Abstract
In the Huh-7.5 hepatoma cell line, replication of the genotype 1a H77 strain of hepatitis C virus (HCV) is attenuated compared to that of the genotype 1b Con1 strain. This study identifies the poorly characterized integral membrane protein, NS4B, as a major determinant for this replication difference. Chimeric H77 subgenomic replicons containing the entire NS4B gene from Con1 in place of the H77 NS4B sequence replicated approximately 10-fold better than the H77 parent and to levels similar to that of the adapted Con1 replicon. An intermediate level of replication enhancement was conferred by H77 chimeras containing the poorly conserved N-terminal 47 residues or the remaining less-divergent C terminus of Con1 NS4B. The replication-enhancing activity within the N terminus of NS4B was further mapped to two Con1-specific amino acids. Experiments to elucidate the mechanism of enhanced H77 replication revealed that Con1 NS4B primarily increased H77 RNA synthesis on a per cell basis, as indicated by the similar capacities of chimeric and parental replicons to establish replication in Huh-7.5 cells and the higher levels of both positive- and negative-strand RNAs for the chimeras than for the H77 parent. Additionally, enhanced H77 replication was not the result of Con1 NS4B-mediated effects on HCV translation efficiency or alterations in polyprotein processing. Expression of Con1 NS4B in trans did not improve the replication of the H77 parental replicon, suggesting a cis-dominant role for NS4B in HCV replication. These results provide the first evidence that allelic variation in the NS4B sequence between closely related isolates significantly impacts HCV replication in cell culture.
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus that has emerged as a serious global health concern, causing chronic liver diseases including cirrhosis and hepatocellular carcinoma. The single-stranded HCV RNA genome of ∼9.6 kb is composed of 5′ and 3′ nontranslated regions (NTRs) flanking one large open reading frame (ORF). Translation of the ORF is mediated by an internal ribosome entry site (IRES) located within the 5′ NTR. The nascent polyprotein is processed co- and posttranslationally into the structural proteins (core and envelope glycoproteins, E1 and E2), the hydrophobic peptide p7, and the nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B. While the structural proteins and p7 are liberated from the polyprotein by a cellular signal peptidase, cleavages within the nonstructural portion of the polyprotein are mediated by two viral proteases; the NS2-3 autoprotease cleaves at the NS2/3 junction, and the remaining four sites, NS3/4A, NS4A/4B, NS4B/5A, and NS5A/5B, are processed by the NS3 serine protease together with its cofactor NS4A (for recent reviews, see references 3 and 9).
HCV RNA replication takes place in a membrane-associated replication complex or replicase (14, 21, 43, 48). The nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B are sufficient for RNA replication in cell culture (40) and are believed to function as the enzymes and accessory factors that catalyze and regulate replication of the HCV genome. Although the details of replication complex assembly and maintenance are essentially unknown, the enzymatic functions of NS3 and NS5B are fairly well defined. In addition to the serine protease activity, NS3 possesses RNA helicase and nucleoside triphosphatase activities (29, 49) that are essential for an unknown step(s) in RNA replication (33, 35). The NS5B protein is the RNA-dependent RNA polymerase (4, 39). Within the replication complex, NS5B synthesizes complementary negative-strand RNA intermediates of the HCV genome, which in turn act as templates for the production of an excess of positive-strand HCV RNA. In contrast, the role of NS5A in RNA replication is unknown. NS5A is a serine phosphoprotein with basally phosphorylated and hyperphosphorylated variants (25, 50) and is a major target for adaptive mutations that dramatically enhance genotype 1 replication in cell culture (5, 6, 20, 34, 38, 54).
In addition to NS5A, the function of the NS4B protein in RNA replication is not well understood. NS4B, a 27-kDa hydrophobic protein that associates with membranes of the endoplasmic reticulum (ER) (23, 30), contains at least four predicted transmembrane domains with the N and C termini located in the cytoplasm, although the N-terminal tail of NS4B has been suggested to posttranslationally translocate to the ER lumen (41, 42). A putative amphipathic helix within the N-terminal 26 residues of NS4B also contributes to membrane association, and this domain is critical for HCV replication in cell culture (13). The NS4B protein reorganizes intracellular membranes into distinct membranous structures that represent a site of viral replication in Huh-7 cells (11, 18, 44). Thus, NS4B has been proposed to play a structural role in RNA replication by serving as the scaffold for replication complex assembly. Despite significant sequence variations in NS4B between different HCV isolates, single amino acid substitutions in NS4B can greatly impact RNA replication efficiency in cell culture, and critical residues are found throughout the NS4B protein (37). Moreover, several replication-enhancing adaptive mutations map to NS4B (20, 38). In addition to its apparent role in providing a platform for RNA replication, NS4B has been implicated in nucleotide binding (12), modulation of HCV and host translation (16, 22, 26), modulation of NS5A hyperphosphorylation (31, 45), and negative regulation of NS5B by inhibition of NS3 function (47). However, the importance of these properties in HCV replication is unknown.
The closely related hepatitis C genotype 1 strains, Hutchinson (H77) (1a) and Con1 (1b), require adaptive mutations in order to productively replicate in cell culture. Adaptive mutations that permit efficient RNA replication, albeit to varying degrees, have been identified in the NS3, NS4A, NS4B, NS5A, and NS5B proteins, and specific combinations of these replication-enhancing mutations can further increase RNA replication (reviewed in reference 8). Previous experiments have revealed that adapted replicons derived from H77 and Con1 replicate at different rates in the highly permissive Huh-7.5 cell line (6). Specifically, H77 replicons carrying the adaptive mutations P1496L (NS3) and S2204I (NS5A) have lower replication capacities than Con1-derived replicons harboring only the S2204I adaptive change in the NS5A protein. This observation suggests that innate genetic differences between Con1 and H77 govern replication efficiency in Huh-7.5 cells. In this study, chimeric H77 replicons were created to explore the genetic basis for the attenuated H77 replication phenotype. This experimental approach identified the NS4B sequence as a major determinant for the lower replication rate of H77 in cell culture and implicated NS4B in the maintenance of RNA synthesis.
MATERIALS AND METHODS
Cell culture.
Huh-7.5 cells (7) were cultured in Dulbecco's modified Eagle medium (DMEM; Mediatech) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 0.1 mM nonessential amino acids (HyClone) (DMEM-10% FBS).
Plasmid construction.
Standard recombinant DNA technologies were used to construct and purify all plasmids. PCR was performed using Klentaq LA DNA polymerase (Wayne Barnes, Washington University, St. Louis, MO), and all regions subjected to PCR amplification were sequenced to ensure that no additional mutations were introduced. Primers used in this study are shown in Table 1. Plasmid DNA was purified by CsCl gradient centrifugation prior to linearization and RNA transcription.
TABLE 1.
Oligonucleotides used for constructing chimeric and mutant H77 replicons
| Name | Sequence (5′-3′)a |
|---|---|
| 42 | (−) CCTCAGCTAGCGCACTCTTCCATCTCATC |
| 43 | (+) CCACAGCTAGCCACCTCCCTTACATCGAAC |
| 44 | (−) GCTTCGCCCAGAAGGCTTCGA |
| 45 | (+) CGCGGGGTAACCACGTCTCC |
| 46 | (−) CCACGGATCCGGAGCATGGCGTGGAGC |
| 47 | (+) CCACGGATCCTGGCTAAGAGATGTTTGG |
| 48 | (−) GTGGTGTACGCGTTAATGGG |
| 51 | (+) ATGCATGTCGGCCGACCTGG |
| 52 | (−) CCTCGCTAGCGCACTCTTCCATCTCATCG |
| 53 | (+) CCTCGCTAGCCACTTACCGTACATCGAGCA |
| 54 | (−) AGAAGACCTCGAGTTTCTGC |
| 55 | (+) AGCTCCTGAGGCGACTGCATCAGTGGATAAGCTCGGAGTGTACCACTCCATGCTCCGGATCC |
| 56 | (−) AGTCGCTCAGCACCTCGCATATCCAGTCCCAGATGTCCCTTAGCCAGGATCCGGAGCATGGA |
| 61 | (−) CCTACGCCCAGAAGGCCTCGAGGGTCCGCCACTTGG |
| 61-S | (+) TACTGATGACCCATTTCTTTA |
| 71 | (+) CTAGGTCATGAGCGGCGAGATGCCCTCCACCGAGGACCTGGTGAACCTACTCCCTGCTATCCTCTC |
| 125 | (+) CATCATGACATGCATGTCGG |
| 133 | (−) CCATCTCGAGTTTCTGCCAGTTGGTCTGGACAGCAGGGGTGATAACCTCTGCTTGCTTGGTCGCGGTCTGCAGGAGGCCG |
| 135 | (−) CCACTCTAGATTAGCATGGCGTGGAGCAGTCCT |
| 159 | (+) GAGTGCGCTAGCCACCTCCCTTACATCGAACAGGGAATGATGCTCGCCGAACAATTCAAACAGAAGGCACTCGGGTTGCTGCAAACAGCCACCAAGCAAG |
| 190 | (−) CACACTCGAGGGTCCGCCACTTGGATTCCACCACGGGAGCAGCAGCCTCTGCTTGGCGGGACGCG |
| 191 | (−) CACACTCGAGTTTCTGCCAGTTGGTCTGGACAGCAGGGGTGATAACCTCCGCTTGCTTGGTGGCT |
| 209 | (−) CACACTGCAGGAGGCCGAGGGCCTTCTGCTTGAACTGCTCAGCGAGCTGCATCCCTTGCTCGATGTACG |
| 210 | (−) CACACTGCAGGAGGCCTATGGCCTTCTGCTTGAACTGC |
| 218 | (+) CCATGGATCCACCATGGCCTCACACCTCCCTTACA |
| 221 | (+) CCACGCTAGCCACCTCCCTTACATCGAACAGGGAATGATGCTCGCCGAACAATTCAAAC |
| 222 | (+) CCACGAATGCAGCTCGCCGAACAATTCAAACAGAAGGCACTCGGGTTGCTGCAAACAGCC |
| 223 | (−) CCACCTGCAGGAGGCCGATGGCCTTCTGCTTGAACTGCTCAGCGAGCTGCATCCCTTGCTCGATGTACG |
| 227 | (−) CCACTCTAGATTATCGGTTGGGGAGGAGGTAG |
| 228 | (+) CCACGGATCCACCATGGCGCCCATCACGGCGTAC |
| 2420 | (−) GGAGTTGTCCGTGAAC |
Restriction sites used for cDNA cloning are underlined, and the polarity of each oligonucleotide is indicated as either genome RNA (+) or its complement (−).
