Abstract
Respiratory syncytial virus (RSV) is a common cause of severe lower respiratory tract infection in children. Severe RSV disease is related to an inappropriate immune response to RSV resulting in enhanced lung pathology which is influenced by host genetic factors. To gain insight into the early pathways of the pathogenesis of and immune response to RSV infection, we determined the transcription profiles of lungs and lymph nodes on days 1 and 3 after infection of mice. Primary RSV infection resulted in a rapid but transient innate, proinflammatory response, as exemplified by the induction of a large number of type I interferon-regulated genes and chemokine genes, genes involved in inflammation, and genes involved in antigen processing. Interestingly, this response is much stronger on day 1 than on day 3 after infection, indicating that the strong transcriptional response in the lung precedes the peak of viral replication. Surprisingly, the set of down-regulated genes was small and none of these genes displayed strong down-regulation. Responses in the lung-draining lymph nodes were much less prominent than lung responses and are suggestive of NK cell activation. Our data indicate that at time points prior to the peak of viral replication and influx of inflammatory cells, the local lung response, measured at the transcriptional level, has already dampened down. The processes and pathways induced shortly after RSV infection can now be used for the selection of candidate genes for human genetic studies of children with severe RSV infection.
The severity of respiratory syncytial virus (RSV) infection in young children varies from a nonclinical or mild upper respiratory tract infection to severe lower respiratory tract infection that may lead to hospitalization and occasionally to death. Some children are more prone to a severe course of disease, such as premature-born children, children younger than 3 months of age, children with chronic lung disease or congenital heart disease, and immunocompromised children (27, 35). However, the biological mechanisms underlying the highly variable disease course in children are still poorly understood. The current belief is that children with severe RSV disease suffer from enhanced inflammatory lesions rather than from virus-induced cytopathology (25). In line with this, naturally occurring polymorphisms in genes affecting the inflammatory immune response influence the severity of RSV-induced disease (5, 11, 12, 15).
Immune responses to viral pathogens are initiated among others via the recognition of pathogen-associated molecular patterns by various Toll-like receptors (TLR), leading to the induction of innate immune responses, proinflammatory cytokines, and the Th1 pathway (reviewed in references 18 and 26). Innate immunity to RNA viruses is initiated by TLR3 and murine TLR7 or human TLR8, which are important for the responses to double-stranded and single-stranded RNAs, and through intracellular RNA recognition molecules, such as RIG-I and Mda5 (reviewed in reference 21). Both TLR3 and RIG-I have also been shown to be involved in the response to RSV, which is a single-stranded RNA virus (24, 33). In addition, a role for TLR4 in the initiation of an RSV-specific immune response has been postulated (23, 39). RSV, on the other hand, can interfere with the induction of the host response by modulating TLR3 and TLR7 signaling and virus-induced IFN responses (31, 32, 34, 36, 37).
Murine models for studying RSV-induced pathology have been developed, and these models have shown that RSV induces a complex immune response. Priming of mice with various RSV proteins and subsequent RSV challenge can lead to various degrees of lung pathology (6, 8, 16, 38). Although primary RSV infection induces proinflammatory and Th1 responses, models of RSV-induced pathology have implicated Th2 cytokines, such as interleukin-4 (IL-4), in this process. In accordance with this, polymorphisms in IL-4 and the IL-4 receptor have been associated with severe RSV disease in children (5, 13, 30).
To gain more insight into the early pathways of the pathogenesis of and immune response to RSV, we determined the transcription profiles of the lungs of RSV-infected mice at 1 and 3 days after infection by using microarray analysis. Mice are rather resistant to RSV infection and clear the virus quite rapidly. Studying early responses to such a self-limiting RSV infection in the lung may give insight into processes necessary for viral clearance and protection from RSV-induced pathology. In addition, genes and biological pathways which are regulated at early time points in infection can serve as candidates for studying susceptibility to RSV infection in children. Subsequent analysis of polymorphisms in selected candidate genes in human genetic association studies, together with the here-described murine studies, may help to elucidate the mechanisms that underlie an adequate response to acute primary RSV infection.
MATERIALS AND METHODS
Virus.
Human RSV type A2 (RSV A2) was obtained from the ATCC (Rockville, MD). The virus was cultured on HEp-2 cells (ATCC, Rockville, MD) in medium (RPMI 1640; Gibco BRL, Life Technologies, Rockville, MD) containing 10% heat-inactivated fetal calf serum (Greiner, Frickenhausen, Germany), 2 mM glutamine, 100 IU/ml penicillin, and 100 U/ml streptomycin, as described elsewhere (4). The infectivity of the virus stock (PFU RSV/ml) was assessed by a quantitative plaque-forming assay (4). As a control, mock was prepared using HEp-2 cells that were not infected with RSV.
