Abstract
Studies have shown that human immunodeficiency virus type 2 (HIV-2) is less pathogenic than HIV-1, with a lower rate of disease progression. Similarly, plasma viral loads are lower in HIV-2 infection, suggesting that HIV-2 replication is restricted in vivo in comparison to that of HIV-1. However, to date, in vivo studies characterizing replication intermediates in the viral life cycle of HIV-2 have been limited. In order to test the hypothesis that HIV-2 has a lower replication rate in vivo than HIV-1 does, we quantified total viral DNA, integrated proviral DNA, cell-associated viral mRNA, and plasma viral loads in peripheral blood samples from groups of therapy-naïve HIV-1-infected (n = 21) and HIV-2-infected (n = 18) individuals from Dakar, Senegal, with CD4+ T-cell counts of >200/μl. Consistent with our previous findings, total viral DNA loads were similar between HIV-1 and HIV-2 and plasma viral loads were higher among HIV-1-infected individuals. Proportions of DNA in the integrated form were also similar between these viruses. In contrast, levels of viral mRNA were lower in HIV-2 infection. Our study indicates that HIV-2 is able to establish a stable, integrated proviral infection in vivo, but that accumulation of viral mRNA is attenuated in HIV-2 infection relative to that in HIV-1 infection. The differences in viral mRNA are consistent with the differences in plasma viral loads between HIV-1 and HIV-2 and suggest that lower plasma viral loads, and possibly the attenuated pathogenesis of HIV-2, can be explained by lower rates of viral replication in vivo.
Two closely related human lentiviruses, human immunodeficiency virus type 1 (HIV-1) and HIV-2, have been shown to cause AIDS. However, it is now well recognized that the in vivo pathogenicity of HIV-2 is attenuated relative to that of HIV-1, with significantly lower rates of disease progression and transmission (20, 27, 28, 43). Consistent with these observations, plasma viral loads are significantly lower in people infected with HIV-2 (1, 2, 36, 37, 42). Interestingly, studies have shown that quantities of total viral DNA in peripheral blood mononuclear cells (PBMCs) are similar in people infected with HIV-1 and people infected with HIV-2 (5, 32, 36).
Within an infected cell, HIV DNA is found in a number of forms, including integrated proviral DNA, unintegrated linear viral DNA, and unintegrated one- and two-long-terminal-repeat (LTR) episomal viral DNA (31). The integration of proviral DNA is necessary for viral replication (10, 11, 21, 44). In addition, the integration of proviral DNA can result in a stable long-term infection. HIV-1 has been shown to establish latent infection in vivo within resting memory CD4+ T cells, a reservoir in which the integrated proviral genome can persist for decades (reviewed in references 22 and 35). Although integration is necessary for viral replication, studies of HIV-1 have shown that the majority of in vivo viral DNA exists in the linear unintegrated form (8), while integrated proviral DNA accounts for a minor fraction of the total DNA load in vivo (6, 8, 9, 15, 16).
While higher plasma viral loads in HIV-1 infection suggest higher replication rates, no studies have clearly shown a difference in replication in vivo between HIV-1 and HIV-2. Additionally, previous studies comparing viral DNA in HIV-1 and HIV-2 infection have focused on total viral DNA and, to date, no studies have quantified integrated proviral DNA in people infected with HIV-2. In this study, we measured viral life cycle intermediates (integrated proviral DNA and viral mRNA) in conjunction with total viral DNA loads and plasma viral loads in people infected with HIV-1 or HIV-2 to determine whether quantitative differences that can explain the lower plasma viral loads observed in HIV-2 infection exist at a specific point of the viral life cycle.
MATERIALS AND METHODS
Sample acquisition.
PBMC samples were obtained from a cohort of female sex workers in Dakar, Senegal, that have been followed since 1985. Epidemiologic and clinical aspects of this cohort have previously been described elsewhere (19). All subjects signed informed consents and participated in protocols approved by the Counseil National de Lutte Contre le Sida Comite Ethique et Juridique and the Harvard School of Public Health Human Subjects Committee. CD4+ T-cell counts were determined, and serum samples were diagnosed for HIV-1- and HIV-2-specific antibodies as previously described (19). All subjects enrolled in this study were antiretroviral therapy naïve and had CD4+ T-cell counts above 200/μl at the time of sample acquisition.
DNA and mRNA extraction.
