virus (21). Recent reports by others (13, 38, 49) support our conclusions. On the other hand, other studies suggest that NF-κB activity may not serve such a central role in viral gene regulation (8, 22, 28, 32). The reason for these differences is unknown but probably reflects differences in the systems and viruses used. Nevertheless, our data argue that NF-κB activity is required for efficient gene expression and viral replication.
To this end, we recently expanded our efforts into understanding the upregulation of NF-κB activity following infection. Specifically, we focused on the mechanisms responsible for the rapid induction of NF-κB activity following infection. We previously hypothesized that HCMV possessed a kinase that could directly phosphorylate the IκBs (74). The possibility was further supported by our observation that the activities of IKKα and IKKβ, although responsible for much of the induced NF-κB activity, were not responsible for all of the induced NF-κB activity (20). The question then became what was responsible for this remaining activity—was it a virion-associated kinase?
The identification of the cellular and viral proteins present in the HCMV virion in the mass spectrometry study by Varnum et al. (67) provided us with a clue as to the identity of this virus-associated kinase. Casein kinase II (CKII) was detected in HCMV virions (67). CKII is a ubiquitous tetrameric serine-threonine protein kinase (3, 41) that was identified as having multiple target substrates, including IκBα (5, 40, 52, 58). Could cellular CKII packaged with the virion be responsible for the additional level of NF-κB activation that our previous studies demonstrated (20)? In this study, we examined that possibility and now report that active CKII can be found in the virion and that this virion CKII possessed true IκBα kinase activity. When CKII activity was blocked in infected cells with a pharmacological inhibitor, viral IE transcription was significantly decreased, as was IE protein production, suggesting that CKII activity supports maximal MIEP transactivation. Our observation that the HCMV virion has bona fide IKK-like activity is, to our knowledge, the first report of a virus having this sort of enzymatic activity in a viral particle, underscoring the seminal role that NF-κB activation likely plays in HCMV-infected cells.
In light of these data, we propose the following model: when HCMV infects a new cell, the packaged cellular CKII enters the cell and rapidly directs the phosphorylation of IκBα, resulting in additional levels of NF-κB activation above that initiated by viral receptor/ligand interactions (24, 69, 70, 73, 74). Because of the diverse array of distinct cell types that HCMV infects in vivo and the unique biological signaling pathways that each cell type utilizes, the existence of multiple mechanisms to induce NF-κB would not be redundant. Rather, it would be a requirement to ensure that sufficient threshold levels of NF-κB activity are reached in each cell type infected.
MATERIALS AND METHODS
Virus and cell culture.
The Towne/E strain of HCMV was used in these studies (20, 21, 73, 74). Virus was passaged in primary human embryonic lung (HEL) fibroblasts grown in Eagle's minimal essential medium (Cellgro Mediatech, Inc., Herndon, VA) supplemented with 4% heat-inactivated fetal bovine serum (Gemini, Woodland, CA), penicillin, and streptomycin at 37°C in a 5% CO2 incubator. All experiments utilized gradient-purified virus. Virus-containing supernatant was harvested and gradient purified by high-speed centrifugation (36,000 × g for 90 min) through a sucrose cushion (72, 73). For in vitro kinase assays (20, 21), the virus pellet was resuspended in phosphate-buffered saline (PBS) and then sonicated to lyse the virus. For IE gene and protein expression assays, gradient-purified virus was utilized to infect HEL fibroblasts at a multiplicity of infection of 3 to 5 (20, 21, 73, 74).
Antibodies and reagents.
For Western blot analyses, CKII was detected using a goat polyclonal CKIIα (C-18) antibody (catalog no. sc-6479) obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The secondary antibody used was a horseradish peroxidase-conjugated donkey anti-goat immunoglobulin G antibody (catalog no. sc-2056; Santa Cruz Biotechnology, Inc.). The same anti-CKIIα antibody was used in the immunoprecipitation experiments. In experiments in which CKII activity was inhibited, TBB (4,5,6,7-tetrabromo-1H-benotriazole; Calbiochem, La Jolla, CA), a specific CKII inhibitor was utilized (7, 54, 55). TBB is a halogenated benzimidazole derivative that is cell permeable and acts to selectively block ATP binding to CKII due its high-affinity binding to the unique hydrophobic pocket of CKII (7, 54, 55). TBB was dissolved in dimethyl sulfoxide (DMSO). In all experiments, a DMSO solvent-alone control was performed. An antibody to IKKβ (catalog no. sc-8014; Santa Cruz Biotechnology, Inc.) was also employed in our system.
In vitro kinase assay.
Briefly, gradient-purified virions were lysed (by sonication), and the extract was mixed with a 10× kinase buffer (200 nM HEPES, 100 mM MgCl2, 1 mg/ml bovine serum albumin, 30 mM β-mercaptoethanol) supplemented with 1.5 μl of 1 mM cold ATP, 5 μl of a 1 mg/ml concentration of substrate (various glutathione S-transferase [GST]-tagged IκBα substrates), and 1 μl of [γ-32P]ATP (0.25 μCi/μl; ICN Biochemicals, Inc., Irvine CA) (20). The equivalent of 5 × 105 infectious viral particles was analyzed in each sample of the kinase assays. The assay mix was incubated for 30 min at 30°C, after which samples were boiled and subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE). Gels were stained with a Coomassie blue solution (to control for equal loading of the substrate) and then visualized by autoradiography (20). Densitometry was performed on the autoradiograms as discussed in Results. As a positive control and a comparison test reagent, human recombinant wild-type activated full-length CKII (Upstate USA, Inc., Charlottesville, VA) was used. Specifically, 0.2 μg of recombinant CKII (rCKII) was used in each test group in the kinase assays. For one of the experiments, proteinase K was utilized following the protocol of Yao and Courtney (71). Briefly, gradient-purified virions were collected and then treated with proteinase K for 10 min either prior to sonication or after sonication. Following the inactivation of the protease, samples were analyzed by kinase assay.
GST-IκBα substrate induction and purification.
Competent Escherichia coli BL21 cells were transformed with plasmids encoding the GST fusion proteins to be used as substrates in our in vitro kinase assays (20). The following GST-IκBα constructs used in our experiments were obtained as a generous gift from John Hiscott, Institut Lady Davis de Recherches Medicales, Montreal, Quebec, Canada (1, 2, 19, 39, 40): IκBα, a full-length wild-type IκBα; IκBα(2NΔC), a truncated IκBα mutant containing only the first 54 NH2-terminal amino acids and in which Ser32 and Ser36 were mutated to alanines; IκBα(3C), a full-length IκBα mutant in which Ser283 and Thr291 and Thr299 were mutated to alanines; IκBα(Δ2), a truncated IκBα mutant in which the COOH-terminal amino acids 269 to 317 were deleted; and IκBα(2NΔ4), a truncated IκBα mutant in which COOH-terminal amino acids 297 to 317 were deleted and in which both Ser32 and Ser36 were mutated to alanines. We also utilized the following constructs: IκBα(269-296), a truncated IκBα mutant containing only the residues of IκBα from 269 to 296; IκBα(283-296), a truncated IκBα mutant containing only the residues of IκBα from 283 to 296; IκBα(269-282), a truncated IκBα mutant containing only the residues of IκBα from 269 to 282; and IκBα(283-296ΔST), a truncated IκBα mutant containing only the residues of IκBα from 283 to 296 and that had Ser283, Ser288, Thr291, Ser293, and Thr296 mutated to aspartic acid, aspartic acid, alanine, aspartic acid, and alanine, respectively. These additional GST-IκBα constructs were constructed using short oligonucleotides (Integrated DNA Technologies, Inc., Coralville, IA) encompassing the wild-type regions of IκBα stated above or that had specific point mutations incorporated into the oligonucleotide sequence. Restriction sites specific for BamHI and EcoRI were engineered into the 5′ and 3′ termini of the oligonucleotides, respectively, to facilitate cloning of the fragments into the pGEX2T expression vector. Annealed oligonucleotides were ligated into the BamHI and EcoRI restriction sites of a pGEX2T vector (downstream of the GST-coding sequence). Constructs were sequenced to confirm authenticity.
