Abstract
The stable signal peptide (SSP) of the lymphocytic choriomeningitis virus surface glycoprotein precursor has several unique characteristics. The SSP is unusually long, at 58 amino acids, and contains two hydrophobic domains, and its sequence is highly conserved among both Old and New World arenaviruses. To better understand the functions of the SSP, a panel of point and deletion mutants was created by in vitro mutagenesis to target the highly conserved elements within the SSP. We were also able to confirm critical residues required for separate SSP functions by trans-complementation. Using these approaches, it was possible to resolve functional domains of the SSP. In characterizing our SSP mutants, we discovered that the SSP is involved in several distinct functions within the viral life cycle, beyond translocation of the viral surface glycoprotein precursor into the endoplasmic reticulum lumen. The SSP is required for efficient glycoprotein expression, posttranslational maturation cleavage of GP1 and GP2 by SKI-1/S1P protease, glycoprotein transport to the cell surface plasma membrane, formation of infectious virus particles, and acid pH-dependent glycoprotein-mediated cell fusion.
The Arenaviridae comprise a group of enveloped RNA viruses that include several causative agents of hemorrhagic fevers in the New World and Africa. Among these are Lassa fever virus (LASV) and Junín virus (JUNV). The prototypic arenavirus, lymphocytic choriomeningitis virus (LCMV), has a bisegmented, single-stranded, negative-sense RNA genome. Each of the two segments uses an ambisense coding strategy to direct the synthesis of two polypeptides. The large (L) segment (7.2 kb) encodes a small RING finger protein, Z, and the RNA-dependent RNA polymerase, the L protein, while the small (S) segment (3.4 kb) encodes the nucleoprotein, NP, and the glycoprotein (GP) precursor pGPC (5). The 498-amino-acid pGPC of LCMV consists of three domains (Fig. 1A) that are produced as independent polypeptides by posttranslational processing: residues 1 to 58 comprise the stable signal peptide (SSP), which is cotranslationally cleaved by signal peptidase and followed by the GP precursor GPC (residues 59 to 498), which is further processed into GP1 (residues 59 to 265) and GP2 (residues 266 to 498) (5, 7, 26) by the cellular protease, SKI-1/S1P (3). GP1 is heavily glycosylated, contains the receptor binding site and antibody neutralization sites, and is noncovalently associated with GP2. GP2 contains a transmembrane region and anchors the GP complex in the lipid bilayer of the cell membrane and virus envelope (6). LCMV cell entry involves a fusion event that requires exposure to acidic pH (9) to trigger GP1 dissociation from GP2 and to induce irreversible conformational changes in GP2 that mediate membrane fusion (10).
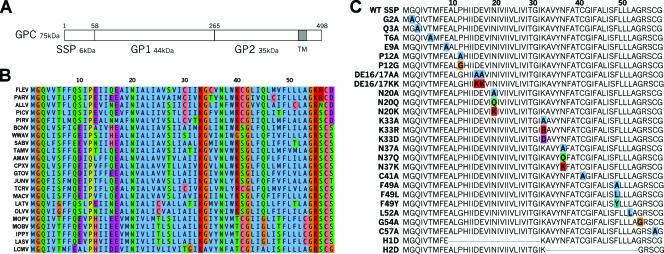
FIG. 1.
(A) LCMV GPC. Overview of pGPC protein, with cleavage sites for the SSP and GP1/GP2 indicated. (B) Alignment of arenavirus SSP sequences. Sequence alignment of arenavirus SSPs was done using Jalview software (8). Coloring is patterned after ClustalX conventions, with aliphatic and aromatic residues in blue, amines and hydroxyls in green, acidic residues in purple, positively charged residues in red, proline in yellow, glycine in orange, and cysteine and histidine in pink. (C) SSP mutant panel. The LCMV SSP amino acid sequence is shown, with locations of SSP mutations noted in bold and with color coding as described for panel B. Arenavirus nomenclature follows standard International Committee on Taxonomy of Viruses abbreviations. Accession numbers for the aligned sequences are as follows: ALLV, AAG42529; AMAV, AAN32967; BCNV, AAX99345; CPXV, AAN32963; FLEV, AAN09937; GTOV, NP_899210; IPPY, YP_516230; JUNV, NP_899218; LASV, AAL13212; LATV, AAN09940; LCMV, NP_694851; MACV, NP_899212; MOBV, YP_516226; MOPV, YP_170709; OLVV, AAC54654; PARV, AAN09944; PICV, AAC32279; PIRV, YP_025080; SABV, YP_089665; TAMV, AAN09948; TCRV, NP_694849; and WWAV, AAK60497.
Signal peptides (SPs) mediate protein translocation into the lumen of the endoplasmic reticulum (ER), promoting proper folding and posttranslational modifications of these polypeptides. Following translocation, an SP is typically cleaved and released into the ER membrane (23). Typical SPs range in size from 15 to 25 amino acids and contain a hydrophobic core of approximately seven residues, a polar C terminus of approximately five residues, and a net positive N terminus of variable length (24). Notably, the highly conserved SSPs of LCMV and LASV contain 58 amino acids (11, 16), which is unusually long for a canonical SP. Arenavirus SSPs contain a conserved N-terminal myristoyl moiety acceptance site (G2), an N-terminal extension, two hydrophobic domains (H1 [residues 10 to 32] and H2 [residues 34 to 53]) separated by a single positive charge (K33), and a C-terminal SP cleavage site (residues 54 to 58). Arenavirus SSPs are present as stable polypeptides in infected cells and are also found in viral particles (16). LASV SSP cleavage is a prerequisite for GPC posttranslational cleavage into GP1 and GP2 (11). LASV SSP replacement with a heterologous mammalian or viral SP does not affect pGPC expression, translocation, and SP release, but GPC cleavage into GP1 and GP2 requires trans-complementation with a bona fide LASV SSP (12). Likewise, myristoylation of the SSP of the New World arenavirus JUNV was shown to be strictly required for GP-mediated cell fusion (27). The conserved lysine at position 33 of JUNV SSP (K33) is also critical for GP-mediated cell fusion, and the charge at that position affects the pH at which fusion occurs (28). Moreover, the JUNV SSP associates specifically with the cytoplasmic domain of GP2, which appears to mask an ER retention signal, thus ensuring that only fully formed GP complexes leave the ER (2).