Cryopreserved PBMC samples were thawed and rested overnight in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum and 1% antibiotics to ensure sample viability and remove dead cells. The following day, PBMC samples were divided equally, with half used for the extraction of DNA and half used for the extraction of mRNA. DNA was extracted (blood and cell culture DNA mini kit; QIAGEN), and DNA concentrations were determined by an optical density reading at 260 nm. Polyadenylated mRNA was extracted (Oligotex mRNA direct mini kit; QIAGEN) and immediately converted to cDNA (TaqMan reverse transcription reagents; Applied Biosystems) by using an equal-volume mixture of random hexamer and poly(T) primers.
Quantification of total viral DNA and viral mRNA.
HIV-1 total viral DNA and mRNA were quantified using a previously described real-time PCR assay (25) that amplifies a highly conserved region corresponding to the HIV-1 LTR-gag junction. For the quantification of HIV-2 total viral DNA and mRNA, we developed a novel HIV-2 real-time PCR assay that amplifies a highly conserved region corresponding to the HIV-2 LTR-gag junction. To construct a quantification standard, a fragment of HIV-2 LTR-gag was amplified by PCR from a previously cloned HIV-2 DNA sample using primers AM2gag1f (AGACCCTGGTCTGTTAGGACCCTT) and AM2gag1r (CAATTCATTCGCTGCCCACAC) cloned into an expression vector (pCR2.1; Invitrogen) and transformed into competent cells (TOP10; Invitrogen). Cloned plasmid was purified (SNAP MiniPrep kit; Invitrogen), and the concentration of purified plasmid was determined by an optical density reading at 260 nm. For the quantification of HIV-2 samples, real-time PCRs were performed by using primers HIV2gagF (CCAACCACGACGGAGTGCTC) and HIV2gagR (CTCTCAAGACGGAGTTTCTCGC) and detected by using the probe HIV2gagP (AGGCCTCCGGGTGAAGGTAAG), which contained a 5′ 6-carboxyfluorescein fluorescent reporter and a 3′ MGB nonfluorescent quencher. For HIV-1 and HIV-2 real-time PCR assays, real-time PCR reagents and reaction conditions were as previously described (25). Viral mRNA was standardized to cellular beta-actin mRNA. Beta-actin mRNA was quantified using commercially available reagents (TaqMan B-actin detection reagents; Applied Biosystems) according to manufacturer recommendations.
Quantification of integrated proviral DNA.
Quantities of integrated proviral DNA were determined for HIV-1 and for HIV-2 by using an endpoint dilution Alu-PCR approach as previously described (9). A series of six threefold serial dilutions were performed in duplicate from a starting quantity of 1 μg genomic DNA, and Alu-PCR was performed using primers Alu1 (GCCTCCCAAAGTGCTGGGATTACAG) (34) and AM1C1r (CTTAATACTGACGCTCTCGCACCC) for HIV-1 samples and Alu1 and AM2C1r (AAGGGTCCTAACAGACCAGGGTCT) for HIV-2 samples in a total volume of 100 μl under the following reaction conditions: denaturation at 94° for 5 min; 35 PCR cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 3 min; and a final extension at 72°C for 7 min. The amplification of integrated proviral DNA was assayed by real-time PCR with 5 μl of the first-round Alu-PCR product by using previously described HIV-1 and HIV-2 real-time PCR assays (25). Real-time PCR assays were performed for each sample in parallel with equivalent concentrations of nonamplified genomic DNA from that respective sample to control for nonamplified viral DNA carryover from the Alu-PCR. Integrated proviral DNA copy numbers were determined by using the QUALITY program (38).
Quantification of plasma viral loads.
HIV-1 plasma viral loads were determined by using a commercially available assay (Amplicor HIV-1 monitor test, version 1.5; Roche). To determine HIV-2 viral loads, we developed an in-house real-time PCR viral load assay. Briefly, we amplified a fragment of HIV-2 LTR-gag from a previously cloned HIV-2 DNA sample using primers HIV2gagF and AM2gag1r and synthesized a T7 promoter-driven in vitro transcription construct by using commercially available reagents (BLOCK-iT RNAi TOPO transcription kit; Invitrogen) and primer AM2gag1r. HIV-2 RNA was generated by in vitro transcription (MEGAshortscript; Ambion). In vitro-transcribed HIV-2 RNA was purified (MEGAclear; Ambion), treated with DNase (New England Biolabs), and repurified, and the concentration of in vitro-transcribed HIV-2 RNA was determined by an optical density reading at 260 nm. To create a full-length HIV-2 genomic RNA standard, full-length HIV-2 genomic RNA from a previously described in vitro infection (25) was extracted (QIAamp viral RNA mini kit; QIAGEN), treated with DNase (New England Biolabs), and repurified (MEGAclear; Ambion). Quantities of full-length HIV-2 genomic RNA were determined by using the in vitro-transcribed HIV-2 RNA as a quantitative standard in a two-step, real-time reverse transcription-PCR (TaqMan reverse transcription reagents and TaqMan universal PCR master mix; Applied Biosystems) using the primer HIV2gagR for the cDNA synthesis reaction and the HIV-2 LTR-gag real-time PCR primers and probe described above. For the quantification of plasma viral loads, RNA standard curves were generated from the full-length HIV-2 genomic RNA standard, spiked to noninfected lysis buffer, and extracted in parallel with 280 μl of plasma from HIV-2-infected subjects (QIAamp viral RNA mini kit; QIAGEN). HIV-2 plasma viral loads were quantified relative to the full-length HIV-2 genomic RNA standard curve in a two-step, real-time reverse transcription-PCR, as described above.