The transformed bacteria were cultured overnight in 2X-YTG medium (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl) supplemented with 2% glucose. Following overnight incubation, the first culture was expanded in 2X-YT medium, and then protein synthesis was induced by the addition of 0.5 M isopropyl-β-d-thiogalactopyranoside (IPTG) (Novagen, San Diego, CA). Cultures were then grown for 5 h, and bacteria were pelleted by centrifugation. Aliquots of uninduced and induced cultures were subjected to SDS-PAGE and stained with a Coomassie blue solution to visualize the protein bands and to confirm protein induction. Bacterial pellets, in which GST fusion protein expression was induced, were resuspended in the Bugbuster reagent (Novagen) to lyse the cells. Benzonase nuclease (Novagen), lysozyme (Sigma), and a protease inhibitor cocktail (catalog no. p-8340; Sigma) were added to the lysed bacteria. Insoluble material was removed following centrifugation, and the supernatant was harvested. The supernatant was applied to a chromatography column containing GST bind resin (Novagen) and washed with 1× GST bind/wash buffer (Novagen). After washing, the bound protein was eluted with glutathione elution buffer (Novagen), and the eluted protein was collected. Purified protein was concentrated using a Centricon centrifugal filter device (Millipore, Bedford, MA) and analyzed by SDS-PAGE followed by Coomassie blue staining.
Immunoprecipitation (IP) experiments.
Purified and sonicated virus was incubated with the CKII antibody or a control antibody overnight at 4°C. Then protein G plus protein A agarose suspension beads (Oncogene Science, Cambridge, MA) were added and incubated for an additional hour at 4°C. The samples were centrifuged at 15,000 × g for 5 min, and the supernatant was collected. The agarose bead precipitates were washed with buffer A (1% NP-40, 0.5% sodium deoxycholate, 100 mM sodium chloride, 10 mM Tris, 1 mM EDTA; a final pH of 7.2) and then subsequently with buffer B (1 M sodium chloride, 0.1% NP-40, 10 mM Tris; a final pH of 7.2) and buffer ST (150 mM sodium chloride, 50 mM Tris-HCl, 50 mM Tris base; a final pH of 7.2). After the Sepharose beads were washed, the beads were diluted in the kinase buffer. Both the supernatant and precipitate containing the beads were used in the in vitro kinase assays. Last, a CKII blocking peptide (catalog no. bp300-198; Bethyl Laboratories Inc., Montgomery, TX) was utilized to examine the specificity of the anti-CKII antibody. For the experiments using the blocking peptide, the peptide was incubated with the appropriate target antibody for 2 h prior to use in IP experiments.
Western blot analyses.
Infected fibroblast cultures incubated for various times postinfection were harvested for Western blot analysis in Laemmli sample buffer (Bio-Rad Laboratories, Inc., Hercules, CA) supplemented with β-mercaptoethanol. Samples were boiled, subjected to SDS-PAGE, and then transferred to immunoblot polyvinylidene difluoride membranes overnight (Bio-Rad Laboratories). Equal amounts of protein were added per lane, where indicated. Membranes were incubated in a blocking buffer (5% skim milk, 0.1% Tween 20, 1× PBS). Primary anti-CKIIα, IE1-72, or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were diluted in blocking buffer and incubated with the membranes. Blots were then washed in a 1× PBS-0.1% Tween 20 solution and incubated with the secondary antibody, washed again, and developed using the ECL+ system (Amersham Biosciences, Piscataway, NJ), according to the manufacturer's protocol (21).
RNA harvesting and real-time PCR.
Briefly, total cellular RNA from a time course of infected fibroblasts was harvested utilizing the QIAGEN RNeasy kit (QIAGEN, Inc., Valencia, CA) (20). Isolated RNA samples were treated with DNase RQ1 (Promega, Madison, WI) according to the manufacturer's protocol. Reverse transcription was performed on 600 ng of RNA in a reaction mixture containing 500 μM of each deoxynucleoside triphosphate, 2.5 μM random hexamers, 0.3 U RNase inhibitor, and 150 U Moloney murine leukemia virus reverse transcriptase (Invitrogen Corporation, Carlsbad, CA). Reaction mixtures were incubated at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min. Quantitative real-time PCR was performed with specific primers for the HCMV IE1-72 (UL123) gene (National Center for Biotechnology Information GenBank accession no. M21295). The forward primer used was 5′-AGTGACCGAGGATTGCAACG-3′, and the reverse primer was 5′-CCTTGATTCTATGCCGCACC-3′. Human GAPDH mRNA levels were also measured, using the forward primer 5′-GAAGGTGAAGGTCGGAGT-3′ and the reverse primer 5′-GAAGATGGTGATGGGATTTC-3′. All primers were made by Integrated DNA Technologies, Inc. The IE1-72 PCR was validated to our GAPDH normalization control to ensure equal PCR efficiency of both primer sets (data not shown). We used the ΔΔct method for quantitation as described by Applied Biosystems, Inc. (Foster City, CA). Amplification and detection were performed with the iCycler IQ real-time PCR detection system in reaction mixtures containing the transcription reaction mixture (100 ng cDNA), IQ SYBR green supermix, and 15 μM primers. The incubation conditions consisted of 3 min at 95°C for polymerase activation and 40 cycles of 15 s at 95°C and 1 min at 60°C. Results are presented as the mean values of three separate independent experiments (run in triplicate) with standard errors of the means. The HCMV-infected sample was normalized to a value of 1, with the decrease observed following TBB treatment shown as a percentage of that normalized value.
RESULTS
The HCMV virion possesses IKK-like activity.
Previously, we documented that rapid NF-κB activation occurred through activation of the canonical IKKα/IKKβ pathway (20). Our study also suggested that at least one additional pathway exists, because although a majority of the rapid activation of NF-κB occurred through the activity of the IKKs, they alone were not responsible for all of the induced activity. The possibility that the virus uses additional pathways for the rapid induction of NF-κB was put forth in an earlier study on transcription factor regulation following HCMV infection (74): we hypothesized that the HCMV virion had the potential to target NF-κB activation through the direct phosphorylation of IκBα either by possessing a virus-encoded kinase with IKK-like activity or by capturing a cellular kinase with IKK-like activity. In either case, such a protein with IKK-like activity would allow the virus to bypass the IKK signaling pathway and mediate NF-κB activation by a distinct mechanism.
