Abstract
Only few of the human endogenous retrovirus (HERV) sequences in the human genome can produce proteins. We have previously reported that (i) patients with germ cell tumors often make antibodies against proteins encoded by HERV-K elements, (ii) expression of the HERV-K rec gene in transgenic mice can interfere with germ cell development and induce carcinoma in situ, and (iii) HERV-K np9 transcript is overproduced in many tumors including breast cancers. Here we document that both Np9 and Rec physically and functionally interact with the promyelocytic leukemia zinc finger (PLZF) tumor suppressor, a transcriptional repressor and chromatin remodeler implicated in cancer and the self-renewal of spermatogonial stem cells. Interaction is mediated via two different central and C-terminal domains of Np9 and Rec and the C-terminal zinc fingers of PLZF. One major target of PLZF is the c-myc proto-oncogene. Coexpression of Np9 and Rec with PLZF abrogates the transcriptional repression of the c-myc gene promoter by PLZF and results in c-Myc overproduction, altered expression of c-Myc-regulated genes, and corresponding effects on cell proliferation and survival. Thus, the human endogenous retrovirus proteins Np9 and Rec may act oncogenically by derepressing c-myc through the inhibition of PLZF.
Approximately 8% of the human genome consists of sequences of retroviral origin (23). These stem from infections and reinfections of the germ line by exogenous retroviruses and from amplifications through retrotransposition in the past 40 million years (4). It is estimated that 2,000 proviruses (DNA copies of the retroviral RNA genomes) and more than 30,000 solitary sequences related to the viral regulatory long terminal repeats are present, of which the vast majority have accumulated multiple nonsense mutations. However, up to 50 provirus copies may contain intact open reading frames for the viral proteins Gag, Prt, Pol, and Env (25, 29). Some of these proteins have important functions; for instance, human endogenous retrovirus W (HERV-W) Env is specifically expressed in trophoblasts of the primate and human placenta, where it induces cell fusion and thereby contributes to the generation of the syncytiotrophoblast cell layer (31). Proteins encoded by the HERV-K family have been implicated in pathological changes: patients with germ line tumors frequently produce antibodies directed against HERV-K proteins (35, 36), HERV-K rec expression interferes with germ cell development and causes carcinoma in situ in transgenic mice (16), and HERV-K Np9 is preferentially expressed in transformed cells and interacts with ligand of Numb protein X, an important player in the Notch signal transduction pathway (2, 3). rec transcript is produced from the env gene by alternative splicing and encodes the 14.5-kDa Rec relative of the regulatory Rev/Rex proteins of the human immunodeficiency and T-cell leukemia viruses (16, 26, 27, 41). Due to a 292-bp deletion and the generation of a specific splice donor site in a HERV-K provirus subtype, the env open reading frame in this provirus produces np9 instead of rec transcript, which codes for the 9-kDa nuclear Np9 protein (2). Np9 and Rec share only the N-terminal 14 amino acid residues.
The promyelocytic leukemia zinc finger (PLZF) protein was first identified in patients with acute promyelocytic leukemia as part of a fusion protein containing, as the second partner, the retinoic acid alpha receptor (RARα) (10, 11). Acute promyelocytic leukemia cells expressing the PLZF-RARα fusion protein are nonresponsive to all-trans retinoic acid and come with a poor prognosis (18). Functionally, PLZF is a 673-amino-acid transcriptional repressor with nine C-terminal Krüppel-like C2H2 zinc fingers and an N-terminal POZ (pox virus and zinc finger) domain. The repression of target genes such as cyclin A2 (42) and c-myc (30) requires DNA binding via the zinc finger domain and is mediated through association of the POZ domain with the transcriptional corepressors N-CoR (nuclear receptor corepressor), SMRT (silencing mediator of retinoid and thyroid receptor), Sin3A, and HDAC1 (histone deacetylase 1) (14, 17, 21, 22, 24). PLZF seems to act primarily as a guardian of stem cell pluripotency in the hematopoietic system (34) and the germ line (8, 12). We demonstrate here that the tumor-associated HERV proteins Np9 and Rec can bind to the PLZF tumor suppressor and stem cell regulator and inhibit it as a transcriptional repressor.
MATERIALS AND METHODS
Plasmids.
pGEX-rec-1.exon, pGEX-rec-47/105, and pGEX-rec-21/75 were constructed by EcoRI and XhoI digestion of pJG4-5. The fragments were inserted in frame into pGEX-4T-1. pGEX-rec was described previously (7). pGEX-rec-47/89 was constructed by amplification of a rec fragment with the primers GGCAGAATTCCCAACTTGGGCACAACTAAA (A), and GGATACCTCGAGTCACACCATTGATACAATCATC (B) that introduced restriction sites for EcoRI and XhoI. pGEX-np9, pGEX-np9ΔC, and pGEX-np9N were previously described (3). pGEX-np9 was constructed by amplification of EGFP-np9 (where EGFP is enhanced green fluorescent protein) (3) with primers CGCGCGGATCCATGAACCCATCGCAGATGCAA (C) and CGCGCGGATCCAACAGAATCTCAAGGCAGAAG (D). pGEX-np9-55mut1 was constructed by amplification of pGEX-np9-NLSmut1 (3) with primers C and D. Plasmids pSG5-PLZF(245/399) and pSG5-PLZF(395) were constructed by BamHI digestion of pJG4-5; pJG4-5-PLZF(245/543) was digested with BamHI and XhoI. The digested fragments were inserted into pSG5. pSG5-PLZF(543) was constructed by amplification of the relevant fragment from vector pSG5-PLZF using the primers GCGCCGGAATTCCCACCATGGACCCCTACGAGT (E) and GGCGCAGATCTTCACACATAGCACAG (F). pSG5-np9 was previously described (3). pSG5-np9-55mut1 was constructed by inserting the BamHI fragment from pGEX-np9-55mut1 into pSG5 vector. pSG5-rec-47/89 was constructed by amplification from pGEX-rec-47/89 with primers GGGGGGGAATTCATGGCACAACTAAAG (G) and GGGGGATCCTCAGACACCATTGATACAATCATC (H). The c-myc promoter reporter constructs cmyc2.5 and cmyc2,5ΔPLZF were described previously (30).