Statistical analyses.
Statistical analyses were performed using SAS 9.1. For analytical purposes, HIV-2-infected subjects with plasma viral loads of <400/ml were assigned values of 400/ml in accordance with the detection limit of the HIV-1 plasma viral load assay (Amplicor HIV-1 monitor test, version 1.5; Roche). The Wilcoxon rank sum test was used for an unadjusted comparison of CD4+ T-cell counts, age, total viral DNA, integrated proviral DNA, viral mRNA, plasma viral load, and percentage of total viral DNA integrated between HIV-1- and HIV-2-infected subjects; two-sided P values are reported. Because of the limited number of study subjects available, we were unable to directly match HIV-1-infected and HIV-2-infected individuals on the basis of CD4+ T-cell count and age. Alternatively, to adjust for CD4+ T-cell counts and age in the comparison of HIV-1-infected and HIV-2-infected individuals, regressions were performed using the ROBUSTREG procedure, which provides stable results in the presence of outlying values, and values of total viral DNA, integrated proviral DNA, viral mRNA, and plasma viral loads were log transformed. Correlations of total viral DNA, integrated proviral DNA, percentage of total DNA integrated, and viral mRNA with plasma viral loads were examined using the Pearson's correlation coefficient (for HIV-1) and Spearman's correlation coefficient (for HIV-2) by using log-transformed values of total viral DNA, integrated proviral DNA, viral mRNA, and plasma viral loads.
RESULTS
For this study, we examined 21 subjects infected with HIV-1 and 18 subjects infected with HIV-2 from Dakar, Senegal. All had CD4+ T-cell counts above 200/μl and were antiretroviral therapy naïve. Although all subjects were in the asymptomatic phase of infection, CD4+ T-cell counts were lower (P = 0.0384) in HIV-1-infected subjects (median and interquartile range values were 560 and 404 to 614 cells/μl, respectively [for HIV-2-infected subjects, median and interquartile range values were 946 and 534 to 1,503 cells/μl, respectively]). In addition, HIV-1-infected subjects (median and interquartile range values were 34.9 and 27.4 to 42.2 years, respectively) were younger (P = 0.0039) than HIV-2-infected subjects (median and interquartile range values were 44.2 and 40.1 to 48.9 years, respectively).
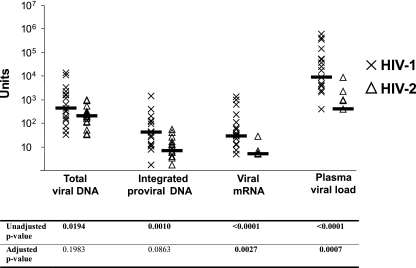
Previous studies have indicated that plasma viral loads are higher in people infected with HIV-1 than in people infected with HIV-2 (1, 2, 36, 37, 42), despite their having similar amounts of total viral DNA (5, 32, 36). For this study, we quantified total viral DNA by using real-time PCR assays for HIV-1 and HIV-2 that amplified highly conserved regions of the viral genomes that correspond to the LTR-gag junction. Twenty of 21 HIV-1-infected and 17 of 18 HIV-2-infected individuals had detectable total viral DNA. Among study subjects, total viral DNA loads were slightly higher in HIV-1-infected subjects than in HIV-2-infected subjects (Fig. 1). However, after adjusting for CD4+ T-cell counts and age, there was no statistical difference in total viral DNA levels between HIV-1- and HIV-2-infected subjects. In contrast, in both unadjusted and adjusted analyses, plasma viral loads were significantly higher in HIV-1-infected subjects than those in HIV-2-infected subjects.
