Abstract
The coxsackievirus and adenovirus receptor (CAR) is both a viral receptor and homophilic adhesion protein. The extracellular portion of CAR consists of two immunoglobulin (Ig)-like domains, each with a consensus sequence for N-glycosylation. We used chemical, genetic, and biochemical studies to show that both sites are glycosylated and contribute to the function of CAR. Although the glycosylation of CAR does not alter cell surface levels or junctional localization, it affects both adhesion and adenovirus infection in unique ways. CAR-mediated adhesion appears to require at least one site of glycosylation since cells expressing CAR without glycosylation do not cluster with each other. In contrast, glycosylation of the Ig-like domain proximal to the membrane is key to the cooperative behavior of adenovirus binding and infection. Contrary to the hypothesis that cooperativity improves viral infection, our data show that although glycosylation of the D2 domain is required for adenovirus cooperative binding, it has a negative consequence upon infection. This is the first report dissecting the adhesion and receptor activities of CAR, revealing that factors other than the binding interface play a significant role in the function of CAR. These data have important implications for both cancers with altered glycosylation states and cancer treatments using oncolytic adenovirus.
Protein glycosylation plays important roles in protein folding, conformation, localization, stability, and cellular interactions (17, 19). Additionally, many physiological activities are affected by glycosylation, such as cell adhesion, migration, ligand recognition, and binding to receptors (9, 20, 24). Thus, it is not surprising that protein glycosylation levels are implicated in many disease states. In particular, the effects of N- and O-linked glycosylation on viral-host cell interactions have been studied, focusing predominantly on the glycosylated viral coat proteins of enveloped viruses or viruses utilizing sialic acid as a receptor (7, 12, 22, 23, 29). Adenovirus (Ad) is a nonenveloped virus that has been reported to contain glycosylated proteins (15, 30, 31) including a mono-GlcNac addition to the fiber knob protein (6). The effect of this glycosylation on Ad infection is unclear. In contrast, glycosylation has never been described for any coxsackievirus proteins.
The coxsackievirus and Ad receptor (CAR) is the primary receptor for most Ads and coxsackie B viruses (2). CAR is also involved in many biological processes such as homophilic adhesion, neutrophil transmigration, protein trafficking, and modulation of cellular growth and motility (4, 5, 8, 10, 14, 27, 28, 32). CAR is often called a glycoprotein and contains two putative glycosylation sites, one on each of its two extracellular immunoglobulin (Ig)-like domains (N106 in D1 and N201 in D2) (1, 2). Detection of CAR by Western blotting reveals two bands, one at approximately 40 kDa (the predicted molecular mass) and one at 46 kDa. Honda et al. demonstrated that an overnight incubation in the presence of peptide N-glycosidase F (PNGase F) reduces the 46-kDa band to 40 kDa, suggesting that this shift in molecular size is due to deglycosylation (14). Additionally, the solved cryoelectron microscopy structure of full-length CAR interacting with coxsackievirus shows an irregular mass on the D1 loop, consistent with glycosylation (13). Together, these data suggest that CAR may be a glycoprotein. However, whether only one or both predicted sites are glycosylated and whether there are additional cryptic sites remain unclear. Moreover, the relevance of glycosylation to the biology of CAR, either as a viral receptor or in its endogenous function as a homophilic adhesion protein, remains to be tested.
MATERIALS AND METHODS
Antibodies, cells, and viruses.
FLAG M2 antibody (Ab) was purchased from Sigma (F3165; St. Louis, MO); Alexa-488- and Alexa-568-conjugated goat anti-mouse or anti-rabbit Abs were from Molecular Probes (Eugene, OR). The CAR Ab RmcB (CRL-2379; ATCC, Manassas, VA) was produced by the University of Iowa Hybridoma Core. Rabbit anti-CAR-1605 was produced in rabbits immunized with a glutathione S-transferase fusion to the intracellular C terminus (amino acids 261 to 365). COS-7 cells (ATCC) were maintained under standard culture conditions (Dulbecco's modified Eagle's medium [DMEM] with 10% fetal calf serum [FCS], penicillin, and streptomycin). CHO-K1 cells (BD Biosciences, Franklin Lakes, NJ) were maintained under standard culture conditions (DMEM and FCS supplemented with tetracycline l-glutamine, penicillin, and streptomycin). Ad serotype 5 containing the β-galactosidase (Ad-β-Gal), green fluorescent protein (peGFP-N1; Clontech, Mountain View, CA), or CAR gene has previously been described (10, 11). All recombinant Ads were generated by the University of Iowa Gene Transfer Vector Core.
Cell transfection.
COS-7 cells were electroporated as previously described (10). Briefly, 10 million cells were mixed with 20 μg of plasmid DNA, 400 μl of cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4, 10 mM KH2PO4, 25 mM HEPES, 2 mM EGTA, 5 mM MgCl2, 2 mM ATP, and glutathione) and put in an electroporation cuvette (Bio-Rad Laboratories, Hercules, CA) for 30 min on ice. After electroporation, cells were seeded onto 10-cm dishes or collagen-coated glass chamber slides and studied 2 days later. CAR-deficient CHO-K1 cells were seeded into six-well or 24-well dishes (n = 6 replicates) and transfected using Lipofectamine 2000 according to the manufacturer's standard protocol (Invitrogen). We routinely achieve approximately 70 to 90% transfection efficiency with these protocols (10).