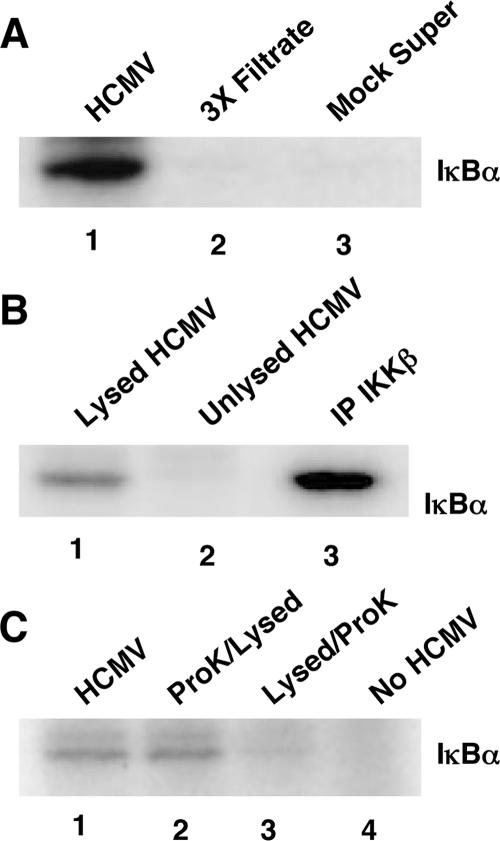
To test our hypothesis, we initially examined whether the HCMV virion contained IKK-like enzymatic activity. We harvested extract from gradient-purified HCMV and performed in vitro kinase assays using wild-type GST-IκBα as a substrate (Fig. 1A). The results documented that IκBα was phosphorylated by the gradient-purified viral extract (Fig. 1A, lane 1), but not by filtrate from gradient-purified virus run three times through a 0.2-μm filter (Fig. 1A, lane 2) or supernatant collected from mock-infected fibroblasts (Fig. 1A, lane 3). Further results (Fig. 1B) showed that IκBα phosphorylation occurred only when the virus was lysed (Fig. 1B, lane 1) prior to use in a kinase assay. No IκBα phosphorylation occurred in unlysed viral samples (Fig. 1B, lane 2). For a positive control, cellular IKKβ was immunoprecipitated (Fig. 1B, lane 3) from infected fibroblast lysate (20).
FIG. 1.
Purified HCMV virions possess IKK-like kinase activity. Kinase assays using a wild-type GST-IκBα substrate (labeled as IκBα in the figures) were performed with different test samples. (A) Lane 1 contains gradient-purified, sonicated HCMV extract (Towne/E); lane 2 contains extract from purified HCMV filtered three times through a 0.2-μm filter prior to sonication (3X Filtrate); and lane 3 contains supernatant from mock-infected cells (Mock Super). (B) Lane 1 contains lysed gradient-purified virus (Towne/E); lane 2 contains unlysed gradient-purified virus; and lane 3 contains a positive control (IP cellular IKKβ activity). (C) Lane 1 contains gradient-purified, sonicated HCMV extract (Towne/E). Lanes 2 and 3 show the results of kinase assays performed on viral extract that had either been pretreated with proteinase K prior to virion lysis (ProK/Lysed) or treated with protease K after virion lysis (Lysed/ProK). Lane 4 contains the substrate-alone control. Representative experiments are shown (from at least three independent experiments).
Next, to provide confirmation that the enzymatic activity was present within the virion and not stuck on the outside of the viral envelope, we employed the strategy that Yao and Courtney used to demonstrate that the herpes simplex virus 1 ICP0 protein localized to the virion (71). This strategy involved examining viral extract that had either been pretreated with proteinase K prior to virion lysis (Fig. 1C, lane 2) or treated with proteinase K after virion lysis (Fig. 1C, lane 3). The results presented in Fig. 1C, using these different treatment groups in a kinase assay showed that enzymatic activity was observed only when the viral extract was pretreated with protease K prior to virion lysis (Fig. 1C, compare lanes 2 and 3). Similar kinase activity was observed when the virus was lysed only (Fig. 1C, lane 1). Together, these data suggest that an enzyme that directly phosphorylates IκBα exists within the HCMV virion.
CKII is packaged within the HCMV virion.
We began using a biochemical approach to purify and identify the protein with IKK-like enzymatic activity. At the time these studies were under way, Varnum et al. published the HCMV AD169 proteome in which they utilized mass spectrometry to identify the viral and cellular proteins in the HCMV virion (67). A host protein of interest to our work was the identification of CKII in the HCMV proteome (67) because of the following molecular traits of this enzyme. CKII is a serine-threonine kinase that appears to be constitutively active (3, 41, 55) that, like the well-characterized IKK proteins (33, 35), can phosphorylate IκBα and regulate its turnover (5, 40, 58) and induce the activation of NF-κB (52). This finding suggested that CKII could be the kinase our molecular data suggested existed in the Towne/E HCMV virion.
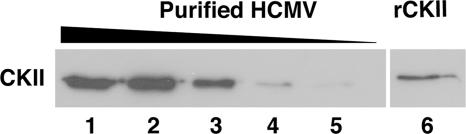
To examine whether CKII was found in the HCMV Towne/E virion, we looked for the presence of CKII in gradient-purified virus by Western blot analysis (Fig. 2). The results from this experiment performed on a titration of gradient-purified virion extract (∼7.5 × 106, ∼4.5 × 106, ∼2.2 × 106, ∼9 × 105, and ∼4.5 × 105 infectious viral particles) show that bona fide CKII was present in the virion of the Towne/E strain of HCMV, consistent with our hypothesis that this virus contains an enzyme with IKK-like activity. Full-length human rCKII (0.1 μg) was used as a positive control. In the study by Varnum et al. (67), the laboratory strain AD169 was utilized to examine the HCMV proteome; thus, with our data from Fig. 2, CKII can be observed in multiple strains of HCMV. In addition, CKII was detected in a third laboratory strain, TB40-fibroblast, and a clinical-like isolate, TB40-endothelial, by Western blot analysis (data not shown). On the other hand, we could not detect IKKα or IKKβ in the purified Towne/E virion extract (data not shown), consistent with the results observed in AD169 (67).
FIG. 2.
Cellular CKII is found in purified HCMV virions. A titration of gradient-purified HCMV (Towne/E, ∼7.5 × 106 [lane 1], ∼4.5 × 106 [lane 2], ∼2.2 × 106 [lane 3], ∼9 × 105 [lane 4], and ∼4.5 × 105 [lane 5] infectious viral particles) was examined by Western blot analysis using an antibody specific for human CKII. Human rCKII (0.1 μg) is shown as a positive control (lane 6). The results from a representative experiment a43 shown. The experiment was repeated.
The enzyme within the HCMV virion shows an IκBα phosphorylation pattern identical to that of purified human CKII.