FIG. 1.
Total viral DNA, integrated proviral DNA, viral mRNA, and plasma viral loads for HIV-1-infected and HIV-2-infected subjects. Units refer to copies per 106 PBMCs (total viral DNA and integrated proviral DNA), copies per 105 copies beta-actin (viral mRNA), and viral load per milliliter of plasma (plasma viral load). Unadjusted P values refer to Wilcoxon rank sum two-sided P values. Adjusted P values refer to robust regression P values adjusted for CD4+ T-cell count and age. Differences between HIV-1 and HIV-2 that reach statistical significance are in bold.
In order to compare amounts of viral mRNA between HIV-1 and HIV-2, viral transcripts isolated though the extraction of polyadenylated cellular mRNA were quantified using the same real-time PCR assays that were used to quantify total viral DNA. Unlike for HIV-1-infected subjects, in which viral transcripts were detectable for the majority of subjects, amounts of viral mRNA were below the limit of detection for most HIV-2-infected individuals. The difference in quantities of viral mRNA between HIV-1 and HIV-2 were statistically significant in both unadjusted and adjusted analyses.
In order to compare the amounts of integrated proviral DNA in PBMCs of subjects infected with HIV-1 with those of subjects infected with HIV-2, we used a previously described endpoint dilution Alu-PCR approach (9) that selectively amplifies integrated proviral DNA by using a virus-specific primer and a primer specific to conserved sequences in Alu elements. Alu elements are retrotransposons, and approximately 1.2 million copies of Alu elements are found in the human genome, accounting for 10 to 11% of the entire genome (18). To avoid unequal amplification between HIV-1 samples and HIV-2 samples, HIV-1 and HIV-2 primers were approximately equidistant from the 5′ end of the respective viral genomes, and have previously been shown to amplify integrated proviral DNA (25). Twenty of 21 HIV-1-infected subjects and 16 of 18 HIV-2-infected subjects had detectable integrated proviral DNA. Amounts of integrated proviral DNA were larger for HIV-1 than HIV-2, and values were slightly different after adjusting for CD4+ T-cell counts and age.
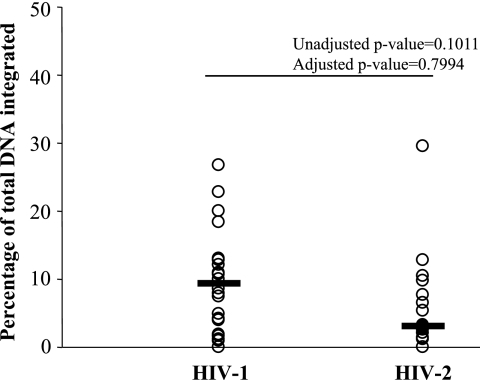
Previous studies of HIV-1 indicate that only a small proportion of total viral DNA is integrated in vivo (6, 8, 9, 15, 16). Among study subjects with detectable quantities of integrated proviral DNA, we observed similar results (Fig. 2) for both HIV-1 (the median percentage of total DNA integrated was 9.2%) and HIV-2 (the median percentage of total DNA integrated was 3.1%). There was no statistically significant difference in the percentage of DNA integrated between HIV-1-infected subjects and HIV-2-infected subjects.
FIG. 2.
Percentage of total DNA in the integrated form for HIV-1-infected and HIV-2-infected subjects. Values are plotted for all subjects who had detectable integrated proviral DNA (n = 20 for HIV-1; n = 16 for HIV-2). Unadjusted P values refer to Wilcoxon rank sum two-sided P values. Adjusted P values refer to robust regression P values adjusted for CD4+ T-cell count and age.
We examined the relationship of total viral DNA, integrated proviral DNA, percentage of total DNA integrated, and viral mRNA to plasma viral loads for HIV-1-infected subjects and HIV-2-infected subjects (Table 1). As expected, for HIV-1, quantities of total viral DNA, integrated proviral DNA, and viral mRNA all significantly correlated with plasma viral loads. In contrast, for HIV-1, there was no correlation between the percentage of total viral DNA integrated and plasma viral loads. Among HIV-2-infected subjects, we observed no significant correlation between plasma viral loads and total viral DNA or integrated proviral DNA. However, despite the small number of HIV-2-infected subjects having detectable mRNA, quantities of viral mRNA positively correlated with plasma viral loads among HIV-2-infected subjects. As with HIV-1, there was no relationship between the percentage of total viral DNA integrated and plasma viral loads.
TABLE 1.