Tunicamycin, PNGase F, biotinylation, immunoprecipitation, and Western blotting.
Twenty-four hours after electroporation, COS cells were either treated with 0.5 μg/ml tunicamycin for 24 h at 37°C (catalog no. T7765; Sigma) or placed on ice, washed once with ice-cold phosphate-buffered saline ([PBS] catalog no. 14287; Invitrogen, Carlsbad, CA), biotinylated with 3 ml of 1 mg/ml EZ-Link sulfo-NHS-SS-biotin ([sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate] catalog no. 21331; Pierce, Rockford, IL) for 45 min, quenched with 100 mM glycine, and washed three times with PBS prior to lysis; alternatively, PNGase F enzyme solution (P7367; Sigma) was added directly to the lysates and incubated for 1 h at 37°C. For lysis, cells were placed on ice, washed once with ice-cold PBS, lysed (50 mM Tris-HCl, pH 7.5, 137 mM NaCl, 1% Triton X-100, 5 mM EDTA, and 1 mM EGTA with protease inhibitors [10 μg/ml] leupeptin, aprotinin, and pepstatin and 1 mM phenylmethylsulfonyl fluoride), sonicated (5 s), and spun in a microcentrifuge (16,000 × g) for 10 min. For immunoprecipitation, supernatant, normalized to equal protein, was incubated with CAR Ab RmcB or FLAG Ab and immunoprecipitated with protein G-Sepharose (catalog no. 17061801; GE Healthcare Life Sciences, Pittsburgh, PA) or incubated with Ultralink Immobilized NeutrAvidin Biotin Binding Protein (catalog no. 53150; Pierce, Rockford, IL) or CAR Ab 1605 cross-linked to protein G Dynabeads (Invitrogen) using the standard manufacturer's protocol (23). Beads were suspended in loading buffer (4% sodium dodecyl sulfate, 100 mM dithiothreitol, 20% glycerol, 65 mM Tris, pH 6.8, 0.005% bromophenol blue), and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA), blocked with 5% bovine serum albumin, washed, and probed with primary FLAG Ab, followed by washing and incubation with anti-mouse or rabbit conjugated to horseradish peroxidase (Pierce). Bands were detected with enhanced chemiluminescence reagents (catalog no. 34080; Pierce).
Site-directed mutagenesis.
Site-directed mutagenesis of CAR was performed according to the manufacturer's standard protocol (Stratagene, Cedar Creek, TX) with the following primers: N115Q, 5′-AATCTGGTGATGCATCAATACAGGTAACGAATTTACAACTGTCAG; N210Q, 5′-CATCTGTTATATCTGTAAAACAGGCCTCTTCTGAGTACTCTGGG.
Immunofluorescence staining.
Electroporated COS cells grown on collagen-coated chamber slides were washed, fixed with methanol plus 1% paraformaldehyde at −20°C, and blocked with 2% bovine serum albumin in SuperBlock (Pierce). Cells were incubated with primary Ab, washed extensively, and then incubated with goat anti-mouse Alexa-568 secondary Ab. Nuclei were stained with TO-PRO3 (Molecular Probes), and samples were cover-slipped with Vectashield mounting medium (Vector Laboratories, Inc., Burlingame, CA). Images were acquired with a Bio-Rad MRC-1024 laser scanning confocal microscope (Hercules, CA) mounted on a Nikon E600 microscope (Melville, NY) using a 60× oil immersion lens.
Adhesion.
Transfected cells were washed once with Ca2+-free Hanks balanced salt solution ([HBSS] catalog no. 14170-112; Invitrogen) and incubated with 1 mM EDTA in HBSS until they were released from the plate. Cells were pelleted and washed twice with HBSS, resuspended at 2 × 106 cells/ml in HBSS supplemented with HBSS-dialyzed FCS (2%), and placed on a rotor for 1 h at 37°C. Equal volumes of cells were then gently plated out and supplemented with a small volume of growth medium. Clumps consisting of more than 6 cells were counted after 1 h at 37°C in at least 10 separate fields of view at a magnification of ×10 on a light microscope (Nikon Eclipse TS100).
Labeling of Ad with [methyl-3H]thymidine.
Ad5GFP (where Ad5 is Ad serotype 5) was labeled with [methyl-3H]thymidine as previously described (18). Briefly, 150-mm2 dishes were seeded with 2.5 × 107 HEK 293 cells in 15 ml of DMEM-10% FCS. Twenty-four hours later, these cells were infected with recombinant Ad at a multiplicity of infection (MOI) of 50 or higher. Ten hours postinfection, 1 mCi of [methyl-3H]thymidine (TRK758, 3.15 TBq/mmol, 85.0 Ci/mmol; GE Healthcare) was added to the medium, and cells were further incubated at 37°C until ∼30 h postinfection. Cells were then harvested, pelleted, washed once with PBS, and resuspended in 2 ml of PBS. Virus was released from the cells by three freeze-thaw cycles. Cell debris was removed by centrifugation, and viral material was subjected to ultracentrifugation in two sequential CsCl gradients and subsequent dialysis against PBS-3% sucrose. Virion-specific radioactivity, measured by liquid scintillation (Packard Tri-Carb 1500 liquid scintillation analyzer), ranged from 6 × 10−6 to 4 × 10−5 cpm per virion.