Cell culture and transfection.
All cells were maintained at 37°C in a 5% CO2 atmosphere. 293T cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum and 1% sodium pyruvate. The U937T:PLZF45 inducible PLZF system was previously described (30) and is based on the U937T autoregulatory Tet-off system, in which withdrawal of tetracycline (Tet) leads to gene expression. U937T cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Paisley, United Kingdom), 1 mg/ml of G418 (PAA, Linz, Austria), 0.5 μg/ml puromycin (Calbiochem, Nottingham, United Kingdom), and 0.1 μg/ml Tet (Sigma, St. Louis, MO). U937:PLZF45-Rec was maintained in 300 μg/ml hygromycin B (PAA). PLZF expression was induced by washing the cells three times in phosphate-buffered saline (PBS) and replating them in the medium described above. The stably Rec-expressing U937:PLZF45 cells were developed by electroporating cells with the pCMV-Rec (where CMV is cytomegalovirus) and pcDNA3.1/Hygro(−) vectors and subsequent selection with hygromycin B (PAA). Cos-1 cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum and 1% sodium pyruvate. 293T cells (3 × 105) were plated in 24-well dishes at 16 h before transfection with Nanofectin I (PAA), according to the manufacturer's recommendations. Cos-1 and Tera-1 cells were plated in six-well dishes at 24 h before transfection with FuGene-6 (Roche, Mannheim, Germany), as specified by the supplier.
GST pull-down assays.
Gluthatione S-transferase (GST) proteins were generated by transforming Escherichia coli with plasmids pGEX, pGEX-rec, pGex-rec-1.exon, pGEX-rec-PES, pGEX-rec-47/105, pGEX-rec-21/75, pGEX-rec-47/89, pGEX-np9, pGEX-np9ΔC, pGEX-np9-55, pGEX-np9-55mut1, or pGEX-np9N. Exponentially growing cultures were induced with isopropyl-1-thio-β-galactopyranoside (final concentration, 100 nM) for 4 h at 37°C, and cell pellets were resuspended in GST lysis buffer (10 mM Tris-HCl, pH 7.5, 0.14 M NaCl, 3 mM MgCl2, 0.5% NP-40, 2 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 20 μg/ml aprotinin, and lysozyme in a final concentration of 50 mg/ml). The extracts were sonicated for 1 min on ice and cleared by centrifugation; 200 μl of supernatant was then added to glutathione-Sepharose beads (80 μl, diluted 1:1 in PBS) (Amersham Biosciences, Freiburg, Germany) and incubated for 4 h at 4°C with gentle shacking. Beads were collected by centrifugation and washed five times in GST linking buffer (500 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM EDTA, 0.1% NP-40, 2 mM DTT, 0.2 mM PMSF, 20 μg/ml aprotinin). PLZF, PLZF carrying amino acid residues 245 to 399 [PLZF(245/399)], PLZF carrying amino acid residues 395 to 673 [PLZF(395)], PLZF(245/543), or PLZF(543) labeled with 35S-translabeled methionine-cysteine were synthesized in vitro employing pSG5 as a vector in the rabbit reticulocyte lysate-based TNT T7-coupled in vitro transcription and translation system (Promega, Madison, WI), following the manufacturer's protocol. Protein-coated glutathione-Sepharose beads were incubated overnight at 4°C with GST linking buffer and 15 μl of radiolabeled protein. Pellets were then washed five times with GST linking buffer and boiled for 5 min in sodium dodecyl sulfate (SDS) gel loading buffer. The supernatants were loaded onto SDS-polyacrylamide gels. The gels were fixed with (50% [vol/vol] methanol, 10% [vol/vol] acetic acid) for 30 min, washed three times for 15 min in H2O, and incubated for 1 h in 1 M Na-salicylate before drying. Ten percent of the in vitro translated protein input (1.5 μl), diluted in 18.5 μl of SDS gel loading buffer, was used as input control.
In vitro coimmunoprecipitation.
GST, GST-Np9, PLZF, or PLZF(245/543) labeled with 35S-translabeled methionine-cysteine was synthesized in vitro using the pSG5 constructs in the rabbit reticulocyte lysate-based TNT T7-coupled in vitro transcription and translation system (Promega). A total of 10 μl of each preparation was mixed and incubated for 1 h on ice. The monoclonal GST antibody 6G9 was added to 80 μl of protein G-Sepharose (Amersham Biosciences) and linking buffer (radioimmunoprecipitation assay buffer: 150 mM NaCl, 0.1% NP-40, 0.5% deoxycholate, 0.1% SDS, 2 mM DTT, 0.2 mM PMSF, 20 μg/ml aprotinin) and incubated for 4 h at 4°C with gentle shacking. Beads were collected by centrifugation and washed three times in linking buffer. Protein-coated protein G-Sepharose beads were incubated overnight at 4°C with linking buffer and 20 μl of radiolabeled protein. Pellets were then washed three times with linking buffer and boiled for 5 min in SDS gel loading buffer. The supernatants were separated on SDS-polyacrylamide gels. The gels were fixed 50% (vol/vol) methanol-10% (vol/vol) acetic acid for 30 min, washed three times for 15 min in H2O, and incubated for 1 h in 1 M Na-salicylate before drying. Ten percent of in vitro translated protein input (1.5 μl) in 18.5 μl of SDS gel loading buffer was used as an input control.
SDS-polyacrylamide gel electrophoresis gradient, Western blot analysis, and antibodies.