The H77-derived replicon plasmids pH/ΔE1-p7(L+I) (H/WT) (Fig. 1) and pH/SG-Neo(L+I) (HNeo/WT), which carry two adaptive mutations (P1496L in NS3 and S2204I in NS5A), as well as pH/SG-Neo/pol− (HNeo/pol−), have been described previously (6). The polymerase-defective derivatives (pol−) used in this study contain a mutation of Gly-Asp-Asp to Ala-Ala-Gly in the active site of the NS5B RNA-dependent RNA polymerase. The amino acid positions in NS4B refer to the first amino acid residue of the NS4B protein.
FIG. 1.
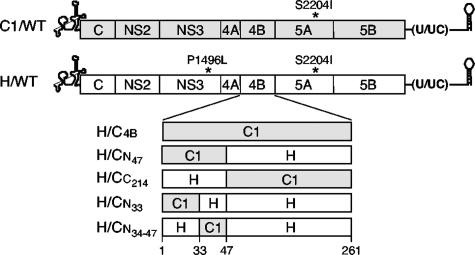
Schematic diagram of the Con1 and H77 subgenomic replicons used to measure transient replication efficiency in the Huh-7.5 cell line. The 5′ and 3′ NTR structures are illustrated, and the Con1 and H77 ORFs are depicted as shaded and open boxes, respectively, with the positions of the cell culture-adaptive mutations indicated above the diagrams. Chimeric NS4B proteins are shown below, with open boxes representing H77 NS4B sequences (H) and shaded boxes representing Con1-derived NS4B sequences (C1). The nomenclature used for each replicon is given on the left, and amino acid positions 1, 33, 47, and 261 are indicated below.
To facilitate the construction of NS4B chimeras, NheI and BamHI restriction sites were engineered in NS4B and NS5A, respectively, in pH/ΔE1-p7(L+I) and in the Con1 replicon plasmid pC1/ΔE1-p7(I) (K. J. Blight, unpublished data) containing the NS5A adaptive change S2204I. Plasmid pH/ΔE1-p7(L+I)/NB was constructed in two stages. First, the NheI site was introduced to make the intermediate plasmid pH/ΔE1-p7(L+I)/4B_NheI by inserting the EagI-NheI fragment of a PCR product amplified from pH/ΔE1-p7(L+I) with primers 51 and 52, the NheI-XhoI portion of a second PCR product amplified from pH/ΔE1-p7(L+I) with primers 53 and 54, and the XhoI-PinAI fragment from pH/ΔE1-p7(L+I) between the PinAI and EagI sites of pH/ΔE1-p7(L+I). Introduction of the NheI site changed the first two amino acids of H77 NS4B from Ser-Gln to Ala-Ser (SQ→AS). Subsequently, a BamHI site in NS5A was created by introducing a silent T-to-A nucleotide change at position 4409 in pH/ΔE1-p7(L+I)/4B_NheI to produce pH/ΔE1-p7(L+I)/NB. Briefly, the Bsu36I-BlpI portion of a PCR product amplified from pH/ΔE1-p7(L+I)/4B_NheI with primers 55 and 56 was ligated with the BlpI-EcoRI and EcoRI-Bsu36I fragments from pH/ΔE1-p7(L+I)/4B_NheI.
Similarly, NheI and BamHI sites were also introduced into pC1/ΔE1-p7(I) via a multistep process. First, the SphI-EcoRI fragment corresponding to the NS4A-4B-5A coding region was subcloned into similarly digested pGEM3Zf(+) (Promega) to create pGEM3Zf(+)/C1_4A(Sph)-5A(EcoRI). To insert the NheI site, two PCR products were amplified from pC1/ΔE1-p7(I) with primer pairs 41 plus 42 and 43 plus 44. These gel-purified products were mixed with primers 41 and 44 for mutually primed synthesis to complete the junction, and this product was subsequently digested with SphI and BglI. A BamHI site was introduced by a similar strategy. PCR products initially amplified from pC1/ΔE1-p7(I) using primer pairs 45 plus 46 and 47 plus 48 were combined in a reaction with the external primers 45 and 48, and the final product was digested with BstEII and MluI. SphI-BglI- and BstEII-MluI-digested PCR products and the BglI-BstEII fragment from pC1/ΔE1-p7(I) were ligated between the SphI and MluI sites of pGEM3Zf(+)/C1_4A(Sph)-5A(EcoRI) to create pGEM3Zf(+)/C1_4A-5A/NB. Finally, C1/ΔE1-p7(I)/NB was constructed via a 4-piece ligation of the XbaI-EagI, EagI-SphI, and EcoRI-XbaI fragments from pC1/ΔE1-p7(I) and the SphI-EcoRI fragment from pGEM3Zf(+)/C1_4A-5A/NB. Additionally, a XhoI site was inserted into pC1/ΔE1-p7(I)/NB to create pC1/ΔE1-p7(I)/NBX by ligating the BsrGI-NheI, BglI-EcoRI, and EcoRI-BsrGI fragments from pC1/ΔE1-p7(I)/NB with the NheI-BglI portion of a PCR product amplified from pC1/ΔE1-p7(I)/NB with primers 43 and 61.
In order to retain HpaI as the runoff site for RNA transcription of H77 replicons that contain Con1 NS4B, a silent T-to-G nucleotide substitution was first engineered at position 4127 in the Con1 NS4B sequence of pC1/ΔE1-p7(I)/NBX. This plasmid, designated pC1/ΔE1-p7(L+I)/4B_Hpa(−) (C1/WT) (Fig. 1), was constructed via a 4-piece ligation of the BsrGI-BspHI, BstEII-EcoRI, and EcoRI-BsrGI fragments from pC1/ΔE1-p7(I)/NBX and the BspHI-BstEII portion of a PCR product amplified from pC1/ΔE1-p7(I)/NBX using primers 71 and 48. The silent mutations creating the NheI, XhoI, and BamHI sites or removing the HpaI site did not alter the replication capacity of Con1 RNA (data not shown).
The H77 replicon plasmid containing the Con1 NS4B gene, pH/ΔE1-p7(L+I)/C1_4B (H/C4B) (Fig. 1), was constructed by cloning the NheI-BamHI fragment from pC1/ΔE1-p7(I)/4B_Hpa(−) together with the NsiI-NheI and BamHI-BstEII fragments from pH/ΔE1-p7(L+I)/NB into pH/ΔE1-p7(L+I)/NB digested with BstEII and NsiI.
Plasmid pH/ΔE1-p7(L+I)/C1_4BN47 (H/CN47) (Fig. 1), harboring the first 47 amino acids of Con1 NS4B, was created by ligating the BglII-NheI, XhoI-MluI, and MluI-BglII fragments from pH/ΔE1-p7(L+I)/NB and the NheI-XhoI fragment from pC1/ΔE1-p7(L+I)/NBX.
Plasmid pH/ΔE1-p7(L+I)/C1_4BC214 (H/CC214) (Fig. 1), containing the C-terminal 214 amino acids from Con1 NS4B (residues 48 to 261 of Con1 NS4B), was constructed in a 4-piece ligation reaction containing the NsiI-XhoI fragment from pH/ΔE1-p7(L+I), the XhoI-BamHI fragment from pC1/ΔE1-p7(I)/4B_HpaI(−), and the BamHI-BstEII and BstEII-NsiI fragments from pH/ΔE1-p7(L+I)/NB.
The chimeric construct pH/ΔE1-p7(L+I)/C1_4BN33 (H/CN33) (Fig. 1), containing the N-terminal 33 amino acids of Con1 NS4B, was constructed by ligating the XhoI-EcoRI and EcoRI-NsiI fragments from pH/ΔE1-p7(L+I) with the NsiI-XhoI portion of a PCR product amplified from pH/ΔE1-p7(L+I)/C1_4B with primers 125 and 191.
The replicon plasmid pH/ΔE1-p7(L+I)/C1_4BN34-47 (H/CN34-47) (Fig. 1), containing amino acids 34 to 47 from Con1 NS4B, was created by ligating the NsiI-XhoI fragment of a PCR product amplified from pH/ΔE1-p7(L+I) using primers 125 and 190 and the XhoI-EcoRI and EcoRI-NsiI fragments from pH/ΔE1-p7(L+I).
To introduce the NS4B mutation M12Q, L22I, or both into pH/ΔE1-p7(L+I), PCRs were first performed using pH/ΔE1-p7(L+I) as a template with forward primer 125 and one of the following reverse primers: 209 (M12Q), 210 (L22I), or 223 (M12Q and L22I). PCR products were digested with EagI and PstI and combined in a ligation reaction with the Bsu36I-BsrGI, BsrGI-EagI, and PstI-Bsu36I fragments from pH/ΔE1-p7(L+I) to create plasmids pH/ΔE1-p7(L+I)/4B_M12Q (M12Q), pH/ΔE1-p7(L+I)/4B_L22I (L22I), and pH/ΔE1-p7(L+I)/4B_ML→QI (ML→QI).
Plasmid pH/ΔE1-p7(L+I)/4B_SR→TK (SR→TK), carrying the double mutation S29T and R30K in NS4B, was constructed by digesting a PCR fragment amplified from pH/ΔE1-p7(L+I) with primer 125 and mutant primer 133 with NsiI and XhoI and ligating into EcoRI-NsiI-digested pH/ΔE1-p7(L+I) together with the XhoI-EcoRI fragment also from pH/ΔE1-p7(L+I).