Correlation of total viral DNA, integrated proviral DNA, percentage of total DNA integrated, and viral mRNA, with plasma viral loads for HIV-1-infected subjects and HIV-2-infected subjects
| Virus | Correlation statistics for viral load anda:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Total viral DNA
|
Integrated proviral DNA
|
% of total DNA integrated
|
Viral mRNA
|
|||||
| r | P | r | P | r | P | r | P | |
| HIV-1 | 0.56 | 0.0088 | 0.53 | 0.0130 | −0.16 | 0.4900 | 0.75 | <0.0001 |
| HIV-2 | 0.39 | 0.1091 | 0.22 | 0.3837 | −0.02 | 0.9371 | 0.49 | 0.0389 |
Correlations were examined using log-transformed total viral DNA, integrated proviral DNA, viral mRNA, and plasma viral loads. Correlation coefficients (r) and P values refer to Pearson's correlation coefficient (for HIV-1) and Spearman's correlation coefficient (for HIV-2). Correlations that reach statistical significance are in bold.
DISCUSSION
In HIV-1 infection, higher plasma viral loads are predictors of more rapid disease progression (3, 23, 29, 30, 33). Plasma viral loads have been clearly demonstrated to be lower in HIV-2 infection than in HIV-1 infection (1, 2, 36, 37, 42), possibly explaining the lower rate of disease progression of HIV-2 (12). This observation, in conjunction with similar total viral DNA loads in people infected with these viruses (5, 32, 36), suggests that HIV-2 maintains a lower rate of replication in vivo. However, host factors, such as differences in adaptive immune responses, could also account for the difference in plasma viral loads between these viruses. To address the hypothesis that HIV-2 has a lower replication rate in vivo than HIV-1 does, we quantified replication intermediates in the viral life cycles of HIV-1 and HIV-2 in vivo.
Consistent with previous reports, we observed significantly higher plasma viral loads in people infected with HIV-1 than in people infected with HIV-2, and after an adjustment for CD4+ T-cell counts and age, we observed no difference in total viral DNA. The interpretation of total viral DNA with regard to viral replication is difficult, as studies have shown that only a minority of HIV-1 DNA is actually integrated in vivo (6, 8, 9, 15, 16). Furthermore, this measurement includes one- and two-LTR circles, which are dead-end products. To better compare the nature of viral DNA between HIV-1 and HIV-2 in vivo, we quantified levels of integrated proviral DNA. We observed only modestly higher levels of integrated proviral DNA in HIV-1-infected individuals, and after comparing the percentage of viral DNA in the integrated form, we observed no difference between HIV-1 and HIV-2. While integrated proviral DNA clearly represents a minor proportion of the overall viral DNA load in HIV-2 infection, our data indicate that the integration of viral DNA is not overtly impaired in HIV-2 infection relative to that in HIV-1.
The HIV-1 and HIV-2 real-time PCR assays used in this study to quantify viral mRNA amplify a fragment of the LTR-gag junction, which is encoded in processive unspliced viral mRNA. Levels of viral mRNA in HIV-2-infected individuals were significantly lower than those in HIV-1-infected individuals. This difference is consistent with the lower plasma viral loads among the HIV-2-infected individuals and implies that lower plasma viral loads can be accounted for by lower viral mRNA levels in HIV-2 infection in vivo. Hermankova et al. (14) reported extremely little accumulation of processive viral mRNA in latently infected resting CD4+ T cells during HIV-1 infection in vivo. Given the observations that HIV-2 is able to establish integrated proviral infection, yet transcription remains limited, HIV-2 may have a much higher propensity for establishing latent infection in vivo than HIV-1 does. In support of this notion, the Nef protein of HIV-2 has been shown to downmodulate T-cell receptor-CD3 within infected cells and block responsiveness to T-cell activation, whereas the HIV-1 Nef protein fails to do so, contributing to increased cell death in HIV-1-infected cells and possibly increased persistence in HIV-2-infected cells (40).
Among HIV-1-infected individuals, total viral DNA, integrated proviral DNA, and viral mRNA all correlated with plasma viral loads. Of these measures, viral mRNA had the strongest correlation with plasma viral load. Interestingly, despite the small number of individuals with detectable mRNA, viral mRNA also correlated with plasma viral load for HIV-2. These findings support the notion that viral mRNA is a relevant marker of viral replication. For both HIV-1 and HIV-2, no correlation was observed between the percentage of total viral DNA integrated and plasma viral load, indicating that the proportion of integrated proviral DNA is not a pertinent measure for viral pathogenesis.
