Abstract
Previously, we reported that treatment of cells with sphingomyelinase inhibits human immunodeficiency virus type 1 (HIV-1) entry. Here, we determined by measuring fluorescence recovery after photobleaching that the lateral diffusion of CD4 decreased 4-fold following sphingomyelinase treatment, while the effective diffusion rate of CCR5 remained unchanged. Notably, sphingomyelinase treatment of cells did not influence gp120 binding, HIV-1 attachment, or fluid-phase and receptor-mediated endocytosis. Furthermore, sphingomyelinase treatment did not affect the membrane disposition of the HIV receptor proteins CD4, CXCR4, and CCR5, as determined by Triton X-100 extraction. Restriction of CD4 diffusion by antibody cross-linking also inhibited HIV infection. We therefore interpret the decrease in CD4 lateral mobility following sphingomyelinase treatment in terms of clustering of CD4 molecules. Examination of fusion intermediates indicated that sphingomyelinase treatment inhibited HIV at a step in the fusion process after CD4 engagement. Maximal inhibition of fusion was observed following short coculture times and with target cells that express low levels of CD4. As HIV entry into cells requires the sequential engagement of viral envelope protein with CD4 and coreceptor, we propose that sphingomyelinase inhibits HIV infection by inducing CD4 clustering that prevents coreceptor engagement and HIV fusion.
Human immunodeficiency virus (HIV) fusion is initiated following the engagement of CD4 by gp120, the receptor binding subunit of the HIV envelope protein (Env) (25). This interaction triggers conformational changes in the Env, allowing for the engagement of the second HIV receptor, generally either CXCR4 or CCR5 (1, 5). Coreceptor engagement is preceded by a lag time of several minutes following gp120-CD4 binding (10). This allows for the spatial recruitment of coreceptor molecules (32), generating close proximity to each other and to the CD4-Env complex. A trimolecular complex of Env-CD4-coreceptor then forms, eliciting additional conformational changes in the Env. This triggers the refolding of gp41, the fusogenic moiety, into a six-helix bundle and the merging of viral and cellular membranes (reviewed in reference 9).
The lipid content of the cell membrane is composed primarily of glycerophospholipids, sphingolipids, and cholesterol. Sphingolipids and cholesterol segregate from glycerophospholipids, creating a more ordered phase in the cell membrane termed “rafts” (reviewed in reference 41). The lipid composition of the target cell plays an important role in the HIV fusion process (35, 39). Receptor recruitment, a prerequisite for fusion, is sensitive to lipid modulation (32). Cholesterol depletion inhibits the ability of gp120 to induce the colocalization of CD4 and the coreceptor (26). In primary cells where receptor molecules are expressed in low numbers, cholesterol depletion inhibits fusion and infection. However, overexpression of Env and the receptors in many model fusion systems obscures this requirement of receptor recruitment (44). It has been demonstrated that cholesterol depletion inhibits receptor recruitment by decreasing the diffusion rate of CCR5, implicating receptor restriction as one possible mechanism by which modulation of cellular lipids can inhibit HIV fusion (42).
The application of sphingomyelinase (Smase) to cells alters the lipid content of the plasma membrane by generating ceramide upon cleaving sphingomyelin. Ceramide is extremely hydrophobic and, upon formation, promotes the coalescence of membrane domains into what have been termed “membrane platforms” (16). This property of ceramide to facilitate large domain formations has been exploited by a variety of microbes to facilitate entry and infection. Neisseria gonorrhoeae infection is entirely dependent on the activity of an acid Smase at the cell surface, which triggers phagocytosis of the bacterium into mucosal epithelial cells (13). Likewise, ceramide formation due to Smase activity induces the formation of large raft signaling platforms that have been implicated in facilitating the internalization of Pseudomonas aeruginosa (14), Sindbis virus (20), Plasmodium falciparum (17), and rhinovirus (15).
We showed previously that Smase activity can have adverse effects on HIV infection (6). In the current work, we probe the mechanistic details of how ceramide modulation inhibits HIV fusion. Here, we demonstrate that Smase activity significantly restricts the lateral diffusion of CD4, while coreceptor diffusion is unaltered. We show that restricting CD4 diffusion by antibody cross-linking inhibits HIV infection. Smase inhibits HIV infection at a late step in the fusion process, prior to coreceptor engagement. We demonstrate that Smase-mediated inhibition of HIV fusion is overcome when CD4 is expressed at high levels, if fusion occurs over a sufficiently long period. Collectively, these results indicate that Smase inhibits the HIV fusion process by restricting the lateral mobility of CD4, which may be a result of clustering of CD4 molecules. As HIV entry is a highly orchestrated event requiring the sequential interaction of CD4 and the coreceptor, receptor clustering would be expected to have severe consequences for viral fusion.
MATERIALS AND METHODS
Cells and reagents.
The TZM-bl indicator cell line, obtained through the AIDS Research and Reference Reagent Program, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc., is a HeLa cell line derivative that expresses high levels of CD4 and CCR5 along with endogenously expressed CXCR4. TZM-bl cells contain HIV long terminal repeat-driven β-galactosidase and luciferase reporter cassettes that are activated by HIV TAT expression. HeLa cells expressing different levels of CD4 and coreceptor were gifts from David Kabat (Oregon Health Sciences University). Smase derived from Bacillus cereus, streptavidin-horseradish peroxidase (HRP), and HRP were obtained from Sigma (St. Louis, MO). Phycoerythrin (PE)-conjugated anti-CD4, -CXCR4, or -CCR5 monoclonal antibodies (MAbs) were from BD-Pharmingen (San Jose, CA). Gp120-biotin (Bt) was obtained from Intracell (Issaquah, WA), and biotinylated transferrin, anti-transferrin receptor, Alexa-conjugated secondary antibodies, and CellTracker reagents were obtained from Molecular Probes (Eugene, OR).
Analysis of lipid composition.
The sphingolipid analyses were conducted by liquid chromatography and electrospray tandem mass spectrometry using a PE Sciex API 3000 triple quadrupole mass spectrometer equipped with a turbo ion-spray source, as described previously (30). Internal standards for the mass spectrometric analyses were obtained from Avanti Polar Lipids (Alabaster, AL); these were C12-ceramide (N-dodecanoyl-sphingosine, d18:1/12:0), C12-glucosylceramide, and C12-sphingomyelin.
Infectivity assay.
TZM-bl cells (2 × 104 per well) were added to 96-well microtiter plate wells (Falcon, Lincoln Park, NJ) in 100 μl of complete medium (Gibco) and allowed to adhere for 15 to 18 h at 37°C. An equivalent amount of virus stock (NL4-3; multiplicity of infection [MOI] of 0.01) was added to the cell monolayers in the presence of 40 μg/ml DEAE-dextran in Dulbecco's modified Eagle's medium (DMEM) in a final volume of 100 μl. Viral infection was allowed to proceed for 2 h at 37°C, following which, 100 μl of complete DMEM was added. Luciferase activity was measured after 15 to 18 h at 37°C with 5% CO2 in a humidified incubator using a Promega (Madison, WI) luciferase assay system kit. All infectivity assays were performed in triplicate.
Biochemical measurement of fluid-phase uptake.