To construct mutant chimeras pH/ΔE1-p7(L+I)/C1_4BN33_Q12M (Q12M) and pH/ΔE1-p7(L+I)/C1_4BN33_Q12M+I22L (QI→ML), DNA fragments containing the Q12M mutation alone or the Q12M and I22L mutations were amplified by PCR using mutant primer 221 or 159, respectively, primer 54, and the template pH/ΔE1-p7(L+I)/C1_4BN33. The resultant PCR products were digested with NheI and XhoI and ligated together with the NsiI-NheI fragment from pH/ΔE1-p7(L+I)/C1_4B and the XhoI-EcoRI and EcoRI-NsiI fragments from pH/ΔE1-p7(L+I).
The single-amino-acid substitution I22L in NS4B was introduced into a pH/ΔE1-p7(L+I)/C1_4BN33-derived PCR product by use of primers 222 and 54. The PCR product was subsequently digested with BsmI and XhoI before being combined in a 4-piece ligation reaction with the NsiI-BsmI fragment from pH/ΔE1-p7(L+I)/C1_4B and the XhoI-EcoRI and EcoRI-NsiI fragments from pH/ΔE1-p7(L+I) to create plasmid pH/ΔE1-p7(L+I)/C1_4BN33_I22L (I22L).
Plasmids pH/SG-Neo(L+I)/C1_4B (HNeo/C4B) and pH/SG-Neo(L+I)/C1_4BN47 (HNeo/CN47) were constructed by replacing the NsiI-MluI fragment of pH/SG-Neo(L+I) with the corresponding fragment from pH/ΔE1-p7(L+I)/C1_4B or pH/ΔE1-p7(L+I)/C1_4BN47, respectively. Similarly, pH/ΔE1-p7(L+I)/C1_4B/pol− (H/C4B/pol−) and pH/ΔE1-p7(L+I)/C1_4BN47/pol− (H/CN47/pol−) were created by exchanging the NsiI-MluI fragment in pH/ΔE1-p7(L+I)/pol− (H/pol−) (K. J. Blight, unpublished data) with the analogous segment from pH/ΔE1-p7(L+I)/C1_4B or pH/ΔE1-p7(L+I)/C1_4BN47, respectively.
To construct pcDNA3.1/C1_4B (pC4B), which expresses Con1 NS4B under the control of a human cytomegalovirus promoter, the Con1 NS4B gene was amplified by PCR from pC1/ΔE1-p7(I) using primers 218 and 135. The amplification product was digested with BamHI and XbaI and ligated into the similarly digested eukaryotic expression vector pcDNA3.1Zeo (Invitrogen). Plasmids pcDNA3.1/H_NS3-5B (pH3-5B) and pcDNA3.1/H_NS+C1_4B (pH/C4B), which contain the H77 nonstructural coding region with either H77 or Con1 NS4B, respectively, were constructed by inserting the following between the BamHI and XbaI sites of pcDNA3.1Zeo: the BamHI-ApaLI portion of a PCR product amplified from pH/SG-Neo(L+I) with primers corresponding to the N-terminal part of NS3 (primers 228 and 2420), the ApaLI-NotI fragment from either pH/SG-Neo(L+I) (for pcDNA3.1/H_NS3-5B) or pH/ΔE1-p7(L+I)/C1_4B (for pcDNA3.1/H_NS+C1_4B), and the NotI-XbaI portion of a second PCR product containing the C terminus of NS5B amplified from pH/SG-Neo(L+I) using primers 61-S and 227.
For transcription of negative-strand replicon RNA for Northern blot analysis, plasmid pGEM3Zf(+)/H77_NotI-NheI, corresponding to the 5′ NTR-Neo-encephalomyocarditis virus IRES-NS3-4A-4B-5A-5B sequence from pH/SG-Neo(L+I), was constructed by linearizing pH/SG-Neo(L+I) with NotI, filling in the 5′ overhang with Klenow fragment, and digesting the fragment with NheI prior to subcloning between the SmaI and XbaI sites of the transcription vector pGEM3Zf(+).
RNA transcription.
In vitro transcription reaction mixtures containing 40 mM Tris-HCl (pH 7.5), 10 mM NaCl, 12 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol, 0.035 U inorganic pyrophosphatase (Fermentas), 50 U RNasin (Promega), 3 mM each ATP, CTP, GTP, and UTP, 100 U T7 RNA polymerase (Epicentre Biotechnologies), and 1 μg of HpaI-linearized (H77) or ScaI-linearized (Con1) template DNA were incubated for 1.5 h at 37°C. Transcription was terminated by the addition of 5 U RNase-free DNase I (Roche) and a 20-min incubation at 37°C. After extraction with phenol and chloroform, HCV RNA was precipitated with isopropanol and dissolved in RNase-free water. The remaining template DNA was degraded by two serial DNase I digestions for 20 min at 37°C, followed by extraction with phenol and chloroform and precipitation with ethanol. RNA pellets were washed in 80% ethanol before being dissolved in RNase-free water. RNA concentrations were determined by measuring the optical density at 260 nm, and the integrity and concentrations were checked by agarose gel electrophoresis and ethidium bromide staining. For synthesis of negative-strand RNA for Northern blot analysis, plasmid pGEM3Zf(+)/H77_NotI-NheI was linearized with AflII and transcribed using SP6 RNA polymerase (Epicentre Biotechnologies).
RNA transfection of Huh-7.5 cells.
Subconfluent Huh-7.5 cells grown for 24 to 30 h were detached by trypsin treatment, collected by centrifugation (500 × g, 5 min, 4°C), washed twice with ice-cold RNase-free phosphate-buffered saline (PBS), and resuspended at 1.25 × 107 cells/ml in PBS. Unless otherwise indicated, 1 μg of transcribed RNA was mixed with 0.4 ml of the cell suspension, transferred to a cuvette with a gap width of 0.2 cm (BTX), and immediately pulsed (900 V; pulse length, 99 μs; 5 pulses; 1-s intervals) in a BTX ElectroSquarePorator. After electroporation, cells were kept at room temperature for 10 min and then transferred to 10 ml DMEM-10% FBS. For RNA and protein analysis, 0.5 ml of the electroporated cell suspension was seeded into six-well plates and harvested at the time points indicated below and in Results.
To select G418-resistant colonies, electroporated cells (1,250, 2,500, 5,000, 104, or 2.5 × 104 cells) were plated on 100-mm-diameter dishes together with cells transfected with HNeo/pol− RNA, to maintain the total number of cells at 5 × 105 per dish. After 48 h, the medium was replaced with DMEM-10% FBS supplemented with 0.8 mg G418 per ml (Gibco). The medium was replaced every 3 to 4 days, and after about 3 weeks, G418-resistant colonies were fixed with 7% formaldehyde, stained with 1% crystal violet in 50% ethanol, and counted to determine the colony-forming efficiency per μg of transfected RNA (CFU/μg).
To obtain stable population cell lines supporting persistent H77 replication, Huh-7.5 cells were electroporated with G418-selectable replicons (HNeo/WT, HNeo/C4B, or HNeo/CN47), and 1.5 × 106 cells were plated on 150-mm-diameter dishes. Approximately 48 h after plating, the medium was replaced with DMEM-10% FBS supplemented with 1 mg/ml G418, and the G418-resistant colonies selected after 3 weeks were detached by trypsin treatment and subsequently cultured in DMEM-10% FBS containing 0.75 mg/ml G418.
HCV RNA quantitation.
Total cellular RNA was extracted from Huh-7.5 cell monolayers using TRIzol reagent (Invitrogen) and the manufacturer's protocol. HCV-specific RNA levels in 1 μg of total RNA were quantified using an ABI PRISM 7000 sequence detection system (Applied Biosystems). Real-time reverse transcription-PCR (RT-PCR) amplifications were performed using the TaqMan EZ RT-PCR core reagents (Applied Biosystems), primers specific for the HCV 5′ NTR (5′-CCTCTAGAGCCATAGTGGTCT-3′ [sense; 50 μM] and 5′-CCAAATCTCCAGGCATTGAGC-3′ [antisense; 50 μM]), and 10 μM probe (6-carboxyfluorescein-CACCGGAATTGCCAGGACGACCGG-6-carboxytetramethylrhodamine; Applied Biosystems). RT reaction mixtures were incubated for 30 min at 60°C, followed by inactivation of the reverse transcriptase and activation of Taq polymerase at 95°C for 7 min. Forty cycles of PCR were performed with cycling conditions of 15 s at 95°C and 1 min at 60°C. Synthetic HCV RNA standards of known concentrations were run in parallel reactions and used to calculate a standard curve.
Western blot analysis.
Transfected cell monolayers were washed once with ice-cold PBS and lysed in 0.1 M sodium phosphate (pH 7.0) containing 1% sodium dodecyl sulfate (SDS), 80 μg/ml phenylmethylsulfonyl fluoride (PMSF), and 2× Complete protease inhibitor cocktail (Roche). Cellular DNA was sheared by repeated passage through a 27.5-gauge needle, and the lysates were subsequently heated to 70°C for 10 min and clarified by centrifugation. Ten to 12 μg of protein lysate was resolved on an SDS-8% polyacrylamide gel and transferred to nitrocellulose membranes. Blots were blocked for 1 h in 3% goat serum-PBS-0.5% Tween 20, followed by 1 h to overnight in 5% milk-PBS-0.5% Tween 20. For NS5A and NS3 detection, polyclonal antisera, GSK#308 and GSK#367 (kindly provided by Robert Sarisky, GlaxoSmithKline, Collegeville, PA), respectively, were diluted 1:1,500 in PBS-0.5% Tween 20-0.3% goat serum and incubated for 1 h at room temperature with constant rocking. After several washes with PBS-0.5% Tween 20, the secondary antibody (a horseradish peroxidase-coupled goat anti-rabbit antibody) (Pierce) was added at a 1:25,000 dilution in PBS-0.5% Tween 20-0.3% goat serum for 1 h at room temperature with constant rocking. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected by using mouse anti-GAPDH (Imgenex) diluted 1:1,500 in PBS-0.5% Tween 20-0.3% goat serum as a primary antibody and horseradish peroxidase-conjugated goat anti-mouse (Pierce) diluted 1:25,000 in PBS-0.5% Tween 20-0.3% goat serum as a secondary antibody. Proteins were visualized by chemiluminescence using the SuperSignal West Pico chemiluminescent substrate (Pierce). To quantify HCV protein or GAPDH levels, antibody-probed membranes were developed using ECL plus Western blotting detection reagents (Amersham Biosciences), and the signal was measured on a FLA-5000 scanner with a 473-nm laser (Fuji).
