Abstract
The structure of immature West Nile virus particles, propagated in the presence of ammonium chloride to block virus maturation in the low-pH environment of the trans-Golgi network, was determined by cryo-electron microscopy (cryo-EM). The structure of these particles was similar to that of immature West Nile virus particles found as a minor component of mature virus samples (naturally occurring immature particles [NOIPs]). The structures of mature infectious flaviviruses are radically different from those of the immature particles. The similarity of the ammonium chloride-treated particles and NOIPs suggests either that the NOIPs have not undergone any conformational change during maturation or that the conformational change is reversible. Comparison with the cryo-EM reconstruction of immature dengue virus established the locations of the N-linked glycosylation sites of these viruses, verifying the interpretation of the reconstructions of the immature flaviviruses.
West Nile virus, dengue virus, yellow fever virus, and tick-borne encephalitis virus, members of the Flaviviridae family (10), are enveloped viruses with single-stranded RNA genomes. West Nile virus infections in the United States (9) resulted in more than 140 deaths in 2006 (http://www.cdc.gov/ncidod/dvbid/westnile).
Genomic RNA replicated in the cytoplasm associates with the capsid (C) protein during the viral life cycle. Immature flavivirus particles are formed when these nucleocapsid cores acquire their lipid envelope together with these membrane-anchored envelope (E) protein and the precursor membrane (prM) protein during the process of budding into the lumen of the endoplasmic reticulum (ER). The prM and E proteins exist as trimers of heterodimers on the surface of the immature particles (19), which then pass through the trans-Golgi network (TGN). The E and prM proteins become glycosylated in the ER, albeit at different residues for West Nile and dengue viruses. The fusion peptide on the E protein is protected from premature fusion with membranes in the cell by being covered with the amino-terminal precursor peptide pr (19), rendering the immature particles noninfectious. The maturation process is activated by furin cleavage of the prM protein after exposure to acidic pH, which results in the release of the precursor peptide (16).
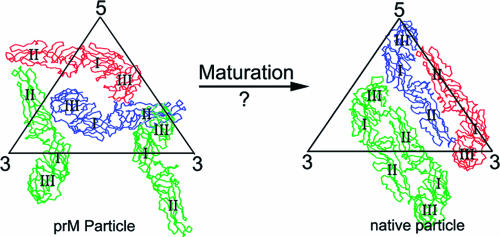
The maturation of flaviviruses can be inhibited by growing the viruses in a medium containing the acidotropic agent NH4Cl, allowing the accumulation of immature virus particles for structural studies (14). The immature dengue virus and yellow fever virus particles, produced by adding NH4Cl to the medium (ammonium chloride-treated immature particles [ACIPs]), have diameters of about 600 Å, with trimers of prM-E heterodimers arranged icosahedrally, forming 60 protruding spikes on the surface of each particle (19). In contrast, mature infectious virus particles contain 90 E-E homodimers that form a much smoother surface and these particles have diameters of only about 500 Å (12, 17). Without visualizing any intermediate states in virus maturation, the large conformational changes that must occur are difficult to comprehend (Fig. 1). It is, therefore, essential to determine that the immature structure, as described by Zhang et al. (19), is, indeed, a true structural intermediate in the life cycle of the virus.
FIG. 1.
Diagram showing the structural rearrangement required for immature particles to become mature particles. The Cα backbones of the three independent E molecules per icosahedral asymmetric unit are colored green, red, and blue. The three domains in each E molecule are labeled I, II, and III. (Reprinted from the EMBO Journal [19] with permission of the publisher.)
Organization of immature West Nile virus.
