Abstract
Analysis of severe acute respiratory syndrome coronavirus (SCoV) by either sucrose gradient equilibrium centrifugation or a virus capture assay using an anti-SCoV S protein antibody demonstrated that the SCoV 6 protein, which is one of the accessory proteins of SCoV, was incorporated into virus particles. Coexpression of the SCoV S, M, E, and 6 proteins was sufficient for incorporation of the 6 protein into virus-like particles. Cells transfected with plasmid expressing the 6 protein released SCoV 6 protein; however, infected cells released SCoV 6 protein only in association with SCoV particles.
Severe acute respiratory syndrome coronavirus (SCoV) is known to be the causative agent of the global severe acute respiratory syndrome epidemic occurring during 2002 and 2003 (1, 8, 9). The 3′ one-third of the SCoV genome encodes the spike (S), membrane (M), and envelope (E) proteins (all of which are envelope proteins); the N protein (which forms a helical nucleocapsid with the genomic RNA); and the eight accessory proteins 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b (10, 15, 19). Expression studies have uncovered some of the biological activities of these accessory proteins (20), and yet their functions in infected cells and hosts are largely unexplored. SCoV accessory proteins are dispensable for SCoV replication in cultured cells (20, 21), and the 3a, 7a, and 7b proteins are incorporated into virus particles (4, 6, 16, 17).
The 63-amino-acid SCoV 6 protein is a membrane protein that is present mainly in the endoplasmic reticulum and Golgi compartments (3, 14) proximal to the virus assembly site. Past studies showed that SCoV lacking the gene encoding the 6 protein is viable (21), and a murine coronavirus expressing SCoV 6 protein has increased virulence in mice (14). The latter observation may be related to a finding that SCoV 6 protein expression accelerates murine coronavirus replication (18). Furthermore, an expression study demonstrated that the SCoV 6 protein, like the SCoV 3b and N proteins, is an interferon antagonist (7), while further studies are needed to establish the biological functions of the 6 protein in SCoV-infected cells.
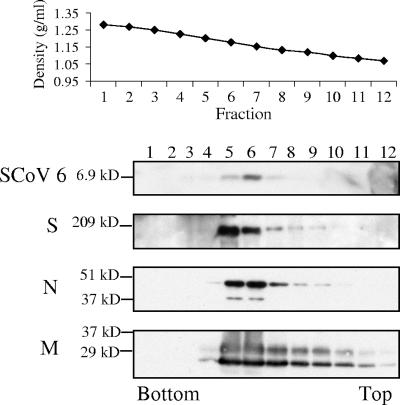
To determine whether the SCoV 6 protein is a virion-associated protein, we investigated the presence of the 6 protein in the purified SCoV particles. Supernatants harvested from SCoV-infected Caco2 cells were irradiated with 2 × 106 rads from a Gammacell 60Co source (model 109A; J. L. Shepherd and Associates, San Fernando, CA) and clarified by low-speed centrifugation and filtration through a 0.45-μm syringe filter. Then, SCoV was purified by ultracentrifugation on a 20 to 60% continuous sucrose gradient at 26,000 rpm for 18 h using a Beckman SW28 rotor (4, 5). Twelve fractions, each consisting of 1 ml, were collected, and each was examined on Western blots using antibodies against the S, M, N, and 6 proteins (Fig. 1). The rabbit anti-SCoV 6 protein peptide antibody was prepared by immunizing rabbits with the synthetic peptide N-ELDDEEPMELDY, corresponding to amino acids 51 to 62 of the 6 protein. Commercially available anti-S antibody (IGM-541; IMGENEX) and anti-SCoV M antibody (AP6008b; Abgent) were used. Mouse anti-N serum was provided by Xiao-Hua Li at The University of Texas Southwestern Medical Center, Dallas, TX. In agreement with our previous studies (4, 5), the strongest signals for the S, N, and M proteins were detected in fractions 5 and 6 (Fig. 1), showing that the buoyant density of SCoV was between 1.18 and 1.20 g/ml. Most of the 6 protein signal was present in fraction 6, strongly suggesting the association of the 6 protein with virions.
FIG. 1.