Ad infection.
Transfected CHO-K1 cells were infected with Ad-β-Gal (MOI as indicated in figures) for 1 h at 37°C and lysed after 24 h. β-Gal expression per milligram of protein was determined, as previously described (10). Binding of Ad was determined by 3H-labeled Ad as previously described (11). Briefly, cells were transfected as above, washed twice with either ice-cold or room temperature (RT) PBS, incubated with 3H-labeled Ad at various MOIs for 1 h on ice or at RT. A sample of inoculum was put directly into scintillation vials to determine unbound virus. Cells were washed two times with ice-cold or RT PBS and lysed with 1% Triton X-100. Lysate (bound virus) was put into scintillation vials and measured in a liquid scintillation counter. Data analysis and nonlinear regression were performed using Origin, version 7.5, software (OriginLab Corp., Northampton, MA).
RESULTS
CAR is an N-glycosylated protein.
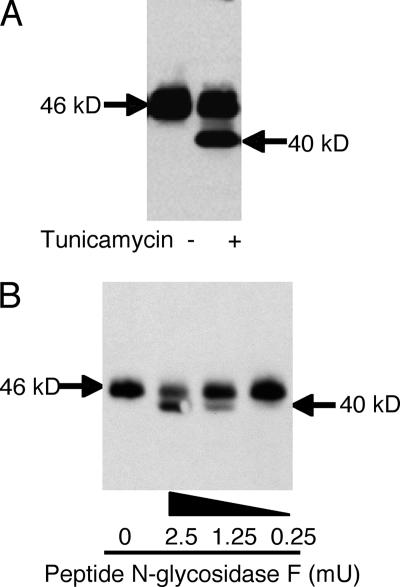
In order to investigate whether CAR is exclusively N-glycosylated, 24 h after transfection with CAR, COS cells were incubated with tunicamycin, an antibiotic that blocks the reaction of UDP-GlcNAc and dolichol phosphate in the first step of glycoprotein synthesis, thus inhibiting the synthesis of all N-linked glycoproteins. In the absence of tunicamycin, CAR-expressing COS cells show a 46-kDa band (Fig. 1A). Tunicamycin treatment 24 h after transfection results in an additional 40-kDa band, consistent with the complete inhibition of glycosylation of newly synthesized CAR. The presence of the 46-kDa band even after 24 h of tunicamycin treatment suggests that CAR has a turnover of more than 24 h. These data suggest that either CAR is glycosylated or a glycosylated protein modifies CAR. To directly evaluate whether CAR itself is glycosylated, lysates from CAR-expressing COS cells were incubated with increasing doses of PNGase F for 2 h (Fig. 1B). When no PNGase F was added, a single band of 46 kDa was observed. The addition of 2.5 mU of PNGase F resulted in a more robust 40-kDa band relative to the 46-kDa band, consistent with the idea that CAR is a glycoprotein. However, these data do not define whether one or both possible N-glycosylation sites are glycosylated.
FIG. 1.
CAR is a glycosylated protein. Panel A shows the presence of an additional lower-molecular-mass band after treatment of CAR-transfected CHO-K1 cells with the glycosylation inhibitor tunicamycin (10 μg/ml for 24 h). Panel B shows a reduction in molecular mass from 46 kDa with mock treatment to the lower, predicted 40-kDa band upon treatment of CAR isolated from transfected COS cells with PNGase F at increasing doses.
CAR has independent glycosylation sites in each extracellular Ig-like domain.
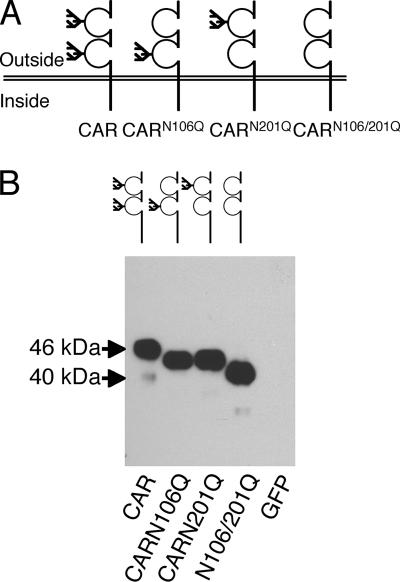
CAR is predicted to be N-glycosylated at amino acids N106 and N201; thus, we created single and double glycosylation-deficient CAR constructs by site-directed mutagenesis (CARN106Q, CARN201Q, and CARN106Q/N201Q). Both of these residues are in the extracellular domain of CAR, with residue 106 in the first D1 domain and 201 in the second or D2 domain (Fig. 2A). To confirm the size and expression of these constructs, we transfected them into COS cells and analyzed cell lysates by Western blotting (Fig. 2B). The resulting bands reflected the loss of glycosylation in a stepwise manner such that wild-type (wt) CAR was larger than either CARN106Q or CARN201Q, which were of equivalent size, but each was larger than CARN106Q/N201Q. This confirms that CAR is glycosylated at both of these two sites to a similar extent and that the 46-kDa form of CAR is the mature glycosylated form of the protein.