Fluid-phase endocytosis was quantitated as described previously (27). In brief, cells were plated on six-well plates 24 h prior to the assay. The cells were washed once with DMEM and incubated at 37°C with 0.5 ml prewarmed medium containing 10 μg/ml HRP (Type 2; Sigma Chemical Co., St. Louis, MO). Following incubation for the specified time, the dishes were placed on ice and washed 10 times with ice-cold phosphate-buffered saline (PBS). Subsequently, the cells were trypsinized, washed an additional two times, and finally resuspended in 0.5 ml of 0.1% Triton X-100. Total cell protein was calculated using bicinchoninic acid (BCA; Pierce Chemical Co.), and all samples were normalized for protein content. HRP activity was determined using 3-3′,5,5′-tetramethylbenzidine, a soluble chromogen substrate for HRP.
Cell fractionation assays.
Subconfluent TZM cells were incubated with Bt-conjugated transferrin (transferrin-Bt) or Bt-conjugated gp120 (gp120-Bt) for 3 h at 37°C. To remove surface-bound protein, the cells were washed three times with ice-cold PBS and trypsinized for 5 min at room temperature. Trypsin activity was quenched by the addition of serum-containing medium, and the cells were then washed twice in ice-cold PBS. The cells were resuspended in 2 ml of hypotonic solution (20 mM Tris-HCl [pH 8], 10 mM KCl, 1 mM EDTA) for 15 min at 4°C and disrupted by Dounce homogenization (15 strokes, 7-ml B pestles). The nuclei and cell debris were pelleted by centrifugation (3,000 rpm for 5 min at 4°C). The postnuclear extracts were centrifuged at 22,000 rpm for 30 min at 4°C. The pellet representing the vesicular fraction, including endosomes, was resuspended in lysis buffer (0.5% Triton X-100). Total cell protein was calculated using BCA, and all samples were normalized for protein content. Transferrin and gp120 content were then quantitated by immunoblot analysis.
Preparation of detergent-resistant membrane fractions.
Plasma membrane rafts were obtained as previously described (44). Briefly, TZM cells were treated at 37°C with 50 mU/ml of Smase for 10 min at 37°C. The enzyme was removed, and the cells were washed with ice-cold PBS, followed by lysis at 4°C in a buffer containing 1.4 ml of 25 mM morpholineethanesulfonic acid (MES) (pH 6.5), 0.15 M NaCl, 1% (vol/vol) Triton X-100, protease inhibitors (0.1 μg of phenylmethylsulfonyl fluoride/ml, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 μg of pepstatin A/ml), and 1 mM sodium orthovanadate. The lysate was then homogenized and adjusted to 40% sucrose. A 5 to 30% linear sucrose gradient was formed above the homogenate. The centrifugation was carried out at 45,000 rpm for 16 to 20 h in an SW60 rotor (Beckman Instruments, Palo Alto, CA). From the top of each gradient, 0.35-ml fractions were collected to yield a total of 12 fractions. Proteins were quantitated using BCA (Pierce Biotechnology), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subjected to immunoblot analysis. Fractions 1 and 2 were generally lacking proteins and thus were not subjected to SDS-PAGE. Fraction 12 represented the nuclear portion. Prototypical raft proteins localize on the top of the gradient in fractions 4 to 7; nonraft marker proteins localize predominantly in fractions 9 to 11.
Immunoblot analysis.
Cellular proteins were resolved by SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes. The blots were incubated for 1 h in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.2% Tween 20) containing 5% powdered skim milk. After four washes with TBST, the membranes were incubated for 1 h with the primary antibody (streptavidin-HRP diluted 1:3,000) in TBST with 3% bovine serum albumin. Immunoreactivity was detected by using an enhanced chemiluminescence detection kit (Amersham, Piscataway, NJ).
Analysis of receptor expression.
Cells were harvested with cell dissociation buffer (Gibco) and resuspended at 0.5 × 106 cells/ml in PBS. Following 20 min of incubation in PBS with 5% fetal bovine serum and 5% normal mouse serum, PE-conjugated mouse immunoglobulin G (IgG) anti-CD4 (RPA-T4), anti-CXCR4 (12G5), anti-CCR5 (2D7), or anti-transferrin receptor MAbs were added at 5 μg/ml. GM1 expression was assayed by incubation with fluorescein isothiocyanate-conjugated cholera toxin B at 5 μg/ml. Following 1 h of incubation at 4°C, the cells were washed three times with ice-cold PBS and samples were split into duplicates. In experiments determining Triton X-100 extraction of proteins, 300 μl of ice-cold Triton X-100 (1%, vol/vol) in PBS was added to one set of samples for 30 min on ice. Control samples received PBS alone. The cells were subsequently pelleted to remove the Triton X-100 and washed twice with PBS on ice. The samples were maintained on ice and analyzed with a FACSCalibur (Becton Dickinson, San Jose, CA) at 10,000 events/sample with respect to unlabeled cells.
gp120 binding assay.
Cells were harvested with cell dissociation buffer and resuspended at 106 cells/ml in PBS. The cells were treated for 10 min at 37°C with 50 mU/ml of Smase or control buffer and then incubated with gp120 (5 μg/ml) for 2 h at 4°C. Cells were washed and directly fixed with 2% paraformaldehyde. The cells were stained with the gp120-specific MAb 2G12 (5 μg/ml). Following staining, samples were resuspended in 1 ml of PBS and read by a FACSCalibur (Becton Dickinson, San Jose, CA) at 10,000 events/sample with respect to unlabeled cells.
Viral binding assays.
The cells were washed twice with PBS and resuspended at 3 × 106 cells in 2 ml of complete medium. Purified HIV type 1 (HIV-1) (40 ng of p24 ADA isolates) was added to the cells for 3 h at 4°C. Following this incubation, the cells were spun down and unbound virus was aspirated. The cells were washed five times with cold PBS to remove unbound material. Cell lysates were prepared by resuspending the pellet in 300 μl of lysis buffer (1% Triton X-100) and freezing at −20°C. Viral attachment was monitored by measuring the amount of p24 in the supernatant of cell lysates by enzyme-linked immunosorbent assay (ELISA). Total cell protein was calculated using BCA, and all samples were normalized for protein content. The average was taken from the results of three separate experiments that were normalized to 100% relative to the results for the untreated control sample.
FRAP acquisition and analysis.
Fluorescence recovery after photobleaching (FRAP) analysis was performed using a Zeiss LSM 510 (Carl Zeiss, Jena, Germany) confocal laser scanning microscope. HeLa cells were plated on 35-mm glass-bottom dishes (MatTek, Ashland, MA) and transfected 24 h prior to confocal analysis with either CD4-green fluorescent protein (GFP) or CCR5-N-terminally tagged GFP (these constructs were generous gifts from W. Popik and S. Manes, respectively, and have been described previously) (12, 38). Most of the protein was expressed on the surface, although some intracellular accumulation was observed in some cells. During FRAP analysis, cells were kept at physiological conditions of 37°C and 5% CO2 in a stage incubation system (Incubator S; PeCon GmbH, Erbach, Germany). A 488-nm Ar+ laser line was used for GFP excitation, and emission light was collected with a 500- to 550-band-pass filter. A 40×/1.3 numerical-aperture oil-immersion objective lens was used with a zoom factor of 4. The detector pinhole was opened slightly more than 1 Airy unit in order to acquire an optical section of 2-μm thickness. This allowed more light to be collected, for better quantification. Three prebleach images were acquired, to determine the rate of nonpurposeful photobleaching, using a nominal laser intensity of 1%. Then photobleaching was performed in a 2-μm, visually uniform region of the cell membrane. Bleaching was performed using 100% laser intensity for a duration of approximately 1 s (20 to 50 scans of the laser) to achieve close to 100% bleaching of the GFP. After photobleaching, images were acquired at 1 s apart for 8 to 10 images and then the time resolution was changed to 10 s to follow the recovery to completion. A total of 20 to 40 data points per image were acquired for image analysis.