Association of the SCoV 6 protein with purified SCoV particles. Culture media from SCoV-infected Caco2 cells were inactivated by irradiation. Samples were clarified and partially purified by centrifugation through 20% sucrose cushions. Pellets were suspended in NTE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA) and purified by ultracentrifugation on a continuous 20 to 60% sucrose gradient by using a Beckman SW28 rotor. Twelve fractions were taken from the bottom. The purified viruses in each fraction were pelleted by centrifugation through a 20% sucrose cushion. Samples were dissolved in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 50 mM dithiothreitol) and separated on a 15% SDS-PAGE gel for detection of the SCoV 6 protein, on a 12% SDS-PAGE gel for the M and N proteins, or on an 8% SDS-PAGE gel for the S protein. Western blotting analysis using anti-SCoV 6 antibody (SCoV 6), anti-SCoV S protein antibody (S), anti-SCoV N antibody (N), and anti-SCoV M protein antibody (M) was performed to detect the SCoV 6, S, N, and M proteins, respectively.
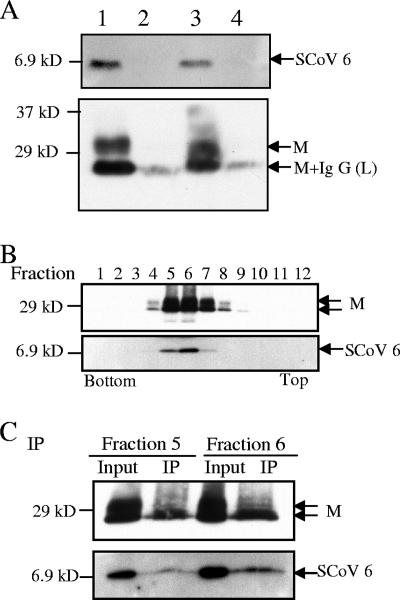
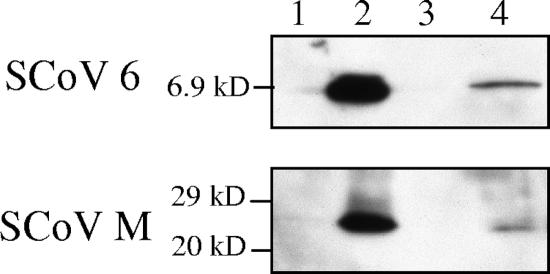
The presence of the 6 protein in virions was further confirmed using a virus capture assay (4, 5). Clarified virus samples were partially purified by centrifugation through 20% sucrose cushions at 26,000 rpm for 3 h in a Beckman SW28 rotor, followed by immunoprecipitation with mouse monoclonal anti-SCoV S protein antibody (NR-617; NIH Biodefense and Emerging Infections Research Resources Repository) or with an unrelated mouse monoclonal anti-H2K antibody in the absence of detergent (11, 12). We collected the pelleted SCoV particles, the immunoprecipitated samples, and the supernatant from the samples after initial anti-SCoV S protein antibody-mediated precipitation (virus capture assay supernatant). About 10% of the pelleted sample (Fig. 2A, lane 1), ca. 10% of the immunoprecipitated samples (Fig. 2A, lanes 2 and 3), and 3.3% of the virus capture assay supernatant (Fig. 2A, lane 4) were applied to the gel electrophoresis. Western blot analysis demonstrated that both the 6 and the M proteins were present in the virions immunoprecipitated by anti-S antibody (Fig. 2A, lane 3) and showed that neither the 6 protein nor the M protein was detected in samples immunoprecipitated by the anti-H2K antibody (Fig. 2A, lane 2). Furthermore, no 6 or M protein was detected in the supernatants of the immunoprecipitated samples (Fig. 2A, lane 4), even after longer exposure of the gel (data not shown), demonstrating that viral particles were efficiently captured by this assay and that most 6 protein was associated with virions. To firmly establish the presence of SCoV 6 protein in the SCoV particle, we performed a virus capture assay using highly purified SCoV particles. SCoV was purified by a 20 to 60% sucrose gradient as described above (Fig. 2B), and the purified SCoV in fraction 5 and that in fraction 6 were separately subjected to the virus capture assay. Western blot analysis showed the presence of the M and 6 proteins in the pelleted samples (Fig. 2C, lanes Input) as well as in the samples obtained after the virus capture assay (Fig. 2C, lanes IP). We found that monoclonal anti-SCoV S protein antibody failed to immunoprecipitate 6 protein in the extracts of 6 protein-expressing cells (data not shown), demonstrating that there was no cross-reactivity between the anti-SCoV S protein antibody and the 6 protein. Taken together, these studies convincingly showed that the SCoV 6 protein was incorporated into viral particles. The stoichiometry of the 6 protein in SCoV particles has not been determined.
