Abstract
Inactivation of gene products by dominant-negative (DN) mutants is a powerful tool to assign functions to proteins. Here, we present a two-step procedure to establish a random screen for DN alleles, using the essential murine cytomegalovirus gene M50 as an example. First, loss-of-function mutants from a linker-scanning library were tested for inhibition of virus reconstitution with the help of FLP-mediated ectopic insertion of the mutants into the viral genome. Second, DN candidates were confirmed by conditional expression of the inhibitory proteins in the virus context. This allowed the quantification of the inhibitory effect, the identification of the morphogenesis block, and the construction of DN mutants with improved activity. Based on these observations a DN mutant of the homologous gene (UL50) in human cytomegalovirus was predicted and constructed. Our data suggest that a proline-rich sequence motif in the variable region of M50/UL50 represents a new functional site which is essential for nuclear egress of cytomegalovirus capsids.
Herpesviruses have the largest genomes among vertebrate viruses. The cloning of herpesvirus genomes as bacterial artificial chromosomes (BACs) (21) paved the way for genome-wide studies by means of both random and targeted mutagenesis (4, 5, 12, 40). Gene-by-gene insertion mutagenesis of the human cytomegalovirus (HCMV), for example, revealed that only 41 of the more than 200 predicted open reading frames (ORFs) are essential for virus growth in tissue culture (12). While the function of nonessential genes can be studied using deletion and loss-of-function mutants (35), this approach is not applicable for essential genes. So far, their study requires the cumbersome establishment and optimization of trans complementation.
trans-dominant alleles and mutants can be used to generate the null phenotype in the presence of the wild-type (wt) gene product and permit functional analyses of essential genes (16). Dominant-negative (DN) mutants of host genes have been proven to be a valuable tool to elucidate viral pathways from entry (9) to exit (34). Naturally selected DN mutants have been shown to contribute to host resistance to retroviruses (10, 22), and the artificial expression of a trans-dominant mutant viral gene can render cells resistant to virus infection, a condition termed intracellular immunization (2, 37).
Knowledge of protein structure, function, or sequence motifs improves the design of trans-dominant mutants. This has been shown for single proteins of human immunodeficiency virus type 1 (for review see references 31 and 37), of a hepadnavirus (28, 29), and of herpes simplex virus type 1 (14, 32, 38). Recently, even a genome-wide screen for DN mutants based on protein structures of poliovirus gene products was conducted (11). To date the information on the majority of herpesvirus proteins is too limited for knowledge-based construction of DN mutants. Therefore, the entire coding sequence of a target gene needs to be subjected to mutagenesis to locate potential DN mutants. An optimal screening approach should be (i) generally applicable, thereby allowing the inclusion of any viral gene of interest; (ii) subtle, to minimize the risk of protein misfolding; (iii) genome based, to study the mutant's function in the relevant biological context; and (iv) suitable for high-throughput analysis because of the high number of mutants which have to be analyzed to cover large ORFs. Here, we propose a combined screening and testing approach for herpesvirus genes which matches these requirements.
MATERIALS AND METHODS
Cells and viruses.
BALB/c murine embryonic fibroblasts (MEFs), human foreskin fibroblasts (HFFs), M2-10B4 bone marrow stromal cells (ATCC CRL-1972), mouse lymphoid endothelial cells immortalized by simian virus 40 (SVEC 4-10 endothelial cells; ATCC CRL-2181), and NIH 3T3 fibroblasts (ATCC CRL-1658) were prepared and treated as described previously (1, 20). Mouse mammary epithelial C127 cells (ATCC CRL-1616) and 293 cells (ATCC CRL-1573) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. The murine CMV (MCMV) mutant GFP-1 bears the regulation cassette expressing the enhanced green fluorescent protein (GFP) under the control of the HCMV-IE/tetO2 promoter (27). All the other MCMV mutants used in this study were reconstituted from the respective MCMV BACs by transfection of MEFs with 2 to 3 μg of purified BAC DNA using SuperFect transfection reagent (QIAGEN) according to the manufacturer's instructions. Virus stocks were propagated in M2-10B4 cells as described previously (20). Virus titers were quantified on MEFs by a standard plaque assay (24).
Growth analysis.
For determination of in vitro growth of MCMV mutants, MEFs or SVEC 4-10 or C127 cells were infected in duplicate at a multiplicity of infection (MOI) of 0.1 PFU per cell. The inoculums were removed after 1 h, and normal medium or medium supplemented with 1 μg/ml doxycycline (Dox) was added. On day 3, Dox was added a second time. Supernatants of infected cells were harvested on days 3 and 5 after infection, and the amounts of the released infectious particles were determined by plaque assay on MEFs.
Plasmid construction. (i) Constructs with constitutive expression.
Loss-of-function mutants from a linker-scanning library of M50 used in an earlier study (6) were used in the primary screen for inhibitory mutants. The mutants were subcloned into the pOriR6K-zeo-ie vector (GenBank accession no. AY700022) under the control of the strong constitutive HCMV immediate-early promoter-enhancer. The sites of 5-amino-acid (aa) insertions of the mutants used in this study are indicated in Fig. 2. The deletion in pM50-del4 (6) (renamed pM50-ΔVR in this study) comprises aa 179 to 276 of the M50 ORF. The hemagglutinin (HA) tag was added to the N terminus of M50 and M50-ΔVR by PCR with primers AB20-03 (CGCGGTACCATGTACCCATACGACGTCCCAGACTACGCTGAGATCGACAAGAATGTGGGC [the underlined oligonucleotide sequences represent restriction sites; sequences in bold code for the HA tag) and AB10-02 (CGCCTCGAGTCACGGATGACC) using the template pOriR6K-zeo-ie-M50 (6) or pM50-ΔVR. The PCR fragments were cut with KpnI/XhoI and inserted into pOriR6K-zeo-ie, giving rise to pM50HA and pM50HA-ΔVR, respectively. In order to destroy the M53 binding in the M50 deletion mutants, pM50-ΔVR and pM50HA-ΔVR were amplified by inverse PCR using primers delmo-NheI-for (GATGCTAGCAGCTATTGGGAGTGTAGGAGC) and delmo2-Nhe-rev (GCTGCTAGCATCGGTGGTCAACATCGCGTCGC). The recircularization of the PCR products via the introduced NheI sites led to the constructs pM50-ΔM-ΔVR and pM50HA-ΔM-ΔVR, in which aa 51 to 59 of M50 were replaced by alanine and serine. In order to delete 29 aa spanning a proline-rich sequence (aa 179 to 207), pOriR6K-zeo-ie-M50 and pM50HA were amplified by inverse PCR using primers del3to4-for (CGCGCTAGCAGCCTGCGCAAACACGGCCC) and del3to4-rev (CGCGCTAGCCCCGCAGTCGGTGGGAACGGAAG). The recircularization of the PCR products via the introduced NheI sites led to the vectors pM50-ΔP and pM50HA-ΔP, respectively. The integrity of every construct was confirmed by DNA sequencing.