FRAP analysis was performed using the Medical Imaging Processing, Analysis, and Visualization (MIPAV; CIT/NIH, Bethesda, MD) (28) software package. The images were preprocessed by correction for background by subtraction and by normalizing to the total fluorescence from the entire cell membrane in order to correct for the (nonpurposeful) photobleaching that occurred during the acquisition of the recovery images. Following preprocessing, the total intensities in the bleached region after photobleaching, I(t), were calculated and fitted to the one-dimensional diffusion FRAP model (26), I(t) = Ifinal{1 − [w2(w2 + 4πDt)−1]1/2}, using Gauss-Newton nonlinear fitting, where Ifinal is the predicted intensity after infinite time, w is the width of the bleached region along the membrane in μm, and D is the diffusion coefficient. The mobile fraction was calculated as: [Ibefore − I(0)]/[Ifinal − I(0)], where Ibefore is the total intensity of the bleached region before photobleaching commenced and I(0) is the intensity immediately following photobleaching.
Infectivity assay.
The infectivity assay was performed as previously described (6). To create temperature-arrested intermediate (TAI) states, virus and cells were cocultured at either 23°C or 4°C for 2 h. Virus was then removed and the cells (with attached virus) were treated with Smase for 10 min at 37°C. Following removal of Smase, the cells were incubated with complete medium and infection was allowed to proceed for 15 to 18 h at 37°C. In control experiments, target cells were treated with Smase for 10 min, the enzyme was removed, virus was then added at 37°C, and infection was allowed to proceed for 15 to 18 h at 37°C.
MAb OKT4 cross-linking and infectivity experiments.
TZM-bl cells (2 × 104 per well) were added to 96-well microtiter plate wells (Falcon, Lincoln Park, NJ) in 100 μl of complete media and allowed to adhere for 15 to 18 h at 37°C. Each well was treated with 100 μl of MAb OKT4 (10 μg/ml) or mouse IgG (10 μg/ml) in combination with goat anti-mouse IgG (40 μg/ml) for 1 h at 4°C in serum-free DMEM. The antibody solutions were then removed, and virus (NL4-3; MOI of 0.01) was added to the cell monolayers in the presence of 40 μg/ml DEAE-dextran in DMEM in a final volume of 100 μl. Viral infection was allowed to proceed as described above.
HIV-1 Env glycoprotein-mediated cell-cell fusion.
Target cells were labeled with the cytoplasmic dye CellTracker orange (CMRA) at 10 μM and Env-expressing cells were labeled with 10 μM CellTracker green as described in the manufacturer's protocol. The labeled target cells were treated with Smase for 10 min and then cocultured with the labeled Env cells for 2 h at 37°C. Dye redistribution was monitored microscopically as described previously (10). The extent of fusion was calculated as % fusion = 100 × (number of bound cells positive for both dyes/number of bound cells positive for CMRA).
RESULTS
Smase treatment increases ceramide levels in HeLa cells.
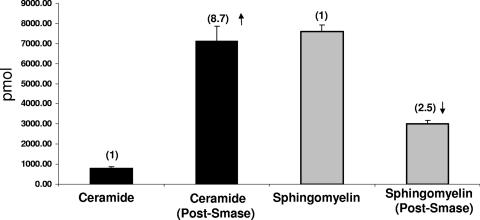
Smase cleaves sphingomyelin at the outer leaflet of the plasma membrane, generating phosphocholine and ceramide. Initially, we determined the effectiveness of this treatment in our assay system following incubation at 37°C for 10 min. Previously, we established that this brief enzymatic treatment inhibits HIV infection (6). As shown in Fig. 1, following lipid extraction and analysis, Smase treatment decreased sphingomyelin from 7,605 to 3,019 pmol and increased ceramide more than eightfold (from 818 to 7,136 pmol). Thus, the increase in ceramide apparently reflects cleavage of sphingomyelin, confirming the activity of this enzyme in our assay system. This suggests that the changes in membrane properties are due to the decrease in sphingomyelin and the increase in ceramide.
FIG. 1.
Smase treatment increases ceramide levels while decreasing sphingomyelin. Smase was added to cells for 10 min at 37°C. Cells were pelleted and frozen, and lipid analysis was performed by liquid chromatography electrospray tandem mass spectrometry as described by Merrill et al. (30). The amounts of the individual subspecies (e.g., d18:1/16:0 ceramide, d18:1/18:0 ceramide, etc.) were summed to give the total amounts shown in the figure. The error bars indicate the standard deviations of three samples.
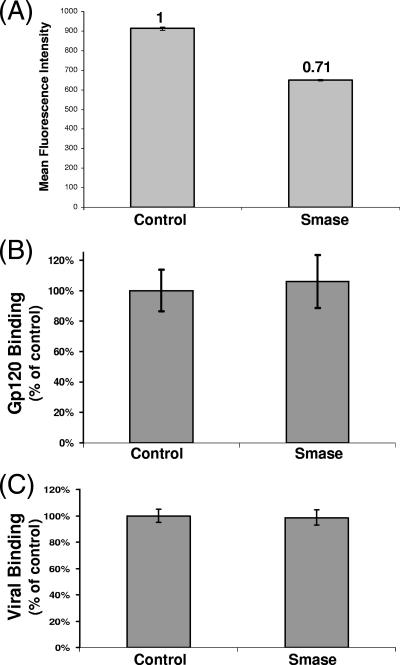
Smase treatment does not influence gp120 or viral binding.
Smase activity generates a large increase in ceramide in the extracellular leaflet of the plasma membrane, possibly affecting HIV-1 receptor expression. To investigate this, we analyzed the expression of the HIV receptors CD4, CXCR4, and CCR5 by flow cytometry following Smase treatment. We observed no change in the expression levels of CCR5 or CXCR4 (data not shown), while CD4 expression was decreased. As shown in Fig. 2A, enzymatic treatment decreases CD4 expression levels approximately 30%. To determine if Smase treatment influences gp120 or viral binding, we incubated treated cells with either the viral Env gp120 or chilled HIV-1 at 4°C. Gp120 engagement was assayed using flow cytometry, by employing the gp120-specific MAb 2G12, while viral binding was determined by a p24 ELISA. As seen in Fig. 2B, Smase activity does not affect gp120 binding to target cells. Quantitative analysis of viral binding also revealed similar values for the HIV antigen p24 (Fig. 2C), indicating that Smase has little effect on this initial step in the fusion cascade.
FIG. 2.