SDS-polyacrylamide gel electrophoresis gradients containing 9.5 to 20% acrylamide were produced following standard procedures. Western blotting was carried out as described elsewhere (7). Np9 was detected with the rabbit polyclonal antiserum K82 (2) diluted at 1:100. The rabbit polyclonal Rec antiserum K3086 was used at a dilution of 1:100. The monoclonal mouse anti-PLZF antibody (Calbiochem) was used at a dilution of 1:50. The monoclonal mouse anti-β-actin antibody (Sigma) was used at 1:2,500; the monoclonal mouse c-Myc antibody sc-40 (Santa Cruz, CA) was used at 1:500; the monoclonal mouse p53 antibody (Calbiochem) was used at 1:2,000, the monoclonal mouse PCNA antibody sc-56 (Santa Cruz) was used at 1:500, and the monoclonal mouse IκBα antibody C-21 (Santa Cruz) was used at 1:500. Immunoprecipitation was performed with 10 μl of the mouse monoclonal GST antibody 6G9 (concentration, 20 μg/ml). The intensities of the immunoblot signals were measured with a Molecular Dynamics Personal Laser Densitometer.
Subcellular protein localization and fluorescence microscopy.
For subcellular localization and colocalization assays, cells were grown to about 20% density on glass coverslips and were transfected with FuGene-6 (Roche), using 1 μg of each DNA construct, according to the supplied protocols. Cells were fixated in paraformaldehyde (4% in PBS) at 48 h after transfection, and DNA was stained with 200 ng/ml DAPI (4′,6′-diamidino-2-phenylindole) in methanol to visualize the nuclei. Intracellular localization and colocalization were studied with a Leica DM IRB/E fluorescence microscope equipped with an Axio Cam color camera (Zeiss) and were analyzed with the Axio Vision 3.0 software.
Reporter gene assays and statistical analysis.
Reporter and effector plasmids were used in a 1:4 ratio, with 100 ng of pEGFP to detect transfected cells in fluorescence-activated cell sorting analyses. 293 cells were transfected with Nanofectin (PAA) as described above. Transfected cells were harvested at 42 to 45 h posttransfection, and lysates were assayed for luciferase activity with a Dual Luciferase kit (Promega) as recommended by the manufacturer. The luciferase activity was normalized to the transfection efficiency measured by fluorescence-activated cell sorting. Each experiment was performed at least three times and in triplicate per transfection. P values were calculated with the Sigma Plot, version 4.01, software (SPSS, Inc., Chicago, IL).
MTT and apoptosis assays.
The relative rate of proliferation was quantified by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) assay. U937T:PLZF45, U937T:PLZF45-CMV, and U937T:PLZF45-Rec cells were grown as specified above in the presence of Tet at the doses indicated in the figure legends. Cells were seeded at a density of 3 × 105 live cells per well on 96-well dishes. At 24 h after Tet exposure, the cells were treated with MTT and analyzed as specified by the supplier (Roche, Mannheim, Germany). To determine the rate of apoptosis, U937T:PLZF45 and U937T:PLZF45-Rec were washed three times in PBS and replated. Tet was added as indicated in the figure legends. Cells were fixed with methanol and incubated with propidium iodide for 15 min according to the supplier's protocol (Boehringer Mannheim, Mannheim, Germany) and were then analyzed by flow cytometry (Beckton Dickinson FACScan) for the number of cells with a sub-2N DNA content.
RESULTS
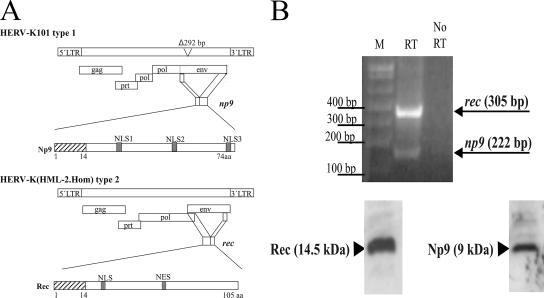
The HERV-K family of human endogenous retroviruses contains intact open reading frames for proteins specifically overproduced in transformed cells and certain cancers. The aim of our work is to examine whether these proteins merely constitute markers for tumors or whether they can actively contribute to the various stages of tumor development. In particular, we paid attention to the nuclear HERV-K proteins Np9 and Rec which are encoded by transcripts spliced from the env reading frames of the HERV-K subtypes 1 and 2, respectively (Fig. 1A). Amplification by reverse transcription-PCR of the np9 transcript from Tera-1 cells gives rise to a 222-bp fragment; the rec transcript can be amplified from the same cells as a 305-bp fragment. The 9- and 14.5-kDa proteins produced by these transcripts can be detected with our specific polyclonal anti-Np9 antibody K82 and anti-Rec antibody K3086 (Fig. 1B; 15 μg of total protein loaded per lane). To find protein interaction partners of Np9 and Rec, yeast two-hybrid screens were employed. An initial search with Rec (formerly called cORF) as bait identified PLZF as an interacting protein (7). Since Np9 and Rec share the N-terminal 14 amino acid residues, we asked whether Np9 can also associate with PLZF. Figures 2 and 3 document that both proteins bind to PLZF, and they summarize the mapping of the interaction domains. Np9 with a mutated nuclear localization signal 1 (NLS1; amino acid positions 22 to 29) can no longer bind to PLZF in GST pull-down assays (Fig. 2A). In contrast, the interaction of Rec with PLZF is dependent upon amino acid positions 21 to 47 in the center and 89 to 105 in the C terminus of Rec (Fig. 2B). PLZF with a deletion of the C-terminal four zinc fingers fails to associate with Np9 (Fig. 2C), whereas deletion of all nine zinc fingers is required to inhibit binding to Rec in GST pull-downs (Fig. 2D). The interaction of Np9 with PLZF (Fig. 2E) and Rec with PLZF (7) was confirmed by direct coimmunoprecipitation of proteins generated from in vitro transcribed and translated material. The extreme instability of Np9 protein in vivo (see reference 3) precluded coimmunoprecipitation from transfected cells.
FIG. 1.