Northern blot analysis.
Total cellular RNA was extracted from Huh-7.5 cells supporting H77 replication with TRIzol reagent and was quantified with a spectrophotometer at 260 nm. Ten micrograms of the total RNA was denatured by treatment with 6 M glyoxal in 50% dimethyl sulfoxide and 10 mM sodium phosphate (pH 7.0) for 1 h at 50°C, separated by sodium phosphate-buffered 1% agarose gel electrophoresis, and transferred overnight to Nytran SuPerCharge nylon membranes using the TurboBlotter system (Schleicher & Schuell) and 20× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate). The RNA was immobilized on the membrane by UV cross-linking. Hybridization was performed for 16 h at 47°C with [α-32P]UTP-labeled positive- or negative-sense riboprobes annealing to the NS5B sequence of the H77 genome (nucleotides 7861 to 8205) in a solution containing 4× SSC, 50% deionized formamide, 8× Denhardt's reagent, 0.1% SDS, 50 mM sodium phosphate (pH 6.8), 10% dextran sulfate, 150 μg of sheared denatured salmon sperm DNA per ml, 75 μg yeast tRNA per ml, and 400 μg torula yeast RNA per ml. The membranes were washed four times in 2× SSC for 5 min at room temperature, twice in 0.2× SSC-0.1% SDS for 30 min at 66°C, and twice in 0.2× SSC for 5 min at room temperature. To correct for total-RNA amounts loaded in each lane, the membrane was subsequently hybridized with a GAPDH-specific antisense riboprobe. Specific HCV and GAPDH signals were quantified by phosphorimaging with a FLA-5000 scanner (Fuji), and the number of replicon molecules was determined by comparison with a serial dilution of in vitro-transcribed RNA corresponding to a known number of positive- or negative-sense H77 RNA strands mixed with 10 μg of total RNA from naïve Huh-7.5 cells.
DNA transfection.
Huh-7.5 cells were electroporated with replicon RNA 36 h prior to transfection with 0.5 or 1 μg of the DNA plasmids pcDNA3.1Zeo (pcDNA), pcDNA3.1/C1_4B (pC4B), pcDNA3.1/HA-C1_4B, pcDNA3.1/H_NS+C1_4B (pH/C4B), pcDNA3.1/H_NS3-5B (pH3-5B), or pcDNA3.1/C1_NS3-5B by using LT1 transfection reagent (Mirus Corporation) and following the manufacturer's instructions. Approximately 60 h after the addition of DNA-LT1 complexes, cells were either washed once with ice-cold PBS, lysed in 1% SDS in 0.1 M sodium phosphate (pH 7.0), and analyzed by Western blotting as described above or used to prepare total RNA for quantitative RT-PCR.
Polyprotein processing was assessed using the vaccinia virus-T7 hybrid system. Briefly, subconfluent Huh-7.5 cells were infected for 1 h at room temperature with vTF7-3, a vaccinia virus expressing the T7 RNA polymerase. Subsequently, cell monolayers were washed once with DMEM-10% FBS and subjected to transfection with 1 μg of plasmid DNA using LT1 transfection reagent. Twenty-four hours later, cells were incubated for 1 h in methionine- and cysteine-deficient MEM containing 5% dialyzed FBS (dMEM); that medium was then replaced with dMEM supplemented with 200 μCi/ml Express 35S protein labeling mix (Perkin-Elmer). Cells were labeled for 1 h, washed once with ice-cold PBS, and harvested in 1% SDS lysis buffer as described above. Sixty micrograms of protein lysate was mixed with 0.5 ml TNA (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.67% bovine serum albumin, 1 mM EDTA, 0.33% Triton X-100, 80 μg/ml PMSF, 1× Complete inhibitor cocktail) and 1 μl of HCV-positive patient serum (kindly provided by Charles Rice, The Rockefeller University, New York, NY) and incubated overnight at 4°C with rocking. Fifty microliters of prewashed Pansorbin cells (Calbiochem) were added and incubated for 2 h at 4°C with rocking, and immune complexes were collected by centrifugation. Immunoprecipitates were washed three times in TNA containing 0.125% SDS and once with 50 mM Tris-HCl (pH 7.5)-150 mM NaCl-1 mM EDTA-80 μg/ml of PMSF-1× Complete inhibitor cocktail before being heated at 80°C for 20 min in protein sample buffer. Metabolically labeled proteins were separated on an SDS-9% polyacrylamide gel and visualized either by fluorography and autoradiography or by phosphorimaging with a FLA-5000 scanner.
RESULTS
The Con1 NS4B sequence enhances H77 replication.
Previously, experiments with the Huh-7.5 cell line showed that the replication ability of H77 replicons carrying adaptive mutations P1496L in NS3 and S2204I in NS5A was significantly lower than that of S2204I-containing Con1 replicons (6); however, the genetic determinants responsible for this replication difference have not been defined. In this study, the relative contribution of the NS4B sequence to H77 replication efficiency in Huh-7.5 cells was addressed by replacing the H77 NS4B gene in the H77 replicon with its counterpart from the Con1 strain. The replicative ability of this chimeric replicon (H/C4B) (Fig. 1) was compared to those of the parental (H/WT) (Fig. 1), Con1 (C1/WT) (Fig. 1), and replication-defective (H/pol−) replicons by quantifying HCV RNA levels at 24-h intervals after RNA transfection of Huh-7.5 cells and by examining HCV protein expression after 96 h in culture. As shown in Fig. 2A, HCV RNA levels for the chimeric H/C4B replicon were ∼10-fold higher than that of the parental H77 replicon and were similar to that of the Con1 replicon. Additionally, the amount of NS3 and NS5A protein expression was significantly higher in H/C4B-transfected cells than in cells electroporated with the H/WT replicon (Fig. 2B). Enhanced replication was not restricted to H77 subgenomic replicons; genome-length H77 RNA containing Con1 NS4B also replicated at a ∼10-fold-higher level than the full-length H77 parent (data not shown). Thus, the NS4B sequence influences the HCV replication rate and is a major determinant for the lower-replication phenotype of H77 replicons.
FIG. 2.
Con1 NS4B enhances H77 replication. (A) Huh-7.5 cells were electroporated with 0.5 μg of replicon RNA. At the indicated times posttransfection, total cellular RNA was harvested, and HCV RNA in 1 μg of total RNA was quantified by real-time RT-PCR. (B) Ninety-six hours after RNA transfection, Huh-7.5 cell monolayers were lysed, and NS3 (top) and NS5A (bottom) expression was analyzed by immunoblotting. The migration of NS3 and NS5A is shown on the right. The differences in mobility between Con1- and H77-derived NS3 and between Con1- and H77-derived NS5A reflect differences in specific amino acid composition between these two strains. Due to slight differences in antibody reactivity, the relative expression levels of Con1 and H77 NS3 or NS5A cannot be directly compared. Similar results were obtained in four independent repetitions of the experiment.
This clear effect of Con1 NS4B on H77 replication levels led to a closer comparison of the NS4B genes of Con1 and H77. Amino acid alignments of the Con1 and H77 NS4B proteins revealed considerable variation in the N-terminal 47 residues (68% identity), while the C-terminal 214 amino acids shared 90.7% identity (residues 48 to 261) (Fig. 3A). To determine which region of Con1 NS4B might be responsible for replication enhancement, two replicons containing chimeric NS4Bs, one with the N-terminal 47 residues of Con1 NS4B fused to the H77 NS4B C terminus (designated H/CN47) (Fig. 1) and the other containing the H77 N terminus followed by the C-terminal 214 amino acids of Con1 NS4B (designated H/CC214) (Fig. 1), were tested. By 96 h posttransfection, the levels of H/CN47 and H/CC214 RNAs in Huh-7.5 cells were about three- to fourfold higher than those of the parental H/WT replicon but remained lower than that of the H77 chimera containing the entire Con1 NS4B sequence (H/C4B) (Fig. 3B). Furthermore, the relative amounts of NS5A expressed at 96 h after electroporation paralleled the HCV RNA levels (Fig. 3C). Hence, both the N- and C-terminal domains of Con1 NS4B enhance H77 replication, but both together are required to achieve the replication levels of the H/C4B chimera.
FIG. 3.
Replication efficiencies of H77 replicons encoding chimeric NS4Bs. (A) Amino acid sequence alignment of Con1 (C1) and H77 (H) NS4Bs. Amino acid differences are boldfaced, and amino acid positions 1, 33, 47, and 261 in the NS4B sequence are indicated. (B) HCV RNA levels in 1 μg of total Huh-7.5 cellular RNA were quantified 24, 48, 72, and 96 h after transfection by real-time RT-PCR. (C) At 96 h postelectroporation, the relative levels of NS5A were measured by Western blotting using a polyclonal antiserum. The results are representative of three independent repetitions of the experiment.
Identification of two divergent N-terminal residues required for efficient RNA replication.