The SSP functional domains and mechanisms responsible for the multiple and distinct roles of the arenavirus SSPs remain largely unknown. We have analyzed the expression and activities of a panel of mutated LCMV SSPs with the aim of gaining a clearer understanding of SSP functions. We show that single amino acid changes in the SSP can selectively interfere with a variety of downstream GP functions, including transport to the cell surface, formation of infectious particles, and pH-dependent fusion, although pGPC expression and cleavage remain relatively unaffected.
MATERIALS AND METHODS
Cell lines.
DBT murine astrocytoma cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, and 1% penicillin-streptomycin. BHK-21 cells were maintained in DMEM supplemented with 10% FBS, 10 mM HEPES, 1% penicillin-streptomycin, and tryptose phosphate broth. 293T cells were maintained in DMEM with 10% FBS.
Plasmids and transfections.
All SSP deletion and point mutants were made from pCMV-WT GP (LCMV Arm53b pGPC) by use of a QuikChange PCR mutagenesis kit (Stratagene), and mutations were confirmed by automated dideoxynucleotide sequencing. Each SSP mutation appears in the context of the full-length pGPC gene. pCMV-SSP-WT was made from pCMV-WT pGPC by adding stop codons in frame at the end of the SSP (residue 58 of pGPC) to allow for expression of wild-type (WT) SSP only. DSP is an SSP deletion construct made from the parent vector, pCMV-WT GP, with the sequence encoding the first 58 amino acids deleted, up to the naturally occurring methionine at position 59. Plasmids expressing the LCMV L (pC-L), NP (pC-NP), and Z (pC-Z) proteins and T7 RNA polymerase (pC-T7) under the control of a modified chicken β-actin promoter, as well as minigenome (MG) 7 under the control of a T7 promoter, have been described previously (19, 21). All transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, using 2.5 μl Lipofectamine/μg plasmid DNA. Transfection medium was removed and replaced with fresh medium at 5 h posttransfection.
Western blots.
293T cells were lysed at 48 h posttransfection in lysis buffer (2% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl, pH 6.8, 15% glycerol, 0.1% bromphenol blue, 0.1 M dithiothreitol). Cells were scraped, collected in lysis buffer, boiled for 10 min, and then passed three times through a 30-gauge needle. Lysates were then separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were then blocked with 5% powdered milk in phosphate-buffered saline (PBS) for 1 h at room temperature, probed with primary anti-GP2 monoclonal antibody 33.6 (25) overnight at 20°C, washed three times with 0.05% NP-40 in PBS, probed with an anti-mouse secondary antibody conjugated to alkaline phosphatase for 2 h at 20°C, and washed an additional three times. Bound antibody was visualized by incubation with Tris-buffered nitro blue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate. Blots were scanned and then quantified using ImageJ software (1).
Immunofluorescence assay.
Transfected 293T cells were collected, washed in cold PBS with 1% bovine serum albumin, and then incubated for 30 min with the mouse monoclonal antibody 33.6. Following primary antibody incubation, cells were washed three times and then incubated for 30 min with anti-mouse-fluorescein isothiocyanate (FITC). Cells were finally washed three times, fixed with 2% normal buffered formalin in PBS, and counted by fluorescence-activated cell sorting (FACS). Data were analyzed using FlowJo and WinMDI software.
VLP infectivity assay.
Virus-like particle (VLP) assays were done as described previously (19, 21). Briefly, 293T cells (1.5 × 106) growing in 35-mm-diameter wells were transfected with pC-L (1 μg), pC-NP (0.8 μg), pC-Z (0.1 μg), pC-T7 (1 μg), and pT7-MG#7 (0.5 μg) together with the indicated WT or mutant pGPC plasmid (0.5 μg). At 72 h posttransfection, VLP-containing supernatants (1.5 ml) were harvested and clarified at 3,000 × g for 10 min at 4°C. Aliquots of the clarified supernatant (600 μl) were used to infect fresh monolayers of BHK-21 cells. After 5 h of adsorption, the inoculum was removed, and LCMV ARM helper virus was added at a multiplicity of infection of 3 to provide the viral trans-acting factors required for RNA replication and expression of the MG RNA brought into the cells by the VLPs present in the transfected cell supernatant. After adsorption for 3 h, the inoculum was removed and fresh medium was added, and at 72 h postinfection, BHK-21 cells were lysed and assayed for chloramphenicol acetyltransferase (CAT) expression to quantify VLP infectivity.
CAT enzyme-linked immunosorbent assay to quantify VLP infectivity.
Cells were collected in cold PBS, and whole-cell extracts were made by resuspending cell pellets in 0.25 M Tris-HCl, pH 7.5, freeze-thawing them three times, and collecting the supernatant. The concentration of CAT, in ng/ml, for each whole-cell extract was measured using a CAT enzyme-linked immunosorbent assay kit from Roche Applied Science, Indianapolis, IN.
DBT cell fusion assay.
DBT cells were transfected with WT pGPC and mutated SSPs, and at 24 h posttransfection, cells were exposed to pH 5 buffered medium for a 1-hour interval, returned to neutral pH for 1 hour, and then fixed in 20% formaldehyde-PBS and stained with 0.1% crystal violet-20% ethanol. The extent of syncytium formation was quantified using representative images with at least 100 visible nuclei and the following equation to calculate the fusion index (FI): FI = [1 − (number of cells/number of nuclei)] (17, 19, 21).
Statistical analysis.
Statistical comparisons were generated using Instat software (Graphpad). A Pearson (r) correlation statistic was calculated to compare the relative power of each functional assay in predicting the results of the other functional assays presented in this study. The mean data presented in Table 1 were entered as a matrix, with each column representing a functional assay and each row relating to one specific mutated construct. The correlation coefficient was then calculated for pairs of columns (functional assays), excluding any rows (mutated constructs) that were not analyzed in both of the assays being tested.
TABLE 1.