Animals.
Female specific-pathogen-free BALB/c mice were obtained from Harlan Olac (Horst, The Netherlands) and were used at 6 to 10 weeks of age. A week before the experiments started, mice were housed per group according to the experimental setup under specific-pathogen-free and temperature-controlled conditions. Mice were kept in a 12-h light/dark cycle and received water and food ad libitum. The study was approved by the National Institute for Public Health and the Environment committee on animal welfare.
Experimental design.
Mice (n = 7 per group) were infected intranasally with 106 PFU RSV or with mock or were not inoculated. Before inoculation, mice were anesthetized with enfluran. At day 1 or 3 after infection, mice were intraperitoneally anesthetized with ketamine, xylazine (Rompun), and atropine (KRA) and sacrificed. After perfusion, the lungs and bronchial lymph nodes were removed. The right lung was kept in RNAlater RNA stabilization reagent (QIAGEN). The left lung was fixed intratracheally using 4% formalin for histological examination. Fixed lungs were embedded in Paraplast (Monoject, Kildare, Ireland). Sections of 5 μm were stained with hematoxylin and eosin. Different lung lesions were scored semiquantitatively as absent (0), minimal (1), slight (2), moderate (3), marked (4), or severe (5), as previously described (2). The lesions were scored blindly.
Microarray analyses.
Tissues kept in RNAlater RNA stabilization reagent (QIAGEN) were stored at 4°C for 2 to 4 days. Subsequently, RNA was extracted using RNeasy kits (QIAGEN). RNA concentrations were measured using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA quality was determined using the Agilent 2100 bioanalyzer (Agilent, Amstelveen, The Netherlands). Microarray slides containing 21,997 oligonucleotides from the Sigma-Compugen Mouse oligonucleotide library (and appropriate controls) were spotted at the Microarray Department of the University of Amsterdam. RNA amplification and labeling were carried out with an Amino Allyl MessageAmp aRNA kit (Ambion) according to the manufacturer's instructions, using 2 to 3 μg of total RNA as starting material. RNA samples from individual mice were labeled and hybridized against a common reference containing an RNA pool of all samples isolated. Arrays were scanned at two wavelengths by using a ScanArray 4000XL microarray scanner (PerkinElmer). Following microarray scanning, median Cy3 and Cy5 signal intensities per spot were determined using Array Vision software (Imaging Research, St. Catharines, Ontario, Canada). Quality control was performed on raw data by means of visual inspection of the scanned images, as well as a check on the scatter and MA (ratio-intensity) plots. All slides (n = 45) met our quality control criteria, i.e., less than 10% of the spots could be flagged as missing data, and the dye ratio did not show a signal-dependent trend exceeding a factor 10. Raw signal data for oligonucleotide-containing spots were normalized with R software by using a three-step approach of (i) natural log transformation, (ii) quantile normalization of all scans, and (iii) correction of the sample spot signal for differences in the corresponding reference spot signal between arrays.
Normalized data for individual genes were compared between all groups by using a one-way analysis of variance. Initially, genes with a P value of <0.001 and a maximum ratio of >1.5-fold (defined as maximum/minimum between groups) were considered sufficiently relevant for further analysis. The false discovery rate (FDR), i.e., the fraction of false positives in the lists of regulated genes, was <0.05, except in the lymph node arrays, where the FDR was <0.1. The resulting gene lists were further refined using additional criteria and stringencies.
Gene expression patterns were visualized by hierarchical clustering (Euclidian distance clustering and Ward linkage) using the GeneMaths program (Applied Maths, St. Martens-Latem, Belgium). Gene Ontology term enrichment was assessed using Expression Analysis Systematic Explorer (EASE) (http://david.abcc.ncifcrf.gov/ease/ease.jsp) (14), and pathway analysis was performed using MetaCore software.
Real-time PCR.