Smase decreases CD4 expression but does not affect gp120 or viral binding. Cells were treated with Smase for 10 min at 37°C. (A) The expression of CD4 was analyzed by the addition of anti-CD4 PE-conjugated antibody in parallel with a matched isotype control. Antibody binding was quantitated using flow cytometry. The reactivity was compared to the reactivity of the control sample, and the mean fluorescence intensity was quantitated. (B) Cells were incubated with gp120 for 2 h at 4°C. Gp120 binding was determined using the anti-gp120-specific MAb 2G12, and the reactivity was detected by flow cytometry. (C) Cells were incubated with prechilled virions for 2 h at 4°C, washed extensively, and lysed. Viral binding was quantitated using a p24 ELISA. The results represent the averages of three independent experiments; the error bars indicate the standard deviations between experiments.
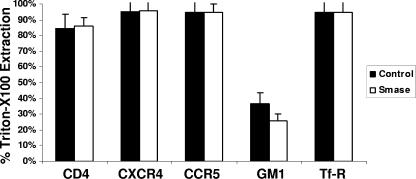
Smase treatment does not influence Triton X-100 extraction of HIV-1 receptor proteins.
Next, we investigated the membrane disposition of the HIV receptors by determining Triton X-100 extraction at 4°C. We compared the extractabilities of CD4, CXCR4, and CCR5 with those of GM1, a characterized raft marker, and transferrin receptor, a well-characterized nonraft marker protein. As seen in Fig. 3, following Triton X-100 treatment, transferrin receptor is predominantly extracted, as expected for this protein. In contrast, GM1, a raft-localized ganglioside, remained predominantly cell associated. In agreement with previous reports, the coreceptors CXCR4 and CCR5 are almost entirely extracted under our assay conditions; notably, the percent extraction remained unchanged following Smase treatment. CD4, which is generally accepted to be a raft-localized protein, is also largely extracted. This result is consistent with results found by others for nonlymphoid cell lines (7). Importantly, there is no difference in the extraction of CD4 following Smase treatment. This result was confirmed by isolating detergent-resistant membrane fractions from sucrose gradients and quantitating CD4 content by Western blotting (data not shown).
FIG. 3.
Smase has minimal effects on detergent extraction of CD4, CXCR4, or CCR5. Cells were treated with Smase for 10 min at 37°C. CD4, CXCR4, CCR5, and transferrin receptor were detected by the addition of the respective PE-conjugated antibodies, while GM1 was detected by the addition of fluorescein isothiocyanate-conjugated cholera toxin B subunit. Following antibody or cholera toxin labeling, the cells were subjected to Triton X-100 extraction at 4°C as described in Materials and Methods. Antibody binding was quantitated using flow cytometry. The reactivity was compared to the reactivity of control samples, and the percent extraction was calculated. The results shown are the averages of three independent experiments. The data are the means ± standard deviations of three independent experiments.
We also investigated the distribution of these receptors by confocal microscopy. However, on our reporter cell line, the HIV receptors are overexpressed, resulting in a continuous plasma membrane staining pattern. The limits of resolution and sensitivity of confocal microscopy cannot resolve any subtle changes in such a staining pattern. Following Smase treatment, no alteration in staining was observed, indicating that no gross membrane effects such as capping are being induced (data not shown).
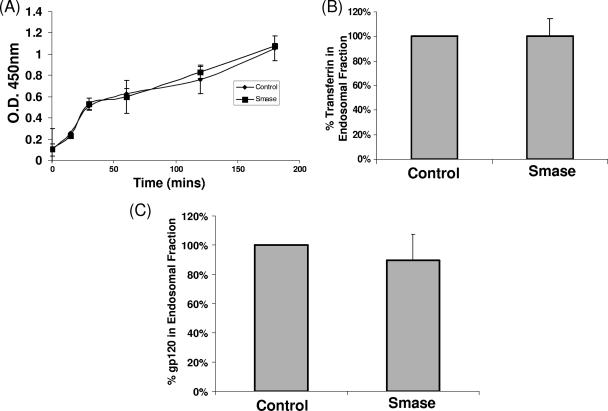
Smase does not affect cellular endocytic pathways.
Smase activity has been shown to influence cellular endocytic pathways (3, 23, 45). Given that HIV infectivity is influenced by modulating cellular endocytic pathways (4, 40), we considered this a possible mechanism resulting in HIV inhibition. Initially, we measured the rate of uptake of a soluble protein, HRP, which enters cells through nonabsorptive endocytosis. As observed in Fig. 4A, Smase activity has no effect on HRP uptake over a 3-h time frame. Notably, it has been shown that Smase activity promotes a rapid stimulation of endocytosis (45). However, even when HRP activity was assayed over the initial 30 min of uptake, no difference was observed.
FIG. 4.
Smase does not affect cellular endocytic pathways. (A) Confluent dishes of TZM-bl cells were incubated in medium containing HRP (10 μg/ml) following control or Smase pretreatment. Internalized HRP was assayed as described in Materials and Methods. The results represent the averages of three independent experiments. O. D., optical density. (B and C) Cells were incubated with transferrin-Bt (B), or gp120-Bt (C) for 3 h at 37°C. The endosomal fraction was isolated as described in Materials and Methods, and the Bt-labeled content was quantitated. The results were normalized to the results for control samples and represent the averages of three independent experiments; the error bars indicate the standard deviations between experiments.
We next investigated the effect of Smase on receptor-mediated endocytosis, as HIV engages specific receptors on the cell surface. We employed transferrin, the soluble ligand for the transferrin receptor, as a classical marker protein for this pathway. Endocytic uptake was analyzed following the incubation of cells with medium containing transferrin-Bt after Smase or control treatment. Following 3 h at 37°C, the cells were trypsinized to remove cell surface ligand and subjected to subcellular fractionation. The vesicular endosome fraction was isolated from the cellular lysate and analyzed for transferrin content by Western blotting. Densitometric analysis of such blots provided the data shown in Fig. 4B. As shown, when internalized transferrin was quantitated, Smase pretreatment appeared to have a minimal effect on endocytic accumulation. Thus, it appears that receptor-mediated endocytosis is unaffected by Smase activity.
Given that productive viral entry is mediated by specific Env-receptor interactions, we also determined if the viral Env gp120 undergoes increased endocytosis following Smase treatment. To test this, we quantitated endocytic uptake of gp120-Bt as described previously for transferrin. As shown in Fig. 4C, the gp120 content of the endocytic fraction is unaltered by Smase treatment. We also performed kinetic analysis of gp120 uptake over a 30-min to 3-h time course, and no difference in the rate of gp120 endocytosis was observed at any time point (data not shown).
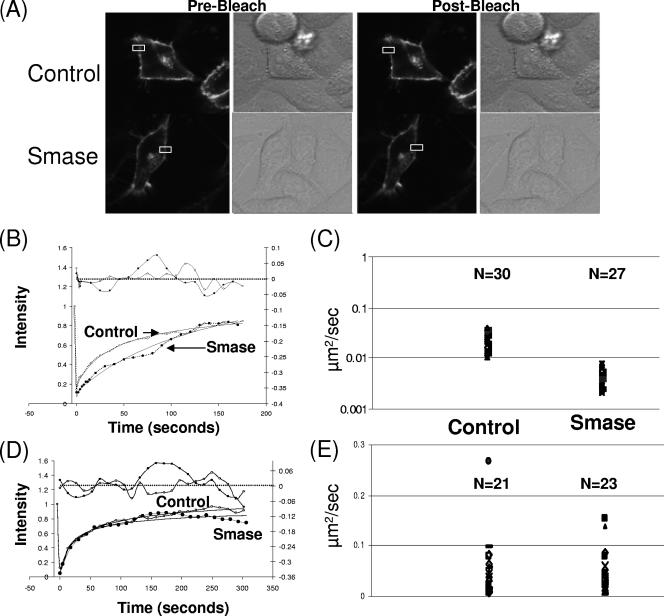
Smase restricts CD4 mobility.