FIG. 2.
CAR is N-glycosylated at two separate sites, with one in each of the extracellular domains. Panel A shows a schematic representation of the glycosylation mutants created by site-directed mutagenesis. Panel B shows the difference in molecular mass by Western blotting of COS-7 cells expressing wt CAR, CARN106Q, CARN201Q, CARN106Q/N201Q, or GFP probed with a CAR-specific Ab.
Loss of glycosylation does not alter the cell surface or junctional localization of CAR.
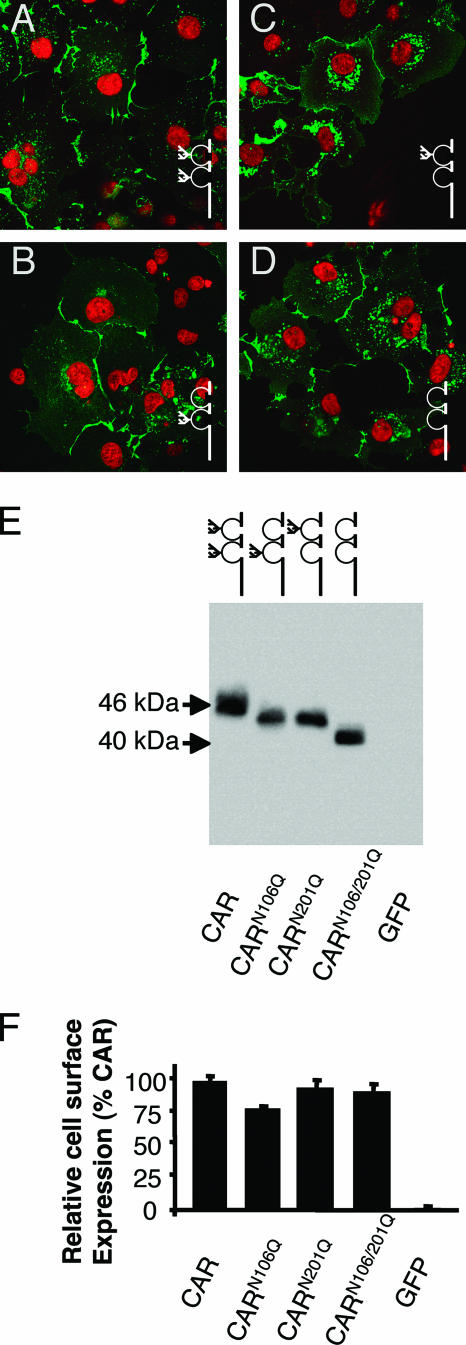
CAR normally localizes to the junctions between cells, where it behaves as a homophilic adhesion molecule (Fig. 3A). To ensure that the glycosylation mutants localized to the cell surface and in particular to cell-cell junctions, COS cells and CAR-deficient CHO-K1 cells (data not shown) were transfected and analyzed by immunocytochemistry. Despite some intracellular differences in localization, all three of the glycosylation mutants localized primarily to the cell-cell junctions in a similar manner to wt CAR (Fig. 3B to D; CARN106Q, CARN201Q, and CARN106Q/N201Q, respectively, are shown in green; nuclei are counterstained in red). This is in contrast to mutations altering the asparagine to alanine, which did not reach the cell surface (data not shown), suggesting that the trafficking and cell surface junctional localization were not altered by the absence of glycosylation provided that the asparagine was replaced by a similar amino acid. To confirm the cell surface expression of these constructs, each was expressed in CHO-K1 cells and subjected to cell surface biotinylation (n = 4 experiments). All biotinylated proteins were isolated using streptavidin-labeled beads and analyzed by Western blotting using a CAR-specific Ab (Fig. 3E). Although there appeared to be a small decrease in the amount of CARN106Q, there were no statistically significant differences in the amount of protein present at the cell surface in comparison to wt CAR (Fig. 3F). In a manner similar to the unbiotinylated lysates shown in Fig. 2B, the molecular mass of the cell surface protein reflected a stepwise size difference in accordance with the amount of glycosylation. In contrast, immunoblotting for GFP showed that this cytoplasmic protein was not present after precipitation using streptavidin (data not shown). These data suggest that loss of glycosylation does not significantly affect cell surface expression and cell-cell junctional localization. Interestingly, Western blotting for endogenous CAR in several cell lines that display junctional CAR localization and support Ad infection (including HeLa, 293T, COS, and A549) reveals CAR-specific bands that run with different electrophoretic mobilities, which is consistent with differential glycosylation (data not shown).