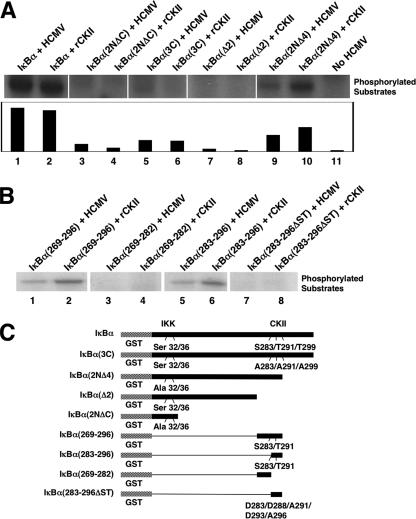
The data above suggested that the HCMV virion possessed an enzyme, possibly CKII, that directly phosphorylated IκBα. To provide evidence for this suggestion, we next examined whether the phosphorylation pattern of the virion-associated kinase activity was consistent with that documented for CKII by comparing the kinase activity of gradient-purified HCMV lysate to that of rCKII against various GST-IκBα substrates. IκBα has three distinct regions: an NH2-terminal region; the ankyrin repeat region; and, the COOH-terminal PEST region. The NH2-terminal region of IκBα is the target of the canonical IKK pathway (29, 68) that we have shown is upregulated in infected cells (20), while the COOH-terminal PEST region is the target of CKII-mediated phosphorylation (40, 58, 68). In vitro kinase assays (Fig. 3A and B) using various IκBα constructs (represented in Fig. 3C) as target substrates demonstrated that the HCMV virion-mediated phosphorylation of IκBα occurred in the COOH-terminal region of the molecule. Furthermore, the HCMV virion-mediated pattern of phosphorylation of the different IκBα substrates mimicked that observed for rCKII. Specifically, as shown in Fig. 3A and B, both the virus and rCKII phosphorylated IκBα to high levels, while the traditional IκBα kinase control construct [IκBα(2NΔC); contains only the first 54 N-terminal amino acids and has Ser32 and Ser36 mutated to alanines (1, 2, 19, 39, 40)] showed no activity. The construct with the known COOH-terminal CKII phosphorylation sites mutated [IκBα(3C); Ser283, Thr291, and Thr299 mutated to alanines (1, 2, 19, 39, 40)] and the construct with the last 48 COOH-terminal amino acids deleted [IκBα(Δ2); missing amino acids 269 to 317 (1, 2, 19, 39, 40)] showed little or no activity with both the purified HCMV extract and rCKII. A low amount of activity was consistently detected in the IκBα(3C) construct with both the purified viral extract and rCKII. This low level of activity probably represents the phosphorylation of one of the additional serine or threonine residues (such as Ser288 and Ser293 or Thr273 or Thr296) in the COOH terminus that are not ordinarily targeted by CKII with a high efficiency when Ser283, Thr291, and Thr299 are available for phosphorylation. A construct with only the last 21 COOH-terminal amino acids deleted (missing amino acids from positions 297 to 317) and Ser32 and Ser36 mutated to alanines [IκBα(2NΔ4) (1, 2, 19, 39, 40)] was also phosphorylated, although to a lower level due to the loss of the threonine at position 299. The similarities between the activity of the virus-associated kinase and rCKII was also seen when additional kinase assays were performed using short COOH-terminal GST-IκBα mutant constructs (Fig. 3B and C). When constructs with only the COOH-terminal residues of IκBα from 269 to 296 [IκBα(269-296)] or 283 to 296 [IκBα(283-296)] were used in kinase assays, similar patterns of phosphorylation were observed between the virus-associated kinase and rCKII. In contrast, no activity was seen in the constructs lacking the CKII target serines or threonines [IκBα(269-282) (representing only the residues of IκBα from 269 to 282) and IκBα(283-296ΔST) (representing only the residues of IκBα from 283 to 296 that had Ser283, Ser288, Thr291, Ser293, and Thr296 mutated to aspartic acid, aspartic acid, alanine, aspartic acid, and alanine, respectively)]. Together, these results suggest that HCMV packages (incorporates) an active enzyme that phosphorylates the COOH-terminal domain of IκBα in a pattern identical to that documented for functional human CKII (40, 58, 68).
FIG. 3.
The virion-associated kinase phosphorylates the COOH terminus of IκBα. (A) Kinase assays comparing purified HCMV (Towne/E) virion extract with that of rCKII were performed on different GST-IκBα mutant substrates. IκBα represents wild-type full-length IκBα (lanes 1 and 2). IκBα(2NΔC) represents only the 54 NH2-terminal amino acids of IκBα (in addition, Ser32 and Ser36 were mutated to alanines) (lanes 3 and 4). IκBα(3C) represents a full-length IκBα in which the dominant COOH-terminal CKII phosphorylation sites were mutated to alanines (lanes 5 and 6). IκBα(Δ2) represents an IκBα COOH-terminal deletion mutant (lanes 7 and 8). IκBα(2NΔ4) represents a mutant IκBα in which Ser32 and Ser36 were mutated to alanines, and the molecule was truncated after amino acid 296 (lanes 9 and 10). Lane 11 corresponds to no HCMV extract added. The results of the phosphorylation pattern of the different GST-IκBα substrates from the same experiment are presented side by side in a composite figure. The results of the kinase assay were analyzed by densitometry (shown in the graph at the bottom of panel A). The data are expressed in arbitrary units. (B) Kinase assays comparing purified HCMV (Towne/E) virion extract with that of rCKII were performed on additional GST-IκBα mutant substrates. IκBα(269-296) represents a COOH terminal deletion mutant of IκBα with only the amino acid residues from 269 to 296 (lanes 1 and 2). IκBα(283-296) represents a COOH terminal deletion mutant of IκBα with only the amino acid residues from 283 to 296 (lanes 5 and 6). IκBα(269-282) represents a COOH terminal deletion mutant of IκBα with only the amino acid residues from 269 to 282 (lanes 3 and 4). IκBα(283-296ΔST) represents a COOH terminal deletion mutant of IκBα with only the amino acid residues from 283 to 296 that had Ser283, Ser288, Thr291, Ser293, and Thr296 mutated to aspartic acids or alanines (lanes 7 and 8). The results from the same experiment are presented side by side in a composite figure. (C) Model of the different IκBα mutants used in the kinase assays. The results are from a representative experiment (three independent experiments performed).
Active CKII is observed within the HCMV virion.
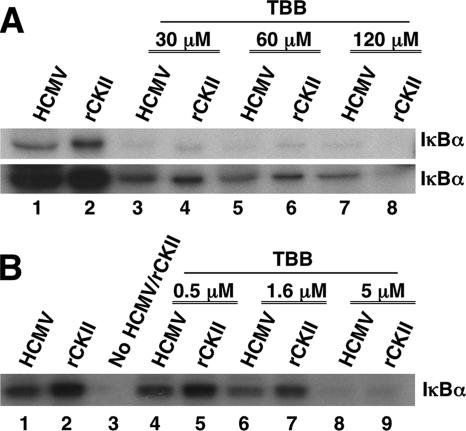
Next, we wanted to molecularly define that the enzyme packaged within the HCMV virion was active CKII. We utilized a specific CKII inhibitor, TBB (7, 54, 55), that fills the unique hydrophobic pocket of CKII, resulting in the targeted inactivation of the enzyme. Gradient-purified HCMV extract was either mock treated or pretreated for 4 h with increasing concentrations of TBB (30, 60, and 120 μM) and then examined in a kinase assay for activity against an IκBα substrate (Fig. 4A). This concentration of TBB initially used is reported to be effective in inhibiting CKII activity in cells (54). rCKII was used as a positive control and as a comparative test variable. Following incubation with TBB, the virus-associated and rCKII-mediated enzymatic activity against GST-IκBα was significantly inhibited (Fig. 4A, top and bottom blots). This inhibition occurred in a dose-dependent manner as demonstrated in the bottom blot, which represents the results of a longer exposure of the same autoradiogram shown in the top blot of Fig. 4A. The solvent-alone control (DMSO) showed no difference compared to the untreated virus in our kinase assays (data not shown). Because the 50% inhibitory concentration of TBB on purified human CKII is 1.6 μM (54), we next performed kinase assays using concentrations of TBB (0.5, 1.6, and 5 μM) around this 50% inhibitory concentration of 1.6 μM (Fig. 4B). We saw ∼45% decrease in substrate phosphorylation by the virion-associated kinase at the 1.6 μM concentration of TBB. A similar decrease was seen with rCKII. These results suggest that an enzyme with a functional equivalent to cellular CKII is packaged by HCMV and that this enzyme phosphorylates IκBα.