The lateral diffusion of HIV receptors is an important requirement in the fusion process (9). Hence, we investigated the effect of Smase on the effective diffusion rates of CD4 and CCR5. FRAP is one technology available to measure the lateral mobility of proteins in membranes. This approach utilizes the confocal laser to focus on a small region of the cell until the fluorescent signal is irreversibly bleached. Following bleaching, the recovery of the fluorescent signal is monitored over time. The rate of this recovery is used to determine the effective diffusion coefficient for the protein of interest in the cell. To investigate receptor mobility, we employed GFP-tagged CD4 and CCR5 constructs which result in a bright fluorescent membrane staining pattern when expressed in target cells (Fig. 5A). Following Smase treatment, we bleached these samples and monitored the recovery of the fluorescent signal. Quantitative analysis of the recovery rate allows for calculation of the effective diffusion coefficient for both of these proteins. Experiments to calculate the mobility of CD4 were performed on the same day under the same FRAP conditions. Three separate experiments with paired samples were performed. A total of 30 control and 27 Smase-treated cells were analyzed, and the average effective diffusion coefficient for CD4 was calculated. A representative image and a recovery curve are shown in Fig. 5A and B for both control and Smase-treated cells. The average effective diffusion coefficient is shown in Fig. 5C. Notably, there was an approximately fourfold change in the effective diffusion rate of CD4 following Smase treatment. The effective diffusion rate decreased from 0.022 μm2/s to 0.005 μm2/s following enzyme treatment. We also determined the mobile fraction of CD4 by calculating the overall extent of signal recovery. By observing the recovery curve of the fluorescent signal, we can determine if the recovery plateaus before 100% of the signal is achieved, indicating a subset of CD4 molecules that are immobile in the membrane (24). In both the presence and absence of Smase treatment, almost 100% recovery was observed (control treatment, 96%; Smase treatment, 98.6%). This indicates that most of the CD4 molecules under these FRAP conditions were mobile in the membrane.
FIG. 5.
Smase restricts CD4 mobility, while CCR5 mobility is unaffected. FRAP analysis was conducted as described in Materials and Methods. Cells were transfected to express CD4-GFP (A, B, and C) or CCR5-GFP (D and E) and treated with control or Smase for 10 min. FRAP was performed within 1 h of treatment using a Zeiss LSM 510 (Carl Zeiss, Jena, Germany) confocal laser scanning microscope. (A) A representative control and a Smase-treated CD4-expressing cell are shown; the bleach box is indicated in white. (B) A representative recovery curve (broken line) and a fit (solid line) for CD4-GFP diffusion following control (square) or Smase (star) treatment are shown. A residual plot is shown at the top of the graph. (C) The average effective diffusion coefficient for CD4 following control treatment is 0.022 μm2/s with a standard deviation of 0.008 μm2/s; following Smase treatment, the average effective diffusion coefficient is 0.005 μm2/s with a standard deviation of 0.002 μm2/s. The number of cells analyzed for each condition is indicated. (D) A representative recovery curve (broken line) and fit (solid line) are shown for CCR5-GFP diffusion following control or Smase treatment. A residual plot is shown at the top of the graph. (E) The average effective diffusion coefficient for CCR5 following control treatment is 0.049 μm2/s with a standard deviation of 0.057 μm2/s; following Smase treatment, the average effective diffusion coefficient is 0.050 μm2/s with a standard deviation of 0.042 μm2/s.
CCR5 mobility is unaffected by Smase.
Using FRAP analysis, we also determined the effective diffusion rate for CCR5. Experiments were performed on the same day under the same FRAP conditions. Three separate experiments with paired enzyme and control treatments were performed. A total of 21 control and 23 Smase cells were analyzed, and the average effective diffusion coefficient for CCR5 was calculated. A representative recovery curve is shown in Fig. 5D for both control and Smase-treated cells. As shown in Fig. 5E, there was no change in the effective diffusion rate of CCR5 following Smase treatment. Under these conditions, we calculate the effective diffusion rate of CCR5 to be approximately 0.05 μm2/s. We calculate the mobile fraction in control cells to be 99%, while Smase treatment decreased this fraction slightly to 94%.
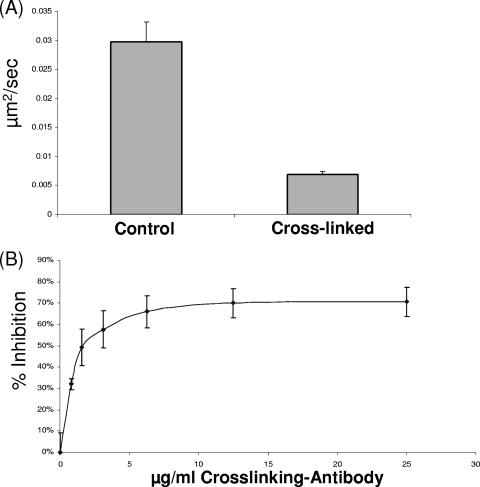
Cross-linking CD4 reduces the effective diffusion rate and inhibits HIV infection.
To further investigate the influence of CD4 diffusion on HIV infection, we cross-linked CD4 using the nonneutralizing anti-CD4 MAb OKT4 in combination with an anti-mouse IgG antibody. MAb OKT4 binds an epitope on CD4 that is not involved in the HIV fusion process and has no inhibitory effects on gp120 engagement (2). We confirmed that cross-linking CD4 with MAb OKT4 does not downregulate CD4 or alter HIV binding under our assay conditions (data not shown). By employing FRAP, we confirmed that we were effectively cross-linking CD4, with a fourfold decrease in the effective diffusion rate of CD4 observed following cross-linking (Fig. 6A). No such effect was detected on cells treated with an isotype control antibody in combination with an anti-mouse IgG antibody (data not shown). We next investigated the effects of cross-linking CD4 on HIV infection by pretreating cells with increasing concentrations of MAb OKT4 in combination with an anti-mouse IgG antibody. Following 1 h of pretreatment at 4°C, the antibody cocktail was removed, virus was added, and infection proceeded at 37°C. Control experiments with an isotype antibody were carried out in parallel. As shown in Fig. 6B, MAb OKT4 cross-linking of CD4 effectively inhibits HIV infection. Dose-dependent inhibition of infection was observed, while control antibody pretreatment resulted in minimal inhibition (data not shown).
FIG. 6.