FIG. 3.
CAR glycosylation mutants are expressed on the cell surface and localize to cell-cell junctions. Cells expressing CAR or each of the glycosylation mutants were analyzed for indirect immunocytochemistry (A to D) using a CAR-specific Ab (green) and a nuclear stain (red) in COS-7 cells: CAR (A), CARN106Q (B), CARN201Q (C), and CARN106Q/N201Q (D). (E) Cell surface expression by biotinylation and streptavidin precipitation followed by Western blotting with a CAR-specific Ab in CHO-K1 cells expressing CAR or each of the glycosylation mutants. (F) Quantification of cell surface expression as a percentage of CAR expression by cell surface biotinylation in CHO-K1 cells (n = 4 experiments). There was no significant difference found between CAR and CARN201Q or CARN106Q/N201Q. CARN106Q showed approximately 20% less cell surface protein compared to wt CAR.
Glycosylation of CAR affects cell adhesion.
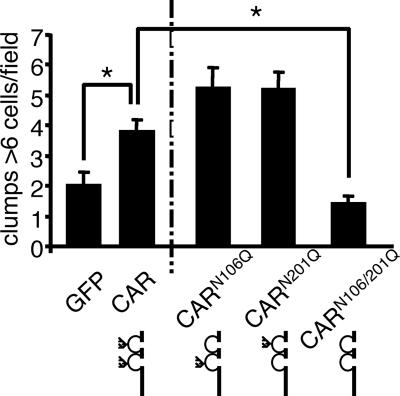
To determine if glycosylation plays a role in CAR homophilic adhesion, CAR-negative CHO-K1 cells were transfected with CAR, CARN106Q, CARN201Q, CARN106Q/N201Q, or GFP. Approximately 48 h after transfection, monolayers of cells were harvested, resuspended as a single cell suspension, and incubated for 1 h to allow cell-cell clumping (Fig. 4; data are from 10 to 15 fields of view in three replicate experiments). GFP-transfected cells contained an average of approximately 2 clusters of 6 cells per clump per field of 10 counted fields, whereas CAR normally contained 3 to 4 large clumps containing 6 or more cells per field. The complete loss of glycosylation in the CARN106Q/N201Q mutant abrogated clumping, suggesting that glycosylation is important for CAR-mediated cell clumping. CARN106Q and CARN201Q behaved similarly and maintained clumping, indicating that glycosylation of either the D1 or D2 domain is sufficient to maintain CAR-mediated cell adhesion. To confirm CAR specificity, clumping was performed in the presence of 0.5 mg/ml of anti-CAR RmcB monoclonal Ab. As predicted, due to Ab bivalency, increased clumping was apparent in CAR-expressing cells but not control cells (data not shown).
FIG. 4.
CAR glycosylation affects CAR-mediated cell clumping. CHO-K1 cells expressing CAR, CARN106Q, CARN201Q, CARN106Q/N201Q, or GFP were made into a single cell suspension and allowed to clump for 1 h at 37C. The average number of clumps with 6 or more cells was counted over 10 fields of view. The elimination of 1 glycosylation site significantly increased cell clustering while deletion of both sites significantly decreased adhesion. *, = P < 0.01 (n = 3 experiments).
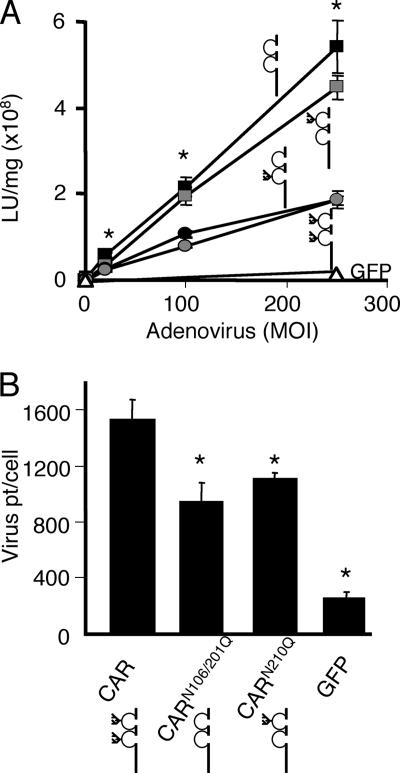
Glycosylation of the D2 domain on CAR decreases Ad infection.