Biochemical properties of mutated SSPsf
| Mutation | % pGPC expressiona | % Surface expressionb | GP1/GP2 cleavage (%)c | FId | VLP infectivity (%)e |
|---|---|---|---|---|---|
| WT GP | 100 | 100 | 100 | 0.80 | 100 |
| No GP | NT | NT | NT | 0.05 | <1 |
| G2A | 124 | 122 | 100 | 0.12 | 35 |
| Q3A | 164 | 88 | 111 | 0.74 | 109 |
| T6A | 134 | 86 | 102 | 0.65 | 110 |
| E9A | 198 | 87 | 105 | 0.71 | 22 |
| P12A | 91 | 112 | 59 | 0.33 | 1 |
| P12G | 80 | 106 | 79 | 0.60 | 4 |
| DE16/17AA | 153 | 138 | 109 | 0.50 | <1 |
| DE16/17KK | 73 | 36 | 20 | 0.20 | <1 |
| N20A | 149 | 143 | 81 | 0.19 | 1 |
| N20Q | 157 | 122 | 77 | 0.28 | 2 |
| N20K | 104 | 88 | 45 | 0.12 | 9 |
| K33A | 69 | 67 | 59 | 0.06 | <1 |
| K33R | 153 | 81 | 81 | 0.80 | 72 |
| K33D | 43 | 35 | 26 | 0.07 | <1 |
| N37A | 52 | 64 | 37 | 0.09 | 3 |
| N37Q | 53 | 76 | 46 | 0.55 | 93 |
| N37K | 132 | 91 | 34 | 0.52 | 8 |
| C41A | 95 | 104 | 65 | 0.69 | 130 |
| F49A | 80 | 99 | 16 | 0.06 | <1 |
| F49L | 87 | 51 | 22 | 0.52 | 2 |
| F49Y | 53 | 85 | 37 | 0.74 | 86 |
| L52A | 63 | 149 | 55 | 0.66 | 74 |
| G54A | 63 | 122 | 4 | 0.30 | <1 |
| C57A | 83 | 13 | 13 | NT | NT |
| H1D | 55 | 15 | 2 | NT | NT |
| H2D | 5 | 8 | <1 | NT | NT |
Relative to WT GPC expression. Mean values were obtained by densitometric analysis of band intensities in two to four replicate Western blots stained with a GP2-specific antibody. Typical standard deviations for this assay fell between 9 and 14% of the values reported here.
Percentage of transfected cells staining for GP2 relative to that for the WT control, as measured by FACS. Mean values were calculated from two to four replicates, with typical standard deviations of 7 to 19% of the means.
Relative intensity of GP2, expressed as a percentage of the cleavage observed in WT control cells. The methodology was duplicated from the percent expression, as described above, with typical standard deviations ranging from 4 to 8% of the means.
Calculated using the formula FI = [1 − (number of cells/number of nuclei)], based on examination of at least four microscopic fields. Standard deviations between 1 and 24% of the means were typical.
Percentage of infectivity in a VLP system relative to the WT control, as measured by MG CAT expression. Standard deviations are shown in Fig. 4C.
NT, not tested.
RESULTS
Generation of SSP mutations.
We designed a panel of mutated SSPs (Fig. 1C) based on previous studies of arenavirus SSPs and SPs in general as well as on common amino acid characteristics revealed by alignment of arenavirus SSP sequences (Fig. 1B). The LCMV SSP contains two hydrophobic domains, yet either one alone would suffice to allow translocation of pGPC into the ER lumen (13). To assess the individual role of each of the two hydrophobic domains present within the SSP, we generated deletion mutants H1D and H2D. To examine the roles affected by potential LCMV SSP myristoylation, we introduced an alanine mutation, G2A, at the conserved myristoylation site. Sequence conservation at the N terminus of SSP led us to construct mutants with the additional mutations Q3A, P12A, P12G, DE16/17AA, and DE16/17KK to assess the respective functional contributions of these residues. The conserved basic residue K33 marks the boundary between the two hydrophobic domains, and it may be required to properly orient SSP in the lipid bilayer (18). To investigate charge requirements at residue K33 within the LCMV SSP, we generated mutants with the mutations K33A, K33R, and K33D. To evaluate the role of the highly conserved asparagine residues at positions 20 and 37 (N20 and N37), we generated mutants with the following mutations: N20A and N37A (alanine substitution), N20Q and N37Q (conservative substitution), and N20K and N37K (polar substitution). Near the C terminus of SSP, there are two highly conserved cysteine residues, C41 and C57. C41 of the LASV SSP has been shown to be involved in SSP dimerization via disulfide bond formation (13). We therefore generated C41A and C57A LCMV SSP mutants to assess the role of these residues in LCMV GP functions. We also generated mutations to alter a highly conserved phenylalanine at position F49 of SSP, namely, F49A, F49L, and F49Y. Finally, we mutated the highly conserved leucine and glycine residues present at positions 52 and 54, respectively, to alanine (L52A and G54A). The full mutated SSP panel is detailed in Fig. 1C.
Effects of SSP mutations on pGPC expression and processing.
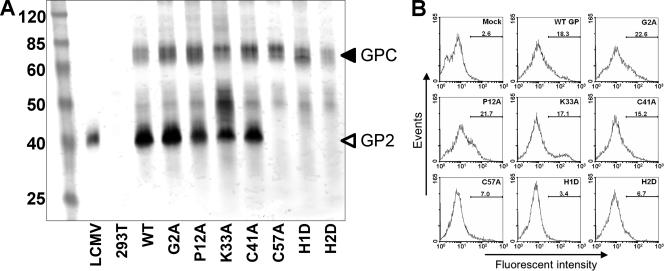
To assess the role of the SSP in pGPC expression and processing, we transfected 293T cells with mutated SSP pGPC constructs and used the corresponding cell lysates in Western blot assays. Membranes were probed with a monoclonal antibody specific for GP2. The presence of the GP2 antigen in both the unprocessed pGPC and processed GP2 allowed us to compare overall levels of pGPC expression and the efficiency of GPC processing into GP1 and GP2. A representative Western blot is shown in Fig. 2A, and the results obtained from the complete set of mutated SSPs are summarized in Table 1. The double banding pattern for GPC may possibly represent uncleaved pGPC and GPC. The intermediate bands seen migrating between GPC and GP2 are unidentified and may be related to aberrant GPC processing or posttranslational modifications. Most mutations did not impair pGPC expression, with the exception of the H2D mutation, which abrogated pGPC expression, and the P12G, DE16/17KK, K33A, K33D, N37A, N37Q, L52A, G54A, and H1D mutations, all of which resulted in reduced pGPC expression levels. Several mutations within the SSP resulted in greater expression levels of pGPC than those of WT pGPC. The SSP is essential for GP1/GP2 cleavage (12), and thereby, in some cases, SSP mutations may have contributed to increased levels of uncleaved GPC in the transfected cells. Table 1 shows the percentage of GP1/GP2 cleavage relative to that of the WT for all SSP mutants tested. Sixteen of the SSP mutations tested resulted in aberrant GPC cleavage, including the P12A, P12G, DE16/17KK, N20A, N20Q, N20K, K33A, K33R, K33D, N37A, N37Q, N37K, C41A, L52A, G54A, and C57A mutations, whereas the H1D mutation resulted in nearly undetectable levels of GPC cleavage. These results highlight the functional importance of absolute conservation of some residues located near the borders of the hydrophobic regions extending from residues 18 to 32 and 41 to 54 and stress the role of the SSP in GPC expression and processing.