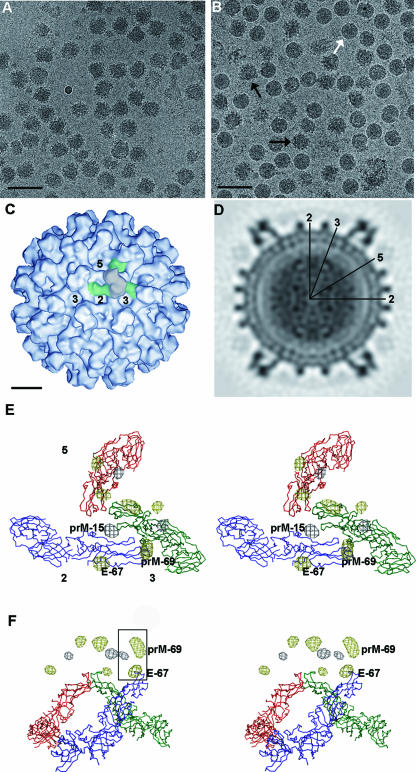
A three-dimensional, 24-Å-resolution, cryo-electron microscopy (cryo-EM) reconstruction of the West Nile virus ACIPs (Table 1; Fig. 2A) showed surface features similar to those of dengue virus ACIPs (19). The surface of the virus is dominated by 60 icosahedrally arranged trimeric glycoprotein spikes, as is the surface of naturally occurring immature particles (NOIPs) (Fig. 2C). Cross-sectional views of the reconstruction show a multilayered structure composed of the prM and E glycoproteins (between 199 and 287 Å radius), a lipid bilayer (167 to 199 Å radius), and the nucleocapsid core (within a radius of 167 Å), as also seen in NOIPs (Fig. 2D). Similar to that of dengue virus, the nucleocapsid core does not show well- ordered structural features on the icosahedrally averaged cryo-EM map.
TABLE 1.
Cryo-EM image reconstruction dataa
| Type of virus particle | No. of micrographs | Defocus (μm) | No. of particles | PFT CCb | Resolution (Å) |
|---|---|---|---|---|---|
| NOIPsc | 45 | 1.3-3.1 | 341 | 0.288 | 24 |
| ACIPsd | 39 | 1.2-3.3 | 720 | 0.261 | 24 |
Images were recorded with an FEI Philips CM300 transmission electron microscope at a calibrated magnification of ×47,400. Micrographs were digitized with a Zeiss-PHODIS scanner to yield a sampling corresponding to 2.95 Å between pixels on the specimen.
PFT CC is the correlation coefficient (CC) between the model projection and the corresponding raw image after polar Fourier transformation (PFT) (1).
West Nile virus NOIPs were centered and classified as described by Furst et al. (6) but using the program SPIDER (5). The self-common line method (2) was then applied to the class averages to generate an initial model for the subsequent reconstruction of the NOIP data as for the reconstruction of the West Nile virus ACIP data.
The reconstruction of West Nile virus ACIPs was performed with the cryo-EM image reconstruction of dengue virus ACIPs as a starting model by using the PFT procedure (1).
FIG. 2.
Cryo-EM micrographs, density maps, and difference density maps showing the positions of carbohydrate sites. (A) Cryo-EM micrograph of West Nile virus ACIPs. Immature particles were produced from cells treated with ammonium chloride. Confluent Vero cells were infected with West Nile virus at a multiplicity of infection of 1.0 in the presence of 2% fetal calf serum. At 22 h after infection, the cell culture medium was exchanged with medium containing 20 mM NH4Cl. This first overlay was discarded at 23 h after infection and replaced with fresh NH4Cl-containing medium. The cell culture supernatant was harvested 48 h after infection, and immature virus was purified as described previously for mature particles (8). Cryo-EM micrographs showed that the ammonium chloride-treated preparation consisted almost exclusively of immature particles (ACIPs). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of these ACIPs showed that the majority of the prM protein was uncleaved. (B) Cryo-EM micrograph of West Nile virus particles produced without NH4Cl. Black arrows point to NOIPs, whereas the white arrow points to mature particles. Scale bars in both panels A and B represent 1,000 Å. Mature West Nile virus (NY99) was propagated and purified as described elsewhere (8), except that the virus was harvested 30 h after infection and concentrated by polyethylene glycol precipitation. The analysis of these particles by SDS-PAGE showed the presence of E, C, and prM proteins, as well as M proteins. Cryo-EM showed that up to one in five particles in the resulting preparations was an NOIP. (C) Surface representation of NOIPs. Icosahedral 5-, 3-, and 2-fold axes are labeled. In one spike, prM protein is colored gray and E protein is colored green. The scale bar represents 100 Å. (D) Central cross-section showing the multilayer organization of West Nile virus NOIPs. Icosahedral symmetry operators are indicated with black lines. (E) Stereo view of a spike showing the difference density between West Nile virus and dengue virus ACIPs. The Cα backbone of the E molecules is shown in green, red, and blue. Positive and negative peaks are colored gray and yellow, respectively. The icosahedral 5-, 3-, and 2-fold axes are labeled. The N-linked glycosylation sites (Asn-15 in the prM protein of West Nile virus [prM-15], Asn-67 in the E protein of dengue virus [E-67], and Asn-69 in the prM protein of dengue virus [prM-69]) associated with the difference densities are labeled. (F) Stereo view showing the same difference map as panel E but from a side view. The three difference density peaks associated with one prM-E heterodimer are outlined with a black rectangle.