We hypothesized that since Ad binds to the D1 domain of CAR, lack of glycosylation of this domain may affect Ad interactions with CAR. Alternatively, since crystal structures and footprint mapping of the Ad-CAR interaction suggest that the glycosylation site is on the opposite side of the structural Ad binding site, it may have no effect on the interaction (27). We further hypothesized that the lack of glycosylation at D2 would have no effect on Ad infection if the role of the D2 domain is strictly to position the D1 domain at an appropriate distance from the membrane for efficient Ad interaction (26). CAR-negative CHO-K1 cells were transfected with GFP, CAR, CARN106Q, CARN201Q, or CARN106Q/N201Q. The cells were infected 48 h later with Ad-β-Gal at various MOIs (n = 6 replicates) and lysed 24 h postinfection to determine the β-Gal expression per milligram of protein (Fig. 5A). Transfection of these cells with GFP resulted in very little Ad-β-Gal infection even at high MOIs. As expected, transient expression of CAR significantly increased the ability of Ad to infect these cells. The CARN106Q mutant behaved identically to wt CAR, suggesting that the D1 glycosylation neither inhibits nor augments Ad binding or infection. Surprisingly, mutation of the D2 domain glycosylation site either on its own (CARN201Q) or in combination with the D1 mutation (CARN106Q/N201Q) significantly increased Ad-mediated gene transfer. We hypothesized that this increase in infection was simply due to an increase in Ad binding. To test this, CHO-K1 cells transfected with CAR, CARN210Q, CARN106Q/N201Q, or GFP were incubated for 1 h at 4°C in the presence of 3H-labeled Ad at an MOI of 100 (i.e., 10,000 particles per cell). Figure 5B shows that the binding of Ad to CARN210Q or CARN115Q/N201Q was reduced significantly in comparison to wt CAR at an MOI where there was roughly twice as much infection in cells expressing CAR D2 mutants (Fig. 5A). These experiments suggest that, contrary to our hypotheses, D1 glycosylation does not alter Ad binding or infection, while the lack of glycosylation of the D2 domain paradoxically decreases Ad binding and increases infection.
FIG. 5.
The lack of glycosylation at D2 augments Ad infection but decreases Ad binding. Panel A shows the infection of CHO-K1 cells expressing CAR (gray circle), CARN106Q (black circle), CARN201Q (gray square), CARN106Q/N201Q (black square), or GFP (▵), with an Ad carrying the β-Gal gene. Whereas cells expressing GFP are not well infected by Ad, cells expressing CARN106Q infect to a similar extent as CAR, and cells expressing CARN201Q or CARN106Q/N201Q mediate Ad infection significantly better than CAR. *, P < 0.01 (n = 3). Panel B shows that the binding of 3H-labeled Ad to CHO-K1 cells expressing CAR is significantly greater than CARN201Q, CARN106Q/N201Q, or GFP. *, P < 0.01 (n = 3 experiments).
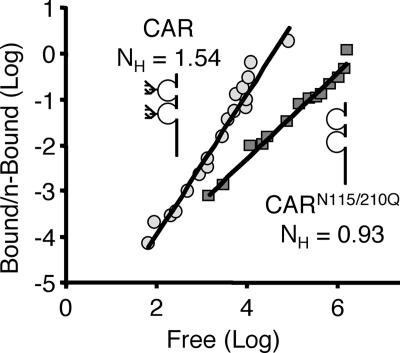
Glycosylation at the D2 domain increases the cooperativity of Ad-CAR binding.
Ad binding reflects both the multivalency of virus attachment points (12 trimeric fiber knobs that can bind up to 36 CAR molecules) and the ability of receptors to migrate within the membrane (21). Thus, we hypothesized that the lack of D2 glycosylation results in increased cooperativity such that more receptors bind a single virion (i.e., fewer virions bound), resulting in improved infection. CHO-K1 cells were transfected with GFP, CAR, or CARN106Q/N201Q. Cells were then incubated with 3H-Ad-GFP over a large range of MOIs at RT, which is a temperature that supports cooperative binding (21). In order to determine unbound and bound virus after 1 h, both the medium and cell lysates were each counted in a scintillation counter. Figure 6 shows one representative experiment plotted as the log of free particles versus the log of bound virus/(number of receptor sites − number of bound particles) (Hill plot). The Hill coefficient was determined by nonlinear regression. As expected, wt CAR showed a Hill coefficient greater than 1 (1.20 to 1.54), indicating positive cooperativity. In contrast, the Hill coefficient for CARN115Q/N201Q was roughly 1 (0.93 to 1.06), indicating no cooperativity. These data suggest that glycosylation of the D2 domain is important for CAR cooperativity and, moreover, that the lack of cooperativity augments Ad infection.
FIG. 6.
Glycosylation is required for the cooperative binding of Ad. CHO-K1 cells expressing either CAR (gray circle) or CARN106Q/N201Q (gray square) were incubated with 3H-labeled Ad at MOIs ranging from 100 to 106 particles per cell. The amount of 3H remaining in the infection medium as well as cell-associated 3H was determined after an incubation of 1 h at RT. Data were analyzed by nonlinear regression and graphed as a Hill plot. CAR showed positive cooperativity with a Hill coefficient greater than 1 (n = 3 experiments; Hill coefficients of 1.54, 1.20, and 1.36) while CARN106Q/N201Q showed no binding cooperativity (n = 2 experiments; Hill coefficients of 0.93 and 1.06). One representative experiment is shown.
DISCUSSION
CAR is a glycoprotein with two N-glycosylation sites, one in each of the two Ig-like extracellular domains. Although glycosylation at each of these sites may not be identical, both are glycosylated to a similar degree. The amount of glycosylation on CAR does not affect either cell surface or junctional localization in nonpolarized cells. Surprisingly, glycosylation affects both the homophilic adhesion as well as viral receptor functions of CAR differentially, thus separating the evolution of these two functions and their dependence on extracellular domain glycosylation.