FIG. 2.
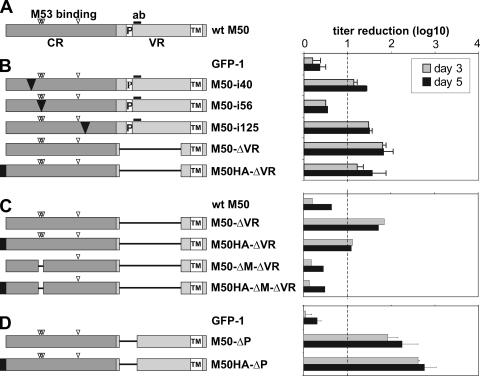
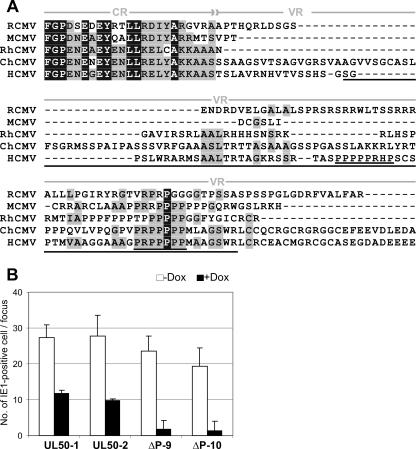
Results of the primary screen for inhibitory mutants of M50. The amino acid sequence encoded by the M50 gene of MCMV is displayed to show the positions of the different mutations which were analyzed. Transposon insertions resulting in the introduction of 5 aa (arrowheads) are indicated along the ORF. Mutants that caused a null phenotype after they were placed into the M50-deleted MCMV genome are indicated by filled symbols; mutants which did not affect the function are indicated by empty ones (6). Black arrowheads indicate the mutants that interfered with virus reconstitution after insertion into the wt genome, and gray arrowheads indicate mutants that did not interfere. The amino acid sequence deleted in M50-ΔVR is indicated by white lettering in a black box. The sequence which is deleted in the ΔP mutant of M50 is indicated by bold lettering. The underlined sequence in the M50 ORF shows the region which is essential for binding to the M53 protein.
(ii) Construction of the entry vectors.
For the generation of a suitable insertion vector for conditional expression of the inhibitory mutants, the enhanced GFP ORF was deleted from vector pO6-IET-gfp (27) with PmeI/ApaI. The synthetic oligonucleotides IET-linker1 (CGCCCCGGGGGCGCGCCGTTAACACGCGTGGGCCCGCG) and IET-linker2 (CGCGGGCCCACGCGTGTTAACGGCGCGCCCCCGGGGCG) were annealed, cut with SmaI and ApaI, and then inserted into the vector, giving rise to pO6-IET-linker. The tetR gene was excised from pcDNA6/TR (Invitrogen) by MluI, filled in, and inserted into pO6-IET-linker at the filled-in SphI site, giving rise to pO6-IET-entry. Essentially the same procedure has been carried out to modify pO6-SVT-gfp to construct an entry vector, pO6-SVT-entry. These plasmids were used as insertion vectors for the different conditional constructs listed below.
(iii) Constructs for conditional expression.
The ORFs for M50-i40, M50-i56, M50-i125, M50-ΔVR, M50HA-ΔVR, M50-ΔM-ΔVR, M50HA-ΔM-ΔVR, M50-ΔP, and M50HA-ΔP were excised from pM50-i40, pM50-i56, pM50-i125, pM50-ΔVR, pM50HA-ΔVR, pM50-ΔM-ΔVR, pM50HA-ΔM-ΔVR, pM50-ΔP, and pM50HA-ΔP, respectively, by KpnI/XhoI and then filled in by T4 DNA polymerase and inserted into the pO6-IET-entry vector, which was linearized by HpaI, giving rise to pO6-IET-M50-i40-TR, pO6-IET-M50-i56-TR, pO6-IET-M50-i125-TR, pO6-IET-M50-ΔVR-TR, pO6-IET-M50HA-ΔVR-TR, pO6-IET-M50-ΔM-ΔVR-TR, pO6-IET-M50HA-ΔM-ΔVR-TR, pO6-IET-M50-ΔP-TR, and pO6-IET-M50HA-ΔP-TR, respectively.
pO6-SVT-UL-50-TR was constructed by cloning of the HCMV UL50 ORF from pO6-ie-UL50 (30) into the pO6-SVT-entry vector at the AscI and HpaI sites. The N-terminal variable region of UL50 (aa 189 to 253) was deleted by inverse PCR using UL50-ΔPfor (CGCGCTAGCCTGTGTCGTTGTGAGGCCTGTATGG) and UL50-ΔPrev (CGCGCTAGCGCCGCTGTGACTGCTTACGGTGACG) primers, resulting in pO6-ie-UL50-ΔP. The mutant ORF from this construct was inserted into pO6-SVT-entry at the AscI and HpaI sites, generating pO6-SVT-UL50-ΔP-TR.
Construction of recombinant MCMV BACs.
To insert the constitutive expression cassettes into the wt MCMV BAC, Escherichia coli strain DH10B (Invitrogen) containing the wt FLP recombination target (FRT) MCMV (wt-FRT-MCMV) BAC (7) and the temperature-sensitive FLP recombinase expression plasmid pCP20 (8) was transformed with pOriR6K-zeo-ie constructs containing the different M50 mutants and treated as described previously (6). The correct recombinations were verified by restriction analysis, and BAC DNA was prepared as described above. MEFs were transfected with purified BAC DNA derived from two independent clones for each mutant, and virus reconstitution was performed by assessing plaque formation for the period of 4 weeks after transfection. Constructs which did not lead to viral plaque formation were newly reisolated from bacteria and retested three times in total.