In addition to microarray analysis, we measured the expression levels of seven genes by real-time reverse transcription-PCR. All reagents and equipment were obtained from Applied Biosystems (Foster City, CA). The following TaqMan gene expression assays were used: Mm00445235_m1 (Cxcl10), Mm00515153_m1 (Ifit1), Mm00801778_m1 (Ifng), Mm00445259_m1 (IL-4), Mm00439646_m1 (IL-5), Mm00446190_m1 (IL-6), and Mm00434204_m1 (IL-13). Assays for hypoxanthine phosphoribosyltransferase 1 and Polr2a were custom-made and included as endogenous controls. The presence of genomic DNA in RNA samples and amplification efficiency for all assays were assessed before the start of the measurements. RNA was converted to cDNA using a High-Capacity cDNA archive kit according to the manufacturer's instructions. For each measured gene, 1 μl of assay was mixed with 10 μl TaqMan Fast universal PCR master mix and added to 33 ng of every cDNA sample in 9 μl Milli-Q in duplicate. The cDNA was amplified in a 96-well plate during 40 cycles of 3 s at 95°C and 30 s at 60°C, preceded by 20 s at 95°C for enzyme activation, using the 7500 Fast real-time PCR system. No-template controls were included in all plates. Threshold cycles were automatically derived from the amplification plots constructed of the ROX-normalized fluorescence signals by 7500 Fast system SDS software v1.3. The expression levels of the endogenous controls were comparable to each other and for all samples. The means of the hypoxanthine phosphoribosyltransferase 1 and Polr2a levels of all samples were therefore used to normalize the expression of the other genes. Relative quantification of the mRNA copies in the RSV/mock-challenged samples compared to that of the controls was performed by the comparative threshold cycle method using Microsoft Excel.
RESULTS
Gene expression in the lung upon RSV infection.
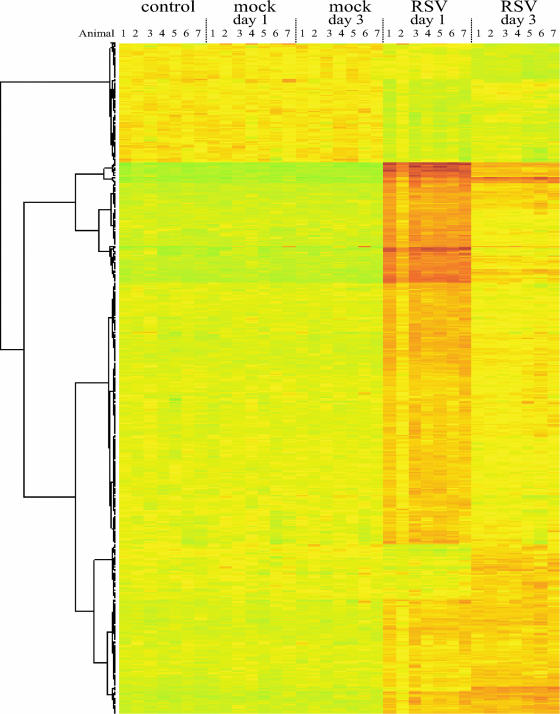
To assess which genes and biological pathways are induced at early time points after RSV infection, the transcriptional profiles of the lungs of RSV-infected BALB/c mice were determined on days 1 and 3 after RSV inoculation by using microarrays (n = 7 per group, 1 lung per array). Cluster analysis of all arrays clearly shows that all mice infected with RSV showed similar responses on day 1 and day 3 after infection, with the exception of mouse 2 on day 1, which displayed a somewhat less pronounced response (Fig. 1). This was, however, not related to less-efficient infection since the amount of viral RNA was similar to that of the other mice in the group (data not shown).
FIG. 1.
Cluster analysis of genes regulated in the lungs of BALB/c mice in response to RSV infection. Genes with a change of >1.5-fold (P < 0.001) are depicted. Each row represents the lung of an individual mouse, and each group (control, mock infection day 1, mock infection day 3, RSV infection day 1, and RSV infection day 3) comprises seven mice.
To ensure that the differences in transcription profiles observed are related to RSV infection, we compared mock-infected mice on days 1 and 3 with uninfected mice. No statistically significant differences in expression profiles could be detected between these groups, indicating that mock infection does not alter gene expression in the lung. RSV infection, however, strongly affected gene expression in the lung, i.e., 584 genes were regulated (>1.5-fold up-regulation by one-way analysis of variance between all groups, P < 0.001, FDR = 0.0081). A total of 475 of these genes were up-regulated, and 109 genes were down-regulated. The strongest response was observed on day 1, and the expression of the majority of up-regulated genes had already faded on day 3. A subset of genes remained highly expressed both on day 1 and on day 3, and a smaller group of genes was induced only on day 3 (Fig. 1). The down-regulated genes were less strongly regulated than the up-regulated genes, i.e., none of them reached a greater-than-twofold down-regulation.