FIG. 2.
Results of the SCoV capture assay. (A) SCoV particles in clarified media from SCoV-infected Caco2 cells were pelleted down by centrifugation on a 20% sucrose cushion. The suspended pellets were immunoprecipitated by a nonspecific monoclonal mouse anti-H2K antibody (lane 2) or by a mouse monoclonal anti-SCoV S antibody (lane 3) to capture virions. Samples were examined with rabbit anti-SCoV 6 protein antibody (top panel) and anti-SCoV M antibody (bottom panel) by Western blot analysis. Lanes 1 and 4 represent a SCoV sample partially purified by a 20% sucrose cushion and supernatant from the SCoV samples after purification with anti-SCoV S protein antibody, respectively. A minor signal detected in lanes 2 and 4 represents the immunoglobulin G light chain, which comigrated with a fast-migrating major M protein signal in lane 3. (B) SCoV samples were purified by ultracentrifugation on a 20 to 60% sucrose gradient. Twelve fractions were collected from bottom to top and subjected to Western blot analysis to detect the SCoV M and 6 proteins. (C) Fractions 5 and 6 from panel B, containing the purified SCoV particles, were further subjected to the virus capture assay to detect virion-associated SCoV M protein and 6 protein. Input, pelleted samples from fractions 5 and 6; IP, sample obtained after immunoprecipitation.
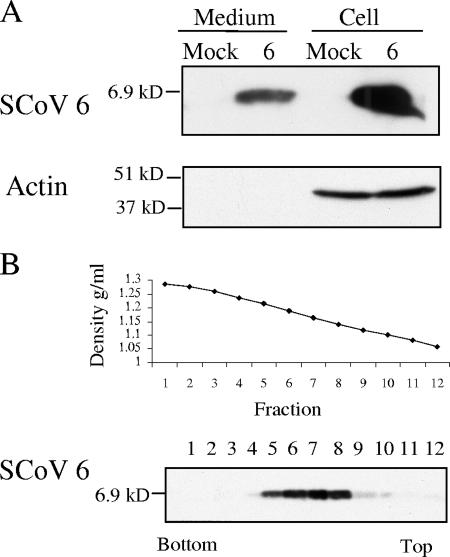
We next tested whether the SCoV 6 protein was released from 6 protein-expressing cells. Cultures of 293T cells were transfected with the control, pCAGGS plasmid, or pCAGGS-6 plasmid expressing the 6 protein. At 48 h posttransfection, culture media were collected and clarified, and the released materials were pelleted through a 20% sucrose cushion (5). Western blot analysis of the pellet demonstrated that the 6 protein was released into the culture media (Fig. 3A). Release of the 6 protein was not cell type dependent, as it was also released from Vero E6 cells (data not shown). Actin was not detected in the supernatants of transfected cells, suggesting that the observed release of the 6 protein was unlikely due to compromised cellular membrane integrity or contamination with cell debris. Although the released 6 protein exhibited a broad buoyant density of approximately 1.12 to 1.21 g/ml, the peak of the 6 protein signal was found in fractions 6, 7, and 8, with a density of approximately 1.14 to 1.19 g/ml (Fig. 3B). As the 6 protein is a membrane protein (3, 14), it was probably released as a membrane-associated form when expressed alone.
FIG. 3.