In the same way, the constructs with regulation cassettes for the identified inhibitory mutants (pO6-IET-M50-i40-TR, pO6-IET-M50-i56-TR, pO6-IET-M50-i125-TR, pO6-IET-M50-ΔVR-TR, pO6-IET-M50HA-ΔVR-TR, pO6-IET-M50-ΔM-ΔVR-TR, pO6-IET-M50HA-ΔM-ΔVR-TR, pO6-IET-M50-ΔP-TR, and pO6-IET-M50HA-ΔP-TR) were inserted into the wt-FRT-MCMV BAC by using the FRT/FLP system, and viruses M50-i40, M50-i56, M50-i125, M50-ΔVR, M50HA-ΔVR, M50-ΔM-ΔVR, M50HA-ΔM-ΔVR, M50-ΔP, and M50HA-ΔP, respectively, were reconstituted by transfection of MEFs.
Construction and analysis of the spread of HCMV mutants.
The plasmids pO6-SVT-UL50-TR and pO6-SVT-UL50-ΔP-TR were flipped into the AD169-derived BAC AD169-FRT carrying an FRT site at the UL45 locus as described earlier (1). The mutants were reconstituted to virus by transfection of BAC DNA into HFFs using the MBS transfection kit (Stratagene) according to the instructions of the manufacturer. Two independent clones of the recombinant BACs, AD169FRT-SVT-UL50-1 and AD169FRT-SVT-UL50-2 carrying the wt UL50 and AD169FRT-SVT-UL50-ΔP-9 and AD169FRT-SVT-UL50-ΔP-10 carrying the mutant ORF, were reconstituted to virus in the absence of Dox. After the plaques became apparent, the cells were removed from the transfected dish by trypsinization and fivefold serial dilutions with uninfected HFFs were performed. Cells (2 × 104 per well) were plated onto 96-well plates from each dilution either in the presence or in the absence of Dox in triplicate. Parallel with the plating CMV-positive human serum (1:200) was added to the culture medium to prevent virus spread via the supernatants. At 24 h postplating one of the triplicates of each dilution was stained with an immediate-early 1 antigen (IE1)-specific mouse monoclonal antibody (Perkin-Elmer) and was visualized by a Cy3-coupled goat anti-mouse immunoglobulin G (IgG) serum (Dianova) to determine the density of infected cells in each dilution by fluorescence microscopy. Five days after the plating the remaining two replicates were stained as described above and the sizes of infection foci were determined by counting the number of IE1-positive cell nuclei by fluorescence.
Semiquantitative Western blot analysis of protein expression.
MEFs or SVEC or C127 cells were infected on six-well plates in the absence or presence of 1 μg/ml Dox with M50HA-ΔVR for 3 days at an MOI of 0.1. In experiments done on day 3 postinfection, the supernatants were removed and stored for plaque assays. Cells were washed with phosphate-buffered saline (PBS) and lysed in 150 μl triple detergent lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.01% sodium dodecyl sulfate [SDS], 1% NP-40, 0.5% sodium deoxycholate, freshly supplemented with protease inhibitor cocktail [Roche]) per well. The lysates were diluted in 1:3 steps, separated in a 15% SDS-polyacrylamide gel, and transferred to Hybond-P membranes (Amersham). The membranes were treated with a monoclonal rat antibody to the HA tag (3F10) coupled to horseradish peroxidase (Roche) or a polyclonal rabbit antiserum specific to M50 (23) followed by a horseradish peroxidase-coupled anti-rabbit antiserum (Dianova). The specific signals were detected with ECL-Plus reagent (Amersham). The intensities of the specific bands were quantified by Quantity One software (Bio-Rad). The dilutions where the signals were in the linear range of detection were used to calculate the relative expression levels. The signals obtained with the anti-M50 detection were multiplied with the dilution factor and set as 1 for calculation of the relative amounts of the HA-tagged proteins using the dilution factor-corrected signals obtained by the anti-HA blot assay.
Coimmunoprecipitation.
293 cells were transfected with 5 μg DNA (2.5 μg of each plasmid) per 6-cm dish with SuperFect transfection reagent (QIAGEN) according to the manufacturer's instructions. Cells were harvested 1 day posttransfection and resuspended in 1 ml PBS. NIH 3T3 cells were mock infected or infected with wt-FRT-MCMV or M50HA-ΔP at an effective MOI of 10 with or without addition of 1 μg/ml Dox for 28 h. Cells were resuspended in 1 ml PBS. Fifty microliters of the cell suspensions was lysed directly in loading buffer (62.5 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 6 M urea, 5% β-MetOH, 0.01% bromophenol blue, 0.01% phenol red). The cells in the remaining 950 μl of the cell suspensions were lysed in 1 ml high-salt lysis buffer (400 mM NaCl, 20 mM Tris, pH 8.0, 1% Triton X-10, freshly supplemented with protease inhibitor cocktail [Roche]) for 20 min on ice. The insoluble cell debris was removed by centrifugation (15,000 × g, 4°C, 20 min). The coimmunoprecipitations were performed with anti-HA-Matrix (Roche) according to the manufacturer's instructions or with 5 μl anti-M50 antiserum and 50 μl protein A-Sepharose (Amersham) following a standard protocol by use of the high-salt lysis buffer for the washing (19). The pelleted proteins were separated in a 10% SDS-polyacrylamide gel, transferred to Hybond-P membranes (Amersham), and stained with a peroxidase-conjugated anti-HA antibody (3F10; Roche) or a polyclonal rat antiserum specific to M53 (19) followed by peroxidase-conjugated donkey anti-rat IgG as secondary antibody (Dianova) followed by ECL-Plus immunodetection (Amersham).
Confocal laser scanning microscopy.
NIH 3T3 cells grown on glass coverslips were infected with M50HA-ΔVR or M50HA-ΔP at an MOI of 0.1 for 1 day with or without addition of 1 μg/ml Dox. Cells were washed with PBS and fixed with phosphonoformic acid as described previously (23). The proteins were visualized using a monoclonal rat antibody to the HA tag (3F10) coupled to fluorescein isothiocyanate (FITC; Roche), a polyclonal rabbit antiserum specific to M50 (23) followed by Texas Red-conjugated donkey anti-rabbit IgG (Dianova), or a polyclonal rat antiserum specific to M53 (19) followed by FITC-conjugated donkey anti-rat IgG (Dianova) as a secondary antibody. The confocal fluorescence images were collected with an LSM501Meta system (Zeiss).