The origins of the endogenous retrovirus proteins Np9 and Rec. (A) Schematic presentation of the HERV-K type 1 and 2 proviral sequences. The indicated open reading frames encode the proteins Gag (group-specific antigens and the viral capsid proteins), Prt (protease), Pol (reverse transcriptase/RNase H) and Env (envelope proteins). A 292-bp deletion in the env reading frame of HERV-K type 1 gives rise to the alternatively spliced np9 transcript; the rec transcript is spliced from the full-length env reading frame in the type 2 sequence. Np9 and Rec share the N-terminal 14 amino acid residues (hatched boxes). NES, nuclear export signal; LTR, long terminal repeat. (B) Results of reverse transcription-PCR amplification of the rec and np9 transcripts from Tera-1 cells are shown in the top panel. M, marker; RT, reverse transcriptase. The bottom panel shows the expression of Rec and Np9 in Tera-1 cells. Western blot analysis was performed on 15 μg total protein with the rabbit polyclonal anti-Np9 antibody K82 and anti-Rec antibody K3086 at a 1:100 dilution.
FIG. 2.
Mapping of the interaction of Np9 and Rec with PLZF by GST pull-down assays. (A) GST-Np9 and the indicated GST-Np9 mutants but not GST alone precipitate full-length in vitro translated 35S-labeled PLZF. An outline of the Np9 variants and the PLZF interaction is also shown at bottom. Np9-55mut1 harbors a mutation that incapacitates the NLS1. (B) GST-Rec and the indicated variants bind to radiolabeled PLZF. A scheme summarizing the interactions is given below. NES, nuclear export signal. (C) GST-Np9 but not GST alone precipitates the indicated full-length or truncated variants of in vitro translated 35S-labeled PLZF. (D) GST-Rec precipitates the indicated deletion mutants of radiolabeled PLZF. (E) In vitro coimmunoprecipitation of Np9 and PLZF. The GST-Np9 and PLZF proteins as well as the Np9 binding-defective PLZF(245/543) were transcribed and translated in vitro and coprecipitated with the anti-GST monoclonal antibody 6G9. Note that GST-Np9 fails to bring down PLZF(245/543).
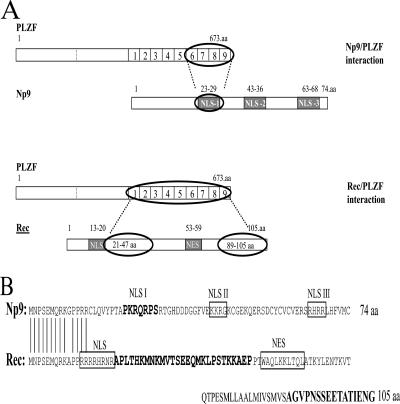
FIG. 3.
Schematic summary of the interaction between Np9 and Rec with PLZF and comparison of the Np9 and Rec domains that interact with PLZF. (A) The NLS1 region between amino acid residues 23 and 29 on Np9 associates with the four C-terminal zinc fingers of PLZF. On Rec, a region between amino acid residues 21 and 47 situated downstream of the NLS and the C-terminal 16 amino acid residues make contact with the PLZF zinc fingers. NES, nuclear export signal. (B) Alignment and comparison of the PLZF-interacting domains on Np9 and Rec (highlighted). NES, nuclear export signal. Note the differences between the PLZF-interacting NLS1 of Np9 and the noninteracting NLS of Rec.
In summary, Np9 and Rec interact with PLZF not through their identical N termini but via distinct domains in the center and the C termini (Fig. 3A). The association of Np9 with PLZF requires amino acid positions 23 to 29 containing NLS1, whereas Rec-PLZF interaction is dependent upon amino acid positions 21 to 47 in the center and 89 to 105 in the C terminus of Rec. Both make contact with the zinc fingers of PLZF but in a distinct manner. The domains on Np9 and Rec do not share any detectable homology or similarities in local hydrophobicity; the PLZF-interacting NLS1 of Np9 is distinct from the other NLSs in Np9 and Rec (Fig. 3B).
Since interaction in vivo would require codistribution of the proteins in cellular compartments, colocalization studies were performed. For this purpose, plasmids expressing fusion proteins consisting of (i) the green or red fluorescent proteins and full-length PLZF and (ii) the green or red fluorescent proteins and Np9 or Rec were employed. All fusion proteins as well as the native proteins localized to the nucleus (Fig. 4A). Cotransfections of the plasmids into Cos-1 cells revealed that both Np9 and Rec partially colocalize with PLZF in dot-like subnuclear structures, of which some appeared to be identical with the nucleoli (Fig. 4B). Similarly, colocalization was observed in Tera-1 cells (Fig. 4C). Combined, these data are compatible with the idea that Np9 and Rec interact with PLZF in the cell nucleus.
FIG. 4.
Intracellular localization of Np9, Rec, and PLZF. (A) Cos-1 cells were transiently transfected with plasmids producing the indicated fusion proteins involving EGFP or the red fluorescent protein (Dsred). (B) Transient transfection of Cos-1 cells with plasmids producing the indicated fusion proteins revealed a partial colocalization of Np9 and Rec with PLZF in dot-like subnuclear structures. Some of these were identical with the nucleoli spared in the DAPI-stained nuclei. (C) Transient transfection of Tera-1 cells with the indicated plasmids.