Our data suggest that HIV-2 is able to establish a stable integrated proviral infection within PBMCs and that replication is attenuated at some point following integration, but prior to the accumulation of late transcripts. The reason for the block remains unknown. HIV-1 and HIV-2 have different regulatory elements within their promoter regions and have been shown to respond differently to cellular stimuli (13), suggesting possible differences in transcriptional initiation between these viruses. In addition, integration within heterochromatin has been shown to be associated with transcriptional inhibition and viral latency during in vitro HIV-1 infection (17, 24) and we have recently reported evidence of proviral integration within heterochromatin in a higher proportion of people infected with HIV-2 than with HIV-1 (25). Differences between HIV-1 and HIV-2 may also exist following transcriptional initiation. For instance, the HIV-2 LTR is significantly larger than the HIV-1 LTR, has been shown to undergo 5′ LTR splicing, and has a complex secondary structure that may affect transcriptional trans-activation (4, 7). While the exact effect this has on mRNA accumulation in vivo remains unknown, it is possible that differences in the LTR structure result in less-efficient transcriptional elongation and/or the switch from early to late transcripts in HIV-2 infection.
To our knowledge, this is the first study to report direct evidence of a difference in the replication rates of HIV-1 and HIV-2 in vivo. In a recent report from our lab, we prospectively examined viral evolution over a decade of infection among eight HIV-2-infected individuals and observed significantly lower rates of viral evolution for HIV-2 than for HIV-1 (26), which also supports the notion that HIV-2 replication is attenuated in vivo in comparison to HIV-1 replication. It should be noted that the results of the current study do not exclude the possibility that differences in adaptive immune responses may also contribute to the lower plasma viral loads observed with HIV-2 infection. Recent data from our laboratory indicate a broad neutralizing response in people infected with HIV-2 (39).
While it is well recognized that disease progression is significantly slower in HIV-2 infection, the virologic explanations for the attenuated pathogenicity of HIV-2 have remained unclear. Studies in vitro imply no difference in cytopathogenicity between HIV-1 and HIV-2 (41). However, cytopathogenicity is partially a function of viral replication. Through characterizing intermediates of the viral life cycles of HIV-1 and HIV-2 in people infected with these viruses, we have found direct evidence that replication rates are lower for HIV-2 than for HIV-1 in vivo. Additionally, we note that viral replication rates for HIV-1 and HIV-2 correspond with relative rates of disease progression for these viruses, implying that the difference in the pathogenicity of HIV-1 and HIV-2 may be explained by differences in viral replication. Since this difference appears to be manifested after the viral integration step, our data suggest that HIV-2 is capable of efficient cellular infection but has a propensity for viral latency. These observations are consistent with in vivo and epidemiologic population data, whereby HIV-2 persists despite a significant impairment in replicative capacity relative to HIV-1.
Acknowledgments
We thank Shaun Rodriguez for helpful suggestions and Beth Chaplin and Christopher Mullins for technical assistance.
This work was supported by grants from the National Institutes of Health (RO1AI46187, AI52734-01A1, and 5 T32 AI007638).
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Alabi, A. S., S. Jaffar, K. Ariyoshi, T. Blanchard, M. Schim van der Loeff, A. A. Awasana, T. Corrah, S. Sabally, R. Sarge-Njie, F. Cham-Jallow, A. Jaye, N. Berry, and H. Whittle. 2003. Plasma viral load, CD4 cell percentage, HLA and survival of HIV-1, HIV-2, and dually infected Gambian patients. AIDS 17:1513-1520. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, S., H. Norrgren, Z. Da Silva, A. Biague, S. Bamba, S. Kwok, C. Christopherson, G. Biberfeld, and J. Albert. 2000. Plasma viral load in HIV-1 and HIV-2 singly and dually infected individuals in Guinea-Bissau, West Africa: significantly lower plasma virus set point in HIV-2 infection than in HIV-1 infection. Arch. Intern. Med. 160:3286-3293. [DOI] [PubMed] [Google Scholar]
- 3.Arduino, J. M., M. A. Fischl, K. Stanley, A. C. Collier, and D. Spiegelman. 2001. Do HIV type 1 RNA levels provide additional prognostic value to CD4+ T lymphocyte counts in patients with advanced HIV type 1 infection? AIDS Res. Hum. Retrovir. 17:1099-1105. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B. 1992. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 20:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry, N., K. Ariyoshi, O. Jobe, P. T. Ngum, T. Corrah, A. Wilkins, H. Whittle, and R. Tedder. 1994. HIV type 2 proviral load measured by quantitative polymerase chain reaction correlates with CD4+ lymphopenia in HIV type 2-infected individuals. AIDS Res. Hum. Retrovir. 10:1031-1037. [DOI] [PubMed] [Google Scholar]