Transmission electron microscopy.
NIH 3T3 cells were grown on carbon-coated sapphire discs and infected with M50HA-ΔP virus at an MOI of 0.5 without or with addition of 1 μg/ml Dox for 48 h. Cells were fixed by high-pressure freezing with an HPF 01 instrument (Engineering Office, M. Wohlwend GmbH, Switzerland), freeze-substituted, and plastic embedded as described previously (36).
RESULTS
Mutant screening of an essential viral gene for inhibitory effects.
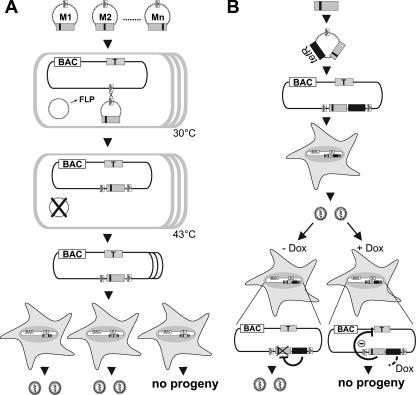
DN mutants of a protein are defined by their ability to fulfill one essential function and, by lacking a second, to inhibit their wt counterpart. We used the MCMV to establish a random screening approach to viral DN mutants. The viral proteins involved in nuclear egress of virus capsids are conserved among all herpesviruses and form the nuclear egress complex (NEC) at the inner nuclear membrane to execute mechanistically still-unresolved but crucial functions (6, 18, 23, 25, 39). The interaction between the MCMV NEC proteins M50 and M53 is essential for function. We have mapped the mutual binding sites of M50 and M53, but no further structure-function relations are known (6, 19). A library of M50 mutants was made available from this study which relies on the random insertion of 5 aa or a stop codon into the protein coding sequence. Longer parts of the M50 coding region in which insertions did not lead to a null phenotype were subjected to deletions until the mutant became nonfunctional. Loss-of-function mutants from this library were now placed under the control of the strong HCMV immediate-early promoter-enhancer and reinserted via an FRT site into the MCMV BAC that still contains the wt M50 gene (Fig. 1A). The resulting recombinants were screened for inhibition of virus reconstitution. To this end the DNAs of mutant MCMV BACs were transfected one by one into primary mouse fibroblasts (MEFs) and viral plaque formation was monitored. For the M50 gene, a total of 32 derivatives, 29 loss-of-function mutants, one nonfunctional deletion mutant, and two functional mutants for control purposes were analyzed (Fig. 2). Four of the tested loss-of-function mutants, namely, the mutants with 5-aa insertions at positions 40, 56, and 125 of the M50 ORF and also the M50-ΔVR deletion mutant, reproducibly interfered with plaque formation after transfection.
FIG. 1.
Screening strategy for DN mutants of essential viral genes. (A) Screening for inhibitory mutants. An essential viral gene, the target gene (gray box, T), is subcloned and subjected to a random and comprehensive mutagenesis in vitro leading to a mutant library, M1, M2, … Mn (small black boxes indicate mutations). Mutated ORFs are placed under the control of a strong constitutive promoter into an insertion plasmid containing an FRT site (open box with gray triangle). The insertion plasmids can be maintained only in a special E. coli strain. Normal E. coli (open boxes) carrying an FRT-site-labeled viral BAC and a temperature-sensitive plasmid expressing FLP recombinase (FLP) are transformed with the insertion plasmids carrying different mutants one by one. The FLP recombinase mediates site-specific recombination between the FRT sites in the BAC and the insertion plasmids. This recombinant can then be isolated under combined antibiotic selection for both the BACs and the insertion plasmid. The FLP-expressing helper plasmid is removed by elevated temperature. Then, BAC DNA is prepared and permissive cells are transfected with each construct. The mutants which are able to inhibit the virus reconstitution can be selected on the basis of the inability to form plaques upon transfection. (B) Validation of DN mutants by conditional gene expression. The inhibitory mutants are subcloned under the control of a promoter regulated by TetR (black box) into an insertion plasmid with an FRT site. These constructs are delivered into the viral BAC as described above. Then, permissive cells are transfected with the recombinants in order to reconstitute viruses carrying the regulation cassettes for the inhibitory mutants. The inhibitory mutants are not expressed during reconstitution because in the absence of Dox (− Dox) the constitutively expressed TetR blocks their transcription. The inhibitory function of the mutants can be analyzed upon Dox administration (+ Dox), which leads to the expression of the inhibitory mutant by releasing the expression cassette from TetR regulation.
Validation of DN mutants by conditional expression.
Reconstitution of viruses from isolated DNA is difficult to standardize and may be affected by inhibitory effects at several stages. Rather than specific inhibition of the wt gene function, the toxicity of an abundantly expressed mutant protein also may impair virus reconstitution upon transfection. Thus, the failure to reconstitute virus in the presence of a mutant of M50 is an indication of but not a sufficient proof for the DN effect of a given mutant. Conditional expression of the mutant protein in the context of the virus replication should allow virus reconstitution. By use of a conditional mutant a fully infectious virus and not only the viral genome enters the cell and the intracellular milieu mirrors a wt infection.
Recently, we reported on a regulated viral expression system which operates independently of the viral replication program. Constitutive viral expression of TetR blocks the transcription of the regulated gene. Induction by Dox exposes the viral replication program to the effect of the mutant protein (27). We now inserted each mutant of M50, which had interfered with virus reconstitution, into this regulation cassette. To study the inhibitory capacity in the context of virus replication, these constructs were then integrated into the MCMV genome (Fig. 1B). Viral growth was analyzed in MEFs under multistep growth conditions using 0.1 PFU of the analyzed virus per cell in both the absence and the presence of Dox. In the absence of Dox all recombinants could be reconstituted to virus and the progeny grew to titers indistinguishable from those of the GFP-1 control virus (data not shown). The number of infectious units was determined by a plaque assay on days 3 and 5 after infection. At least a 90% reduction of virus titers in the presence of Dox was arbitrarily considered as significant inhibition. The presence of Dox had no effect on the replication of the GFP-1 control, whereas the Dox-induced expression of M50-i40, M50-i125, M50-ΔVR, and M50HA-ΔVR proteins reduced growth of the viruses in the range of 1 to 2 orders of magnitude (Fig. 3B). The degrees of regulation were comparable on day 3 and day 5, demonstrating the stability of the regulation cassette under replication conditions in vitro. One mutant, M50-i56, had no effect in response to Dox, showing the advantage of a conditional system over simple genome transfection.