FIG. 2.
(A) Representative Western blot of mutant SSP panel subgroup. At 48 h posttransfection, 293T cell lysates were separated by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with a GP2-specific antibody, 33.6. GPC and GP2 bands are indicated with arrows. (B) Representative immunofluorescence assay of mutant SSP panel subgroup. At 48 h posttransfection, 293T cells were probed for cell surface GP expression, using a GP2-specific antibody with a FITC-labeled secondary antibody. Cells were then fixed and counted using a FACSCalibur flow cytometer. FlowJo and WinMDI software was used for analysis, and data are reported as the percentage of positively stained cells for each mutated SSP.
Effects of SSP mutations on GP cell surface localization.
We next examined the effects of SSP mutations on transport of the GP complex to the cell surface, where viral assembly and budding occur. For this experiment, cells were transfected with the various pGPC constructs, and cell surface expression of GP2 was determined by FACS. Expression levels of GP2 derived from pGPC expression with mutated SSPs were normalized with respect to GP2 levels obtained from WT pGPC. Representative results are shown in Fig. 2B, whereas a summary of the results obtained with all SSP mutants tested is shown in Table 1. Mutations DE16/17KK, K33D, C57A, H1D, and H2D caused >50% reductions in GP2 expression at the cell surface, while several others were less severely impaired. Because the antibody we used for the immunofluorescence assay does not distinguish between GPC and GP2, we cannot rule out that some mutated SSPs with non-WT levels of cleaved GP2 localized to the cell surface, since proper GPC cleavage is not required for cell surface localization. Although some of the mutations near the N terminus of the SSP inhibited expression at the cell surface, GPC transport appeared to be more sharply affected by mutations in the signal peptidase recognition region as well as by charge reversal and alanine scanning mutations flanking the hydrophobic region extending from residues 18 to 32. Charge reversal may have the side effect of affecting SSP membrane topology and, thereby, any SSP functions dependent upon correct SSP topology.
Effects of SSP mutations on the generation of infectious VLPs.
All mutated SSPs with WT or near-WT levels of pGPC expression and processing were tested for the ability to form VLPs, using previously described procedures (19, 21). Reporter gene expression can be driven only by the intracellularly reconstituted viral polymerase, and VLP formation and infectivity closely mimic the corresponding steps by a bona fide virus during its natural course of infection. VLP infectivity was assessed based on levels of reporter gene expression in BHK cells that were coinfected with VLPs produced by transfected cells and a bona fide LCMV helper virus to provide viral trans-acting factors required for RNA replication and expression of the MG RNA associated with infectious VLPs. Results for all SSP mutations tested are shown in Fig. 3 and Table 1. Some of our mutated SSPs were unable to form infectious VLPs at WT levels; these included the G2A, E9A, P12A, P12G, and DE16/17AA mutants, despite their having WT or near-WT levels of pGPC expression, processing, and cell surface localization. These results suggest that the amino-terminal region of the SSP may be essential for virus particle formation and/or fusion with a host cell and that this SSP function is distinct from pGPC expression, processing, and trafficking.
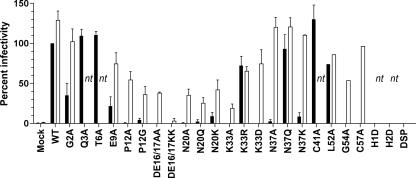
FIG. 3.
Infectivities of mutated SSPs in VLPs. The VLP infectivity for each mutated SSP is expressed as a percentage of WT VLP infectivity. Solid black bars indicate the percentages of WT VLP infectivity for SSP mutations, and white bars indicate the percentages of WT VLP infectivity for SSP mutations complemented with the WT SSP in trans. nt, not tested. Error bars indicate standard errors of the means. DSP is a pCMV-GP vector with the SSP deleted.
Rescue of noninfectious VLPs by trans-complementation with WT SSP.
The WT SSP was added to the six-plasmid VLP system to determine its ability to act in trans to rescue VLP infectivity. Unlike the mutated SSPs, which were expressed as part of the pGPC polypeptide, the WT SSP was expressed alone as only the first 58 amino acids of pGPC. With each SSP mutation that displayed less than WT VLP infectivity, expression of the WT SSP in trans was able to partially or fully restore VLP infectivity (Fig. 3). The G2A, E9A, K33D, L52A, and C57A mutants, which had reduced or no VLP infectivity, were restored to near-WT levels, and the P12A, P12G, DE16/17AA, N20A, N20Q, N20K, K33A, K33D, and G54A mutants, which showed little or no infectivity, exhibited only a partial restoration of infectivity. In selected instances, notably, for the N37A, N37Q, and N37K mutants, infectivity was restored to levels greater than the WT level. Rescue of particle infectivity by the addition of the WT SSP in trans suggests not only that the SSP is involved in creating infectious VLPs but that this activity is distinct from its roles in pGPC expression, processing, and localization to the cell surface.
Effects of SSP mutations on GP-mediated cell fusion.