FIG. 4.
The CKII inhibitor TBB inhibits the activity of the virion-associated kinase. (A) A kinase assay was performed with the GST-IκBα substrate in which an increasing concentration of TBB (0 μM [lanes 1 and 2], 30 μM [lanes 3 and 4], 60 μM [lanes 5 and 6], and 120 μM [lanes 7 and 8]) was incubated for 4 h with the purified, sonicated HCMV (Towne/E) virion extract or rCKII prior to their addition to the kinase assay. The bottom panel of this figure represents a longer exposure of the same experiment. (B) A kinase assay was performed with the GST-IκBα substrate in which an increasing concentration of TBB (0 μM [lanes 1 and 2], 0.5 μM [lanes 4 and 5], 1.6 μM [lanes 6 and 7], and 5 μM [lanes 8 and 9]) was incubated for 4 h with the purified, sonicated HCMV (Towne/E) virion extract or rCKII prior to their addition to the kinase assay. Lane 3 is a negative control in which no kinase (viral or purified rCKII) was added to the kinase assay. Representative titration experiments are shown; titrations were repeated.
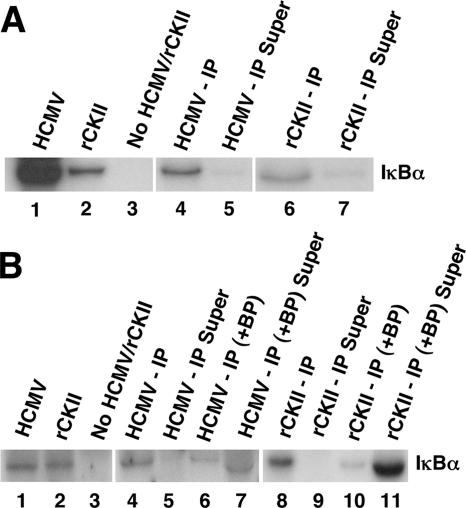
To confirm that the results discussed above represent true CKII activity in the HCMV virion, we performed IP kinase assays. As a control, the activity of rCKII was examined in the kinase assays. As shown in Fig. 5A, phosphorylation of IκBα was seen in the gradient-purified viral and rCKII samples immunoprecipitated with a CKII antibody (Fig. 5A, lanes 4 and 6). Kinase activity was also observed in the gradient-purified virus or rCKII-alone samples. When an isotype control antibody was used, no IP kinase activity was observed (data not shown). The supernatant collected from the IP reaction mixture showed no enzymatic activity (Fig. 5A, lanes 5 and 7).
FIG. 5.
The virion-associated kinase is cellular CKII. (A) An IP kinase assay was performed using a GST-IκBα substrate. Gradient-purified, sonicated HCMV (Towne/E) virion extract was incubated with a CKII antibody overnight prior to use in the kinase assay. rCKII was utilized as a control in this experiment. Lane 1 contains purified, sonicated virion extract. Lane 2 contains rCKII. Lane 3 contains no kinase (viral or rCKII). Lane 4 contains the immunoprecipitated fraction from the purified, sonicated virus (HCMV - IP). Lane 5 contains the supernatant (Super) from lane 4. Lane 6 contains the IP fraction from rCKII. Lane 7 contains the supernatant from lane 6. The results from the same experiment are presented side by side in a composite figure. (B) The IP kinase experiment was performed as stated above for panel A, with the exception that a CKII blocking peptide (BP) was preincubated for 2 h with the CKII-specific antibody in some of the samples prior to their use in the IP kinase assays. GST-IκBα was used as the substrate. Lane 1 contains purified, sonicated virion extract (Towne/E). Lane 2 contains rCKII. Lane 3 contains no kinase (viral or rCKII). Lane 4 contains the IP fraction from the purified, sonicated virus. Lane 5 contains the supernatant (Super) from lane 4. Lane 6 contains the IP fraction from the purified, sonicated virus in which the CKII antibody was preincubated with the BP (+ BP). Lane 7 contains the supernatant from lane 6. Lane 8 contains the IP fraction from rCKII. Lane 9 contains the supernatant from lane 8. Lane 10 contains the IP fraction from the rCKII in which the CKII antibody was preincubated with the BP. Lane 11 contains the supernatant from lane 10. The phosphorylation pattern of the IκBα substrate in the different treatment groups is from the same experiment and is presented side by side in a composite figure. Representative blots are shown from three independent experiments.
To verify the specificity of the IP reaction (with the CKII-specific antibody), a high-affinity CKII peptide that binds to the CKII antibody was utilized. In this experiment, the CKII antibody was preincubated with the blocking peptide for 2 h at 4°C prior to use in IP kinase assays. The results of this experiment are shown in Fig. 5B. The kinase activity of viral extract and rCKII samples in which the CKII antibody was pretreated with the CKII blocking peptide (Fig. 5B, lanes 6 and 10) showed only a limited ability to phosphorylate the IκBα substrate compared to the IP samples alone (Fig. 5B, lanes 4 and 8). Similarly, kinase activity was observed in the positive controls in which no IP was performed (Fig. 5B, lanes 1 and 2). In addition, like the results shown in Fig. 5A, when the supernatant from the IP reactions was collected (Fig. 5B, lanes 5 and 9), no phosphorylation of the IκBα substrate was documented. As expected, when the supernatant was collected from samples in which IP was performed with the CKII antibody pretreated with the blocking peptide (Fig. 5B, lanes 7 and 11), kinase activity was found in the supernatant. The results of Fig. 4 and 5 show that functionally active CKII is found in the virus.
The HCMV virion CKII possesses the molecular characteristics of cellular CKII (data not shown).
For additional molecular controls, we wanted to show that the enzymatic activity of the purified (by IP) virion CKII mimics the enzymatic activity of cellular CKII (rCKII) in regards to IκBα phosphorylation (in kinase assays). We compared the sensitivity of CKII from HCMV virions to that of rCKII to different concentrations of the CKII inhibitor (TBB) and to its targeted phosphorylation of the different IκBα substrates. TBB prevented the phosphorylation of IκBα in a dose-dependent manner by the IP CKII from virions and the IP rCKII. Furthermore, CKII IP from virion extract possessed a pattern of phosphorylation of the different IκBα substrates identical to that observed for IP rCKII. That is, high levels of substrate phosphorylation were observed when both the IκBα and IκBα(2NΔ4) substrates were used. In contrast, only low phosphorylation levels of the IκBα(3C) substrate were seen, and no phosphorylation of the IκBα(Δ2) substrate was seen. Together, these results provide confirmation that the HCMV virion contains genuine CKII.
CKII activity enhances transactivation of the MIEP and production of IE gene products.