FIG. 3.
The effect of conditional expression of the inhibitory M50 mutants on the virus growth. The schematic representation of the analyzed mutants is shown on the left. The wt M50 ORF is shown first. (A) The N-terminal region of the M50 (CR), which is conserved in alpha-, beta-, and gammaherpesvirus families, is indicated by a dark gray box, and the C-terminal variable region (VR) is indicated by a light gray box. The open box labeled with TM indicates the transmembrane domain, and “P” indicates the proline-rich sequence. The HA tag is indicated by a black box. The open arrowheads show the positions of 5-aa insertions affecting M53 protein binding (aa 53 to 57 and aa 114) (6). The sequence used to generate the anti-M50 antiserum (aa 201 to 213) is shown by a thick bar (ab). (B to D) The analyzed M50 mutants carry an insertion (black triangle) of 5 aa after position 40, 56, or 125 of M50 or lack sequences (represented by lines), i.e., a deletion of aa 179 to 276 in the ΔVR constructs (B), of aa 51 to 59 in the ΔM constructs (C), and of aa 179 to 207 in the ΔP constructs (D). The bar diagram on the right side shows the regulation of the virus growth in response to Dox in these mutants. MEFs were infected at an MOI of 0.1 in the absence or presence of 1 μg/ml Dox for 3 (gray bars) or 5 (black bars) days. Cell-free supernatants were taken on the indicated days and were titrated on MEFs to determine the number of the released infectious units. The titer reduction induced by Dox is shown as a ratio between the titer in the absence of Dox and the titer in the presence of Dox. The titer reduction was considered not significant if the ratio was less than 10. Each set was analyzed two times. Shown are means and standard deviations with the exception of the panel shown in the second diagram, which shows results from one experiment.
The inhibitory effect of M50-ΔVR requires binding to M53 and can be improved by targeted mutagenesis.
The M50 protein is predicted to form aggregates with several viral proteins (33). A DN mutant probably loses the interaction with at least one of its partners. The interaction between M50 and M53 is essential for their functionality, and a binding motif of M50 for M53 is located between aa 53 and 56 (6). We predicted that an inhibitory mutant of M50 which does not bind to M53 should lose its DN activity provided that the inhibitory effect is related to an NEC function. Accordingly, we tested the inhibitory potential of M50-ΔVR and M50HA-ΔVR after deleting the M53 binding motif (ΔM). While M50-ΔVR and M50HA-ΔVR showed the expected range of regulation, in the ΔM mutants the inhibitory effect on virus growth in response to Dox was completely lost (Fig. 3C).
An accumulation of proline and arginine residues is consistently found in the variable region of all UL34 homologues of herpesviruses. The DN mutant M50-ΔVR lacks 98 aa including a prominent proline-rich sequence motif shared by all CMVs (see Fig. 7A). To test whether the absence of only this motif would suffice to mediate the DN effect, we deleted only the N-terminal 29 aa of the variable region of M50 (179 to 207, indicated by bold lettering in Fig. 2A, named ΔP). Viruses carrying the regulation cassettes expressing M50-ΔP and M50HA-ΔP were found to be attenuated up to 800-fold in the presence of Dox (Fig. 3D). Thus, a DN mutant found in a first screen can be improved, and this could be documented because the conditional expression provides quantitative information.
FIG. 7.
The deletion of the proline-rich sequence from UL50 of HCMV creates a DN mutant. (A) N-terminal sequence comparison of the CMV UL34 homologues. The amino acid sequence of M50 was aligned with the UL34 family members from CMVs by use of the Vector NTI Align X program (Invitrogen) via the BLOSUM 62 similarity matrix. (RCMV, rat CMV; RhCMV, rhesus monkey CMV; ChCMV, chimpanzee CMV). The alignment of the end of the conserved region (CR) and the N-terminal half of the variable region (VR) is shown. The strictly conserved amino acids are shown by white letters in black boxes. Blocks of similar amino acids are indicated by gray boxes. The region deleted from the ΔP mutant of the HCMV UL50 is underlined. The two proline-rich motifs found in UL50 of HCMV are double underlined. (B) HFFs were transfected with two independent clones of the recombinant BACs, AD169FRT-SVT-UL50-1 and AD169FRT-SVT-UL50-2 expressing the wt UL50 and AD169FRT-SVT-UL50-ΔP9 and AD169FRT-SVT-UL50-ΔP10 expressing the mutant ORF. After the first plaques became apparent, the cells were replated onto 96-well plates together with noninfected HFFs either in the absence (white bars) or in the presence (black bars) of Dox. Five days later the numbers of IE1-positive cell nuclei in infected foci were determined. The mean values for 8 to 10 infected foci obtained in two independent experiments for clones UL50-1, UL50-2, and ΔP10 and three experiments for clone ΔP9 are plotted.
The cell toxicity of M50-ΔVR is not responsible for the inhibitory effect.
Inhibition due to protein toxicity depends on the steady-state concentration of the toxic protein, whereas for a DN effect the ratio of mutant to wt protein is relevant. The necessary ratio depends on the affinity of the mutants for their binding partners and on the number of homologous subunits within the targeted complex. Productive virus infection is characterized by exponentially increasing viral protein levels. For a stable phenotype a critical value of the ratio of wt to mutant protein should be maintained during the entire replication cycle. We studied at which critical threshold the excess of M50-ΔVR mutant protein over wt M50 would provide a stable inhibitory effect. To compare the amounts of wt and DN M50 proteins, we used HA-tagged M50-ΔVR (M50HA-ΔVR) for detection, because it lacks the sequence recognized by the specific antiserum to M50 (23). The HA tagging, probably by affecting the protein folding or stability, weakened the strength of M50HA-ΔVR but did not decrease the improved inhibitory capacity of M50HA-ΔP with the smaller deletion (Fig. 3B and 3C). However, the inhibitory activity of HA-M50HA-ΔVR remained significant.