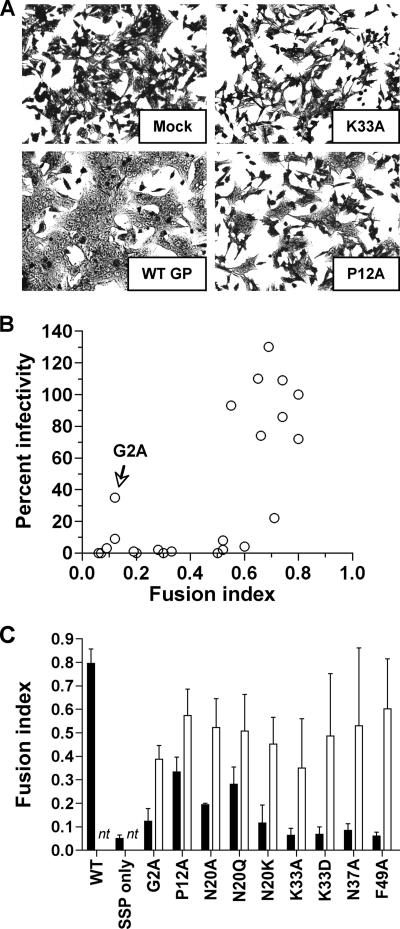
We considered the possibility that SSP mutations affected VLP infectivity by interfering with either the formation of correctly assembled virus particles, the virus fusion event, or both. To distinguish between these possibilities, we tested each mutated SSP with altered VLP infectivity in a cell fusion assay. We transfected each mutated SSP construct into DBT cells and assayed its ability to promote syncytium formation. A representative example of the fusion assay is shown in Fig. 4A, and a summary of the results obtained with all SSP mutations tested is presented in Table 1. The results show that for the G2A, DE16/17KK, N20A, N20K, K33A, K33D, and N37A mutants, loss of VLP infectivity could be explained at least partially by a loss of SSP function in GP-mediated cell fusion. In contrast, the noninfectious VLP E9A, P12G, DE16/17AA, and N37K mutants permitted GP-mediated cell fusion at near-WT levels, whereas the P12A and G54A mutants exhibited partially fusogenic phenotypes. These results suggest that in some cases, the loss of VLP infectivity was not determined by a lack of SSP fusion activity but, instead, by a defect in infectious particle formation. We observed that, with the exception of the G2A mutant, there was no infectivity when the FI fell below 0.5 (Fig. 4B), but GP-mediated fusion alone does not guarantee particle infectivity. Our findings indicated that the SSP is involved in both particle formation and GP-mediated fusion, but our studies did not distinguish between complete inhibition of virion formation and inhibition of GP incorporation into VLPs.
FIG. 4.
(A) DBT cell fusion assay with SSP mutants. pGPC-transfected DBT cells were given a pH 5 pulse at 24 h posttransfection, returned to neutral pH for an additional 1 h, fixed with formaldehyde, and stained with crystal violet. Note that all pictures are shown at the same magnification. (B) Correlation between percent infectivity and FI. Percent infectivity versus FI was plotted for all SSP mutants tested, with the G2A mutant indicated. The overall trend shows no infectivity when the FI is <0.5. (C) Rescue of GP-mediated cell fusion by WT SSP in trans. DBT cells were transfected with mutated SSPs alone (black bars) or with the WT SSP (white bars). nt, not tested. Cells were scored by using the FI for a representative field, calculated as follows: FI = 1 − (number of cells/number of nuclei).
Rescue of fusion by trans-complementation with WT SSP.
We next examined whether a subset of SSP mutants deficient in GP-mediated cell fusion, but with WT levels of pGPC expression, processing, and transport, could be complemented in trans with the WT SSP to restore cell fusion (Fig. 4C). All mutated SSPs tested could be trans-complemented with the WT SSP to WT or near-WT fusion levels, with the exception of the SSP with the putative myristoylation-inhibiting mutation G2A. The mutated G2A SSP could only be restored to just over 50% of WT fusion with trans-complementation, suggesting a requirement for SSP myristoylation in cis.
Effects of SSP F49 mutations on pGPC expression, processing, and cell surface transport.
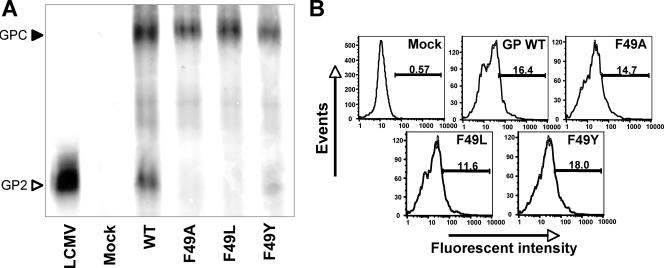
Proteins involved in virus budding often contain late domains. One type of late domain is typified by a YXXL motif, which functions by binding host adaptor proteins AP1 and AP2, which associate with clathrin (20). FXXL is an accepted variant of the YXXL motif. To probe the functionality of the strictly conserved FXXL motif present in the SSP, we generated F49A, F49L, F49Y, and L52A mutants and tested them for pGPC expression and GPC processing into GP1/GP2. As shown in Table 1, the L52A mutant was similar to the WT SSP in each of the functional assays, exhibiting only a slight defect in expression and cleavage. The F49A and F49L mutants had near-WT levels of pGPC expression, whereas the mutated F49Y SSP exhibited reduced pGPC expression (Fig. 5A and Table 1). While none of the F49 mutants exhibited WT levels of GPC cleavage into GP1 and GP2, all three showed intermediate levels of processed GP2 normalized to GPC expression (Table 1).
FIG. 5.
(A) F49 mutant SSP GPC expression and processing. Western blotting was performed under reducing conditions with 293T cell lysates at 48 h posttransfection. The membrane was probed using a GP2-specific monoclonal antibody to visualize GPC expression levels and GP1/GP2 maturation cleavage. (B) F49 mutant SSP cell surface localization. An immunofluorescence assay was performed with 293T cells at 48 h posttransfection. Surface expression was visualized using a GP2-specific monoclonal antibody with a FITC-labeled secondary antibody. Cells were then fixed and counted using a FACSCalibur flow cytometer. FlowJo software was used for analysis, and data are reported as percentages of positively stained cells.