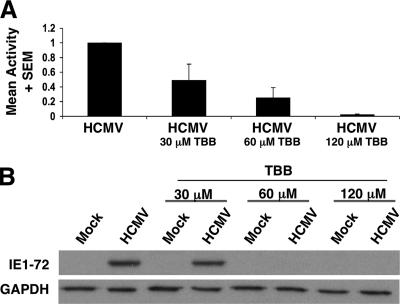
To begin to assess the role that the virion CKII plays in viral infection, we tested whether CKII activity was required for efficient IE gene and protein expression (Fig. 6). For these experiments, different concentrations of TBB were utilized (30, 60, or 120 μM). Cells were pretreated with TBB or the DMSO solvent-alone control for 1 h prior to infection (or mock treatment). At 12 h postinfection, cell lysates were collected, and real-time PCR was performed (Fig. 6A) or Western blot analyses were performed (Fig. 6B) for IE1-72 protein expression. As shown in Fig. 6A, IE1-72 gene expression was decreased when cells were pretreated with increasing concentrations of TBB, demonstrating that CKII activity promotes maximal IE gene expression. There was ∼50% decrease in IE1-72 gene expression when cells were pretreated with a 30 μM concentration of TBB. This inhibition increased to approximately 70% when a 60 μM concentration was used, and nearly 100% inhibition was observed when a 120 μM concentration of TBB was used. The appropriate mock-infected experiments were performed, as well as appropriate comparisons to GAPDH (data not shown) to validate the PCR. When IE1-72 protein expression was examined over the same titration course (Fig. 6B), a ∼40% decrease in IE1-72 protein expression relative to GAPDH was observed at 30 μM; this decrease in IE1-72 expression increased to nearly 100% when a 60 μM concentration was used. For additional controls, we showed that TBB did not induce apoptosis in treated cells in our assay system, nor did it negatively regulate IKKα/β kinase activity. These results provide a biological explanation for the packaging of this constitutively active enzyme into HCMV virions and the first molecular hints for why the virus has evolved to capture this cellular enzyme.
FIG. 6.
CKII activity is required for efficient IE1-72 expression in HEL fibroblasts. HEL fibroblasts were either preincubated with a 30, 60, or 120 μM concentration of TBB (indicated by TBB below the bar) or the DMSO solvent-alone control for 1 h prior to infection (HCMV, Towne/E) or mock infection. At 12 h postinfection, cells were harvested for RNA or protein. (A) Real-time PCR was performed using primers specific for IE1-72. The data were validated as stated in Materials and Methods. Results are presented as means plus standard errors of the means (SEMs) from three independent experiments performed in triplicate. The infected sample was normalized to 1, and the percentage decrease following TBB treatment is shown relative to this normalized value. (B) Western blot analysis using an antibody specific to IE1-72 and GAPDH was performed on the collected samples. Equal amounts of protein were loaded onto all lanes. Representative experiments are shown from three independent experiments.
Pretreatment of cells with TBB would be expected to inhibit cellular and virion-associated CKII activity. Therefore, we cannot yet define the specific contribution of each CKII to the induction of IE gene expression. The data does, however, demonstrate that CKII activity is required for maximal MIEP transactivation. Studies are under way to determine the mechanism(s) of CKII packaging into virions, which in turn should allow us to define the distinction between the contributions of the two pools of CKII to the initiation of viral gene expression. Nevertheless, because CKII from the virion directly phosphorylates IκBα as shown here and all available evidence identifies phosphorylation of IκBα as an essential step in the activation of NF-κB (6, 29, 35, 37), we propose that our data supports the hypothesis that active CKII in the virion contributes to the total pool of activated NF-κB found in infected cells following infection, which in turn contributes to the efficient transactivation of the MIEP.
DISCUSSION
We have focused on the NF-κB pathway and have shown that this transcription factor is needed for efficient transactivation of multiple classes of viral gene products (20, 21). Because of this reported important role for NF-κB in the viral life cycle, we initiated studies to examine the mechanisms for the regulation of NF-κB activation following infection. In this study, we demonstrated that the HCMV virion contains active cellular CKII and that this functionally active CKII phosphorylates IκBα to promote efficient transactivation of the IE genes.
CKII is a cellular serine-threonine kinase that has multiple target substrates, one of which is the NF-κB inhibitor, IκBα (3, 5, 40, 41, 52, 58). Our data shows that this virion-associated CKII directly phosphorylates IκBα in kinase assays, demonstrating that bona fide IKK-like activity exists in the HCMV virion, supporting our earlier model of NF-κB activation following infection (74). Phosphorylation of IκBα is a required regulatory step in the activation of NF-κB (6, 29). There is little evidence that phosphorylation of IκBα normally occurs in the absence of NF-κB activation; therefore, the identification of a kinase in the HCMV virion that phosphorylates IκBα strongly implies functional activation of NF-κB.
CKII regulates IκBα protein levels in a manner distinct from that observed for IKKα and IKKβ. CKII phosphorylates COOH-terminal residues of IκBα and targets the degradation of IκBα via a calpain-mediated pathway (40, 52, 58), in contrast to the IKKs (IKKα and IKKβ) that phosphorylate NH2-terminal residues and target IκBα degradation via a proteasome-mediated pathway (6, 29). In general, CKII is thought of as a constitutively active kinase that lacks the ability to respond to second messengers or kinase-dependent signaling pathways (43), although CK2 activity was recently shown to be altered in response to at least one cellular stimuli, UV irradiation (36). In a cell, CKII mediates constitutive or basal (signal-independent) phosphorylation of IκBα (5, 42, 56). At the molecular level, the presence of multiple pathways to activate NF-κB is likely required for correct differential and/or temporal regulation of gene expression (5, 6, 29, 42, 56). We propose that the virus takes advantage of and manipulates these diverse regulatory pathways to increase the probability of reaching the threshold levels of active NF-κB required in a cell following infection to drive the transactivation of the MIEP (20, 21, 73-75), at least in infected human fibroblasts. Because CKII is generally only a constitutively active kinase (3, 41, 55), one possible mechanism to quickly enhance CKII activity in a cell would be overexpression. Biologically, that process would be expected to occur when the virus deposits active CKII in a cell following infection. On the basis of an examination of the enzymatic activity observed with the viral extracts compared to the enzymatic activity of purified rCKII, we can estimate that between 50 and 500 CKII molecules exist per particle (infectious and noninfectious). Only a few thousand CKII molecules are thought to exist per cell, although the true number is unknown; thus, our data are mathematically consistent with the possibility that virion CKII meaningfully affects cellular substrates in vivo. The most abundant herpesvirus virion proteins, at least for HCMV and Epstein-Barr virus (EBV), from mass spectrometry analyses are capsid and tegument proteins (∼1,000 of the most abundant tegument molecules likely exist per viral particle [34, 67]). Therefore, our estimation of the number of CKII molecules per virion is again mathematically plausible.
It is unknown how this CKII becomes incorporated into the virion. We suggest that a protein-protein interaction of CKII with a predominant viral protein found in the virion allows for incorporation of this cellular molecule into a viral particle. Perhaps a mechanism similar to that documented for how polo-like kinase 1 gets incorporated into the virion exists (26). Future studies will allow for a greater dissection of the molecular role the virion CKII plays following infection.
One of the hallmarks of HCMV is the ability to infect multiple cell types and organ systems (47, 60, 66). Because each cell type infected would have unique signaling requirements for infection and would have unique downstream signaling patterns following receptor ligand engagement, the packaging of a virion-associated cellular kinase would ensure that sufficient levels of NF-κB are reached in a multitude of cell types. Functionally, this NF-κB could have a diverse array of effects, from the activation of the MIEP in cells, such as fibroblasts (20, 21) or other permissive cell types, to the activation of appropriate inflammatory pathways in monocytes (72) that would favor viral dissemination to multiple host organs. Furthermore, with the multiple substrates that CKII is known to phosphorylate (3, 43), such as cell cycle regulatory proteins, enzymes associated with nucleic acid biosynthesis, transcription factors, and even viral proteins (4, 25, 43), it is intriguing to speculate that this factor targets multiple cellular and viral substrates and has many functions in the array of cells that HCMV infects in vivo.