Antibody cross-linking of CD4 restricts mobility and inhibits HIV infection. (A) Cells were treated with MAb OKT4 (10 μg/ml) or control antibody solution for 1 h at 4°C. The antibody solution was removed and FRAP analysis was conducted as described in Materials and Methods. The average effective diffusion rate for CD4 is 0.027 μm2/s with a standard deviation of 0.009 μm2/s. Following cross-linking, the average effective diffusion rate is 0.007 μm2/s with a standard deviation of 0.002 μm2/s. (B) Cells were incubated with the indicated concentration of MAb OKT4 or control IgG for 1 h at 4°C. The antibody solution was removed, virus (isolate NL4-3; MOI = 0.01) was added, and infection was allowed to proceed overnight. The inhibition was calculated relative to the inhibition of control samples with an isotyped antibody, and the average of three independent experiments is shown. The error bars indicate the standard deviations.
Smase inhibits viral fusion after CD4 engagement.
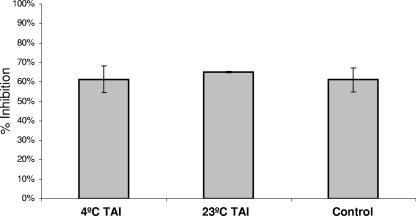
As Smase restricts CD4 diffusion (Fig. 5C), and we determined from cross-linking experiments that CD4 mobility is necessary for HIV infection, we next determined which step in the fusion cascade is targeted by Smase. To do this, we used the approach of creating temperature-arrested fusion intermediates which have well-defined characteristics (10, 29, 31). Previous studies have determined that virus-cell fusion intermediates created by incubation at 4°C are arrested at an early step in the fusion cascade that allows attachment but does not permit further progress. In contrast, intermediates arrested by incubation at 23°C have engaged CD4 and are in close proximity to the coreceptor. Shifting of 23°C-arrested intermediates to 37°C results in rapid fusion without the characteristic lag time that is seen in the fusion of 4°C-arrested intermediates or in the fusion of non-temperature-arrested cocultures (18). By employing specific inhibitors that target different steps in the fusion process, it has been determined that the 23°C-arrested intermediate is arrested at a late step in the cascade, immediately prior to coreceptor engagement and formation of the fusion pore. We created 4°C TAIs and 23°C TAIs by coculturing virus and cells at these temperatures for 2 h. The unabsorbed virus was then removed, and the coculture systems were treated with Smase. In control experiments, target cells were treated with Smase prior to virus coculture. When we analyzed these coculture systems for Smase-mediated inhibition, we observed that Smase was equally effective in inhibiting infection when added to all TAIs (Fig. 7). Thus, HIV fusion is sensitive to Smase inhibition post-CD4 engagement and most likely is sensitive until a late step in the fusion process prior to coreceptor engagement.
FIG. 7.
Smase inhibits fusion at a late step in the fusion cascade. Cells were incubated with virus for 2 h at either 4°C or 23°C. Unbound virus (NL4-3 isolate) was removed, and Smase was added for 10 min at 37°C. After removal of the enzyme, complete medium was added, and infection was allowed to proceed overnight. In control experiments, target cells were treated with Smase for 10 min, the enzyme was removed, virus was then added at 37°C, and infection was allowed to proceed overnight. The inhibition was calculated relative to the inhibition by the control treatment at the respective temperature, and the average of three independent experiments is shown. The data are the means ± standard deviations of three independent experiments. The relative luciferase units obtained are as follows: for the 4°C TAI, 36,463; for the 23°C TAI, 38,854; and for the control, 36,465.
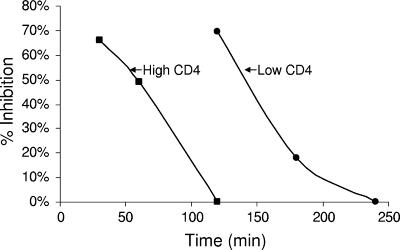
Inhibition of cell-cell fusion by Smase is dependent on CD4 expression levels.
To further examine the hypothesis that Smase inhibits HIV fusion by restricting the lateral diffusion of CD4, we employed a cell-cell fusion system with target cells expressing different CD4 surface densities. These experiments utilize a well-characterized dye transfer system between a stable HeLa cell line expressing IIIb Env and HeLa target cells expressing either low (10 × 103 molecules/cell) or high (150 × 103 molecules/cell) levels of CD4. CXCR4, the coreceptor for the EnvIIIb, is expressed endogenously on HeLa cells. As in the viral fusion assays, we pretreated target cells for 10 min with Smase prior to coculture with Env cells. As shown in Fig. 8, a 10-min treatment of target cells with Smase effectively inhibits cell-cell fusion after a 120-min coculture period, but only with target cells that have a low surface density of CD4. Cells with high CD4 levels demonstrate no inhibition at this time point. When shorter coculture periods are scored for fusion inhibition, high-CD4-expressing target cells do demonstrate susceptibility to Smase-mediated inhibition. As observed in the results shown in Fig. 8, following 30 min of coculture, Smase-treated high-CD4-expressing cells demonstrated 66% fusion inhibition, while after 60 min of coculture, this inhibitory effect decreased to 49%. We reasoned that the lack of inhibition following 2 h of coculture may reflect the high density of CD4 molecules overcoming the mobility restriction imposed by Smase due to closer proximity to the coreceptor. We additionally investigated if cells expressing low CD4 surface densities could overcome Smase-induced fusion inhibition when cultured for extended time periods of 180 to 240 min. This extended coculture time may allow sufficient time for CD4 to engage the coreceptor even when CD4 mobility is restricted due to Smase treatment. As shown in Fig. 8, low-CD4-expressing cells can overcome Smase-mediated inhibition following 180 to 240 min of coculture. At these time points, the inhibitory effects of Smase on fusion decrease from 18% to 0%, respectively. These results indicate that Smase-mediated fusion inhibition is sensitive to CD4 surface densities, highlighting the requirement for receptor mobility in the fusion process.
FIG. 8.
Smase inhibition of cell-cell fusion is dependent on CD4 expression levels and coculture time. HeLa cells expressing HIV-1 IIIb Env, labeled with CellTracker green, were cocultured with HeLa target cells labeled with CellTracker orange. The target cells expressing either low or high CD4 levels were pretreated with Smase for 10 min prior to coculture. Low- and high-CD4-expressing cells were cocultured with Env cells for discrete time periods up to 240 min at 37°C. Images were collected at the indicated time points using a 20× lens objective, and the extent of fusion was scored relative to the extent of fusion of the control cells as described in Materials and Methods. The data are the means of three independent experiments.
DISCUSSION
Ceramide, when generated rapidly upon Smase application, reorganizes the plasma membrane (reviewed in reference 43). In vivo, this plays an important role in many cell signaling events, as the generation of ceramide functions to cluster and activate receptor molecules in rafts (for a review, see reference 16). In our experiments, we determined that Smase, while eliciting the rapid generation of ceramide (Fig. 1), does not alter the membrane raft disposition of CD4 or coreceptors as determined by Triton X-100 extraction. However, we found that the effective diffusion rate for CD4 is reduced fourfold following Smase treatment (Fig. 5C), whereas the CD4 mobile fraction is not reduced. It is important to note that the relationship between CD4 raft localization and susceptibility to HIV-1 infection has been quite controversial. Prior studies have indicated that raft disruption does not affect HIV-1 Env-mediated fusion as long as HIV-1 receptors are in high abundance in the target cells (44). Other studies (37, 38) have also shown that cells expressing CD4 with mutations that placed CD4 in the nonraft domains were perfectly capable of supporting HIV-1 entry. Since these data indicate that HIV-1 fusion/entry is supported mainly by the nonraft (i.e., Triton X-100 extractable) fraction of CD4, we are led to conclude that the Smase effects on CD4 mobility are not raft related. This is consistent with the results of Kenworthy et al. (21), who show that raft association is not the dominant factor in determining long-range protein mobility at the cell surface.