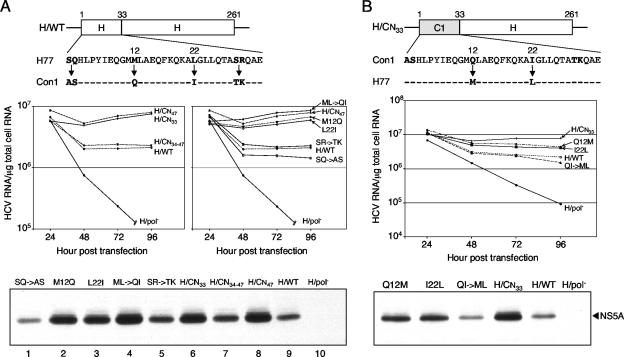
Although the N- and C-terminal domains are both required for optimal NS4B function, initial experimentation focused on defining the replication-enhancing residues within the N terminus of NS4B, since this region is short and poorly conserved and is predicted to reside in the cytoplasm. In order to locate the responsible residue(s), the N-terminal 47 amino acids were first subdivided into two smaller regions based on the degree of amino acid variability between Con1 and H77 NS4B (Fig. 3A). In the H77 replicon, either the first 33 amino acids (H/CN33) or residues 34 to 47 (H/CN34-47) of H77 NS4B were replaced with the corresponding sequence from Con1 NS4B (Fig. 1), and the ability of these chimeras to replicate was measured in Huh-7.5 cells as described above. The levels of HCV RNA and NS5A for H/CN34-47 were equivalent to those for the parental H/WT replicon (Fig. 4A, compare lanes 7 and 9), indicating that residues 34 to 47 of Con1 NS4B were not responsible for increasing H77 replication. In contrast, levels of H/CN33 RNA were three- to fourfold higher than RNA levels for H/WT and similar to those of the H77 chimera carrying the N-terminal 47 amino acids from Con1 NS4B (H/CN47) (Fig. 4A). Similarly, the amounts of NS5A expression in cells supporting H/CN33 and H/CN47 replication were comparable at 96 h after electroporation and were significantly greater than the levels of NS5A in H/WT-transfected cells (Fig. 4A, compare lanes 6, 8, and 9). These results indicate that one or more of the six amino acid differences within the first 33 residues are responsible for the enhanced replication phenotype of the H77 chimera containing the N terminus of Con1 NS4B.
FIG. 4.
Enhancing residues map to the N-terminal 33 amino acids of Con1 NS4B. (A) Replicative abilities of H77 replicons expressing hybrid or mutant NS4B proteins. (Top) Schematic representation of H77 NS4B, with the first 33 amino acids of H77 and Con1 NS4B aligned below. Divergent residues are boldfaced, and arrows indicate the amino acid substitutions tested. (Center) Huh-7.5 cells were electroporated with the parental H/WT replicon, H77 chimeric RNAs (H/CN47, H/CN33, and H/CN34-47) (Fig. 1), and H77 replicons containing the S1A and Q2S (SQ→AS), M12Q, L22I, M12Q and L22I (ML→QI), or S29T and R30K (SR→TK) mutations. At the indicated times postelectroporation, HCV RNA levels were measured as described above. At the 96-h time point, 9 × 104 RNA copies per μg of total cellular RNA were detected in H/pol−-transfected cells. (Bottom) Western blot analysis of NS5A expression levels at 96 h posttransfection. Similar results were obtained in four independent experiments. (B) HCV RNA and NS5A levels of the H77 chimeric replicon H/CN33 (Fig. 1), containing the Q12M or the I22L mutation or both (QI→ML). (Top) Diagram of the hybrid NS4B protein containing the first 33 amino acids of Con1 NS4B fused to the remaining H77 NS4B sequence. Shown below is the alignment of the N-terminal 33 residues of Con1 and H77 NS4B, with the divergent residues boldfaced; the amino acid replacements analyzed are indicated by arrows. (Center) HCV RNA levels quantified at 24 to 96 h after transfection of Huh-7.5 cells. (Bottom) NS5A expression at 96 h after RNA transfection. These results are representative of two repetitions of the experiment.
To identify which residue(s) in the N-terminal 33 amino acids of Con1 NS4B enhanced H77 replication, the six nonconserved amino acids in the H77 parental replicon were replaced either individually or in pairs with the Con1-specific amino acids (Fig. 4A, top), and the replicative ability of mutant replicons was measured in Huh-7.5 cells. Changing the first two residues in H77 NS4B from Ser-Gln to the Con1 sequence (Ala-Ser) led to a slight reduction in HCV RNA (∼1.5-fold) and NS5A levels compared to those for the parental H/WT sequence (Fig. 4A, compare lanes 1 and 9), while the replicon carrying the S29T plus R30K double mutation (SR→TK) replicated in a manner similar to that of H/WT (Fig. 4A, compare lanes 5 and 9). Conversely, both the M12Q and L22I substitutions independently increased RNA (∼3-fold) and NS5A protein levels relative to those for H/WT (Fig. 4A, compare lanes 2, 3, and 9), and when both substitutions were combined (ML→QI), replication levels were slightly higher than those for the H77 chimeric replicon, H/CN47 (Fig. 4A, compare lanes 4 and 8). Thus, Gln-12 and Ile-22 are the only residues within the N terminus of Con1 NS4B that are responsible for the enhanced replication phenotype of the H77 chimera H/CN47.
To confirm the importance of the amino acid sequence at positions 12 and 22, Q12M, I22L, and the Q12M plus I22L double mutation (QI→ML), were introduced into the Con1-specific NS4B sequence present in the H77 chimera H/CN33 (Fig. 4B, top). As anticipated, each of the amino acid substitutions independently decreased H/CN33 replication, and in combination they further reduced replication to levels close to that of the unmodified H/WT replicon (Fig. 4B). Hence, Met at position 12 and Leu at position 22 of NS4B are suboptimal for Con1 NS4B-enhanced H77 RNA replication in Huh-7.5 cells.
H77 replication is enhanced on a per cell basis.
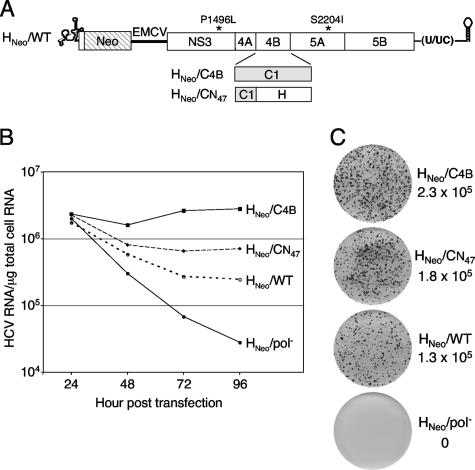
In order to determine whether elevated replication of the H77-Con1 NS4B chimeras occurred via an increase in replication per cell or was the result of replication establishment in a greater frequency of cells, the entire Con1 NS4B gene (HNeo/C4B) or the first 47 amino acids of Con1 NS4B (HNeo/CN47) were introduced into the G418-selectable replicon HNeo/WT (Fig. 5A) (6). HCV RNA levels were quantified for the first 96 h after transfection of Huh-7.5 cells (Fig. 5B), and the number of Huh-7.5 cells transduced to G418 resistance was measured 3 weeks posttransfection (Fig. 5C). As observed for the monocistronic replicon (Fig. 3), HCV RNA levels for the G418-selectable replicons HNeo/C4B and HNeo/CN47 were ∼11- and 3-fold greater, respectively, than that for HNeo/WT (Fig. 5B). However, only slight differences in the number of G418-resistant colonies were observed between these replicons (<2-fold) (Fig. 5C). Furthermore, the levels of HCV RNA in three isolated independent G418-resistant cell clones supporting HNeo/C4B replication were significantly higher than the levels detected in three HNeo/WT-containing cell clones isolated in parallel (3.5- to 24-fold [data not shown]). Taken together, these results suggest that Con1 NS4B primarily enhances H77 RNA replication within individual Huh-7.5 cells.
FIG. 5.
HCV RNA accumulation and G418-resistant colony-forming abilities of H77-derived subgenomic replicons. (A) Structure of the G418-selectable replicon. The 5′ and 3′ NTR structures are shown, and the H77 NS3-5B ORF is depicted as an open box; the polyprotein cleavage products are indicated, along with the positions of the two cell culture-adaptive mutations, P1496L and S2204I. The neomycin phosphotransferase gene (Neo) and the encephalomyocarditis virus (EMCV) IRES are shown. Chimeric NS4B proteins are diagramed below, with the shaded regions representing Con1-derived NS4B sequences and the adopted nomenclature given on the left. (B) Total cellular RNA was harvested at the indicated times postelectroporation, and HCV RNA in 1 μg of total RNA was quantified by real-time RT-PCR. (C) G418 transduction efficiencies of H77 replicons. Immediately after electroporation, 1,250, 2,500, 5,000, 104, or 2.5 × 104 transfected Huh-7.5 cells were coplated with cells transfected with HNeo/pol− RNA as described in Materials and Methods. Following 3 weeks of G418 selection, the resulting colonies were stained with crystal violet and counted for at least three cell densities. The calculated transduction efficiencies (expressed in CFU per μg of transfected RNA) are given to the right of representative dishes that were initially plated with 2.5 × 104 transfected cells. Similar results were obtained in three independent repetitions of the experiment.
Replication of H77 RNA is not enhanced by expressing Con1 NS4B in trans.
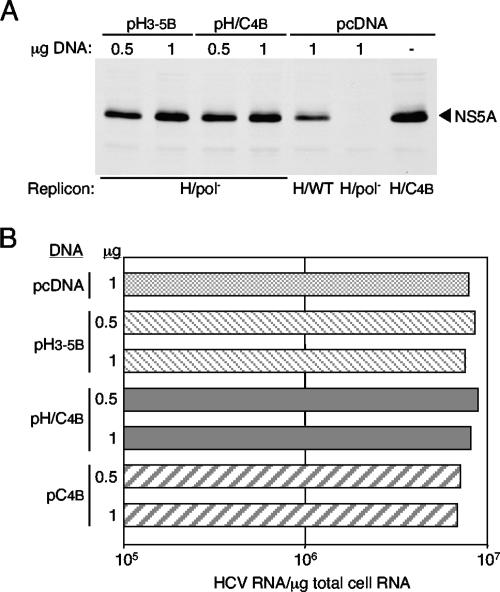
To determine if the Con1 NS4B protein enhanced viral replication in trans, HCV RNA levels in Huh-7.5 cells supporting parental H/WT replication were measured following transfection of NS4B-expressing plasmids. The transfected plasmids expressed either Con1 NS4B (pC4B), N-terminally hemagglutinin (HA) tagged Con1 NS4B (data not shown), the H77 NS3-5B polyprotein containing Con1 NS4B instead of H77 NS4B (pH/C4B), the H77 NS3-5B polyprotein (pH3-5B), or the NS3-5B polyprotein from Con1 (data not shown). First, conditions were established such that >80% of cells received the DNA expression construct (data not shown) and HCV protein levels expressed from the transfected DNA plasmids were comparable to the levels observed in Huh-7.5 cells supporting productive H77-Con1 NS4B chimera replication (Fig. 6A). For example, the amount of NS5A expressed upon transfection of 1 μg of pH3-5B or pH/C4B into cells containing H/pol− RNA was similar to the level of NS5A detected at 96 h after electroporation of the H/C4B chimera (Fig. 6A). When Con1 NS4B was expressed either alone (pC4B) or in the context of the chimeric H77 (pH/C4B) or Con1 polyprotein, H77 RNA levels were similar to those for cells transfected with either the empty expression vector (pcDNA) or a plasmid containing the H77 NS3-5B ORF (pH3-5B) (Fig. 6B; also data not shown). Similarly, plasmid-driven expression of Con1 NS4B (pC4B and pH/C4B) in a stable cell line supporting persistent HNeo/WT replication (Fig. 5A) did not increase RNA replication (data not shown). Thus, under these experimental conditions, Con1 NS4B was not able to enhance the replication of the H77 parental replicon in trans.