Gene Ontology term enrichment using EASE revealed that enriched genes could almost exclusively be categorized into immunological pathways, with the exception of the 109 down-regulated genes, which were involved in metabolism, electron transport, and transcription processes. Enriched categories in the up-regulated genes included defense, immune response, response to pathogen, chemokine activity, chemotaxis, cytokine synthesis, inflammation, acute phase, apoptosis, and cell death. For a complete overview of EASE categories, see Table SA in the supplemental material.
To examine the potential effects of cell influx on transcription profiles, lung pathology was analyzed by histology. Mock infection did not lead to histological changes apart from marginal perivasculitis in two out of seven mock-infected mice both on day 1 and on day 3 after infection (histopathological score [mean ± standard deviation] of 0.3 ± 0.5 for both groups). RSV infection on days 1 and 3 resulted in slight peribronchiolitis (histopathological scores of 1.6 ± 0.5 and 1.1 ± 0.4, respectively), perivasculitis (1.9 ± 0.4 for both time points), and alveolitis (0.9 ± 0.4 and 1.3 ± 0.5, respectively). The minimal to slight inflammatory infiltrates observed were composed of monocytes and lymphocytes. In addition, alveolar macrophages were detected in the alveoli. The mucous cells of the bronchial epithelium displayed some hypertrophy (2.1 ± 0.7 and 2.6 ± 0.5 on days 1 and 3, respectively). Taken together, these data show that RSV-induced inflammation was minimal at these time points and that there were no differences between day 1 and day 3 after RSV infection. This indicates that differences in gene expression between these two time points are not related to differences in inflammatory infiltrate and are most likely the result of altered gene expression in cells that reside in the lung, e.g., epithelial cells and alveolar macrophages.
Pathways activated in RSV infection.
In order to gain more insight into the processes activated by RSV infection and possible interactions between the induced genes in the lung, data were analyzed using the pathway-finding software MetaCore. When all regulated genes were analyzed, several enriched pathways were found that are involved in proinflammatory cytokine production, i.e., the TLR signaling pathway, the IL-1 signaling pathway, and the alpha/beta interferon (IFN-α/β) and IFN-γ signaling pathways. The RhoA regulation pathway, which leads to the inhibition of viral protein synthesis, was also activated. All these pathways were activated both on day 1 and on day 3 after infection. In addition, on day 1 after infection, but not on day 3, several pathways involved in cell death and apoptosis were activated, i.e., caspase cascade activation, the tumor necrosis factor receptor 1 signaling pathway, IAP proteins in apoptosis, and an antiapoptotic tumor necrosis factor/NF-κB/IAP pathway. The identified pathways and the regulated products of genes belonging to these pathways are listed in Table 1. The pathways involved in proinflammatory responses were more highly up-regulated than the pathways involved in apoptosis. In fact, most of the genes involved in apoptosis were not highly up-regulated, i.e., they had a ratio between 1.5- and 2-fold. In addition, closer examination of the genes regulated in these apoptosis pathways revealed that, with the exception of caspase 8 and Bid, the regulated genes (e.g., the survivin, Cdc2, Birc2, Birc3, Arts-1, and Tnfaip3 genes) are involved in inhibiting apoptosis, indicating that the final effect of the activation of these pathways may be cell survival rather than apoptosis.
TABLE 1.
Pathways regulated by RSV infection and genes regulated in these pathwaysa
| Pathway | Products of genes regulated by RSV infection |
|---|---|
| TLR signaling pathway | Tlr1, Tlr2, CD14, IκB, RelB, IL-6, Cxcl10, Cxcl11, Ccl11, Ccl20 |
| IL-1 signaling pathway | IL-1b, I-κBe, RelB, IRF1, F3, Edn1, serpine 1 |
| IFN-α/β signaling pathway | Stat1, Stat2, Irf9, Ifit2, Gip2 |
| IFN-γ signaling pathway | IFN-γ, Socs1, Stat1, CamK2B, IRF1, IRF9, Pkr, Icam1 |
| RhoA regulation pathway | IRF9, Stat1, Stat2, Oas1, Pkr |
| Caspase cascade activation by FADD and RIPK | Bid, Tnfaip33, Birc2, Birc3, RipK1 |
| Tumor necrosis factor receptor 1 signaling in apoptosis | Arts-1, caspase 8, Bid, Birc2, Birc3, RelB, IκBe, RipK1 |
| IAP proteins in apoptosis | Caspase 8, Hsp70, Birc2, Birc3, survivin, Cdc2 |
| Antiapoptotic tumor necrosis factor/NF-κB/IAP pathway | Birc2, Birc3, survivin, RelB, IκBe, RipK1 |
Data were obtained using MetaCore analysis.