The lateral mobilities of viral or receptor proteins have been correlated with their abilities to mediate fusion (19, 22). However, given the low rate of HIV-1 Env-mediated fusion, a reduction of the CD4 diffusion coefficient to 0.005 μm2/s would still provide these molecules sufficient travel time (a distance of 1 μm in about 1 min) to encounter multiple Env and coreceptor molecules on the time scale of fusion (hours). We therefore propose that Smase treatment causes an appreciable fraction of CD4 to aggregate or cluster and these clusters cannot support fusion. The reduction in the lateral mobility of CD4 is consistent with this hypothesis, since it has recently been demonstrated (11) that the dependence of the lateral diffusion coefficient of a membrane protein (D) on the radius R of the protein fits a Stokes-like expression [D (propto) 1/R]. Therefore, clusters of proteins should diffuse much more slowly than dispersed receptors. Based on the inverse relationship between the diffusion coefficient and the radius, for our data, this translates to an average cluster size of approximately 16 CD4 molecules.
While Smase treatment did not alter gp120 or viral binding, it did lead to a reduction in the fluorescence of a secondary Ab against an anti-CD4 MAb (Fig. 2). Based on the clustering hypothesis, we interpret this reduction as self-quenching due to the clustering of CD4 molecules. Our data on the correlation between CD4 cross-linking by MAb OKT4 that reduces its lateral mobility and inhibition of infection (Fig. 6) also support the clustering hypothesis.
HIV fusion can be dissected into discrete steps based on the use of TAIs and specific inhibitors (10, 29, 31). By coculturing virus and cells at 23°C for 2 h, we created fusion intermediates that have been previously well characterized (18). Such intermediates are insensitive to inhibitors that target the CD4-gp120 interaction, but are sensitive to coreceptor- and gp41-targeted inhibitors. Thus, this stable fusion intermediate most likely reflects an accumulation of CD4-engaged viruses that are primed for coreceptor engagement. We determined that this intermediate was sensitive to Smase inhibition, indicating that Smase is targeting a post-CD4 binding step in the fusion process (Fig. 7). As CD4 molecules are clustered following Smase treatment, the likelihood of coreceptor engagement is reduced, resulting in the inhibition of fusion. However, we find about 30% residual fusion after Smase treatment (Fig. 7), presumably due to remaining unclustered CD4.
Our results using a cell-cell fusion assay with various CD4 levels provide additional evidence that Smase, by inducing CD4 clustering, inhibits HIV fusion. Similar to results that were obtained following the depletion of cholesterol from cells (44), we determined that Smase inhibits cell-cell fusion. This is only observed, however, when CD4 is expressed at low levels or following relatively short coculture times if the expression levels are high. It is important to note that the effects of cholesterol depletion and Smase treatment on HIV-1 receptor disposition and function are quite different: in contrast to the effects of Smase treatment, cholesterol depletion has no effect on CD4 mobility (42). However, cholesterol depletion does significantly affect chemokine receptor function (33, 34). By contrast, Smase does not affect ligand binding to coreceptors and had little effect on ligand-induced chemotaxis (36).
Protection of fusion, after Smase treatment, for high CD4 levels follows naturally if only unclustered CD4 can participate in fusion. The level of unclustered CD4 after Smase treatment would be sufficient for fusion. By contrast, in cells with low CD4 levels, the probability of Env-CD4-coreceptor interactions is less favorable and Smase, by clustering CD4 molecules, reduces the frequency of these interactions to a level where fusion is inhibited (Fig. 8). As an extension of this line of reasoning, in cells with low CD4 levels, if sufficient time is allowed to permit complexes to form between unclustered CD4 and Env, allowing for coreceptor engagement, then the restriction imposed by Smase treatment should be overcome.
Collectively, this study shows that Smase treatment of cells inhibits HIV fusion, most likely by altering the topology of the plasma membrane, which results in the clustering of CD4 molecules. Understanding the mechanism by which Smase inhibits HIV infections has important implications for the development of antiretroviral therapeutics. Inhibiting viral infection by inducing receptor clustering (22) is a novel approach targeting the viral entry process. Recently, the concept of targeting sphingolipids as therapeutics has made significant strides (8), with innovative strategies for the local and targeted delivery of sphingolipids emerging. We anticipate that our understanding of Smase-mediated inhibition of HIV infection can be further exploited for the development of antiviral agents.
Acknowledgments
We thank Waldemar Popik (JHU, School of Medicine, Baltimore) for the CD4-GFP construct and Santos Manes (Centro Nacional de Biotecnología/CSIC, Madrid, Spain) for the N-terminally tagged CCR5-GFP construct. We acknowledge Samuel Kelly and Elaine Wang for help with the sphingolipid analyses. We thank the AIDS Research and Reference Reagent Program for the contribution of reagents that were used in this study. We are grateful to Mathias Viard and members of the Blumenthal Lab for expert experimental advice and helpful comments.
This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO56000; and in part (sphingolipid analyses) by NIH grant GM069338 to the Sphingolipid Core of the Lipid Maps Consortium.