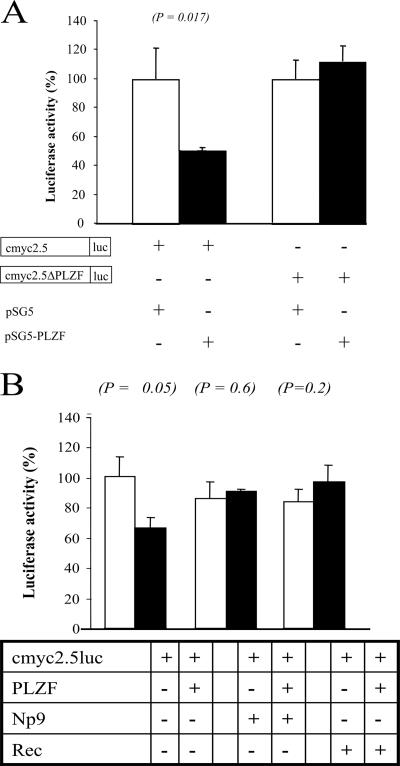
PLZF can suppress cell proliferation, and the PLZF-RARα fusion protein has been associated with neoplastic transformation (10, 11, 18). Previous studies have pointed to PLZF acting as a pleiotropic transcriptional repressor that regulates numerous target genes; however, recent work has indicated that a substantial fraction of the transcriptional changes caused by PLZF may be induced indirectly through the repression of the gene for the cell growth and proliferation-associated transcription factor c-Myc (30). To address whether the PLZF-associating Np9 and Rec proteins can affect PLZF's function as a transcriptional repressor, we first resorted to transient coexpressions of the proteins in the presence of a reporter plasmid carrying the PLZF-responsive fragment of the human c-myc gene promoter or a mutated variant, in front of a luciferase gene. As reported before (30), PLZF was able to significantly suppress luciferase expression driven by the wild-type c-myc promoter fragment in 293T cells whereas, in contrast, the protein exerted no suppressive effect on a construct harboring the promoter with a mutated PLZF recognition site (Fig. 5A), documenting the specificity of the suppressive effect. While expression of Np9 or Rec alone had no effect on the luciferase production by the reporter, coexpression of either protein with PLZF was able to abrogate the suppressive effect of the latter (Fig. 5B). Thus, the PLZF-associating Np9 and Rec proteins can interfere with PLZF's function as a transcriptional repressor in transient reporter assays.
FIG. 5.
Effect of Np9 and Rec on the repression of the c-myc promoter by PLZF. (A) 293T cells were transiently cotransfected with the reporter plasmids cmyc2.5 or cmyc2.5ΔPLZF defective for PLZF binding and with either empty vector or an effector plasmid producing PLZF. The relative luciferase activity was calculated as the percentage of the controls. Error bars indicate the standard deviations from at least three experiments. (B) Transient cotransfection of the cmyc2.5 reporter plasmid with empty vector alone, with empty vector plus Np9- or Rec-producing effector plasmids, and with the PLZF expression plasmid. Note that Np9 and Rec alone do not affect luciferase expression in these PLZF-negative cells. In contrast, PLZF represses luciferase production, and Np9 and Rec can overcome this effect. Luc, luciferase.
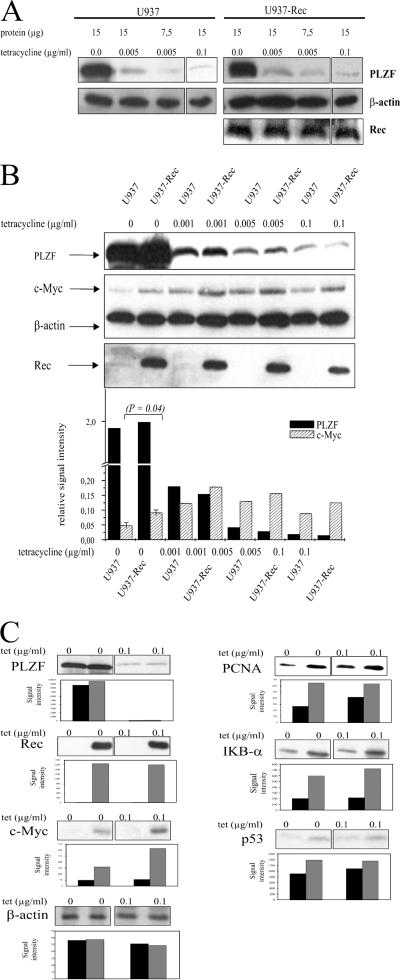
To test whether Rec can regulate the endogenous c-myc gene promoter via PLZF, we employed a U937-derived cell line harboring a PLZF gene under the control of a Tet-responsive promoter. As documented before (30), almost no PLZF was detectable in the presence of Tet (0.1 μg/ml) whereas complete removal of the drug from the culture medium for 24 h resulted in a maximal accumulation of PLZF that was fully reversible. The steady-state levels of PLZF could be fine-tuned by adding increasing amounts of Tet to the culture medium (Fig. 6A). We used this cell line as the basis to develop a new conditionally PLZF-producing cell line that stably expresses Rec from the CMV promoter/enhancer. Stable detectable expression of Np9 could not be achieved due to the very short half-life of this protein (3). The regulation of the PLZF expression levels by Tet was similar in the Rec-deficient and -proficient cell lines (Fig. 6A). In accord with the results by McConnell and colleagues (30) showing that PLZF can directly repress the c-myc gene promoter, the PLZF levels were inversely correlated with the levels of c-Myc at 24 h of incubation with different concentrations of Tet (Fig. 6B). The difference in c-Myc expression in the presence of PLZF and in the presence or absence of Rec was statistically significant (P = 0.04) (Fig. 6B, first two lanes). Furthermore, quantities of Tet that elicited equal PLZF levels in the Rec-negative and -positive cells (0.0 and 0.001 μg/ml, respectively) induced a higher steady-state level of c-Myc in the Rec-producing cells (Fig. 6B). Concomitantly, the Rec-positive cells showed an increase in the expression of the c-Myc-responsive PCNA, IκBα, and p53 genes but not the Myc-independent β-actin gene (Fig. 6C). Thus, the PLZF-interacting HERV-K proteins Np9 and Rec can inhibit the PLZF-mediated repression of the c-myc gene promoter in transient reporter assays, and Rec can counteract the repression of the endogenous c-Myc production by PLZF.
FIG. 6.