Since the hinge angle between domains I and II of the E protein is a little different in mature and immature dengue virus particles (20), the fitting of each domain into the cryo-EM map should be performed independently. However, this approach was not reasonable because of the relatively low resolution of the current West Nile virus reconstruction. Hence, the atomic structure of the West Nile virus E protein (7, 13) was fitted, as a single rigid body, into the cryo-EM density map of immature West Nile virus by using the program EMfit (15) (Table 2). Three E monomers could be positioned into each icosahedral asymmetric unit. Like that of the E proteins in immature dengue virus particles, the organization of the E proteins in West Nile virus does not conform to classical T=3 quasisymmetry (3). The three E molecules within each spike of the immature particles are tilted with respect to the viral surface, with their fusion peptides pointing toward the tip of the spike. The fusion peptides are covered by extra density, attributed to the prM protein.
TABLE 2.
Fitting of the E glycoprotein X-ray structure into the cryo-EM mapa
| Moleculeb | sumf1c (%) | sumf2c (%) | sumf3c (%) | −dend (%) | clashe (%) | centxf (Å) | centyf (Å) | centzf (Å) |
|---|---|---|---|---|---|---|---|---|
| Green | 50.6 | 59.0 | 51.2 | 0.0 | 0.2 | 33.1 | −84.5 | 201.5 |
| Red | 48.5 | 56.4 | 49.3 | 0.0 | 0.0 | 92.2 | −46.6 | 196.5 |
| Blue | 54.3 | 57.6 | 50.6 | 0.2 | 0.0 | 33.2 | −4.4 | 220.5 |
The Cα backbone of the West Nile virus E molecule (Protein Data Bank accession number, 2HG0) was used to fit into the NOIP and ACIP electron density maps with a procedure described by Zhang et al. (19) and Rossmann et al. (15). The pixel size of the ACIP and NOIP West Nile virus maps was adjusted with respect to the cryo-EM map of immature dengue virus by minimizing the root mean square difference in density within the external protein layer (between 190 and 290 Å radius). The cryo-EM densities on the two magnification-adjusted maps were then put onto the same relative scale by minimizing the function ∑(ρden − aρWN − b)2 by using density values within the external protein region, where a and b are scale factors and ρden and ρWN are the density values for the dengue and West Nile virus immature particles, respectively. After having established both the magnification and density scale factors, the difference map between the dengue and West Nile virus immature particles was computed.
sumf1, sumf2, and sumf3 are the average densities at Cα atoms for domains I, II, and III, respectively, expressed as percentages of the highest density in the map.
−den is the percentage of atoms in negative density.
clash is the percentage of atoms that sterically interfere between neighboring molecules.
centx, centy, and centz are the coordinates of the center of mass of the E monomer in the map.
Glycosylation sites in immature particles.