FIG. 6.
Effect of NS4B expression in trans on H77 replication in Huh-7.5 cells. Approximately 36 h following electroporation of Huh-7.5 cells with RNA of the H/pol−, H/WT, or H/C4B replicon, cell monolayers were transfected either with 0.5 or 1 μg of the indicated DNA expressing NS4B under the control of the cytomegalovirus promoter (pC4B, pH/C4B, or pH3-5B) or with 1 μg of the empty vector (pcDNA). After an additional 60 h in culture, cells were lysed, and NS5A (A) and H77 RNA (B) levels were determined as described in Materials and Methods. For reasons that remain unclear, H77 RNA levels in cells transfected with any plasmid DNA were typically higher than RNA levels in cells that were never subjected to DNA transfection. These data are representative of two independent experiments.
Con1 NS4B enhances H77 RNA synthesis.
To determine if Con1 NS4B preferentially enhanced the synthesis of positive-strand RNA, negative-strand RNA, or both, HCV RNA levels in transiently transfected Huh-7.5 cells were analyzed by Northern blot hybridization. At 96 h postelectroporation, a significant increase in positive-strand RNA levels was observed for the chimeric replicons H/C4B (∼8-fold), H/CN47 (∼3.0-fold), and H/CN33 (∼3.0-fold) relative to the level for the parental H/WT replicon (Fig. 7A, lanes 5 to 8). Likewise, negative-strand RNA synthesis was greater for the chimeric replicons (Fig. 7A, lanes 14 to 17). However, the level of negative-strand RNA synthesized in cells supporting transient H/WT replication was at the limit of detection (Fig. 7A, lane 14), making it difficult to quantify the differences in negative-strand synthesis between the H77-Con1 NS4B chimeras and the H77 parental sequence.
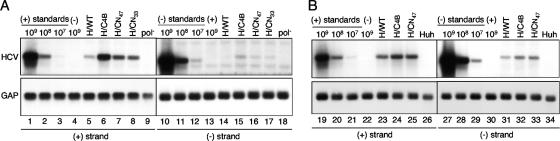
FIG. 7.
Northern blot analysis of positive- and negative-strand HCV RNA levels in Huh-7.5 cells supporting H77 parental and chimera replication. A total of 10 μg of cellular RNA isolated 96 h after electroporation of H77 replicons (A) or 72 h after seeding of G418-resistant Huh-7.5 cells supporting persistent H77 replication (B) was subjected to Northern blot hybridization with radiolabeled riboprobes specific for the detection of H77 positive-strand (+) or negative-strand (−) RNA (top) or GAPDH mRNA (bottom). Signals were quantified by phosphorimaging, compared with serially diluted in vitro-transcribed replicon RNAs of positive (109 to 107 [lanes 1 to 3] and 2 × 109 to 2 × 107 [lanes 19 to 21]) and negative (109 to 107 [lanes 10 to 12] and 2 × 109 to 2 × 107 [lanes 27 to 29]) polarity mixed with 10 μg of total cellular RNA from naïve Huh-7.5 cells, and normalized to the GAPDH message (GAP) to account for variable loading. Note that the negative-strand replicon RNA standard migrates faster due to the lack of the poly(U/UC) and 3′ NTR sequences. Strand specificity for each riboprobe was demonstrated by using 109 (lanes 4 and 13) or 2 × 109 (lanes 22 and 30) molecules of the opposite polarity replicon RNA [(−) or (+)]. Huh in panel B represents RNA isolated from naïve Huh-7.5 cells.
To better assess relative increases in negative-strand synthesis, Northern blot analysis was performed on G418-selected Huh-7.5 cells supporting persistent H77 replication. For this purpose, G418-selected cell populations were used in order to avoid studying potentially nonrepresentative clonal phenotypes of individual G418-resistant colonies. As observed in transient replication, synthesis of both positive- and negative-strand RNA increased for the H77 chimeras containing the entire Con1 NS4B sequence (HNeo/C4B) or its N-terminal 47 residues (HNeo/CN47) relative to that for the parental replicon (HNeo/WT) (Fig. 7B, lanes 23 to 25 and 31 to 33). The ability to detect HNeo/WT negative-strand RNA revealed that all replicons in G418-selected cell lines maintained a ∼22:1 positive- to negative-strand ratio regardless of total HCV RNA production. However, unlike the transient assay, which showed an eightfold increase in the level of positive-strand RNA for the chimera containing Con1 NS4B relative to that for the parental sequence (Fig. 7A, lanes 5 and 6), only a threefold increase was detected in G418-resistant cell lines (Fig. 7B, lanes 23 and 24). This was not an unexpected result, since the stringent G418 selection conditions used to derive the stable cell lines might enrich for cells supporting efficient HNeo/WT replication, while cells containing established but lower replication levels might succumb to selection. Another possibility is that during the >3-week period of G418 selection, mutations that improve H77 replication efficiency may arise. However, sequence analysis of uncloned RT-PCR products amplified from the G418-selected cell populations supporting HNeo/WT, HNeo/C4B, and HNeo/CN47 replication did not identify additional mutations within the NS3-5B coding region. Nevertheless, the data obtained from both transient and stable replication assays support the conclusion that expression of Con1 NS4B from the H77 replicon increases the synthesis of both positive- and negative-strand H77 RNA.
Con1 NS4B does not affect H77 IRES-directed translation.
Given that viral RNA synthesis is dependent on polyprotein production and NS4B has been suggested to modulate the translational activity of the HCV IRES (22, 26), amplification of H77 chimeric RNA levels could be explained by an increase in RNA translation efficiency. To assess the effect of Con1 NS4B on HCV IRES-mediated translation independent of RNA replication, NS5A expression was analyzed 10 to 20 h after electroporation of polymerase-defective H77 replicons expressing either H77 NS4B (H/pol−), the complete Con1 NS4B (H/C4B/pol−), or a hybrid protein containing the N-terminal 47 amino acids of Con1 NS4B (H/CN47/pol−). To achieve HCV protein levels similar to those observed for replicating H77-Con1 NS4B chimeras (Fig. 8A, lanes 1 and 2), Huh-7.5 cells were electroporated with 8 μg of the pol− replicon RNA. At 10 h (Fig. 8A, lanes 3 to 5) and 20 h (data not shown) following transfection, similar amounts of NS5A were translated from the parental and chimeric replicons, indicating that the Con1 NS4B sequence does not appear to stimulate translation from the HCV IRES and that enhanced RNA replication of chimeric H77 replicons is not due to an increased rate of viral protein synthesis.
FIG. 8.
Effect of NS4B on HCV IRES-driven translation and polyprotein processing. (A) Eight micrograms of the polymerase-defective replicons (pol−) expressing H77 NS4B (H/pol−), Con1 NS4B (H/C4B/pol−), or a hybrid NS4B containing the first 47 amino acids from Con1 NS4B (H/CN47/pol−) was electroporated into Huh-7.5 cells. Lysates collected at 10 h postelectroporation were subjected to immunoblot analysis with a polyclonal antiserum to NS5A (top). Relative NS5A levels measured by phosphorimaging were normalized to GAPDH expression (bottom) and to HCV RNA quantified at the same time point (data not shown). Samples collected 96 h after transfection of the replication-competent chimeric replicons, H/C4B and H/CN47, were separated in parallel (lanes 1 and 2). The negative control (Huh) (lane 6) represents Huh-7.5 cells transfected with 8 μg of cellular RNA isolated from naïve Huh-7.5 cells. The migration of NS5A and GAPDH is shown on the left. These results are representative of two independent experiments. (B) Huh-7.5 cells infected with a vaccinia virus expressing T7 RNA polymerase were transfected with 1 μg of the indicated plasmid DNA and incubated for 1 h in the presence of [35S]methionine-cysteine, as described in Materials and Methods. The labeled cells were lysed, and NS3, NS4B, and NS5A were analyzed by immunoprecipitation using a patient serum, followed by SDS-9% polyacrylamide gel electrophoresis, autoradiography, and phosphorimaging. The 35S-labeled NS4B signal was corrected for DNA transfection efficiency by normalizing to NS3 expressed from the corresponding replicon DNA. Values below the gel represent the percentage of normalized NS4B expressed relative to the level of NS4B encoded by the H/WT replicon, which has been set at 100%. The negative control (Huh) represents vaccinia virus-infected Huh-7.5 cells transfected with an unrelated plasmid DNA that was immunoprecipitated with the same antiserum as described above. The mobilities of the molecular mass standards (in kilodaltons) are given on the left, and the migration of the HCV-specific proteins is indicated on the right. Con1 NS4B migrated more slowly than H77 NS4B, presumably reflecting differences in amino acid composition between the proteins.
Chimeric polyproteins are processed in a manner similar to the parental sequence.