The conserved FXXL motif could act as an internalization signal, and therefore mutations within this domain might result in an increased cell surface localization of GP (14, 15). We therefore examined cell surface GP expression of F49 mutants (Fig. 5B; Table 1). Our results showed that cell surface GP expression of the F49L mutant was significantly reduced compared to the WT level of cell surface localization, while the F49A and F49Y mutant expression levels were similar to that of the WT. These findings suggest that it is highly unlikely that the conserved FXXL motif functions as a canonical internalization signal. Another possibility is that residue F49 is part of a potential neutral lipid binding domain (NLBD) involved in SSP membrane association and positioning. Sequence comparison with an NLBD showed that a region of the LCMV SSP beginning with F49 is very similar to an NLBD motif, although direct observation of mutational effects on SSP membrane association was not performed in the present study.
Effects of SSP F49 mutations on VLP infectivity.
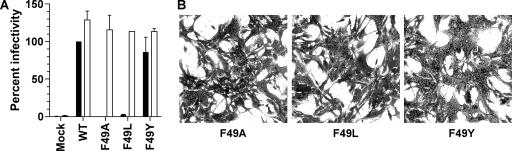
To further assess the function of the conserved F49 residue in the SSP, we examined the effects of SSP F49 mutations on the formation of infectious VLPs. The F49A and F49L mutants yielded essentially no VLP infectivity, while the F49Y mutant gave nearly WT VLP activity. Notably, the conservative F49Y mutation restored VLP infectivity despite the mutant having reduced GPC processing into GP1/GP2. All three F49 mutant SSPs exhibited WT VLP infectivity levels upon trans-complementation with the WT SSP (Fig. 6A; Table 1).
FIG. 6.
(A) F49 mutant SSP infectivity and rescue by WT SSP in trans. The graph shows a summary of F49 mutant GPC expression, GP processing, GP cell surface localization, VLP infectivity, and rescue of VLP infectivity by expression of the WT SSP in trans. Black bars denote mutated SSPs alone, and white bars indicate mutated SSPs complemented with the WT SSP in trans. (B) DBT cell fusion of F49 mutants. GP-transfected DBT cells were given a pH 5 pulse at 24 h posttransfection, returned to neutral pH for an additional 1 h, fixed with formaldehyde, and stained with crystal violet. Note that all pictures are shown at the same magnification.
Effects of F49 mutations on GP-mediated cell fusion.
To further dissect which aspect of the infectious process was inhibited by the SSP mutations in the VLP assay, we tested individual F49 mutants in a pH-dependent cell fusion assay. The F49A mutant was defective in syncytium formation, while the F49L and F49Y mutants showed slightly reduced and WT cell fusion activities, respectively (Fig. 6B; Table 1). This result was expected for the F49A mutant, given that its GP cell surface localization and GP1/GP2 processing were 39% and 13% those of the WT, respectively. In contrast, the fusion results for the F49L and F49Y mutants were unexpected. The F49L mutant showed only 51% of the WT GP cell surface localization, and both the F49L and F49Y mutants exhibited <30% of WT GPC cleavage, yet both were highly active in the GP-mediated cell fusion assay.
DISCUSSION
The work presented here supports the view that the LCMV SSP plays multiple roles in the virus life cycle and that its different functions are mediated by distinct domains and residues within the SSP. We fully characterized 20 different pGPC SSP mutants (Table 1). Correlation analysis (Table 2) revealed that altered GP transport to the cell surface did not predict mutation-induced changes in other SSP functions with statistical significance, possibly because our assay did not distinguish between properly and improperly formed GP complexes. Our results showed a correlation between pGPC processing and cell fusion as well as infectivity. A particularly strong correlation between cell fusion and infectivity was observed. The unique aspect of this analysis is in the establishment of a functional hierarchy for SSP (i.e., pGPC expression is a prerequisite for processing, processing is a requirement for fusion, etc.). An expression of this hierarchy would be as follows: expression > GP1/GP2 cleavage > fusogenicity > infectivity. A defect in one function accurately predicts defects in subsequent SSP activities, while the ability to perform one function does not guarantee the ability to perform other downstream SSP activities. It is anticipated that as future studies uncover novel functions for SSP, this hierarchy can be elaborated.
TABLE 2.
Phenotypic correlation between mutated SSPsa
| Phenotypic characteristic |
r value
|
|||
|---|---|---|---|---|
| Surface transport | Processing | FI | Infectivity | |
| Expression level | 0.56 | 0.81 | 0.34 | 0.08 |
| Surface transport | 0.64 | 0.14 | 0.05 | |
| Processing | 0.41 | 0.34 | ||
| FI | 0.69 | |||
Paired results for 23 to 26 mutated GPC constructs were considered. Pearson correlation coefficients (r values) were calculated as means for comparing the degrees of similarity between two entire panels of results from different functional assays. The data used for this calculation are presented in Table 1.
Our two most comprehensive mutations are H1D and H2D, with each deleting a region including one of the two conserved hydrophobic domains of SSP, H1 (18-32) and H2 (41-54). In a previous study using the LASV SSP, it was shown that either the H1 or H2 hydrophobic domain was sufficient for pGPC translocation into the lumen of the ER but that deletion of the second hydrophobic domain led to reduced SP cleavage from pGPC (13), which is a prerequisite for GP1/GP2 maturation cleavage (12). We observed that our somewhat larger H1D and H2D mutations abrogated virtually all SSP functions. Both H1D and H2D deletion mutations included previously unidentified critical residues for other SSP functions, which explains the loss of activity of the mutants. These results indicate that a fully functional SSP requires additional signals outside both the H1 and H2 domains for native activity.
Myristoylation of the JUNV SSP was shown to be required for GP-mediated cell fusion (27), and the site is conserved across the arenaviruses, including the LCMV SSP. The G2A mutation designed to disrupt the conserved potential myristoylation site caused reduced VLP infectivity and blocked GP-mediated fusion but left all other SSP functions relatively intact. This suggests that the myristoylation site at position G2 of SSP is important for LCMV fusion upon entry into host cells. The Q3A and T6A mutations, despite affecting highly conserved residues, showed little or no inhibition in any of our tests of SSP function. The E9A mutant SSP showed normal GPC expression, processing, and cell surface localization and GP-mediated cell fusion but failed to yield WT levels of VLP infectivity. We observed the trend that infectivity is severely impaired when the FI is below 0.5 (Fig. 4B). However, the E9A mutant had an FI of 0.73, demonstrating that while GP-mediated fusion is required for efficient infectivity, it is a separate and distinct function from infectious particle formation. The E9A mutant's reduced VLP infectivity could be restored by trans-complementation with the WT SSP. The E9A mutation likely causes a loss of particle formation or GP incorporation.