With our work documenting that NF-κB activity promotes efficient viral gene expression (20, 21), we proposed that CKII activity contributes to the transactivation of viral gene expression. The data showing decreased IE gene expression and protein production when CKII activity was inhibited support this proposal. We are continuing to investigate the role CKII plays in viral gene expression. To date, our data provides strong indication of the role the virion-associated CKII plays following viral infection, providing glimpses into a new mechanism that HCMV utilizes to alter an infected cell. Because the mass spectrometry analysis of EBV did not identify CKII in the EBV virion (34), it appears that CKII is only incorporated into HCMV virions and, thus, important only in the infection cycle of this herpesvirus. Discovery of a virion-associated CKII with a possible role in the transactivation of viral gene expression points to the elaborate mechanisms that the virus has evolved to promote and activate cellular players needed for survival and persistence.
Acknowledgments
We thank A. M. Tilley, R. S. Scott, A. H. Yurochko, and T. D. Abita for assistance, helpful discussions, and careful reading of the manuscript.
This work was supported in part by grants from the American Heart Association (0365207B and 0415120B), the March of Dimes Foundation (1-FY01-332), and the National Institutes of Health (P20-RR018724 and R01-AI56077).
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Algarté, M., H. Kwon, P. Genin, and J. Hiscott. 1999. Identification by in vivo genomic footprinting of a transcriptional switch containing NF-κB and Sp1 that regulates the IκBα promoter. Mol. Cell. Biol. 19:6140-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Algarté, M., H. Nguyen, C. Heylbroeck, R. Lin, and J. Hiscott. 1999. IκB-mediated inhibition of virus-induced beta interferon transcription. J. Virol. 73:2694-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allende, J. E., and C. C. Allende. 1995. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313-323. [DOI] [PubMed] [Google Scholar]
- 4.Barrasa, M. I., N. Y. Harel, and J. C. Alwine. 2005. The phosphorylation status of the serine-rich region of the human cytomegalovirus 86-kilodalton major immediate-early protein IE2/IEP86 affects temporal viral gene expression. J. Virol. 79:1428-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barroga, C. F., J. K. Stevenson, E. M. Schwarz, and I. M. Verma. 1995. Constitutive phosphorylation of IκBα by casein kinase II. Proc. Natl. Acad. Sci. USA 92:7637-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates, P. W., and S. Miyamoto. 2004. Expanded nuclear roles for IκBs. Sci. STKE 254:pe48. [DOI] [PubMed] [Google Scholar]
- 7.Battistutta, R., E. De Moliner, S. Sarno, G. Zanotti, and L. A. Pinna. 2001. Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole. Protein Sci. 10:2200-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benedict, C. A., A. Angulo, G. Patterson, S. Ha, H. Huang, M. Messerle, C. F. Ware, and P. Ghazal. 2004. Neutrality of the canonical NF-κB-dependent pathway for human and murine cytomegalovirus transcription and replication in vitro. J. Virol. 78:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehme, K. W., J. Singh, S. T. Perry, and T. Compton. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boldogh, I., M. P. Fons, and T. Albrecht. 1993. Increased levels of sequence-specific DNA-binding proteins in human cytomegalovirus-infected cells. Biochem. Biophys. Res. Commun. 197:1505-1510. [DOI] [PubMed] [Google Scholar]
- 11.Boshart, M. F., F. Weber, G. Jahn, K. Dorsch-Hasler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate-early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 12.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caposio, P., M. Dreano, G. Garotta, G. Gribaudo, and S. Landolfo. 2004. Human cytomegalovirus stimulates cellular IKK2 activity and requires the enzyme for productive replication. J. Virol. 78:3190-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobbs, C. S., L. Harkins, M. Samanta, G. Y. Gillespie, S. Bharara, P. H. King, L. B. Nabors, C. G. Cobbs, and W. J. Britt. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62:3347-3350. [PubMed] [Google Scholar]
- 16.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courtois, G. 2005. The NF-κB signaling pathway in human genetic diseases. Cell. Mol. Life Sci. 62:1682-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crumpacker, C. S. 2000. Cytomegalovirus, p. 1586-1599. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practices of infectious diseases, vol. 2. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 19.DeLuca, C., L. Petropoulos, D. Zmeureanu, and J. Hiscott. 1999. Nuclear IκBβ maintains persistent NF-κB activation in HIV-1-infected myeloid cells. J. Biol. Chem. 274:13010-13016. [DOI] [PubMed] [Google Scholar]
- 20.DeMeritt, I. B., L. E. Milford, and A. D. Yurochko. 2004. Activation of the NF-κB pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 78:4498-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeMeritt, I. B., J. P. Podduturi, A. M. Tilley, M. Nogalski, and A. D. Yurochko. 2006. Prolonged activation of NF-κB by human cytomegalovirus promotes efficient viral replication and late gene expression. Virology 346:15-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eickhoff, J. E., and M. Cotten. 2005. NF-κB activation can mediate inhibition of human cytomegalovirus replication. J. Gen. Virol. 86:285-295. [DOI] [PubMed] [Google Scholar]
- 23.Epstein, S. E., E. Speir, Y. F. Zhou, E. Guetta, M. Leon, and T. Finkel. 1996. The role of infection in restenosis and atherosclerosis: focus on cytomegalovirus. Lancet 348(Suppl.):s13-s17. [DOI] [PubMed] [Google Scholar]
- 24.Feire, A. L., H. Koss, and T. Compton. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. USA 101:15470-15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fish, K. N., C. Soderberg-Naucler, and J. A. Nelson. 1998. Steady-state plasma membrane expression of human cytomegalovirus gB is determined by the phosphorylation state of Ser900. J. Virol. 72:6657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallina, A., L. Simoncini, S. Garbelli, E. Percivalle, G. Pedrali-Noy, K. S. Lee, R. L. Erikson, B. Plachter, G. Gerna, and G. Milanesi. 1999. Polo-like kinase 1 as a target for human cytomegalovirus pp65 lower matrix protein. J. Virol. 73:1468-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghazal, P., M. Messerle, K. Osborn, and A. Angulo. 2003. An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis. J. Virol. 77:3217-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustems, M., E. Borst, C. A. Benedict, C. Perez, M. Messerle, P. Ghazal, and A. Angulo. 2006. Regulation of the transcription and replication cycle of human cytomegalovirus is insensitive to genetic elimination of the cognate NF-κB binding sites in the enhancer. J. Virol. 80:9899-9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-κB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 30.Hummel, M., Z. Zhang, S. Yan, I. DePlaen, P. Golia, T. Varghese, G. Thomas, and M. M. Abecassis. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isomura, H., and M. F. Stinski. 2003. The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J. Virol. 77:3602-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isomura, H., T. Tsurumi, and M. F. Stinski. 2004. Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J. Virol. 78:12788-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jimi, E., and S. Ghosh. 2005. Role of nuclear factor-κB in the immune system and bone. Immunol. Rev. 208:80-87. [DOI] [PubMed] [Google Scholar]
- 34.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karin, M., and F. R. Greten. 2005. NF-κB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5:749-759. [DOI] [PubMed] [Google Scholar]
- 36.Kato, T., Jr., M. Delhase, A. Hoffmann, and M. Karin. 2003. CK2 is a C-terminal IκB kinase responsible for NF-κB activation during the UV response. Mol. Cell 12:829-839. [DOI] [PubMed] [Google Scholar]
- 37.Kim, H. J., N. Hawke, and A. S. Baldwin. 2006. NF-κB and IKK as therapeutic targets in cancer. Cell Death Differ. 13:738-747. [DOI] [PubMed] [Google Scholar]
- 38.Lee, Y., W. J. Sohn, D. S. Kim, and H. J. Kwon. 2004. NF-κB- and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7. Eur. J. Biochem. 271:1094-1105. [DOI] [PubMed] [Google Scholar]
- 39.Le Page, C., O. Popescu, P. Genin, J. Lian, A. Paquin, J. Galipeau, and J. Hiscott. 2001. Disruption of NF-κB signaling and chemokine gene activation by retroviral mediated expression of IKKγ/NEMO mutants. Virology 286:422-433. [DOI] [PubMed] [Google Scholar]
- 40.Lin, R., P. Beauparlant, C. Makris, S. Meloche, and J. Hiscott. 1996. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol. Cell. Biol. 16:1401-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Litchfield, D. W. 2003. Protein kinase CK2: structure, regulation and the role in cellular decisions of life and death. Biochem. J. 369:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McElhinny, J. A., S. A. Trushin, G. D. Bren, N. Chester, and C. V. Paya. 1996. Casein kinase II phosphorylates IκBα at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol. Cell. Biol. 16:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 44.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melnick, J. L., E. Adam, and M. E. DeBakey. 1993. Human cytomegalovirus and atherogenesis, p. 80-91. In Y. Becker, G. Darai, and E. S. Huang (ed.), Molecular aspects of human cytomegalovirus diseases. Springer-Verlag, Berlin, Germany.