Both the CAR-CAR and CAR-Ad adhesion interfaces have been elucidated through crystal structure and genetic mutation studies (3, 16, 25-27). Although not identical in residue usage, the footprints of both interactions overlap each other and are roughly on the opposite side to the predicted D1 N-glycosylation site. This suggests that the presence of glycosylation on the D1 domain may not directly affect these interactions. Our data indicate that this is true for the Ad interaction but not for adhesion.
In cell monolayers, CAR localizes to the adhesion junctions regardless of glycosylation; however, this is in the presence of many other adhesion proteins. We tested the ability of the glycosylation mutants to mediate adhesion under dynamic conditions which limit the effects of major adhesion proteins such as cadherins (i.e., calcium free). Although simplistic, these conditions may expose the effect of altered CAR glycosylation during initiation of the cellular adhesion complex or, alternatively, during the alteration, modulation, or deregulation of that complex (i.e., metastasis, migration, blood flow, or transmigration). In the absence of glycosylation, cell-cell clumping is attenuated. There are at least two possible interpretations: the lack of glycosylation decreases CAR-CAR affinity, resulting in decreased adhesion, or the lack of glycosylation increases CAR-CAR affinity, resulting in cis interactions and thus preventing trans interactions. Conclusive determination of affinity for these interactions requires further investigations using plasmon resonance or sedimentation.
In contrast to the CAR-mediated cell-cell interaction, glycosylation of the D2 domain is key in the Ad-CAR interaction. We have previously shown that the D2 domain of CAR is important for Ad infection and that deletion of this domain reduces Ad binding and infection significantly (11). Additionally, we hypothesized that this domain played a role only in spacing the D1 domain appropriately from the membrane to facilitate an efficient interaction with the Ad fiber knob. In these experiments we could not rule out cis interactions or steric effects of the D2 domain on the Ad-CAR interaction. Here, we show that in spite of decreased binding and a loss of cooperativity, the lack of glycosylation at N201 results in increased infection. Thus, the D2 domain is not simply a structural “spacer”; it plays a key role in cooperativity, and this role is mediated by glycosylation. Cooperative binding, as shown by Persson et al., is thought to require the trimeric nature of the 12 fiber knobs on the Ad particle that can theoretically bind and cluster up to 36 CAR molecules, causing perturbations in the membrane and allowing more (or less) efficient Ad internalization (21). Our data, however, indicate that whereas cooperativity increases binding, it has the opposite effect on subsequent infection steps. Several potential mechanisms exist including the following: the cooperative retention of virus at the cell surface or endosomal compartment; stronger wt CAR-fiber knob interactions resulting in reduced receptor recycling; or loss of glycosylation that may alter coreceptor interactions, improving the efficiency of endocytosis or endosomal escape. Interestingly, although D2 glycosylation reduces Ad infection, the coxsackievirus interaction with CAR may actually require glycosylation since cell-expressed CAR successfully binds coxsackievirus while bacterially expressed (i.e., CAR lacking glycosylation) does not (13).
In summary, CAR is a glycosylated protein, and the sites of glycosylation differentially affect the CAR-CAR and CAR-Ad interactions, revealing an inherent difference between homophilic adhesion and Ad binding. These data have important implications both for cancers with altered glycosylation states and for cancer treatments using oncolytic Ad. It will be important to determine if Ad infection depends on not only the presence of CAR but also the specific degree of glycosylation in vivo.
Acknowledgments
We thank Lisa Jorgensen for assistance with manuscript preparation, Michael Welsh for discussions, the Gene Transfer Vector Core (supported by the Roy J. Carver Charitable Trust, the NHLBI, CFF, and NIDDK), the In Vitro Cell Models Core (supported by the National Heart, Lung and Blood Institute and NIDDK [DK54759]), and the Hybridoma/Tissue Culture Facility.
K.E. is supported by a fellowship from the Parker B. Francis Foundation. This work was supported by a PPG grant from the NIH (HL51670-11).