Release of the SCoV 6 protein from cells expressing the 6 protein. (A) 293T cells were transfected with empty-vector pCAGGS (mock) or pCAGGS-6 (6). At 48 h posttransfection, supernatants from transfected cells were clarified, applied onto a 20% sucrose cushion, and centrifuged at 26,000 rpm for 3 h at 4°C. The pellets were suspended in 1× SDS-PAGE loading buffer (Medium). Cell lysates were prepared with 1× SDS-PAGE loading buffer (Cell). Samples were separated on a 15% SDS-PAGE gel and subjected to Western blot analysis with anti-SCoV 6 protein antibody (6). The membranes were reprobed with anti-actin antibody (Actin). (B) The supernatant of 293T cells expressing the 6 protein was collected 48 h after transfection and partially purified by centrifugation through a 20% sucrose cushion. The pellets were suspended in NTE buffer, loaded on a 20 to 60% continuous sucrose gradient, and centrifuged at 26,000 rpm for 18 h using a Beckman SW28 rotor. Twelve fractions were taken from the bottom, and the densities for each action were measured (upper panel). Each fraction was then diluted at least threefold with NTE buffer and was then centrifuged through a 20% sucrose cushion. The recovered pellets were suspended in 1× SDS-PAGE loading buffer and subjected to Western blot analysis with anti-SCoV 6 protein antibody (lower panel).
A plasmid-based, SCoV virus-like particle (VLP)-producing system (4) was used to learn which other SCoV proteins were required for incorporation of the 6 protein into VLPs. Cultures of 293T cells were cotransfected with pCAGGS-based plasmids expressing the SCoV S, M, E, and 6 proteins. Released VLPs were purified by using the virus capture assay to exclude non-VLP-associated 6 protein that was also present in the sample. Western blot analysis of the captured VLPs showed the presence of the 6 protein in the VLPs (Fig. 4), demonstrating that the S, M, and E proteins were sufficient for the assembly of the 6 protein into VLPs. Because we could not clearly separate the VLPs from the released 6 protein by using sucrose gradient centrifugation (data not shown) and because the virus capture assay relied on the presence of S protein in the VLPs, we were unable to determine whether S protein was required for incorporation of the 6 protein into VLPs.
FIG. 4.
Assembly of SCoV 6 protein into VLPs. A mixture of plasmids, containing 9 μg of pCAGGS-S, 6 μg of pCAGGS-M, 4 μg of pCAGGS-E, and 0.5 μg of pCAGGS-6, was transfected into 293T cells (lanes 2, 3, and 4). As a control, cells were transfected with 19.5 μg of pCAGGS (lane 1). At 48 h posttransfection, culture media were harvested, and the released VLPs were pelleted by centrifugation through a 20% sucrose cushion. SCoV VLPs were resuspended in NTE buffer containing 0.3% of bovine serum albumin and immunoprecipitated with nonspecific monoclonal mouse anti-H2K antibody (lane 3) or mouse anti-SCoV S protein monoclonal antibody (NR-617) (lane 4). The captured SCoV VLPs (lanes 3 and 4) and cell lysates (lanes 1 and 2) were analyzed using Western blot analysis with anti-SCoV 6 protein antibody (top panel) and anti-M protein antibody (bottom panel).