- 6.Calcaterra, S., G. Cappiello, A. Di Caro, A. R. Garbuglia, and A. Benedetto. 2001. Comparative analysis of total and integrated HIV-1 DNA in peripheral CD4 lymphocytes and monocytes after long treatment with HAART. J. Infect. 43:239-245. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, P., A. Garzino-Demo, P. Swinney, and S. K. Arya. 1993. Human immunodeficiency virus type 2 multiply spliced transcripts. AIDS Res. Hum. Retrovir. 9:331-335. [DOI] [PubMed] [Google Scholar]
- 8.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 9.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Englund, G., T. S. Theodore, E. O. Freed, A. Engelman, and M. A. Martin. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlieb, G. S., P. S. Sow, S. E. Hawes, I. Ndoye, M. Redman, A. M. Coll-Seck, M. A. Faye-Niang, A. Diop, J. M. Kuypers, C. W. Critchlow, R. Respess, J. I. Mullins, and N. B. Kiviat. 2002. Equal plasma viral loads predict a similar rate of CD4+ T-cell decline in human immunodeficiency virus (HIV) type 1- and HIV-2-infected individuals from Senegal, West Africa. J. Infect. Dis. 185:905-914. [DOI] [PubMed] [Google Scholar]
- 13.Hannibal, M. C., D. M. Markovitz, N. Clark, and G. J. Nabel. 1993. Differential activation of human immunodeficiency virus type 1 and 2 transcription by specific T-cell activation signals. J. Virol. 67:5035-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermankova, M., J. D. Siliciano, Y. Zhou, D. Monie, K. Chadwick, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2003. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J. Virol. 77:7383-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibanez, A., T. Puig, J. Elias, B. Clotet, L. Ruiz, and M. A. Martinez. 1999. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. AIDS 13:1045-1049. [DOI] [PubMed] [Google Scholar]
- 16.Izopet, J., M. Cazabat, C. Pasquier, K. Sandres-Saune, E. Bonnet, B. Marchou, P. Massip, and J. Puel. 2002. Evolution of total and integrated HIV-1 DNA and change in DNA sequences in patients with sustained plasma virus suppression. Virology 302:393-404. [DOI] [PubMed] [Google Scholar]
- 17.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurka, J. 2004. Evolutionary impact of human Alu repetitive elements. Curr. Opin. Genet. Dev. 14:603-608. [DOI] [PubMed] [Google Scholar]
- 19.Kanki, P., S. M'Boup, R. Marlink, K. Travers, C. C. Hsieh, A. Gueye, C. Boye, J. L. Sankale, C. Donnelly, W. Leisenring, et al. 1992. Prevalence and risk determinants of human immunodeficiency virus type 2 (HIV-2) and human immunodeficiency virus type 1 (HIV-1) in West African female prostitutes. Am. J. Epidemiol. 136:895-907. [DOI] [PubMed] [Google Scholar]
- 20.Kanki, P. J. 1999. Human immunodeficiency virus type 2 (HIV-2). AIDS Rev. 1:101-108. [Google Scholar]
- 21.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassen, K., Y. Han, Y. Zhou, J. Siliciano, and R. F. Siliciano. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10:525-531. [DOI] [PubMed] [Google Scholar]
- 23.Lefrère, J. J., F. Roudot-Thoraval, M. Mariotti, M. Thauvin, J. Lerable, J. Salpetrier, and L. Morand-Joubert. 1998. The risk of disease progression is determined during the first year of human immunodeficiency virus type 1 infection. J. Infect. Dis. 177:1541-1548. [DOI] [PubMed] [Google Scholar]
- 24.Lewinski, M. K., D. Bisgrove, P. Shinn, H. Chen, C. Hoffmann, S. Hannenhalli, E. Verdin, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacNeil, A., J. L. Sankale, S. T. Meloni, A. D. Sarr, S. Mboup, and P. Kanki. 2006. Genomic sites of human immunodeficiency virus type 2 (HIV-2) integration: similarities to HIV-1 in vitro and possible differences in vivo. J. Virol. 80:7316-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacNeil, A., J. L. Sankale, S. T. Meloni, A. D. Sarr, S. Mboup, and P. Kanki. 2007. Long-term intrapatient viral evolution during HIV-2 infection. J. Infect. Dis. 195:726-733. [DOI] [PubMed] [Google Scholar]
- 27.Marlink, R., P. Kanki, I. Thior, K. Travers, G. Eisen, T. Siby, I. Traore, C. C. Hsieh, M. C. Dia, E. H. Gueye, et al. 1994. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science 265:1587-1590. [DOI] [PubMed] [Google Scholar]
- 28.Marlink, R. G., D. Ricard, S. M'Boup, P. J. Kanki, J. L. Romet-Lemonne, I. N′Doye, K. Diop, M. A. Simpson, F. Greco, M. J. Chou, et al. 1988. Clinical, hematologic, and immunologic cross-sectional evaluation of individuals exposed to human immunodeficiency virus type-2 (HIV-2). AIDS Res. Hum. Retrovir. 4:137-148. [DOI] [PubMed] [Google Scholar]
- 29.Mellors, J. W., A. Munoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo, Jr. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 30.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 31.Meyerhans, A., T. Breinig, J. Vartanian, and S. Wain-Hobson. 2004. Forms and function of intracellular HIV DNA, p. 14-21. In T. Leitner, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. W. Mellors, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 2003. Theoretical Biology and Biophysics Group, Los Alamos, NM.