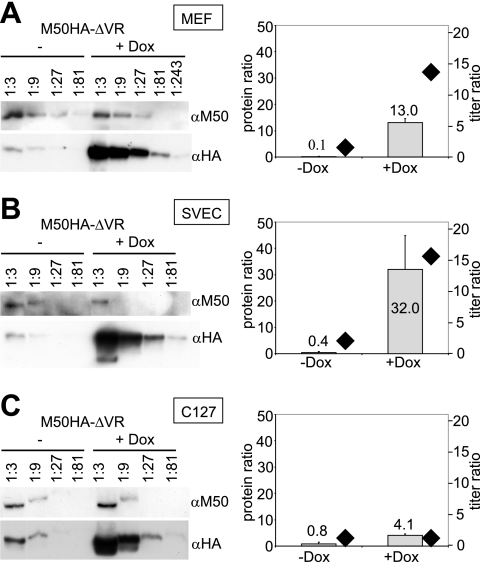
For tet-regulated systems a cell-line-specific degree of regulation and protein expression was reported elsewhere (17, 27). Here, we studied the inhibitory potential of DN M50HA-ΔVR in MEFs (Fig. 4A), in the endothelial cell line SVEC (Fig. 4B), and in the epithelial cell line C127 (Fig. 4C) in order to evaluate the growth phenotype at different mutant/wt protein ratios. The replication of the M50HA-ΔVR virus was attenuated after Dox induction in MEFs and SVEC cells but not in C127 cells (depicted as rhombuses in the panels on the right in Fig. 4). Clearly, the three cell lines could be induced to express comparable total amounts of DN protein, and this excluded cell toxicity as an explanation for the inhibitory effect. However, the determination of protein ratios (Fig. 4, depicted as bars) revealed that in the presence of Dox the amount of M50HA-ΔVR protein exceeded the amount of basal wt M50 protein expression by 13- or 32-fold in MEFs and SVEC cells, respectively, but only by fourfold in C127 cells. A DN effect was apparently achieved after expression of the DN protein beyond a certain ratio of DN mutant to wt protein. The results for the C127 line indicated that the fourfold excess of DN over wt proteins did not yet reach the critical ratio or threshold level of the inhibitor necessary to prevent virus replication.
FIG. 4.
The inhibitory effect is defined by the ratio of DN to wt protein. MEFs (A) and SVEC (B) and C127 (C) cells were infected with M50HA-ΔVR at an MOI of 0.1 in the absence or presence of 1 μg/ml Dox for 3 days. The expression of the wt and the mutant protein was analyzed by Western blotting (left). Lysates of infected cells were diluted serially, and proteins were separated in an SDS-polyacrylamide gel, transferred to a membrane, and stained with specific antibodies to M50 (αM50) or HA (αHA). The M50 antiserum and the HA antibody have comparable affinities for their epitopes (data not shown), allowing the relative quantification of the signals. The bar diagrams (to the right of each blot) show the ratio of the expression of the HA-tagged M50HA-ΔVR protein to that of the wt M50 gene product. The protein ratios were determined in at least two independent experiments and are depicted as gray bars referring to the left ordinate. In parallel, the amounts of the released infectious units were determined in each supernatant by plaque assay. The ratios of the titers in response to Dox (division of the titers obtained in the absence of Dox by the titers in the presence of Dox) are shown as black rhombuses (to be read according to the right ordinate).
Analysis of the DN phenotype.
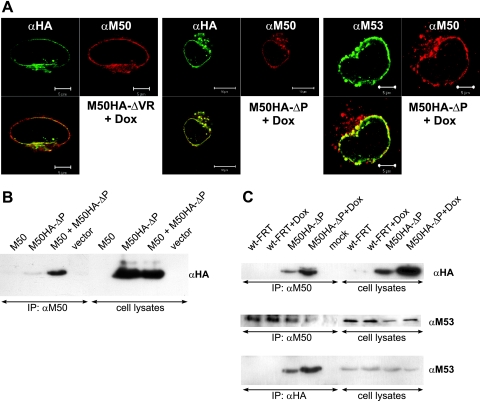
Conditional DN mutants enable the analysis of the null phenotype that they cause in the presence of the wt gene during virus infection. The lack of DN activity of M50-ΔM-ΔVR and M50HA-ΔM-ΔVR, which fail to interact with M53, indicated that the inhibitory effect is dependent on NEC formation. Therefore, we studied the subcellular distribution of the DN mutant proteins in cells infected with M50HA-ΔVR or the functionally improved M50HA-ΔP virus. To this end infected NIH 3T3 fibroblasts were analyzed by immune fluorescence microscopy 24 h postinfection. The mutants M50HA-ΔVR and M50HA-ΔP, which lack the recognition site for the M50 antiserum, were detected by an anti-HA monoclonal antibody. The inhibitory HA-tagged DN protein colocalized with the wt M50 protein at the nuclear rim. Also the M53 protein, which is recruited by the M50 protein (23), was found to colocalize properly in the presence of the DN mutant (Fig. 5A). Therefore, the DN effect of M50HA-ΔP is not due to delocalization of M53 or the wt complex. Next, we asked if the DN protein enters into the complex formed by the wt proteins. Thus, we examined the protein-protein interactions in the presence and absence of M50HA-ΔP. Interestingly, M50HA-ΔP protein already coprecipitated with M50 when expressed in isolation (Fig. 5B) and also during virus replication (Fig. 5C, top), indicating that M50 protein forms homo-oligomers. M53 protein was also present in the complex with both wt M50 and M50HA-ΔP (Fig. 5C, middle and bottom, respectively). Since the complex kept the wt distribution and stability (data not shown), we concluded that the DN function affected the interaction of the NEC with other, not-yet-identified important protein partners.
FIG. 5.
Intracellular distribution of the M50-M53 complex in the presence of the DN M50 mutants. (A) Indirect immunofluorescence images of NIH 3T3 cells infected at an MOI of 0.2 with M50HA-ΔVR or M50HA-ΔP virus for 1 day in the presence of 1 μg/ml Dox were taken. The M50 mutants were stained with an anti-HA antibody coupled to FITC. wt M53 protein was stained with an anti-M53 antibody followed by treatment with an FITC-labeled anti-rat secondary antiserum. The wt M50 protein was stained with an anti-M50 antiserum followed by treatment with Texas Red-labeled anti-rabbit secondary antiserum. (B and C) Coimmunoprecipitation of wt M50, M50HA-ΔP, and M53 proteins. The M50 protein was precipitated from cell lysates by anti-M50/protein A-Sepharose (B and top and middle in panel C), and the M50HA-ΔP protein was precipitated by anti-HA coupled to beads (bottom in panel C). Samples were separated in a 10% SDS-polyacrylamide gel, transferred to a membrane, and stained with antisera to HA or M53. (B) 293 cells were transfected for 1 day with pOriR6K-zeo-ie-M50, pM50HA-ΔP, or pOriR6K-zeo-ie, either alone or in combination. (C) NIH 3T3 cells were infected with wt-FRT-MCMV or M50HA-ΔP virus at an MOI of 10 in the absence or presence of 1 μg/ml Dox for 28 h. IP, immunoprecipitation.