Replacing H77 NS4B with the corresponding Con1 sequence could alter the recognition of the NS4A/4B and NS4B/5A cleavage sites by the H77 NS3-4A serine protease complex. It is therefore possible that alterations in processing at the modified polyprotein junctions account for the increased replication of the H/C4B and H/CN47 chimeras (Fig. 1). Given that chimeric and parental H77 replicons replicate at different rates (Fig. 3), polyprotein processing was examined independently of HCV replication by using a transient vaccinia virus expression system. Small differences in the levels of NS4B expressed from the chimeric and mutant replicon DNAs compared to the parental NS4B sequence (H/WT) were observed (Fig. 8B), but these differences did not correlate with replication efficiency. Given that no uncleaved NS4A-4B precursor was detected and the patient serum used for immunoprecipitation has a greater reactivity to H77 NS4B than to Con1 NS4B (Fig. 8B, compare the percentages of NS4B expressed by H/WT [100%] and C1/WT [61%]), it was concluded that the observed variation in NS4B expression resulted from differences in antibody recognition rather than from slower processing at the NS4A/4B junction. Furthermore, no obvious changes in polyprotein processing were seen for the H77 replicon containing Con1-specific residues in the first two positions of H77 NS4B (SQ→AS) (Fig. 8B). Thus, changing the NS4A/4B and NS4B/5A junctions in H77-Con1 NS4B chimeric replicons does not appear to dramatically alter the biogenesis of the HCV polyprotein.
DISCUSSION
The primary goal of this study was to identify the genetic determinants responsible for the replication difference previously observed between replicons derived from the HCV genotype 1 strains Con1 and H77 (6) (Fig. 2). To determine whether genetic variation within NS4B contributed to genotype 1 replication efficiency in Huh-7.5 cells, a chimeric H77 replicon expressing the entire Con1 NS4B protein, which is ∼87% identical to the NS4B protein of H77, was generated. This chimera had accelerated replication kinetics relative to those of the parental H77 sequence and had essentially acquired the replicative ability of the Con1-derived replicon. In contrast, a lethal phenotype in transient replication assays was observed for a Con1 chimeric replicon containing the NS4B gene from H77 in place of the Con1 NS4B sequence (data not shown). Thus, taken together, the specific sequence of the NS4B protein is critical for robust RNA replication in cell culture.
H77-Con1 NS4B chimeras containing either the N-terminal 47 amino acids (H/CN47) or the C-terminal 214 residues (H/CC214) from Con1 NS4B also replicated with greater efficiency than the parental H77 replicon (Fig. 3). However, compared to the chimeric replicon expressing the entire Con1 NS4B protein, the H/CN47 and H/CC214 chimeras exhibited lower replication rates. The reason these hybrid NS4Bs are less effective remains unclear. One possibility is that the N- and C-terminal regions of NS4B carry out separate functions during RNA replication, perhaps by mediating distinct contacts either with the other HCV replication proteins or with specific host factors essential for robust RNA replication in cell culture. A nucleotide binding motif mediating GTP binding and hydrolysis was recently identified within the C-terminal domain of NS4B (12). Although the exact role of the GTPase in RNA replication has not been resolved, the sequence of the nucleotide binding motif is critical for efficient HCV RNA replication. Within this motif the amino acid at position 132 of NS4B differs between H77 (Val) and Con1 (Ile); however, the impact of this conservative substitution on the GTPase activity of NS4B and RNA replication remains to be tested. Alternatively, optimal NS4B function may depend on intramolecular interactions involving specific sequences in both the N- and C-terminal domains of NS4B. In support of the latter hypothesis, overexpressed NS4B has been shown to form oligomers in cultured cells, and the N-terminal half of NS4B (amino acids 1 to 135) harbors the major determinants mediating NS4B oligomerization (55). However, it is not yet known whether NS4B oligomerization is essential for HCV RNA replication and whether the enhancing amino acids identified in this study are involved in oligomerization.
The N-terminal 47 amino acids of H77 and Con1 NS4B differ at 15 positions; however, only 2 of these residues significantly impact replication (Fig. 4). Specifically, the Con1-derived amino acids, Gln-12 and Ile-22 in NS4B, act in concert to enhance H77 replication to levels comparable to that of the H/CN47 chimera and to achieve optimal replication of the H/CN33 chimera containing the N-terminal 33 amino acids from Con1 NS4B. These observations suggest that Gln at position 12 and Ile at position 22 enhance replication by directly altering replication complex activity, possibly via membrane-protein or protein-protein interactions. Consistent with this hypothesis, these two amino acids reside in a putative amphipathic helix that is important for membrane association and RNA replication, and this domain has also been proposed to have a direct function in replication complex assembly (13). Interestingly, positions 12 and 22 in NS4B are the only residues within this amphipathic helix domain that are not conserved between Con1 and H77. Furthermore, both of these sites are variable among different HCV genotypes: 1a and 3b (Met-12, Leu-22), 1b and 6a (Gln-12, Leu-22), 2a and 2b (Arg-12, Gln-22), 3a (Ala/Val-12, Leu-22), 4a (Gln-12, Leu/Ile/Val-22), and 5a (Ala-12, Leu-22). Although Leu was the most common amino acid observed at position 22 in NS4B in most genotypes, it was surprising to discover that Con1 was the only strain among the genotype 1b sequences analyzed (>100 isolates) that encoded Ile at position 22. It would be interesting to test how other amino acid replacements at these two positions in NS4B impact genotype 1 replication. Although further experimentation is needed, this natural variation may govern strain-specific interactions pivotal for HCV RNA replication.
The H77-Con1 NS4B chimeric replicons consistently produced positive- and negative-strand RNA levels greater than those of the parental H77 replicon (Fig. 7). This suggests that Con1 NS4B (and the N terminus of Con1 NS4B) directly influences the rate of RNA synthesis. Since the steps of the viral replication cycle are tightly coupled, enhanced RNA levels could result from increased translation from the HCV IRES. Previous studies yielded conflicting results: NS4B was reported to stimulate (22) or to repress (26) HCV IRES-directed translation. In the current study, genetic variation in NS4B did not appear to influence viral protein expression (Fig. 8), although the contribution of subtle effects of NS4B on gene expression cannot be entirely ruled out. Furthermore, no significant differences between the cleavage profiles of the parental and chimeric polyproteins were observed (Fig. 8B), and the stabilities of the chimeric NS4B proteins H/CN47, H/CN33, and H/CN34-47 and the L22I mutant were similar to that of NS4B expressed from the H/WT and H/C4B replicons (data not shown). Taken together, it appears that the Con1 NS4B-mediated increase in H77 replication was not due to altered translation efficiency, polyprotein processing, or NS4B stability.
NS4B is postulated to play a central role in mediating the formation of membranous structures that represent a site of viral replication (11, 18). Sequence differences in NS4B have the potential to influence the capacity of NS4B to associate with membranes, induce membrane alterations more favorable for replication complex assembly, or facilitate the formation of productive replication complexes, any of which could significantly impact the ability of HCV replicons to establish RNA replication. In this study, enhanced replication of chimeric replicons was predominantly associated with increased RNA replication per cell, and only minor differences in the frequency of cells able to support chimeric and parental replication were observed (Fig. 5). Thus, our working model is that Con1 NS4B primarily enhances the processes involved in sustaining efficient RNA replication. At this point, it is not clear whether increased RNA synthesis arises from highly efficient replication factories, a greater number of active replication complexes, or both.
If NS4B-induced membrane remodeling was a major mechanism for increased HCV RNA replication, then Con1 NS4B might have the capacity to act in trans to enhance H77 replication. Under the experimental conditions used in this study, expression of Con1 NS4B in trans had no effect on H77 replication, and although this suggests that Con1 NS4B is required in a cis configuration to increase H77 replication, it is still possible that the timing and mode of NS4B expression are critical in order to observe an effect. This observation is consistent with previous reports that NS4B from HCV (1, 52) and other members of the Flaviviridae, including Kunjin virus (28) and bovine viral diarrhea virus (19), cannot trans-complement replicons carrying lethal mutations, insertions, or deletions in NS4B.
How does Con1 NS4B enhance H77 RNA synthesis in Huh-7.5 cells? One possibility is that the Con1 NS4B protein functions more efficiently in the context of the H77-encoded replication machinery than the native H77 NS4B. NS4B has been reported to interact with virtually all the other HCV nonstructural proteins (10, 17, 36, 47), and so far a functional interaction between NS3 and NS4B has implicated NS4B as a regulator of the replication complex. Specifically, an interaction between HCV genotype 1b NS4B and NS3 partially suppresses the nucleoside triphosphatase activity of NS3 in vitro (47). In the case of the flavivirus dengue virus, NS4B interacts with NS3, resulting in the displacement of NS3 from single-stranded RNA (53). However, these NS4B-mediated effects on NS3 were demonstrated in vitro and need to be confirmed in cell cultures supporting RNA replication, where all the components of the replication complex are operative. It should be noted that the H77 replicons used in this study were derived from an infectious molecular clone of the H77 strain that replicates to high titers in chimpanzees (32). While Con1 NS4B may have the capacity to further increase H77 replication in chimpanzee liver, an alternative explanation is that Con1 NS4B functions more efficiently in the Huh-7.5 cellular environment via interactions with specific host components that play a role in the assembly of the viral replication complex. At this stage, very few interactions between NS4B and host cell proteins have been identified (51). While it is not entirely clear whether RNA replication in cell culture depends on NS4B-mediated interactions with cellular proteins, identification of host-interacting partners of NS4B may represent an important step toward a better understanding of NS4B function.
Robust H77 replication in the Huh-7 cell line was previously achieved by combining five cell culture-adaptive mutations in the NS3-4A protease complex and the NS5A protein (54). In Huh-7 cells, the replication efficiency of an H77 replicon carrying these mutations was significantly greater than that of a replicon harboring the adaptive changes used in this study (P1496L in NS3 and S2204I in NS5A) and slightly higher than that of a replicon derived from the robustly replicating genotype 1b HCV-N strain. The mechanism by which these five adaptive mutations promote efficient H77 RNA replication remains unknown. Given the enhanced replication phenotype of the H77-Con1 NS4B chimeras reported here, it is intriguing to speculate that some of these mutations compensate for the less efficient H77 NS4B protein by mediating critical interactions with Huh-7 cell proteins, thereby permitting the assembly of functional replication complexes. Interestingly, replication of the H/C4B chimera reported here requires both P1496L in NS3 and S2204I in NS5A, indicating that in Huh-7.5 cells, Con1 NS4B cannot substitute for these specific adaptive mutations.