The West Nile and dengue virus glycoproteins have 46% sequence identity. West Nile virus has two potential glycosylation sites (at E protein residue 154 [E-154] and prM protein residue 15 [prM-15]) on each prM-E heterodimer that differ from the glycosylation sites of immature dengue virus (at E-67, E-153, and prM-69). A 24-Å-resolution difference map, computed as the immature West Nile virus density minus the immature dengue virus density, showed three strong positive and six strong negative well-defined density peaks per icosahedral asymmetric unit, with magnitudes roughly 1.5 times as large as any other positive or negative peaks. The three positive peaks obey the symmetry of the three prM-E heterodimers within each icosahedral asymmetric unit and can be attributed to the carbohydrate moieties associated with the prM protein (at prM-15) in West Nile virus. The six negative peaks within each icosahedral asymmetric unit can be divided into two sets of three peaks. The spatial relationship among the three peaks of each set is consistent with that among the three E monomers (Fig. 2E and F). Thus, there are two negative peaks associated with each prM-E heterodimer, presumably corresponding to the glycosylation sites at E-67 and prM-69 in dengue virus. The centers of one set of negative difference density peaks are about 17 Å from the corresponding Cα atoms of the glycosylated asparagines (at E-67) and well outside the density of the fitted E proteins. This distance is comparable to distances found in a study of carbohydrate sites in Sindbis virus (18), confirming the correct handedness of the cryo-EM map of immature West Nile virus and dengue virus particles, as well as the fitting of the E molecules into the cryo-EM map (Table 3). The other negative glycosylation site must correspond to the carbohydrate moiety at prM-69 of dengue virus. The West Nile virus glycosylation site at E-154 (which should give a positive peak in the difference map) is structurally aligned with that at E-153 in dengue virus (which should produce a negative peak). As there is no significant peak in the difference map near these residues, it appears that these two peaks have cancelled each other out.
TABLE 3.
Positions of the highest difference map density peaks corresponding to carbohydrate moieties
| Virus | Glycosylation site | Moleculea | xb (Å) | yb (Å) | zb (Å) | Distancec (Å) |
|---|---|---|---|---|---|---|
| West Nile | prM-15 | Green | 47.3 | −77.7 | 260.1 | |
| Red | 86.1 | −45.9 | 258.3 | |||
| Blue | 43.7 | −39.4 | 269.7 | |||
| Dengue | prM-69 | Green | 61.3 | −54.2 | 271.6 | |
| Red | 70.1 | −32.4 | 276.3 | |||
| Blue | 30.9 | −64.0 | 273.3 | |||
| E-67 | Green | 61.1 | −76.9 | 244.4 | 15.5 | |
| Red | 94.5 | −30.1 | 247.0 | 17.3 | ||
| Blue | 19.9 | −32.3 | 261.5 | 17.6 |
The x, y, and z coordinates of the centers of difference density peaks between West Nile virus and dengue virus immature particles are shown.
The distance between the center of the difference density peak and the corresponding Cα atom of the fitted E X-ray structure.
Naturally occurring immature particles resemble immature particles produced by NH4Cl.
The preparation of West Nile virus in Vero cells is a mixture of both mature (smaller and smoother)- and immature (larger and spikier)-looking particles, as observed in cryo-EM micrographs (Fig. 2B), which could not be separated by additional purification steps. About 20% of the manually selected spiky particles converged into a structure described here. The rejected particles probably have an intermediate conformation between those of mature and immature virus particles. The 24-Å-resolution cryo-EM reconstruction of these NOIPs (Fig. 2C) is similar to that of the ACIPs, with a root mean square deviation of 2.9 Å between equivalent Cα atoms of the fitted E molecules. The biggest peaks on the NOIP or ACIP cryo-EM map were three times as large as the biggest peak on the (NOIP-ACIP) difference map. Furthermore, the biggest density peak in the (NOIP-ACIP) difference map was only about three times the root mean square deviation of the density from the mean for the difference map as a whole. Thus, there was no significant difference between these two types of particles at the resolution attained in this study. In addition, the radially averaged correlation between the densities in the two maps, computed as a function of the distance from the center of the particles, showed that the glycoprotein and lipid layers are essentially identical. Thus, the structure of ACIPs is likely to be the true state of the immature particles and is not the result of an artifact created by the NH4Cl treatment.