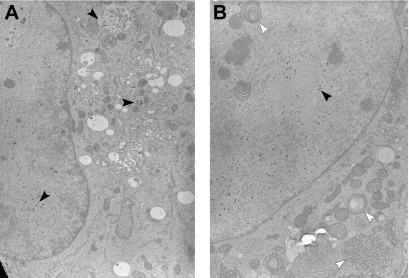
Nuclear export of herpesvirus capsids is a crucial step during herpesvirus replication (15, 23, 26, 39). Thus, DN inhibitors of M50 function might interfere with nuclear export of capsids. To this end, NIH 3T3 cells were infected with M50HA-ΔP virus or wt MCMV (data not shown) and analyzed by transmission electron microscopy of high-pressure frozen and freeze-substituted samples. wt MCMV capsids were found in the nucleus and the cytoplasm irrespective of the presence of Dox. The same capsid distribution was found with M50HA-ΔP virus in the absence of Dox (Fig. 6A). However, in Dox-treated cells all viral capsids were confined to the cell nucleus (Fig. 6B). Moreover, the cell morphology was altered profoundly after infection with M50HA-ΔP virus in the presence of Dox. Stacks of cisternae accumulated in both the cytoplasm and nucleus. The cytoplasmic membranes were decorated with ribosomes and thus represent endoplasmic reticulum (ER), while the nuclear membrane layers were continuous with the inner nuclear membrane. However, we saw this alteration of ER membranes also after infection with an inducible M50HA virus which is not blocked in capsid export (data not shown). Thus, overexpression of M50HA and that of M50HA-ΔP by Dox have similar effects on the cellular membranes. Yet, accumulations of the inner nuclear membrane were seen only in the presence of M50HA-ΔP; the lack of capsid export confirmed the specificity of DN M50HA-ΔP as an inhibitor of the nuclear capsid egress.
FIG. 6.
DN mutants of M50 prevent viral capsid egress from the cell nucleus. NIH 3T3 cells were infected at an MOI of 0.5 with M50HA-ΔP virus for 48 h in the absence (A) or presence (B) of 1 μg/ml Dox. Cells were fixed by high-pressure freezing, freeze-substituted, plastic embedded, and analyzed by electron microscopy. Black arrowheads show viral capsids; white arrowheads show accumulated membranes in the nucleus and cytoplasm after induction of overexpression of the M50HA-ΔP protein.
The proline-rich motif of M50/UL50 is associated with DN function in CMVs.
In order to investigate the functional importance of the proline-rich motif we deleted the corresponding sequences from the subcloned UL50 ORF of HCMV (Fig. 7A). Then, the wt and the mutant UL50 coding sequences were transferred into the regulation cassette (27). This regulation cassette had already been used to insert genes into HCMV BAC AD169-FRT carrying an FRT site at the UL45 locus (1). Two independent clones of the recombinant BACs, AD169FRT-SVT-UL50-1 and AD169FRT-SVT-UL50-2 carrying two copies of the wt UL50 and AD169FRT-SVT-UL50-ΔP9 and AD169FRT-SVT-UL50-ΔP10 carrying the mutant ORF, were reconstituted to virus in HFFs in the absence of Dox. The reconstitution of viruses with the UL50-ΔP-containing regulation cassette was found delayed compared to the wt UL50 constructs. After the plaques became apparent, the infected cells were removed and a focus expansion assay was performed at day 5 after replating (see Materials and Methods). The experiments with each clone were repeated at least two times. There was already a moderate inhibition of spread of recombinants expressing wt UL50 from the second gene copy in the presence of Dox. However, the expression of UL50-ΔP inhibited virus spread by a factor of 10 (Fig. 7B). Thus, the results found in MCMV correctly predicted the function of the HCMV mutant protein.
DISCUSSION
DN mutants, which modulate virus functions, have a broad field of application. They allow a detailed characterization of essential viral functions by the induction of a specific null phenotype and should provide insights into interactions between viral and cellular proteins.
Here, we present a random mutagenesis-based approach to identify DN mutants for a gene of interest. The Tn7-based linker-scanning mutagenesis introduces 5-aa insertions into coding sequences and provides mutant libraries harboring a comprehensive set of subtle insertion mutants of the ORF of interest (3). Such libraries have been applied for mapping functional domains of the MCMV egress proteins by screening for both functional and nonfunctional mutants (6, 19). For M50, 680 mutants were screened by PCR, and among 104 mutants tested for function by complementing an M50-deficient MCMV genome 44 loss-of-function mutants at 34 different positions were found (6). This showed that the mutagenesis is subtle and that in most cases the overall protein structure remained functional upon mutagenesis. Now, the insertion of the nonfunctional mutants of M50 into the wt MCMV genome by the FRT/FLP recombination system allowed a screen for mutants with inhibitory potential. Remarkably, only 4 out of 32 tested nonfunctional mutants revealed an inhibitory function in the virus reconstitution assay. This supports the prediction that a wide screen is necessary to define a DN function by a random approach.
Transfection of the viral nucleic acid is error prone because (i) deregulation of viral gene expression is expected due to the overload of viral genomes, (ii) transgenes under the control of strong promoters will be overproduced and related toxicity issues (response to misfolded proteins) may occur, and (iii) the innate host immune response to nucleic acid transfection induces an antiviral state. This may lead to an overestimation of inhibitory mutants. The conditional expression of the inhibitory mutants in the context of the wt genome allowed the analysis of their inhibitory phenotype. Thus, the usage of regulation cassettes for conditional expression (27) adds to the work load but provides access to quantification, to the analysis of cell toxicity, and to the improvement of DN function.
Here, the random screen in combination with conditional expression identified a new essential motif at the N terminus of the variable region of MCMV M50. The DN mutant lacking this motif still required binding to M53 for its inhibitory activity, suggesting that the proline-rich motif identifies a function of the NEC. This proline-rich sequence is likely to control a binding site for an unknown interaction partner which is required for CMV capsid export from the nucleus. Strong expression of M50 and its derivatives resulted in a characteristic reorganization of ER membranes seen before (13, 15). However, this effect is not identical with the DN phenotype, which includes also the block of capsid export and accumulation of membrane stacks within the nucleus.