Classes of genes regulated in the lung in response to RSV infection.
A limitation of pathways-finding software is that most pathways include all signaling genes but do not include all genes regulated by a given pathway. This is also exemplified by the fact that the pathways induced by RSV infection comprise only 42 of the 584 regulated genes. The other genes were not present in the MetaCore pathways that were enriched upon RSV infection as indicated above. As indicated above, Gene Ontology term enrichment using EASE revealed that most of the genes induced by RSV infection were involved in immunological processes. Although pathway analysis did not reveal other enriched pathways, Gene Ontology term enrichments indicates that specific functional categories can be identified. Therefore, we also subdivided all regulated genes based on their (putative) function into several, more-specific immunological (and other) functional categories listed in Table 2 (for a complete list of regulated genes, see Table SA in the supplemental material). For this analysis, only the highly up-regulated genes (>2-fold change, P < 0.001) were selected. Using these criteria, 182 genes were up-regulated by RSV infection, whereas no genes were down-regulated by infection. Of these genes, 116 were up-regulated on day 1 but not on day 3 after infection, 18 genes were up-regulated on day 3 but not on day 1, and 48 genes were up-regulated at both time points after infection.
TABLE 2.
Classification of genes induced in the lung upon primary RSV infection
| Category | No. of genes up-regulated on:
|
||
|---|---|---|---|
| Day 1 only | Days 1 and 3 | Day 3 only | |
| Acute phase | 1 | 5 | |
| Antigen processing | 7 | 3 | 7 |
| Apoptosis | 3 | 2 | |
| Cell cycle | 1 | 2 | 2 |
| DNA/RNA binding | 4 | 2 | |
| Chemoattraction | 10 | 2 | 1 |
| IFN response | 20 | 14 | |
| Inflammation | 29 | 6 | 5 |
| Metabolism | 3 | 1 | |
| Various functions | 18 | 4 | 2 |
| Unknown function | 20 | 7 | 1 |
| Total | 116 | 48 | 18 |
A large category of genes comprised those that are involved in the IFN response. This group contained both IFN-α/β-regulated genes and IFN-γ-regulated genes. Surprisingly, IFN-α and IFN-β themselves were not up-regulated. There are 14 known IFN-α genes, and 8 of these genes were present on the array. None of these genes was up-regulated. IFN-γ was significantly up-regulated (1.7-fold, P < 0.001) but also did not reach a factor 2. Another large category of induced genes is that of the chemokine genes. Altogether, 11 chemokine genes, 1 chemokine receptor gene, and 1 gene involved in chemokine signaling were up-regulated upon infection. Two other important groups of up-regulated genes are those involved in antigen processing (n = 18) and inflammation (n = 40). The latter set of genes displayed great diversity and included genes involved in or induced by TLR and IL-1 signaling, genes encoding complement components, genes encoding adhesion molecules, and cytokine genes. The products of genes of the four largest categories and their n-fold changes are listed in Table 3.
TABLE 3.