The association of the 6 protein with SCoV particles was tested using sucrose gradient purification of SCoV particles and the more stringent virus capture assay. Initial sucrose gradient centrifugation analysis of SCoV demonstrated cosedimentation of the 6 protein with the S, N, and M proteins, suggesting the association of the 6 protein with SCoV particles. We noted that the abundances of the M and N proteins were similar in fractions 5 and 6, while the SCoV 6 signal in fraction 6 was stronger than that in fraction 5 (Fig. 1). These data may indicate that the abundance of the 6 protein differed among each SCoV particle. A very weak 6 protein signal was detected in the sucrose fractions containing less than 1.18 g/ml of sucrose (Fig. 1), suggesting that a majority of the 6 protein was more likely released in association with SCoV particles from infected cells. The virus capture assay clearly supported this notion; we did not detect any amounts of the 6 protein that were not associated with SCoV particles (Fig. 2A, lane 4), yet we detected the presence of 6 protein in the gradient-purified SCoV particles (Fig. 2B and C). The virus capture assay efficiently excludes various cellular contaminants from the samples and allows preparation of highly purified virions from virally infected cells developing cytopathic effects; this assay is used to demonstrate the specific incorporation of viral RNA and proteins into coronavirus particles (2) and host proteins into retrovirus particles (13). VLP incorporation of the 6 protein also supported the data that suggested that the SCoV 6 protein was virion associated. These data highlight how unlikely the possibility is that the bulk of the SCoV 6 protein in SCoV-infected cell culture media was merely a part of the cellular debris; rather, it was part of the virion. Because those experiments using SCoV were performed after irradiation of samples with 60Co, one might suspect that irradiation of the samples may have artificially caused an association of SCoV particles with released 6 protein, which was not incorporated into SCoV particles. However, this possibility is highly unlikely, because nearly all of the released 6 protein must have artificially bound to SCoV particles by irradiation to generate the data shown in Fig. 1 and 2. In summary, sucrose gradient purification of SCoV particles and the virus capture assay both convincingly pointed to the possibility that most of the 6 protein was released in association with SCoV particles from SCoV-infected cells.
The present study and past studies revealed that SCoV accessory proteins 3a, 6, 7b, and 7a are incorporated into SCoV particles (4, 6, 16, 17), although these accessory proteins are secreted from infected cells and from expressing cells in different ways. The 3a protein is released from both SCoV-infected cells and 3a-expressing cells (5). The 7a protein is not released from infected cells or from 7a-expressing cells (4, 5). The 7b protein is not released from 7b-expressing cells (16), while release of 7b protein from infected cells has not been examined yet. The 6 protein was released from 6 protein-expressing cells (Fig. 3), and yet sucrose gradient purification of SCoV (Fig. 1) and the virus capture assay (Fig. 2) both suggested that, unlike the 3a protein, the 6 protein alone was not released from SCoV-infected cells. We speculate that some viral envelope proteins may interact with the 6 protein, thereby preventing its release and mediating its incorporation into SCoV particles in infected cells.
The report that an attenuated murine coronavirus expressing the SCoV 6 protein exhibits an enhanced virulence in mice (14) and promotes murine coronavirus replication in infected cells (18) points to the possible involvement of the 6 protein in SCoV pathogenesis and replication. A recent expression study reported that the SCoV 6 protein is an interferon antagonist (7). How the 6 protein affects SCoV virulence is unclear, yet the present study and reports by others (7, 14, 18) suggest that the incoming virion-associated 6 protein may alter the cellular environment to facilitate SCoV replication early in infection and inhibits host innate immune functions. Also, the 6 protein may play some role in virus assembly, release, stability, or infectivity or other aspects of pathogenesis.
Acknowledgments
We thank Chien-Te K. Tseng and Eric Mossel for preparation of inactivated virus samples.
This work was supported by Public Health Service grant AI29984 to S.M. and contract N01-AI25489 from the National Institutes of Health. C.H. was supported by the James W. McLaughlin Fellowship Fund.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 2.Escors, D., C. Capiscol, and L. Enjuanes. 2004. Immunopurification applied to the study of virus protein composition and encapsidation. J. Virol. Methods 119:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geng, H., Y. M. Liu, W. S. Chan, A. W. Lo, D. M. Au, M. M. Waye, and Y. Y. Ho. 2005. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 579:6763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang, C., N. Ito, C. T. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 80:7287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang, C., K. Narayanan, N. Ito, C. J. Peters, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. J. Virol. 80:210-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 79:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 9.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 11.Narayanan, K., C. J. Chen, J. Maeda, and S. Makino. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77:2922-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278:52347-52354. [DOI] [PubMed] [Google Scholar]
- 14.Pewe, L., H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 16.Schaecher, S. R., J. M. Mackenzie, and A. Pekosz. 2007. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 81:718-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen, S., P. S. Lin, Y. C. Chao, A. Zhang, X. Yang, S. G. Lim, W. Hong, and Y. J. Tan. 2005. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 330:286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tangudu, C., H. Olivares, J. Netland, S. Perlman, and T. Gallagher. 2007. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 81:1220-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 20.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]