Effect of Rec on the regulation of the endogenous c-myc gene by PLZF. (A) U937T cells that conditionally produce PLZF and a derived cell line which, in addition, expresses Rec from the CMV promoter/enhancer, give rise to equal levels of PLZF upon exposure to low levels of Tet. Western immunoblottings were performed on the indicated quantities of total cell protein with the anti-PLZF monoclonal antibody at a dilution of 1:50, the anti-β actin monoclonal antibody at a dilution of 1:2,500, and the anti-Rec antiserum K3086 at a dilution of 1:100. (B) Western blot analysis shows that Rec expression causes overproduction of endogenous c-Myc despite equally high levels of PLZF. The monoclonal c-Myc antibody 9E10 was used at a dilution of 1:500. The bar diagram indicates the relative signal intensities of the PLZF and c-Myc bands, normalized to β-actin and measured by laser densitometry. The standard deviations of the levels of c-Myc depending on Rec were determined from three blots. (C) Western blots documenting the elevated expression of c-Myc and of known c-Myc-responsive genes in the presence of Rec. The anti-p53 monoclonal antibody DO-1 was used at 1:2,000, the anti-PCNA monoclonal antibody SC56 was used at 1:200, the anti-IκBα antibody was used at 1:500, and the anti-actin antibody was used at 1:2,500. The bar diagrams show the signal intensities.
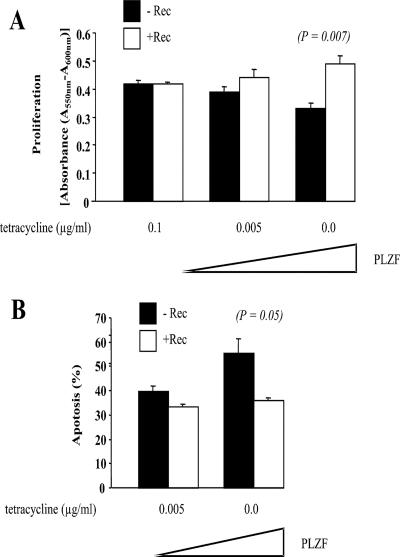
Finally, we asked whether Rec's interference with PLZF-mediated c-myc repression can cause specific phenotype alterations. Previous work determined that ectopic expression of PLZF leads to cell cycle arrest and apoptosis, that this effect is predominantly caused by the repression of c-myc expression, and that the effect is reversible by ectopic c-Myc (30). In this previous study, U937 cells that conditionally produced PLZF ceased to proliferate upon stimulation and instead showed signs of cell cycle arrest and apoptosis. For our studies we employed two derivatives of this cell line that were stably transfected to either harbor empty vector or vector producing Rec. A standard MTT proliferation assay revealed that Rec− and Rec+ cells grew equally well in the absence of PLZF. In contrast, low levels of PLZF (induced by 0.005 μg/ml Tet) (compare with Fig. 6B) and, to an even greater extent, large PLZF levels (induced in the absence of Tet) led to a significantly reduced proliferation of the Rec-deficient cells (Fig. 7A), in accord with the earlier findings (30) and in agreement with the idea that Rec counteracts the suppressive effects of PLZF. Standard apoptosis assays measuring, by flow cytometry, the number of cells with a sub-2N DNA content were performed in parallel and showed that PLZF-expressing cultures experience more apoptosis in the absence of Rec (Fig. 7B), again in agreement with the capacity of Rec to counteract the antiproliferative effects of PLZF. Thus, HERV-K proteins may function as oncoproteins by activating c-myc in cells that express PLZF.
FIG. 7.
Cell proliferation and apoptosis in dependence of Rec expression. (A) U937 cells that conditionally produce PLZF and a derived cell line which, in addition, expresses Rec from the CMV promoter/enhancer, were exposed to the indicated doses of Tet for 24 h. Cell proliferation was determined by measuring absorbance in a standard MTT assay. Error bars denote standard deviations from three experiments. (B) Flow cytometry was employed to determine the number of cells with a sub-2N DNA content indicative of apoptosis, after 24 h of weak (0.005 μg/ml Tet) or strong (no Tet) expression of PLZF. Error bars show standard deviations from three experiments.
DISCUSSION
Our analysis has revealed that Np9, a 9-kDa nuclear protein whose transcript is spliced from the env gene of the endogenous type 1 HERV-K101 retroviral sequences, and Rec, a 14.5-kDa HERV-K type 2-encoded endogenous relative of the Rev/Rex RNA transporters from the exogenous retroviruses human immunodeficiency virus and human T-cell leukemia virus, can directly bind to the zinc finger domain in the C terminus of the PLZF tumor suppressor and stem cell regulator. The interaction abrogates the function of PLZF as a transcriptional repressor of the c-myc proto-oncogene promoter and results in overproduction of this pleiotropic transcription factor and inducer of cell growth and proliferation. Previous studies had already established links between (i) HERV protein expression and the development of germ cell tumors in humans (35, 36), (ii) expression of Rec and the development of carcinoma in situ-like lesions in the testes of transgenic mice (16), and (iii) deficiency of PLZF in knockout mice and the severe testicular dysfunction associated with dysregulated self-renewal of spermatogonial stem cells (5, 8, 12). It is therefore tempting to speculate that Rec and possibly also Np9 can act as oncoproteins through the inhibition of PLZF.
Among the many genes that are downregulated in response to PLZF expression, only those coding for cyclin A2 (42), HoxD11 (5), and c-Myc (30) have so far been documented to be directly targeted by PLZF. In accord with the ideas that c-Myc itself is a transcription factor and that PLZF directly inhibits c-myc expression, more than one-fourth of the genes in a B-lymphoid lineage altered in response to PLZF turned out to be known targets of c-Myc (30). The details of the function of c-Myc in cells are still not fully understood; however, it seems clear that c-Myc is critical for the cell mass increase associated with the entry of cells from G0 phase into the cell cycle. In addition, c-Myc can drive cells into proliferation and control cell survival and differentiation (33). Even without a complete picture of c-Myc function, it is doubtlessly true that the protein is critically involved in tumorigenesis, in particular, leukemogenesis (13, 32). This is thought to depend primarily, if not exclusively, on c-Myc acting as a transcription factor, and a large number of screens have identified more than 600 putative, direct and indirect target genes (33). The findings that testicular germ cell tumors frequently express Rec (35, 36) and that Rec physically associates with and inhibits the c-Myc antagonist and regulator of germ cell differentiation, PLZF (this paper), may point to c-Myc's having a role in germ cell tumorigenesis. However, to date, c-Myc has not been implicated in this lesion. A clinical study has revealed that the levels of c-Myc were unaltered in eight seminomas and elevated in only one of 13 nonseminoma germ cell tumors (37). It should be noted, though, that in contrast to the c-myc gene amplifications and translocations observed in many tumors, overexpression of c-myc through a putative Rec/Np9/PLZF pathway may be transient. Transient overproduction of c-Myc has been implicated, for instance, in the development of genomic instability through the generation of reactive oxygen species, a hallmark of many cancers (38).