Genes up-regulated by infection with RSV that can be classified as being involved in chemoattraction, the IFN response, and inflammation
| Category and gene productc | Description | Fold change on:
|
|
|---|---|---|---|
| Day 1a | Day 3b | ||
| Chemoattraction | |||
| Cxcl10* | Chemokine (C-X-C motif) ligand 10 | 9.66 | 1.50 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 9.26 | 1.94 |
| Cxcl11* | Chemokine (C-X-C motif) ligand 11 | 7.94 | 1.96 |
| Cxcl1 | Chemokine (C-X-C motif) ligand 1 | 7.67 | 2.46 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 5.47 | 1.63 |
| Ccl4* | Chemokine (C-C motif) ligand 4 | 2.86 | 1.73 |
| Cxcl2 | Chemokine (C-X-C motif) ligand 2 | 2.66 | 1.22 |
| Ccl9 | Chemokine (C-C motif) ligand 9 | 2.30 | 2.67 |
| Cxcl9* | Chemokine (C-X-C motif) ligand 9 | 2.29 | 1.23 |
| Cxcl5 | Chemokine (C-X-C motif) ligand 5 | 2.03 | 1.50 |
| Rgs1 | Regulator of G-protein signaling 1 | 2.03 | 1.41 |
| Ccr7 | Chemokine (C-C motif) receptor 7 | 2.01 | 1.57 |
| Ccl8 | Chemokine (C-C motif) ligand 8 | 1.92 | 2.84 |
| Interferon response | |||
| G1p2 | Interferon-alpha-inducible protein | 17.33 | 4.69 |
| Ifi44 | Interferon-induced protein 44 | 16.67 | 4.47 |
| Ifit1 | Interferon-induced protein with tetratricopeptide repeats 1 | 13.36 | 3.15 |
| Ifi202b | Interferon-activated gene 202B | 9.79 | 4.19 |
| Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | 9.02 | 2.63 |
| Ifi204 | Interferon-activated gene 204 | 8.61 | 2.97 |
| Herc5 | Hect domain and RLD5 | 6.84 | 2.02 |
| Ifi1* | Interferon-inducible protein 1 | 6.77 | 2.25 |
| Irf7 | Interferon regulatory factor 7 | 6.72 | 4.95 |
| Stat1* | Signal transducer and activator of transcription 1 | 6.70 | 2.48 |
| Ifih1 | Interferon induced with helicase C domain 1 | 6.47 | 2.28 |
| Ifit2 | Interferon-induced protein with tetratricopeptide repeats 2 | 5.95 | 1.25 |
| Gbp2 | Guanylate nucleotide binding protein 2 | 5.82 | 1.48 |
| Rtp4 | RIKEN cDNA 5830458K16 gene | 5.58 | 3.01 |
| Isg20 | Interferon-stimulated protein | 4.81 | 1.71 |
| Ifi203 | Interferon-activated gene 203 | 4.27 | 1.91 |
| Iigp2 | Interferon-inducible GTPase 2 | 4.22 | 1.64 |
| Ifi27 | Interferon alpha-inducible protein 27 | 4.20 | 6.93 |
| Igtp* | Interferon-gamma-induced GTPase | 4.20 | 1.44 |
| Stat2* | Signal transducer and activator of transcription 2 | 4.02 | 1.95 |
| Usp18 | Ubiquitin-specific protease 18 | 4.01 | 1.80 |
| Oas1a | 2′-5′ Oligoadenylate synthetase 1A | 3.68 | 2.24 |
| Daxx | Fas death domain-associated protein | 3.67 | 1.39 |
| Ifi35* | Interferon-induced protein 35 | 3.07 | 1.44 |
| Nmi* | N-myc (and STAT) interactor | 2.96 | 1.37 |
| Ifi47* | Interferon-gamma-inducible protein 47 | 2.75 | 1.34 |
| Gbp1 | Guanylate nucleotide binding protein 1 | 2.71 | 1.31 |
| Prkr | Protein kinase, interferon-inducible double-stranded RNA dependent | 2.60 | 1.32 |
| Ifitm3 | Interferon-induced transmembrane protein 3 | 2.49 | 1.87 |
| Cd69* | CD69 antigen | 2.38 | 1.14 |
| Ube1l | Ubiquitin-activating enzyme E1-like | 2.31 | 1.31 |
| Isgf3g | Interferon-dependent positive-acting transcription factor 3 gamma | 2.17 | 1.60 |
| Fcgr1* | Fc receptor, IgG, high-affinity I | 2.05 | 1.66 |
| Aif1* | Allograft inflammatory factor 1 | 2.04 | 1.79 |
| Inflammation | |||
| Tgtp | T-cell-specific GTPase | 10.60 | 2.65 |
| Gbp4 | Guanylate nucleotide binding protein 4 | 9.79 | 2.76 |
| Tyki | Thymidylate kinase family LPS-inducible member | 7.90 | 1.66 |
| Il6 | Interleukin-6 | 4.77 | 1.22 |
| Lcn2 | Lipocalin 2 | 4.59 | 6.08 |
| Il18bp* | Interleukin-18 binding protein | 4.07 | 1.63 |
| Lilrb4 | Leukocyte Ig-like receptor, subfamily B, member 4 | 3.80 | 2.52 |
| Il1rn | Interleukin-1 receptor antagonist | 3.25 | 1.37 |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | 3.14 | 1.93 |
| Irg1 | Immunoresponsive gene 1 | 3.04 | 1.38 |