Immature virus particles pass from an environment of neutral pH in the ER to one of acidic pH in the TGN (4, 11) and then, after maturation, back to neutral pH upon exiting the host cell (Table 4, pathways A to D). The large conformational change that occurs during maturation might be caused when entering the low-pH environment of the TGN, when exiting from the TGN (14), or by a combination of both these transitions. The ACIPs are presumably produced because the NH4Cl blocks the maturation of virus particles by raising the pH of the TGN to inhibit either a putative conformational change (16) upon entrance into the TGN and/or the subsequent cleavage of the prM protein. The inhibition of the pH-induced, putative conformational change might stop the exposure of the cleavage site (Table 4, pathway G), or the inhibition of the prM protein cleavage might alter the subsequent conformational changes that are required for maturation (Table 4, pathway F). Both the ACIPs and NOIPs contain uncleaved prM protein, suggesting that the NOIPs must have bypassed the cleavage process. Furthermore, the structures of the ACIPs and the NOIPs are very similar. The NOIPs might not have undergone any conformational change upon entering the TGN (Table 4, pathway B), might have gone through a reverse conformational change upon leaving the TGN (Table 4, pathway D), or might have been released by cytopathic effects without passing the acidic TGN (Table 4, pathway E). However, a low-pH-induced conformational change has been shown to be irreversible for tick-borne encephalitis virus (16). In contrast, particles that are missing the precursor peptide will continue on the pathway to maturation (Table 4, pathways A and C). A lack of cleavage or a lack of conformational change, whichever might be the reason for the production of NOIPs, might be the result of insufficient time of residence in the acidic-pH environment of the TGN, or local variability of pH in the TGN, to permit the complete maturation process.
TABLE 4.
Possible maturation pathways of flavivirus particles
| Growth condition | Pathway, starting with assembled particles in the ER at neutral pH | Conformational change → | Cleavage event in TGN at acidic pH | Conformational change → | Type of particles released into neutral-pH environment |
|---|---|---|---|---|---|
| No NH4Cl | A | No | prM cleavage | Yes | Mature virus |
| B | No | No prM cleavage | No | NOIPs | |
| C | Yes | prM cleavage | Yes | Mature virus | |
| D | Yes | No prM cleavage | Reverse change | NOIPs | |
| E | NAa | TGN bypassed | NA | NOIPs | |
| NH4Cl added | F | No | Blocked cleavage | No | ACIPs |
| G | Blocked | No cleavage | No | ACIPs |
NA, not applicable.
Accession numbers.
The electron density map of West Nile virus NOIPs was deposited with the Electron Microscopy Data Bank (accession number EMD-1314). The fitted model of E molecules was deposited with the Protein Data Bank (accession number 2OF6).
Acknowledgments
We thank Jue Chen for many helpful discussions and Cheryl Towell, Sheryl Kelly, and Sharon Wilder for help in preparing the manuscript. Figures were made using the programs DINO: Visualizing Structural Biology (2002; http://www.dino3d.org) and RobEM (http://cryoem.ucsd.edu/programDocs/runRobem.txt).
The work was supported by a National Institutes of Health Program Project grant (AI 55672) to M.G.R. and R.J.K. and an award from the Keck Foundation for the purchase of the CM300 transmission electron microscope used in this study.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caspar, D. L. D., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27:1-24. [DOI] [PubMed] [Google Scholar]
- 4.Demaurex, N., W. Furuya, S. D'Souza, J. S. Bonifacino, and S. Grinstein. 1998. Mechanism of acidification of the trans-Golgi network (TGN). J. Biol. Chem. 273:2044-2051. [DOI] [PubMed] [Google Scholar]
- 5.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 6.Furst, J., R. B. Sutton, J. Chen, A. T. Brunger, and N. Grigorieff. 2003. Electron cryomicroscopy structure of N-ethyl maleimide sensitive factor at 11 Å resolution. EMBO J. 22:4365-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanai, R., K. Kar, K. Anthony, L. H. Gould, M. Ledizet, E. Fikrig, W. A. Marasco, R. A. Koski, and Y. Modis. 2006. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 80:11000-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufmann, B., G. E. Nybakken, P. R. Chipman, W. Zhang, M. S. Diamond, D. H. Fremont, R. J. Kuhn, and M. G. Rossmann. 2006. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. USA 103:12400-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 10.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 11.Llopis, J., J. M. McCaffery, A. Miyawaki, M. G. Farquhar, and R. Y. Tsien. 1998. Measurement of cytosolic, mitochondrial and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA 95:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 13.Nybakken, G. E., C. A. Nelson, B. R. Chen, M. S. Diamond, and D. H. Fremont. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 80:11467-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174:450-458. [DOI] [PubMed] [Google Scholar]
- 15.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136:190-200. [DOI] [PubMed] [Google Scholar]
- 16.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, Y., W. Zhang, S. Ogata, D. Clements, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]