Several studies have linked endogenous retrovirus proteins, in particular, polypeptides encoded by env-related sequences, to the development and progression of neoplasia. For instance, expression of HERV-K10 gag has been linked to leukemia (15), and env expression has been correlated with human breast cancer (39). Production of Moloney murine leukemia virus full-length Env protein or the Env transmembrane domain allowed immune escape of murine MCA205 cells in immunocompetent mice (28). Interestingly, the presence of serum antibodies directed against the transmembrane domain of HERV-K Env was perfectly correlated with germ cell tumors in humans, whereas antibodies against HERV-K Gag or Env in general were occasionally also observed in patients with other diseases (35). Although Env and the Rec protein studied here share the N-terminal 87 amino acid residues including a domain that interferes with PLZF, it is likely that both proteins contribute to tumorigenesis via distinct pathways as Env is exclusively cytoplasmic while Rec and PLZF are nuclear proteins.
The splice variant Np9, apart from being an interaction partner of PLZF (this paper), can in contrast to Rec also bind to and functionally interfere with the ligand of Numb protein X, a RING-type E3 ubiquitin ligase that regulates the transcription factor Notch via degradation of the Notch-antagonist Numb (3). As an essential part of proproliferative Ras signaling (40), the Numb/Notch pathway has so far been linked primarily to breast cancers, leukemias, and germ cell cancers (1, 6, 9, 19, 20). Recent work has suggested that Notch signaling may be involved in the development of germ cell tumors by causing dysfunction of the mitotic-meiotic switch and, as a consequence, genetic instability (1). Leukemias and germ cell tumors have also been associated with dysregulated PLZF function (16, 43), and both Notch and PLZF have been implicated in stem cell maintenance (8, 12, 19). Np9 and Rec may thus constitute pleiotropic HERV factors that can affect tumorigenesis through their effects on stem cell regulatory PLZF pathways.
Acknowledgments
We thank Martin Jung (Medical Biochemistry) for his help with densitometry and Elisabeth Kremmer (GSF Munich) for the anti-GST antibody.
J.D.L. was supported by grant CA 59936. K.R. and N.M.-L. were supported by grants Ro 1201/10-1 and Mu 452/5-3 from the German Research Foundation.
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Adamah, D. J., P. J. Gokhale, D. J. Eastwood, E. Rajpert De-Meyts, J. Goepel, J. R. Walsh, H. D. Moore, and P. W. Andrews. 2006. Dysfunction of the mitotic:meiotic switch as a potential cause of neoplastic conversion of primordial germ cells. Int. J. Androl. 29:219-227. [DOI] [PubMed] [Google Scholar]
- 2.Armbruester, V., M. Sauter, E. Krautkraemer, E. Meese, A. Kleiman, B. Best, K. Roemer, and N. Mueller-Lantzsch. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800-1807. [PubMed] [Google Scholar]
- 3.Armbruester, V., M. Sauter, K. Roemer, B. Best, S. Hahn, A. Nty, A. Schmid, S. Philipp, A. Mueller, N. Mueller-Lantzsch, E. Krautkraemer, E. Meese, and A. Kleiman. 2004. Np9 protein of human endogenous retrovirus K interacts with ligand of Numb protein X. J. Virol. 78:10310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 5.Barna, M., N. Hawe, L. Niswander, and P. P. Pandolfi. 2000. PLZF regulates limb and axial skeletal patterning. Nat. Genet. 25:166-172. [DOI] [PubMed] [Google Scholar]
- 6.Beverly, L. J., and A. J. Capobianco. 2003. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3:551-564. [DOI] [PubMed] [Google Scholar]
- 7.Boese, A., M. Sauter, U. Galli, B. Best, H. Herbst, J. Mayer, E. Kremmer, K. Roemer, and N. Mueller-Lantzsch. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328-4336. [DOI] [PubMed] [Google Scholar]
- 8.Buaas, F. W., A. L. Kirsh, M. Sharma, D. J. McLean, J. L. Morris, M. D. Griswold, D. G. de Rooij, and R. E. Braun. 23 May 2004, posting date. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36:647-652. [DOI] [PubMed] [Google Scholar]
- 9.Callahan, R., and A. Raafat. 2001. Notch signaling in mammary gland tumorigenesis. J. Mammary Gland Biol. Neoplasia 6:23-36. [DOI] [PubMed] [Google Scholar]
- 10.Chen, S. J., A. Zelent, J. H. Tong, H. Q. Yu, Z. Y. Wang, J. Derre, R. Berger, S. Waxman, and Z. Chen. 1993. Rearrangements of the retinoic acid receptor alpha and promyelocytic leukemia zinc finger genes resulting from t(11;17) (q23;q21) in a patient with acute promyelocytic leukemia. J. Clin. Investig. 91:2260-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Z., N. J. Brand, A. Chen, S. J. Chen, J. H. Tong, Z. Y. Wang, S. Waxman, and A. Zelent. 1993. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 12:1161-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costoya, J. A., R. M. Hobbs, M. Barna, G. Cattoretti, K. Manova, M. Sukhwani, K. E. Orwig, D. J. Wolgemuth, and P. P. Pandolfi. 23 May 2004, posting date. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 36:653-659. [DOI] [PubMed] [Google Scholar]
- 13.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David, G., L. Alland, S. H. Hong, C. W. Wong, R. A. DePinho, and A. Dejean. 1998. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16:2549-2556. [DOI] [PubMed] [Google Scholar]
- 15.Depil, S., C. Roche, P. Dussart, and L. Prin. 2002. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia 16:254-259. [DOI] [PubMed] [Google Scholar]