| Alox12 | Arachidonate 12-lipoxygenase | 2.74 | 3.43 |
| Asb15 | Ankyrin repeat and SOCS box-containing protein 15 | 2.73 | 1.14 |
| Mpa2 | Macrophage activation 2 | 2.73 | 1.49 |
| Samhd1 | SAM domain and HD domain 1 | 2.66 | 1.37 |
| IL-15 | Interleukin-15 | 2.63 | 1.17 |
| Cd19 | CD19 antigen | 2.49 | 1.17 |
| IL-1b | Interleukin 1 beta | 2.43 | 1.52 |
| Casp11 | Caspase 11, apoptosis-related cysteine protease | 2.36 | 1.40 |
| Tlr2 | Toll-like receptor 2 | 2.36 | 1.41 |
| Ear4** | Eosinophil-associated, RNase A family, member 4 | 2.35 | 1.13 |
| Fcer1g** | Fc receptor, IgE, high-affinity I, gamma polypeptide | 2.32 | 2.17 |
| Il1rl1 | Interleukin 1 receptor-like 1 | 2.32 | 1.38 |
| Cd14 | CD14 antigen | 2.29 | 1.91 |
| Gzmb | Granzyme B | 2.18 | 1.36 |
| Ankrd1 | Ankyrin repeat domain 1 (cardiac muscle) | 2.15 | 1.33 |
| Tlr3 | Toll-like receptor 3 | 2.11 | 1.06 |
| Gata3** | GATA binding protein 3 | 2.10 | 1.29 |
| Cd274 | CD274 antigen | 2.06 | 1.06 |
| Osmr | Oncostatin M receptor | 2.05 | −1.01 |
| Clec4e | C-type lectin domain family 4, member e | 2.04 | 1.11 |
| C2 | Complement component 2 (within H-2S) | 2.03 | 1.80 |
| Zc3hav1 | Zinc finger CCCH type, antiviral 1 | 2.02 | 1.41 |
| Cd83 | CD83 antigen | 2.02 | 1.46 |
| Ig region | Ig region | 2.01 | 1.36 |
| Lrg1 | Leucine-rich alpha-2-glycoprotein 1 | 2.01 | 1.78 |
| C3 | Complement component 3 | 1.79 | 2.06 |
| Ly64 | Lymphocyte antigen 64 | 1.71 | 2.03 |
| C3ar1** | Complement component 3a receptor 1 | 1.48 | 3.05 |
| Cd72 | CD72 antigen | 1.43 | 3.90 |
| Clec7a | C-type lectin domain family 7, member a | 1.39 | 2.71 |
| Antigen processing and presentation | |||
| Psmb10* | Proteasome (prosome, macropain) subunit, beta type 10 | 2.39 | 1.30 |
| Psmb8* | Proteasome (prosome, macropain) subunit, beta type 8 | 3.05 | 1.93 |
| Psme1* | Proteasome (prosome, macropain) 28 subunit alpha | 2.35 | 1.41 |
| Psme2* | Proteasome (prosome, macropain) 28 subunit beta | 2.51 | 1.48 |
| Tap2 | Transporter 2, ATP-binding cassette, subfamily B (MDR/TAP) | 2.56 | 1.21 |
| n = 7 genes | MHC class I molecule/genes | 2.42 | 2.42 |
| n = 5 genes* | MHC class II molecule/genes | 2.46 | 2.94 |
Change (n-fold) in regulation compared to that for mock infection on day 1.
Change (n-fold) in regulation compared to that for mock infection on day 3.
Gene products with * are products of Th1-associated genes, and those with ** are products of Th2-associated genes.
Where possible, the 182 up-regulated genes were also designated Th1 genes, i.e., genes regulated by IFN-γ or genes involved in IFN-γ-mediated responses, and Th2 genes, i.e., genes that have been associated with Th2 responses or genes that were shown to be up-regulated in two murine models for allergic asthma (22). Based on this subdivision, primary RSV infection induced 25 (of a total of 182 regulated genes) known Th1-associated genes (13.7%) and 4 known Th2-associated genes (2.2%) (Table 4). The Th1-associated genes were found mainly in the group of antigen-processing genes and included seven major histocompatibility complex (MHC) class II molecules and four immunoproteasome subunits, i.e., Psmb8, Psmb10, Psme1, and Psme2. These immunoproteasome subunits replace the constitutive proteasome subunits upon stimulation with IFN-γ. In addition, several of the chemokines that were induced are regulated by IFN-γ. The Th2 genes included the high-affinity receptor for immunoglobulin E (IgE) (Fcerg1), the complement receptor involved in anaphylatoxin binding (C3aR), an eosinophil-associated RNase (Ear4), and a transcription factor involved in Th2 cell development (Gata3). The genes encoding the Th2 cytokines IL-4, IL-5, and IL-13 were not up-regulated by RSV infection, and IL-10 was up-regulated (1.7-fold) but did not reach a factor 2.