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Bairoch, A., and R. Apweiler. 1997. The SWISS-PROT protein sequence database: its relevance to human molecular medical research. J. Mol. Med. 75:312-316. [PubMed] [Google Scholar]
- 2.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 3.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 4.Bruning, A., and I. B. Runnebaum. 2004. The coxsackie adenovirus receptor inhibits cancer cell migration. Exp. Cell Res. 298:624-631. [DOI] [PubMed] [Google Scholar]
- 5.Carson, S. D., and N. M. Chapman. 2001. Coxsackievirus and adenovirus receptor (CAR) binds immunoglobulins. Biochemistry 40:14324-14329. [DOI] [PubMed] [Google Scholar]
- 6.Cauet, G., J. M. Strub, E. Leize, E. Wagner, A. Van Dorsselaer, and M. Lusky. 2005. Identification of the glycosylation site of the adenovirus type 5 fiber protein. Biochemistry 44:5453-5460. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. C., M. Baylor, and D. M. Bass. 1993. Murine intestinal mucins inhibit rotavirus infection. Gastroenterology 105:84-92. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, C. J., J. T. Shieh, R. J. Pickles, T. Okegawa, J. T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dube, D. H., and C. R. Bertozzi. 2005. Glycans in cancer and inflammation-potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 4:477-488. [DOI] [PubMed] [Google Scholar]
- 10.Excoffon, K. J., A. Hruska-Hageman, M. Klotz, G. L. Traver, and J. Zabner. 2004. A role for the PDZ-binding domain of the coxsackie B virus and adenovirus receptor (CAR) in cell adhesion and growth. J. Cell Sci. 117:4401-4409. [DOI] [PubMed] [Google Scholar]
- 11.Excoffon, K. J., G. L. Traver, and J. Zabner. 2005. The role of the extracellular domain in the biology of the coxsackievirus and adenovirus receptor. Am. J. Respir. Cell Mol. Biol. 32:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna, S. L., T. C. Pierson, M. D. Sanchez, A. A. Ahmed, M. M. Murtadha, and R. W. Doms. 2005. N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J. Virol. 79:13262-13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full-length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda, T., H. Saitoh, M. Masuko, T. Katagiri-Abe, K. Tominaga, I. Kozakai, K. Kobayashi, T. Kumanishi, Y. G. Watanabe, S. Odani, and R. Kuwano. 2000. The coxsackievirus-adenovirus receptor protein as a cell adhesion molecule in the developing mouse brain. Brain Res. Mol. Brain Res. 77:19-28. [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi, M., and J. V. Maizel, Jr. 1974. The polypeptides of adenovirus. VI. Early and late glycopolypeptides. Virology 58:345-361. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, S., A. Jacobs, T. M. Laue, and M. Caffrey. 2004. Solution structure of the coxsackievirus and adenovirus receptor domain 1. Biochemistry 43:1847-1853. [DOI] [PubMed] [Google Scholar]
- 17.Jones, J., S. S. Krag, and M. J. Betenbaugh. 2005. Controlling N-linked glycan site occupancy. Biochim. Biophys. Acta 1726:121-137. [DOI] [PubMed] [Google Scholar]
- 18.Law, L. K., and B. L. Davidson. 2002. Adenovirus serotype 30 fiber does not mediate transduction via the coxsackie-adenovirus receptor. J. Virol. 76:656-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra, N., S. Sinha, T. N. Ramya, and A. Surolia. 2006. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 31:156-163. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsubo, K., and J. D. Marth. 2006. Glycosylation in cellular mechanisms of health and disease. Cell 126:855-867. [DOI] [PubMed] [Google Scholar]
- 21.Persson, R., C. Wohlfart, U. Svensson, and E. Everitt. 1985. Virus-receptor interaction in the adenovirus system: characterization of the positive cooperative binding of virions on HeLa cells. J. Virol. 54:92-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers, G. N., G. Herrler, J. C. Paulson, and H. D. Klenk. 1986. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 261:5947-5951. [PubMed] [Google Scholar]
- 24.Samulski, R. J., L. S. Chang, and T. Shenk. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 63:3822-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seiradake, E., H. Lortat-Jacob, O. Billet, E. J. Kremer, and S. Cusack. 2006. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fibre heads in complex with the D1 domain of CAR. J. Biol. Chem. 281:33704-33716. [DOI] [PubMed] [Google Scholar]
- 26.Tomko, R. P., C. B. Johansson, M. Totrov, R. Abagyan, J. Frisen, and L. Philipson. 2000. Expression of the adenovirus receptor and its interaction with the fiber knob. Exp. Cell Res. 255:47-55. [DOI] [PubMed] [Google Scholar]
- 27.van Raaij, M. J., E. Chouin, H. van der Zandt, J. M. Bergelson, and S. Cusack. 2000. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 Å resolution. Struct. Fold. Des. 8:1147-1155. [DOI] [PubMed] [Google Scholar]
- 28.Walters, R., P. Freimuth, T. Moninger, I. Ganske, J. Zabner, and M. Welsh. 2002. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 110:789-799. [DOI] [PubMed] [Google Scholar]
- 29.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 30.Windheim, M., and H. G. Burgert. 2002. Characterization of E3/49K, a novel, highly glycosylated E3 protein of the epidemic keratoconjunctivitis-causing adenovirus type 19a. J. Virol. 76:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wold, W. S., C. Cladaras, S. L. Deutscher, and Q. S. Kapoor. 1985. The 19-kDa glycoprotein coded by region E3 of adenovirus. Purification, characterization, and structural analysis. J. Biol. Chem. 260:2424-2431. [PubMed] [Google Scholar]
- 32.Zen, K., Y. Liu, I. C. McCall, T. Wu, W. Lee, B. A. Babbin, A. Nusrat, and C. A. Parkos. 2005. Neutrophil migration across tight junctions is mediated by adhesive interactions between epithelial coxsackie and adenovirus receptor and a junctional adhesion molecule-like protein on neutrophils. Mol. Biol. Cell 16:2694-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]