- 46.Mocarski, E. S., Jr., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 47.Myerson, D., R. C. Hackman, J. A. Nelson, D. C. Ward, and J. K. McDougall. 1984. Widespread presence of histologically occult cytomegalovirus. Hum. Pathol. 15:430-439. [DOI] [PubMed] [Google Scholar]
- 48.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 49.Prosch, S., C. Priemer, C. Hoflich, C. Liebenthaf, N. Babel, D. H. Kruger, and H. D. Volk. 2003. Proteasome inhibitors: a novel tool to suppress human cytomegalovirus replication and virus-induced immune modulation. Antivir. Ther. 8:555-567. [PubMed] [Google Scholar]
- 50.Prosch, S., K. Staak, J. Stein, C. Liebenthal, T. Stamminger, H. D. Volk, and D. H. Kruger. 1995. Stimulation of the human cytomegalovirus IE enhancer/promoter in HL-60 cells by TNFα is mediated via induction of NF-κB. Virology 208:197-206. [DOI] [PubMed] [Google Scholar]
- 51.Prosch, S., R. Wuttke, D. H. Kruger, and H. D. Volk. 2002. NF-κB-a potential therapeutic target for inhibition of human cytomegalovirus (re)activation? Biol. Chem. 383:1601-1609. [DOI] [PubMed] [Google Scholar]
- 52.Romieu-Mourez, R., E. Landesman-Bollag, D. C. Seldin, and G. E. Sonenshein. 2002. Protein kinase CK2 promotes aberrant activation of NF-κB, transformed phenotype, and survival of breast cancer cells. Cancer Res. 62:6770-6778. [PubMed] [Google Scholar]
- 53.Sambucetti, L. C., J. M. Cherrington, G. W. Wilkinson, and E. S. Mocarski. 1989. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarno, S., H. Reddy, F. Meggio, M. Ruzzene, S. P. Davies, A. Donella-Deana, D. Shugar, and L. A. Pinna. 2001. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (“casein kinase-2”). FEBS Lett. 496:44-48. [DOI] [PubMed] [Google Scholar]
- 55.Sarno, S., M. Ruzzene, P. Frascella, M. A. Pagano, F. Meggio, A. Zambon, M. Mazzorana, G. Di Maira, V. Lucchini, and L. A. Pinna. 2005. Development and exploitation of CK2 inhibitors. Mol. Cell. Biochem. 274:69-76. [DOI] [PubMed] [Google Scholar]
- 56.Schwarz, E. M., D. Van Antwerp, and I. M. Verma. 1996. Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IκBα. Mol. Cell. Biol. 16:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen, C. Y., M. S. Ho, S. F. Chang, M. S. Yen, H. T. Ng, E.-S. Huang, and C. W. Wu. 1993. High rate of concurrent genital infections with human cytomegalovirus and human papillomaviruses in cervical cancer patients. J. Infect. Dis. 168:449-452. [DOI] [PubMed] [Google Scholar]
- 58.Shen, J., P. Channavajhala, D. C. Seldin, and G. E. Sonenshein. 2001. Phosphorylation by the protein kinase CK2 promotes calpain-mediated degradation of IκBα. J. Immunol. 167:4919-4925. [DOI] [PubMed] [Google Scholar]
- 59.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sinzger, C., and G. Jahn. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39:302-319. [DOI] [PubMed] [Google Scholar]
- 61.Soderberg-Naucler, C. 2006. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J. Intern. Med. 259:219-246. [DOI] [PubMed] [Google Scholar]
- 62.Stenberg, R. M. 1993. Immediate-early genes of human cytomegalovirus: organization and function, p. 330-359. In Y. Becker, G. Darai, and E. S. Huang (ed.), Molecular aspects of human cytomegalovirus diseases. Springer-Verlag, Berlin, Germany.
- 63.Stinski, M. F., D. R. Thomsen, R. M. Stenberg, and L. C. Goldstein. 1983. Organization and expression of the immediate-early genes of human cytomegalovirus. J. Virol. 46:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798-2804. [DOI] [PubMed] [Google Scholar]
- 65.Thomsen, D. R., R. M. Stenberg, W. F. Goins, and M. F. Stinski. 1984. Promoter-regulatory region of the major immediate-early gene of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 81:659-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toorkey, C. B., and D. R. Carrigan. 1989. Immunohistochemical detection of an immediate early antigen of human cytomegalovirus in normal tissues. J. Infect. Dis. 160:741-751. [DOI] [PubMed] [Google Scholar]
- 67.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. I. Camp, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viatour, P., M. P. Merville, V. Bours, and A. Chariot. 2005. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 30:43-52. [DOI] [PubMed] [Google Scholar]
- 69.Wang, X., D. Y. Huang, S. M. Huong, and E.-S. Huang. 2005. Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat. Med. 11:515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, X., S. M. Huong, M. L. Chiu, N. Raab-Traub, and E.-S. Huang. 2003. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 424:456-461. [DOI] [PubMed] [Google Scholar]
- 71.Yao, F., and R. J. Courtney. 1992. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J. Virol. 66:2709-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yurochko, A. D., and E.-S. Huang. 1999. HCMV binding to human monocytes induces immuno-regulatory gene expression. J. Immunol. 162:4806-4816. [PubMed] [Google Scholar]
- 73.Yurochko, A. D., E.-S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E.-S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 71:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yurochko, A. D., T. F. Kowalik, S.-M. Huong, and E.-S. Huang. 1995. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J. Virol. 69:5391-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yurochko, A. D., M. W. Mayo, E. E. Poma, A. S. Baldwin, Jr., and E.-S. Huang. 1997. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J. Virol. 71:4638-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]