- 16.Galli, U. M., M. Sauter, B. Lecher, S. Maurer, H. Herbst, K. Roemer, and N. Mueller-Lantzsch. 2005. Human endogenous retrovirus Rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223-3228. [DOI] [PubMed] [Google Scholar]
- 17.Grignani, F., S. De Matteis, C. Nervi, L. Tomassoni, V. Gelmetti, M. Cioce, M. Fanelli, M. Ruthardt, F. F. Ferrara, I. Zamir, C. Seiser, M. A. Lazar, S. Minucci, and P. G. Pelicci. 1998. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391:815-818. [DOI] [PubMed] [Google Scholar]
- 18.Guidez, F., W. Huang, J. H. Tong, C. Dubois, N. Balitrand, S. Waxman, J. L. Michaux, P. Martiat, L. Degos, Z. Chen, et al. 1994. Poor response to all-trans retinoic acid therapy in a t(11;17) PLZF/RAR alpha patient. Leukemia 8:312-317. [PubMed] [Google Scholar]
- 19.Hayashi, T., Y. Kageyama, K. Ishizaka, G. Xia, K. Kihara, and H. Oshima. 2001. Requirement of Notch 1 and its ligand jagged 2 expressions for spermatogenesis in rat and human testes. J. Androl. 22:999-1011. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi, T., T. Yamada, Y. Kageyama, K. Kihara, K. Ishizaka, G. Xia, and H. Oshima. 2004. Expression failure of the Notch signaling system is associated with the pathogenesis of testicular germ cell tumor. Tumour Biol. 25:99-105. [DOI] [PubMed] [Google Scholar]
- 21.He, L. Z., F. Guidez, C. Tribioli, D. Peruzzi, M. Ruthardt, A. Zelent, and P. P. Pandolfi. 1998. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nat. Genet. 18:126-135. [DOI] [PubMed] [Google Scholar]
- 22.Hong, S. H., G. David, C. W. Wong, A. Dejean, and M. L. Privalsky. 1997. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 94:9028-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 24.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391:811-814. [DOI] [PubMed] [Google Scholar]
- 25.Lower, R., K. Boller, B. Hasenmaier, C. Korbmacher, N. Muller-Lantzsch, J. Lower, and R. Kurth. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. USA 90:4480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lower, R., R. R. Tonjes, C. Korbmacher, R. Kurth, and J. Lower. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magin, C., R. Lower, and J. Lower. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangeney, M., and T. Heidmann. 1998. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc. Natl. Acad. Sci. USA 95:14920-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer, J., M. Sauter, A. Racz, D. Scherer, N. Mueller-Lantzsch, and E. Meese. 1999. An almost intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257-258. [DOI] [PubMed] [Google Scholar]
- 30.McConnell, M. J., N. Chevallier, W. Berkofsky-Fessler, J. M. Giltnane, R. B. Malani, L. M. Staudt, and J. D. Licht. 2003. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol. Cell. Biol. 23:9375-9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 32.Nesbit, C. E., J. M. Tersak, and E. V. Prochownik. 1999. MYC oncogenes and human neoplastic disease. Oncogene 18:3004-3016. [DOI] [PubMed] [Google Scholar]
- 33.Patel, J. H., A. P. Loboda, M. K. Showe, L. C. Showe, and S. B. McMahon. 2004. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer 4:562-568. [DOI] [PubMed] [Google Scholar]
- 34.Reid, A., A. Gould, N. Brand, M. Cook, P. Strutt, J. Li, J. Licht, S. Waxman, R. Krumlauf, and A. Zelent. 1995. Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood 86:4544-4552. [PubMed] [Google Scholar]
- 35.Sauter, M., K. Roemer, B. Best, M. Afting, S. Schommer, G. Seitz, M. Hartmann, and N. Mueller-Lantzsch. 1996. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 56:4362-4365. [PubMed] [Google Scholar]
- 36.Sauter, M., S. Schommer, E. Kremmer, K. Remberger, G. Dolken, I. Lemm, M. Buck, B. Best, D. Neumann-Haefelin, and N. Mueller-Lantzsch. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt, B., R. Ackermann, M. Hartmann, and T. Strohmeyer. 1997. Alterations of the metastasis suppressor gene nm23 and the proto-oncogene c-myc in human testicular germ cell tumors. J. Urol. 158:2000-2005. [DOI] [PubMed] [Google Scholar]
- 38.Vafa, O., M. Wade, S. Kern, M. Beeche, T. K. Pandita, G. M. Hampton, and G. M. Wahl. 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell 9:1031-1044. [DOI] [PubMed] [Google Scholar]
- 39.Wang-Johanning, F., A. R. Frost, B. Jian, L. Epp, D. W. Lu, G. L. Johanning, M. B. Khazaeli, A. F. LoBuglio, D. R. Shaw, and T. V. Strong. 2003. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 22:1528-1535. [DOI] [PubMed] [Google Scholar]
- 40.Weijzen, S., P. Rizzo, M. Braid, R. Vaishnav, S. M. Jonkheer, A. Zlobin, B. A. Osborne, S. Gottipati, J. C. Aster, W. C. Hahn, M. Rudolf, K. Siziopikou, W. M. Kast, and L. Miele. 19 August 2002, posting date. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8:979-986. [DOI] [PubMed] [Google Scholar]
- 41.Yang, J., H. P. Bogerd, S. Peng, H. Wiegand, R. Truant, and B. R. Cullen. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. USA 96:13404-13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeyati, P. L., R. Shaknovich, S. Boterashvili, J. Li, H. J. Ball, S. Waxman, K. Nason-Burchenal, E. Dmitrovsky, A. Zelent, and J. D. Licht. 1999. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene 18:925-934. [DOI] [PubMed] [Google Scholar]
- 43.Zelent, A., F. Guidez, A. Melnick, S. Waxman, and J. D. Licht. 2001. Translocations of the RARα gene in acute promyelocytic leukemia. Oncogene 20:7186-7203. [DOI] [PubMed] [Google Scholar]