The content of this publication does not necessarily reflect the views of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Burkly, L., N. Mulrey, R. Blumenthal, and D. S. Dimitrov. 1995. Synergistic inhibition of human immunodeficiency virus type 1 envelope glycoprotein-mediated cell fusion and infection by an antibody to CD4 domain 2 in combination with anti-gp120 antibodies. J. Virol. 69:4267-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. S., A. G. Rosenwald, and R. E. Pagano. 1995. Ceramide as a modulator of endocytosis. J. Biol. Chem. 270:13291-13297. [DOI] [PubMed] [Google Scholar]
- 4.Daecke, J., O. T. Fackler, M. T. Dittmar, and H. G. Krausslich. 2005. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J. Virol. 79:1581-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 6.Finnegan, C. M., S. S. Rawat, A. Puri, J. M. Wang, F. W. Ruscetti, and R. Blumenthal. 2004. Ceramide, a target for antiretroviral therapy. Proc. Natl. Acad. Sci. USA 101:15452-15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foti, M., M. A. Phelouzat, A. Holm, B. J. Rasmusson, and J. L. Carpentier. 2002. p56Lck anchors CD4 to distinct microdomains on microvilli. Proc. Natl. Acad. Sci. USA 99:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, T. E., C. M. Finnegan, R. Blumenthal, and M. Kester. Cell. Mol. Life Sci., in press. [DOI] [PMC free article] [PubMed]
- 9.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 10.Gallo, S. A., A. Puri, and R. Blumenthal. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40:12231-12236. [DOI] [PubMed] [Google Scholar]
- 11.Gambin, Y., R. Lopez-Esparza, M. Reffay, E. Sierecki, N. S. Gov, M. Genest, R. S. Hodges, and W. Urbach. 2006. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. USA 103:2098-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Mouton, C., R. A. Lacalle, E. Mira, S. Jimenez-Baranda, D. F. Barber, A. C. Carrera, A. Martinez, and S. Manes. 2004. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J. Cell Biol. 164:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grassme, H., E. Gulbins, B. Brenner, K. Ferlinz, K. Sandhoff, K. Harzer, F. Lang, and T. F. Meyer. 1997. Acidic sphingomyelinase mediates entry of N. gonorrhoeae into nonphagocytic cells. Cell 91:605-615. [DOI] [PubMed] [Google Scholar]
- 14.Grassme, H., V. Jendrossek, A. Riehle, G. Von Kurthy, J. Berger, H. Schwarz, M. Weller, R. Kolesnick, and E. Gulbins. 2003. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat. Med. 9:322-330. [DOI] [PubMed] [Google Scholar]
- 15.Grassme, H., A. Riehle, B. Wilker, and E. Gulbins. 2005. Rhinoviruses infect human epithelial cells via ceramide-enriched membrane platforms. J. Biol. Chem. 280:26256-26262. [DOI] [PubMed] [Google Scholar]
- 16.Gulbins, E., S. Dreschers, B. Wilker, and H. Grassme. 2004. Ceramide, membrane rafts and infections. J. Mol. Med. 82:357-363. [DOI] [PubMed] [Google Scholar]
- 17.Hanada, K., T. Mitamura, M. Fukasawa, P. A. Magistrado, T. Horii, and M. Nishijima. 2000. Neutral sphingomyelinase activity dependent on Mg2+ and anionic phospholipids in the intraerythrocytic malaria parasite Plasmodium falciparum. Biochem. J. 346:671-677. [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson, H. I., and T. J. Hope. 2006. The temperature arrested intermediate of virus-cell fusion is a functional step in HIV infection. Virol. J. 3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henis, Y. I., B. Herman-Barhom, B. Aroeti, and O. Gutman. 1989. Lateral mobility of both envelope proteins (F and HN) of Sendai virus in the cell membrane is essential for cell-cell fusion. J. Biol. Chem. 264:17119-17125. [PubMed] [Google Scholar]
- 20.Jan, J. T., S. Chatterjee, and D. E. Griffin. 2000. Sindbis virus entry into cells triggers apoptosis by activating sphingomyelinase, leading to the release of ceramide. J. Virol. 74:6425-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenworthy, A. K., B. J. Nichols, C. L. Remmert, G. M. Hendrix, M. Kumar, J. Zimmerberg, and J. Lippincott-Schwartz. 2004. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 165:735-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leikina, E., H. Delanoe-Ayari, K. Melikov, M. S. Cho, A. Chen, A. J. Waring, W. Wang, Y. Xie, J. A. Loo, R. I. Lehrer, and L. V. Chernomordik. 2005. Carbohydrate-binding molecules inhibit viral fusion and entry by crosslinking membrane glycoproteins. Nat. Immunol. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 23.Li, R., E. J. Blanchette-Mackie, and S. Ladisch. 1999. Induction of endocytic vesicles by exogenous C(6)-ceramide. J. Biol. Chem. 274:21121-21127. [DOI] [PubMed] [Google Scholar]
- 24.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 25.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 26.Manes, S., G. Del Real, R. A. Lacalle, P. Lucas, C. Gomez-Mouton, P. Sánchez-Palomino, R. Delgado, J. Alcamí, E. Mira, and C. Martinez-A. 2000. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 1:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh, M., and A. Helenius. 1980. Adsorptive endocytosis of Semliki Forest virus. J. Mol. Biol. 142:439-454. [DOI] [PubMed] [Google Scholar]
- 28.McAuliffe, M. J. 2005. Medical image processing, analysis and visualization in clinical research, p. 381-386. In F. M. Lalonde, D. McGarry, W. Gandler, K. Csaky, and B. L. Trus (ed.), IEEE computer-based medical systems. IEE Press, Piscataway, NJ.
- 29.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrill, A. H., Jr., M. C. Sullards, J. C. Allegood, S. Kelly, and E. Wang. 2005. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 36:207-224. [DOI] [PubMed] [Google Scholar]
- 31.Mkrtchyan, S. R., R. M. Markosyan, M. T. Eadon, J. P. Moore, G. B. Melikyan, and F. S. Cohen. 2005. Ternary complex formation of human immunodeficiency virus type 1 Env, CD4, and chemokine receptor captured as an intermediate of membrane fusion. J. Virol. 79:11161-11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen, D. H., B. Giri, G. Collins, and D. D. Taub. 2005. Dynamic reorganization of chemokine receptors, cholesterol, lipid rafts, and adhesion molecules to sites of CD4 engagement. Exp. Cell Res. 304:559-569. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, D. H., and D. Taub. 2002. Cholesterol is essential for macrophage inflammatory protein 1 beta binding and conformational integrity of CC chemokine receptor 5. Blood 99:4298-4306. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen, D. H., and D. Taub. 2002. CXCR4 function requires membrane cholesterol: implications for HIV infection. J. Immunol. 168:4121-4126. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, D. H., and D. D. Taub. 2004. Targeting lipids to prevent HIV infection. Mol. Interv. 4:318-320. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, D. H., and D. D. Taub. 2003. Inhibition of chemokine receptor function by membrane cholesterol oxidation. Exp. Cell Res. 291:36-45. [DOI] [PubMed] [Google Scholar]
- 37.Percherancier, Y., B. Lagane, T. Planchenault, I. Staropoli, R. Altmeyer, J. L. Virelizier, F. Arenzana-Seisdedos, D. C. Hoessli, and F. Bachelerie. 2003. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J. Biol. Chem. 278:3153-3161. [DOI] [PubMed] [Google Scholar]
- 38.Popik, W., and T. M. Alce. 2004. CD4 receptor localized to non-raft membrane microdomains supports HIV-1 entry. Identification of a novel raft localization marker in CD4. J. Biol. Chem. 279:704-712. [DOI] [PubMed] [Google Scholar]
- 39.Rawat, S. S., B. T. Johnson, and A. Puri. 2005. Sphingolipids: modulators of HIV-1 infection and pathogenesis. Biosci. Rep. 25:329-343. [DOI] [PubMed] [Google Scholar]
- 40.Schaeffer, E., V. B. Soros, and W. C. Greene. 2004. Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J. Virol. 78:1375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons, K., and W. L. Vaz. 2004. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33:269-295. [DOI] [PubMed] [Google Scholar]
- 42.Steffens, C. M., and T. J. Hope. 2004. Mobility of the human immunodeficiency virus (HIV) receptor CD4 and coreceptor CCR5 in living cells: implications for HIV fusion and entry events. J. Virol. 78:9573-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Blitterswijk, W. J., A. H. van der Luit, R. J. Veldman, M. Verheij, and J. Borst. 2003. Ceramide: second messenger or modulator of membrane structure and dynamics? Biochem. J. 369:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viard, M., I. Parolini, M. Sargiacomo, K. Fecchi, C. Ramoni, S. Ablan, F. W. Ruscetti, J. M. Wang, and R. Blumenthal. 2002. Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J. Virol. 76:11584-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zha, X., L. M. Pierini, P. L. Leopold, P. J. Skiba, I. Tabas, and F. R. Maxfield. 1998. Sphingomyelinase treatment induces ATP-independent endocytosis. J. Cell Biol. 140:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]