Although the mechanism is not well understood, NS4B, together with NS3 and NS4A, appears to be involved in NS5A hyperphosphorylation (31, 45). There is also a growing body of evidence suggesting that disruption of NS5A hyperphosphorylation is important for efficient Con1 replication in Huh-7 cells. For example, mutations in NS5A ablating or reducing Con1 NS5A hyperphosphorylation can dramatically enhance RNA replication (2, 5); nonadapted Con1 RNA replicates efficiently when Huh-7 cells are treated with inhibitors that selectively block NS5A hyperphosphorylation (46); and highly adaptive mutations in Con1 NS4B impair the hyperphosphorylation of NS5A (2, 15). These observations are particularly interesting in light of the fact that robustly replicating Con1 replicons carrying S2204I in NS5A do not express hyperphosphorylated NS5A (5), whereas NS5A hyperphosphorylation is detected during H77 replication (6). Thus, it is conceivable that the lower replication kinetics of H77 parental RNA are in part due to the production of a hyperphosphorylated NS5A species and that Con1 NS4B may improve H77 replication by inhibiting NS5A hyperphosphorylation. However, preliminary data show that during replication of the H77 chimera expressing Con1 NS4B, H77 NS5A remains in a hyperphosphorylated state (data not shown), suggesting that modulation of NS5A hyperphosphorylation is unlikely to be a major mechanism by which Con1 NS4B augments H77 replication.
While the favored hypothesis is that Con1 NS4B directly affects the activity of the H77 replication complex via protein-protein interactions, it is also conceivable that Con1 NS4B influences replication by modulating the cellular environment. This possibility is not without precedent, since HCV genotype 1b NS4B has been shown to inhibit host protein synthesis (16, 26), regulate the ER stress response (51, 56), alter the expression of HeLa cell genes involved in host defense, cell homeostasis, and carcinogenesis (57), and induce interleukin-8 gene expression, possibly through activation of NF-κB (24, 27). Given that ectopic expression of Con1 NS4B did not enhance H77 parental replication (Fig. 6), Con1 NS4B is probably not inducing a cellular environment more conducive to efficient RNA replication.
In conclusion, a chimeric approach has identified NS4B as a crucial player in HCV replication in cell culture and has revealed an unexpected role for NS4B in maintaining RNA synthesis once HCV replication has been established. Currently the exact molecular mechanism by which Con1 NS4B amplifies H77 RNA transcription has not been resolved. However, the NS4B chimeras, with enhanced replication phenotypes compared to that of the parental sequence, represent valuable tools in ongoing efforts to further dissect critical NS4B sequences and better define how NS4B functions in RNA replication.
Acknowledgments
I thank Anne Thierauf for helpful discussions and for H/CN33 mutant replicons; Aster Beyene, Anne Thierauf, Henry Huang, and Sondra Schlesinger for critical reading of the manuscript; and Daniel Ader and Elizabeth Norgard for technical support. I am also grateful to Charles Rice for providing pH/ΔE1-p7(L+I), pH/SG-Neo(L+I), pH/SG-Neo/pol−, and pHCVBMFL/S2204I and to Robert Sarisky for supplying HCV-specific antisera.
This work was supported by the Ellison Medical Foundation program for New Scholars in Global Infectious Disease (ID-NS-119-03) and by NIH grant AI065985.
K.J.B. and Washington University may receive income based on a license of related technology by the University to Apath, LLC. Apath, LLC, did not support this work.
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appel, N., T. Pietschmann, and R. Bartenschlager. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S. E., L. Tomei, and R. DeFrancesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., and E. A. Norgard. 2006. HCV replicon systems, p. 311-351. In S. L. Tan (ed.), Hepatitis C viruses: genomes and molecular biology. Horizon Scientific Press, Norwich, United Kingdom.
- 9.Blight, K. J., and E. A. Norgard. 2006. Recent advances in hepatitis C virus replication, p. 45-90. In K. L. Hefferon (ed.), Recent advances in RNA virus replication. Transworld Research Network, Kerala, India.
- 10.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77:5401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Einav, S., M. Elazar, T. Danieli, and J. S. Glenn. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J. Virol. 78:11288-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elazar, M., P. Liu, C. M. Rice, and J. S. Glenn. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 15.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 101:13038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florese, R. H., M. Nagano-Fujii, Y. Iwanaga, R. Hidajat, and H. Hotta. 2002. Inhibition of protein synthesis by the nonstructural proteins NS4A and NS4B of hepatitis C virus. Virus Res. 90:119-131. [DOI] [PubMed] [Google Scholar]
- 17.Gao, L., H. Aizaki, J.-W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grassmann, C. W., O. Isken, N. Tautz, and S.-E. Behrens. 2001. Genetic analysis of the pestivirus nonstructural coding region: defects in the NS5A unit can be complemented in trans. J. Virol. 75:7791-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy, R., J. Marcotrigiano, K. J. Blight, J. E. Majors, and C. M. Rice. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 77:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, Y., W. Yan, C. Coito, Y. Li, M. Gale, Jr., and M. G. Katze. 2003. The regulation of hepatitis C virus (HCV) internal ribosome-entry site-mediated translation by HCV replicons and nonstructural proteins. J. Gen. Virol. 84:535-543. [DOI] [PubMed] [Google Scholar]
- 23.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H.-G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 24.Kadoya, H., M. Nagano-Fujii, L. Deng, N. Nakazono, and H. Hotta. 2005. Nonstructural proteins 4A and 4B of hepatitis C virus transactivate the interleukin 8 promoter. Microbiol. Immunol. 49:265-273. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko, T., Y. Tanji, S. Satoh, M. Hijikata, S. Asabe, K. Kimura, and K. Shimotohno. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320-326. [DOI] [PubMed] [Google Scholar]
- 26.Kato, J., N. Kato, H. Yoshida, S. K. Ono-Nita, Y. Shiratori, and M. Omata. 2002. Hepatitis C virus NS4A and NS4B proteins suppress translation in vivo. J. Med. Virol. 66:187-199. [DOI] [PubMed] [Google Scholar]
- 27.Kato, N., H. Yoshida, S. K. Ono-Nita, J. Kato, T. Goto, M. Otsuka, K. Lan, K. Matsushima, Y. Shiratori, and M. Omata. 2000. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology 32:405-412. [DOI] [PubMed] [Google Scholar]
- 28.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160-166. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. E., W. K. Song, K. M. Chung, S. H. Back, and S. K. Jang. 1999. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch. Virol. 144:329-343. [DOI] [PubMed] [Google Scholar]
- 31.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 33.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam, A. M. I., and D. N. Frick. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80:404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, C., J. W. Wu, K. Hsiao, and M. S. Su. 1997. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J. Virol. 71:6465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindstrom, H., M. Lundin, S. Haggstrom, and M. A. Persson. 2006. Mutations of the Hepatitis C virus protein NS4B on either side of the ER membrane affect the efficiency of subgenomic replicons. Virus Res. 121:169-178. [DOI] [PubMed] [Google Scholar]
- 38.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 41.Lundin, M., H. Lindstrom, C. Gronwall, and M. A. Persson. 2006. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J. Gen. Virol. 87:3263-3272. [DOI] [PubMed] [Google Scholar]
- 42.Lundin, M., M. Monne, A. Widell, G. von Heijne, and M. A. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyanari, Y., M. Hijikata, M. Yamaji, M. Hosaka, H. Takahashi, and K. Shimotohno. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278:50301-50308. [DOI] [PubMed] [Google Scholar]
- 44.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neddermann, P., A. Clementi, and R. De Francesco. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neddermann, P., M. Quintavalle, C. Di Pietro, A. Clementi, M. Cerretani, S. Altamura, L. Bartholomew, and R. De Francesco. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78:13306-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piccininni, S., A. Varaklioti, M. Nardelli, B. Dave, K. D. Raney, and J. E. McCarthy. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 277:45670-45679. [DOI] [PubMed] [Google Scholar]
- 48.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzich, J. A., J. K. Tamura, F. Palmer-Hill, P. Warrener, A. Grakoui, C. M. Rice, S. M. Feinstone, and M. S. Collett. 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 67:6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong, W.-Y., M. Nagano-Fujii, R. Hidajat, L. Deng, Y. Takigawa, and H. Hotta. 2002. Physical interactions between hepatitis C virus NS4B protein and CREB-RP/ATF6B. Biochem. Biophys. Res. Commun. 299:366-372. [DOI] [PubMed] [Google Scholar]
- 52.Tong, X., and B. A. Malcolm. 2006. Trans-complementation of HCV replication by non-structural protein 5A. Virus Res. 115:122-130. [DOI] [PubMed] [Google Scholar]
- 53.Umareddy, I., A. Chao, A. Sampath, F. Gu, and S. G. Vasudevan. 2006. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 87:2605-2614. [DOI] [PubMed] [Google Scholar]
- 54.Yi, M., and S. M. Lemon. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J. Virol. 78:7904-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, G. Y., K. J. Lee, L. Gao, and M. M. Lai. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80:6013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Y., B. Gao, L. Ye, L. Kong, W. Jing, X. Yang, Z. Wu, and L. Ye. 2005. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J. Microbiol. 43:529-536. [PubMed] [Google Scholar]
- 57.Zheng, Y., L. B. Ye, J. Liu, W. Jing, K. A. Timani, X. J. Yang, F. Yang, W. Wang, B. Gao, and Z. H. Wu. 2005. Gene expression profiles of HeLa cells impacted by hepatitis C virus non-structural protein NS4B. J. Biochem. Mol. Biol. 38:151-160. [DOI] [PubMed] [Google Scholar]