The conditional expression allowed a comparison of the strengths of different dominant inhibitors. Whereas the inhibitory effect of M50HA-ΔP on virus replication was in the range between 2 and 3 orders of magnitude, the DN allele of the smallest capsid protein inhibits virus replication by 6 orders of magnitude (27). Nevertheless, the motif identified in the context of MCMV guided the design of an inhibitory mutant of the homologous protein of HCMV from the same herpesvirus subfamily. It would be interesting to probe the applicability of this approach to the other herpesvirus subfamilies. Notably, the sequence motif that we defined for CMVs is not present in the variable region of the members of the UL34 family encoded by alpha- or gammaherpesviruses.
Our studies of DN mutants of HCMV UL50 also pointed to a difficulty of the present approach. The conditional gene expression cassette established for MCMV did not operate satisfactorily in the HCMV context. In MCMV, the low basal expression of M50 mutants did not interfere with virus reconstitution and propagation. In HCMV, the usage of the immediate-early enhancer prevented the reconstitution of the virus from the BAC (data not shown). Reconstitution was just possible using the simian virus 40 early enhancer-based cassette. However, phenotypic reversion rapidly occurred after passage of the inhibitory mutants even in the absence of Dox. Only the first progeny reconstituted directly from the BAC could be used for functional testing of the HCMV UL50 DN phenotype in the focus expansion assay. Thus, the promoter of the regulation cassette has to be carefully selected for low basal expression and efficient response to Dox and the construct has to be adapted to each virus under study.
Altogether, the random screen for inhibitory mutants combined with conditional expression of the identified mutants demonstrated here should aid the systematic dissection of herpesvirus gene functions by providing access to DN mutants of viral proteins with unknown structure and function in the virus context.
Acknowledgments
We thank Sigrid Seelmeir and Simone Boos for excellent technical assistance, Mark Lötzerich for his kind practical help, and Lars Dölken for critical reading of the manuscript.
This work was supported by grants from the DFG (SFB455 and SPP1175) and the Friedrich-Baur-Stiftung (0020/2003).
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Adler, B., L. Scrivano, Z. Ruzsics, B. Rupp, C. Sinzger, and U. Koszinowski. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 87:2451-2460. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore, D. 1988. Gene therapy. Intracellular immunization. Nature 335:395-396. [DOI] [PubMed] [Google Scholar]
- 3.Biery, M. C., F. J. Stewart, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 5.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 6.Bubeck, A., M. Wagner, Z. Ruzsics, M. Lotzerich, M. Iglesias, I. R. Singh, and U. H. Koszinowski. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bubic, I., M. Wagner, A. Krmpoti, T. Saulig, S. Kim, W. M. Yokoyama, S. Jonji, and U. H. Koszinowski. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Cheshenko, N., W. Liu, L. M. Satlin, and B. C. Herold. 2005. Focal adhesion kinase plays a pivotal role in herpes simplex virus entry. J. Biol. Chem. 280:31116-31125. [DOI] [PubMed] [Google Scholar]
- 10.Crawford, S., and S. P. Goff. 1984. Mutations in gag proteins P12 and P15 of Moloney murine leukemia virus block early stages of infection. J. Virol. 49:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowder, S., and K. Kirkegaard. 2005. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat. Genet. 37:701-709. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 79:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman, A. D., S. J. Triezenberg, and S. L. McKnight. 1988. Expression of a truncated viral trans-activator selectively impedes lytic infection by its cognate virus. Nature 335:452-454. [DOI] [PubMed] [Google Scholar]
- 15.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 79:3713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herskowitz, I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329:219-222. [DOI] [PubMed] [Google Scholar]
- 17.Imhof, M. O., P. Chatellard, and N. Mermod. 2002. Comparative study and identification of potent eukaryotic transcriptional repressors in gene switch systems. J. Biotechnol. 97:275-285. [DOI] [PubMed] [Google Scholar]
- 18.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99-106. [DOI] [PubMed] [Google Scholar]
- 19.Lotzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mura, M., P. Murcia, M. Caporale, T. E. Spencer, K. Nagashima, A. Rein, and M. Palmarini. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc. Natl. Acad. Sci. USA 101:11117-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 24.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rupp, B., Z. Ruzsics, T. Sacher, and U. H. Koszinowski. 2005. Conditional cytomegalovirus replication in vitro and in vivo. J. Virol. 79:486-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scaglioni, P., M. Melegari, M. Takahashi, J. R. Chowdhury, and J. Wands. 1996. Use of dominant negative mutants of the hepadnaviral core protein as antiviral agents. Hepatology 24:1010-1017. [DOI] [PubMed] [Google Scholar]
- 29.Scaglioni, P. P., M. Melegari, and J. R. Wands. 1994. Characterization of hepatitis B virus core mutants that inhibit viral replication. Virology 205:112-120. [DOI] [PubMed] [Google Scholar]
- 30.Schnee, M., Z. Ruzsics, A. Bubeck, and U. H. Koszinowski. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 80:11658-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimano, R., R. Inubushi, Y. Oshima, and A. Adachi. 1999. Inhibition of HIV/SIV replication by dominant negative Gag mutants. Virus Genes 18:197-201. [DOI] [PubMed] [Google Scholar]
- 32.Stow, N. D., O. Hammarsten, M. I. Arbuckle, and P. Elias. 1993. Inhibition of herpes simplex virus type 1 DNA replication by mutant forms of the origin-binding protein. Virology 196:413-418. [DOI] [PubMed] [Google Scholar]
- 33.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239-242. [DOI] [PubMed] [Google Scholar]
- 34.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 36.Walther, P., and A. Ziegler. 2002. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 208:3-10. [DOI] [PubMed] [Google Scholar]
- 37.Wolkowicz, R., and G. P. Nolan. 2005. Gene therapy progress and prospects: novel gene therapy approaches for AIDS. Gene Ther. 12:467-476. [DOI] [PubMed] [Google Scholar]
- 38.Yao, F., and E. Eriksson. 1999. A novel tetracycline-inducible viral replication switch. Hum. Gene Ther. 10:419-427. [DOI] [PubMed] [Google Scholar]
- 39.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]