The P12A SSP mutation affects a weakly conserved domain similar to φ-P-X-V, where φ is any aromatic amino acid and X is any amino acid, which was found to be critical for paramyxovirus budding (22). In the LCMV SP, the sequence is LPHI, and the one conserved across the arenaviruses is I/L/V-P-X-hydrophobic residue. We noted but did not test for functionality of a largely conserved YXXL domain (4) near the N terminus of the SP due to the lack of robust conservation. The P12A mutation did not affect pGPC expression or cell surface localization but caused reduced GPC processing, yet the P12A mutant was unable to form infectious VLPs. This could be explained, at least partly, by a reduced level of GP-mediated fusion that correlated with the reduction in GP cell surface localization. The fact that the P12A mutant could still partially function in the acid-induced fusion assay suggests that the mutation also affects some aspect of GP complex or virus particle formation. Since proline is often involved in secondary structure formation, allowing for tight turns, we also tested the P12G mutation to see if using the smallest side chain possible would give the SSP additional flexibility and restore its activity. The P12G mutant was unable to restore VLP infectivity but did have WT or near-WT levels of pGPC expression, GPC processing and cell surface localization, and GP-mediated cell fusion. These results confirmed that P12 is critical for the SSP's role in particle formation. The fact that the P12A and P12G mutants yielded no infectious VLPs yet the P12G mutant could aid in GP-mediated cell fusion, while that of the P12A mutant was reduced, suggests that the roles of SPs in infectious particle formation and GP-mediated fusion are independent of each other. Due to the loss of infectious VLP infectivity with the P12G mutant with maintained fusogenicity, it is possible that P12 is part of a viral budding domain similar to φ-P-X-V.
Asparagine residues at positions 20 and 37 are strictly conserved among arenavirus SSPs. All mutations tested at either N20 or N37 disrupted one or more SSP functions. Given the mutational effects observed at these two residues, N20 and N37 can both be inferred to play critical roles in SSP function. N20 is strictly required for VLP infectivity, with no substitutions tolerated in that assay. N20 and N37 are also necessary for GPC processing, showing little or no rescue with conservative substitutions.
Lysine 33, which resides between the two hydrophobic domains of the SSP, is conserved among all arenaviruses. The K33R mutant achieved WT levels of pGPC expression, cell surface localization, and GP1/GP2 processing. The K33A and K33D mutants both had reduced pGPC expression, processing, and cell surface localization. This result led to the conclusions that a positive charge is favored over no charge and that no charge is favored over a negative charge at position 33 for pGPC expression, processing, and cell surface transport. For VLP infectivity and GP-mediated cell fusion, a positive charge at position 33 is required: both the K33A and K33D mutants showed no VLP infectivity or GP-mediated cell fusion, while the K33R mutant had near-WT levels of both. The JUNV SSP also demonstrated a strong preference for a positive charge at position 33 to achieve optimal acid pH-dependent cell fusion (28).
The central region of the LCMV SSP, from P12 to K33, displayed an interesting phenotype when probed in our VLP assay. All mutants with substitutions in that region, with the exception of the K33R mutant, were unable to efficiently form infectious VLPs, indicating that this region is critical for particle infectivity, as are other elements of the SSP. However, the inability of the WT SSP to fully rescue particle infectivity when combined with central region mutants suggests that it has a cis-acting role in infectious particle formation. Unlike the other elements of the SSP, whose functions can be restored by the addition of the WT SSP in trans, the central region of the SSP is the first to be assigned a cis-acting role in the viral life cycle beyond translocation of a nascent polypeptide into the lumen of the ER. Another possibility is that some of the SSP mutations may cause the SSP to act in a dominant-negative manner, thereby blocking the function of the WT SSP added in trans.
The C41A mutant SSP performed all SSP functions at WT levels, with the exception of reduced GP1/GP2 processing, which is surprising given that C41 is required for LASV SSP dimerization (13). This leads to the conclusion that in the case of the LCMV SSP, either C41 is not required for SSP dimerization or SSP dimerization is not required for the SSP functions observed in the present study. The opposite situation exists for the C57A mutation. C57 was not required for LASV SSP dimerization, but for the LCMV SSP, the C57A mutation rendered the SSP largely nonfunctional. The C57A mutant reached near-WT levels of pGPC expression, but the SSP functions of GP1/GP2 processing and cell surface localization were severely inhibited. C57 is not part of the typical signal peptidase recognition motif of small, uncharged residues at positions −1 and −3 (24) (residues 56 and 58 of SP), so a loss of SSP cleavage is not a likely explanation for the phenotypes observed with the C57A mutation.
F49 also proved to be a key residue for SSP function. Despite strict conservation of a potential FXXL domain at positions 49 to 52, based on our observations, FXXL in the LCMV SSP is not involved in GP internalization. The F49A and F49L mutants were similar to the WT for pGPC expression, with reduced processing and cell surface transport. If the conserved FXXL domain were involved in GP internalization, an increase in cell surface localization would have been expected when the conserved motif was mutated, but this was not the case. Yet F49 is clearly required for SSP function, as the F49A mutant did not form infectious VLPs or facilitate GP-mediated cell fusion. The F49L mutant was also unable to form infectious VLPs but was still capable of GP-mediated cell fusion. The F49Y mutant had reduced GPC expression and processing but near-WT GP cell surface localization, yet despite these reductions, and unlike the F49A or F49L mutant, the F49Y mutant achieved near-WT levels of VLP infectivity and GP-mediated cell fusion. These observations indicate that position 49 of the SSP requires an aromatic side chain and that a hydrophobic substitution is not sufficient to retain SSP function. The fact that VLP infectivity was fully restored to WT levels for all of the F49 mutants, unlike the case for mutations made in the cis-acting region of the SSP, suggests that F49 is a critical residue in a chaperone function of the SSP. Results for other mutants in our study revealed that this chaperone function may map to the C terminus of the SSP, as L52A, C57A, and G54A mutants also exhibited reduced levels of GP processing and/or cell surface localization. A C57A mutation in the JUNV SSP was recently studied, and while no inhibition of SSP cleavage was found, the alanine substitution did prevent SSP association with the GP complex and subsequent trafficking (29). Our results with the LCMV SSP C57A mutant confirm those observations.