- 32.Norrgren, H., S. Marquina, T. Leitner, P. Aaby, M. Melbye, A. G. Poulsen, O. Larsen, F. Dias, D. Escanilla, S. Andersson, J. Albert, and A. Naucler. 1997. HIV-2 genetic variation and DNA load in asymptomatic carriers and AIDS cases in Guinea-Bissau. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 16:31-38. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien, T. R., W. A. Blattner, D. Waters, E. Eyster, M. W. Hilgartner, A. R. Cohen, N. Luban, A. Hatzakis, L. M. Aledort, P. S. Rosenberg, W. J. Miley, B. L. Kroner, and J. J. Goedert. 1996. Serum HIV-1 RNA levels and time to development of AIDS in the Multicenter Hemophilia Cohort Study. JAMA 276:105-110. [PubMed] [Google Scholar]
- 34.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persaud, D., Y. Zhou, J. M. Siliciano, and R. F. Siliciano. 2003. Latency in human immunodeficiency virus type 1 infection: no easy answers. J. Virol. 77:1659-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popper, S. J., A. D. Sarr, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 2000. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: low virus production in vivo. J. Virol. 74:1554-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popper, S. J., A. D. Sarr, K. U. Travers, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 180:1116-1121. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigo, A. G., P. C. Goracke, K. Rowhanian, and J. I. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retrovir. 13:737-742. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez, S., A. D. Sarr, A. MacNeil, S. Thakore-Meloni, A. Gueye-Ndiaye, I. Traoré, M. C. Dia, S. Mboup, and P. J. Kanki. 2007. Comparison of heterologous neutralizing antibody responses of human immunodeficiency virus type 1 (HIV-1)- and HIV-2-infected Senegalese patients: distinct patterns of breadth and magnitude distinguish HIV-1 and HIV-2 infections. J. Virol. 81:5331-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindler, M., J. Munch, O. Kutsch, H. Li, M. L. Santiago, F. Bibollet-Ruche, M. C. Muller-Trutwin, F. J. Novembre, M. Peeters, V. Courgnaud, E. Bailes, P. Roques, D. L. Sodora, G. Silvestri, P. M. Sharp, B. H. Hahn, and F. Kirchhoff. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055-1067. [DOI] [PubMed] [Google Scholar]
- 41.Schramm, B., M. L. Penn, E. H. Palacios, R. M. Grant, F. Kirchhoff, and M. A. Goldsmith. 2000. Cytopathicity of human immunodeficiency virus type 2 (HIV-2) in human lymphoid tissue is coreceptor dependent and comparable to that of HIV-1. J. Virol. 74:9594-9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, F., S. Matheron, C. Tamalet, I. Loussert-Ajaka, S. Bartczak, J. M. Pepin, C. Dhiver, E. Gamba, C. Elbim, J. A. Gastaut, et al. 1993. Cellular and plasma viral load in patients infected with HIV-2. AIDS 7:1411-1417. [DOI] [PubMed] [Google Scholar]
- 43.Whittle, H., J. Morris, J. Todd, T. Corrah, S. Sabally, J. Bangali, P. T. Ngom, M. Rolfe, and A. Wilkins. 1994. HIV-2-infected patients survive longer than HIV-1-infected patients. AIDS 8:1617-1620. [DOI] [PubMed] [Google Scholar]
- 44.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]