We observed that mutations within the LCMV SSP that did not affect the typical SP motif per se, i.e., a hydrophobic region followed by an appropriate cleavage site, did, however, cause a loss of basic SP function. This suggests that the LCMV SSP may function differently from a canonical SP. Thus, the term “signal peptide” may not be the most apt to describe the LCMV SSP. The LCMV SSP is involved in multiple aspects of GP function during the viral life cycle, including GP-mediated cell fusion and the generation of infectious particles. Our data shown here and those by others (12) indicate that the SSP can act independently from other viral proteins. The observation that trans-complementation with the WT SSP can restore normal function for some mutated SSPs further supports the unique features of the arenavirus SSPs compared to canonical signal peptides.
Our results have shown that the LCMV SSP is involved not only in GPC expression, processing, and cell surface localization but also in particle formation and GP-mediated fusion. Moreover, it appears that within the SSP, specific domains and residues direct each of these functions (Fig. 7). Further studies are needed to elucidate the role of the SSP in particle formation and the components of the SSP required for that function.
FIG. 7.
Summary of SSP mutation effects. The WT SSP sequence is shown at the top. Dashed lines indicate affected deletion regions, and solid lines indicate domains affected by point mutations for the listed functions.
Acknowledgments
This work was supported by NIH grants AI-050840 (to M.J.B.), AI-047140 (to J.C.T.), and AI-065359 (Pacific Southwest Center for Biodefense and Emerging Infectious Diseases) (to J.C.T., B.W.N., and M.J.B.).
This is publication 18629-MIND of the Molecular and Integrative Neurosciences Department of The Scripps Research Institute.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Agnihothram, S. S., J. York, and J. H. Nunberg. 2006. Role of the stable signal peptide and cytoplasmic domain of G2 in regulating intracellular transport of the Junin virus envelope glycoprotein complex. J. Virol. 80:5189-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer, W. R., D. Popplau, W. Garten, D. von Laer, and O. Lenz. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 5.Buchmeier, M. J., J. H. Elder, and M. B. Oldstone. 1978. Protein structure of lymphocytic choriomeningitis virus: identification of the virus structural and cell associated polypeptides. Virology 89:133-145. [DOI] [PubMed] [Google Scholar]
- 6.Burns, J. W., and M. J. Buchmeier. 1991. Protein-protein interactions in lymphocytic choriomeningitis virus. Virology 183:620-629. [DOI] [PubMed] [Google Scholar]
- 7.Burns, J. W., and M. J. Buchmeier. 1993. Glycoproteins of the arenaviruses, p. 17-35. In M. Salvato (ed.), The Arenaviridae. Plenum Press, New York, NY.
- 8.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 9.Di Simone, C., and M. J. Buchmeier. 1995. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology 209:3-9. [DOI] [PubMed] [Google Scholar]
- 10.Di Simone, C., M. A. Zandonatti, and M. J. Buchmeier. 1994. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 198:455-465. [DOI] [PubMed] [Google Scholar]
- 11.Eichler, R., O. Lenz, T. Strecker, and W. Garten. 2003. Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett. 538:203-206. [DOI] [PubMed] [Google Scholar]
- 12.Eichler, R., O. Lenz, T. Strecker, M. Eickmann, H. D. Klenk, and W. Garten. 2003. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 4:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichler, R., O. Lenz, T. Strecker, M. Eickmann, H. D. Klenk, and W. Garten. 2004. Lassa virus glycoprotein signal peptide displays a novel topology with an extended endoplasmic reticulum luminal region. J. Biol. Chem. 279:12293-12299. [DOI] [PubMed] [Google Scholar]
- 14.Eng, F. J., O. Varlamov, and L. D. Fricker. 1999. Sequences within the cytoplasmic domain of gp180/carboxypeptidase D mediate localization to the trans-Golgi network. Mol. Biol. Cell 10:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froeschke, M., M. Basler, M. Groettrup, and B. Dobberstein. 2003. Long-lived signal peptide of lymphocytic choriomeningitis virus glycoprotein pGP-C. J. Biol. Chem. 278:41914-41920. [DOI] [PubMed] [Google Scholar]
- 17.Gallagher, T. M., C. Escarmis, and M. J. Buchmeier. 1991. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J. Virol. 65:1916-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higy, M., S. Gander, and M. Spiess. 2005. Probing the environment of signal-anchor sequences during topogenesis in the endoplasmic reticulum. Biochemistry 44:2039-2047. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K. J., M. Perez, D. D. Pinschewer, and J. C. de la Torre. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J. Virol. 76:6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohno, H., J. Stewart, M. C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, and J. S. Bonifacino. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269:1872-1875. [DOI] [PubMed] [Google Scholar]
- 21.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt, A. P., G. P. Leser, E. Morita, W. I. Sundquist, and R. A. Lamb. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Heijne, G. 1998. Life and death of a signal peptide. Nature 396:111, 113. [DOI] [PubMed] [Google Scholar]
- 24.von Heijne, G. 1985. Signal sequences. The limits of variation. J. Mol. Biol. 184:99-105. [DOI] [PubMed] [Google Scholar]
- 25.Weber, E. L., and M. J. Buchmeier. 1988. Fine mapping of an antigenic site conserved among arenaviruses. Virology 164:30-38. [DOI] [PubMed] [Google Scholar]
- 26.Wright, K. E., R. C. Spiro, J. W. Burns, and M. J. Buchmeier. 1990. Post-translational processing of the glycoproteins of lymphocytic choriomeningitis virus. Virology 177:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.York, J., V. Romanowski, M. Lu, and J. H. Nunberg. 2004. The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 78:10783-10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.York, J., and J. H. Nunberg. 2006. Role of the stable signal peptide of Junin arenavirus envelope glycoprotein in pH-dependent membrane fusion. J. Virol. 80:7775-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York, J., and J. H. Nunberg. 2007. Distinct requirements for signal peptidase processing and function in the stable signal peptide subunit of the Junin virus envelope glycoprotein. Virology 359:72-81. [DOI] [PubMed] [Google Scholar]