TABLE 4.
Th1- and Th2-regulated genes induced in the lung during primary RSV infection
| Category | No. of genes regulated by:
|
|
|---|---|---|
| Th1a | Th2b | |
| Chemoattraction | 4 | |
| IFN response | 11 | |
| Antigen processing | 9 | |
| Inflammation | 1 | 5 |
| Metabolism | ||
| Total | 25 | 5 |
Th1 genes are genes regulated by IFN-γ or involved in IFN-γ-mediated responses.
Th2 genes are genes that have been associated with Th2 responses or that were shown to be up-regulated in two models for allergic asthma (22).
Confirmation of microarray data by real-time PCR.
To validate the gene expression changes found by microarray analysis, we performed real-time PCR. For this purpose, three genes were chosen that displayed relatively high up-regulation (the IL-6, Cxcl10, and Ifit1 genes), one gene which displayed slight up-regulation (the IFN-γ gene), and three genes which were not up-regulated (the IL-4, IL-5, and IL-13 genes). The three genes for which no up-regulation could be detected using microarray analysis were also not up-regulated using real-time PCR read-out (relative increases [n-fold] on day 1 after infection were 1.3 ± 0.5, 1.0 ± 0.4, and 2.3 ± 2.0 for IL-4, IL-5, and IL-13, respectively). IFN-γ was up-regulated (n-fold) 7.9 ± 4.6 and 2.9 ± 0.9 on day 1 and day 3 after infection, respectively. Cxcl10, Ifit1, and IL-6 displayed up-regulation (n-fold) of 1.262 ± 686, 61 ± 19, and 198 ± 96, respectively, on day 1 after infection, and 38 ± 11, 9.5 ± 3.2, and 10 ± 3.8, respectively, on day 3 after infection. Although the trends in regulation are similar between microarray analysis and real-time PCR, the level of up-regulation detected by real-time PCR was higher than that observed on the microarray. This phenomenon is often observed when the two techniques are compared (7, 41).
Gene expression in bronchial lymph nodes upon RSV infection.
On day 3 after inoculation, the transcription profiles of the bronchial lymph nodes of the mock-infected and RSV-infected groups were determined. Lymph nodes of five out of seven mice were randomly selected for microarray analysis (one tissue per array). The response here was much less pronounced than that in the lung. Comparison of mock-infected and RSV-infected mice revealed 40 differentially regulated genes (>1.5-fold change, P < 0.001, FDR = 0.094). Thirty-seven of these genes were up-regulated, and three were down-regulated. EASE again revealed that most of these genes were involved in immunological processes. Of these 40 genes, only 9 genes, whose products are listed in Table 5, had a change of >2-fold (P < 0.001, 1 down-regulated and 8 up-regulated genes). The most strongly up-regulated genes were the granzyme A and granzyme B genes. For a complete list of regulated genes, see Table SB in the supplemental material.
TABLE 5.
Genes differentially expressed (>2-fold change) in bronchial lymph nodes on day 3 after RSV infection compared to mock infection
| Category and gene product | Description | Fold changea |
|---|---|---|
| Acute phase | ||
| Saa2b | Serum amyloid A 2 | 2.06 |
| Apoptosis | ||
| Bid | BH3-interacting domain death agonist | 2.39 |
| Cell cycle | ||
| Slfn4b | Schlafen 4 | 2.05 |
| Chemokine | ||
| Cxcl9b | Chemokine (C-X-C motif) ligand 9 | 2.19 |
| Inflammation | ||
| Glycam1 | Glycosylation-dependent cell adhesion molecule 1 | −2.07 |
| Gzmbb | Granzyme B | 4.14 |
| Gzma | Granzyme A | 3.19 |
| Ly6a | Lymphocyte antigen 6 complex, locus A | 2.24 |
| Unknown function | ||
| Plac8b | Placenta-specific 8 | 2.20 |
Change of expression (n-fold) in lymph nodes of RSV-infected mice compared to expression in mock-infected mice.
Products of genes also at least twofold up-regulated in the lung on day 1 or 3 after RSV infection.
DISCUSSION
This study shows that RSV infection results in a rapid transcriptional response which is very strong on day 1 and fades on day 3, although a small subset of genes (n = 18) is first activated on day 3. The kinetics of the response reveals that the peak of the transcription response in the lung apparently precedes the peak of viral replication, which is normally found between days 4 and 6 (1, 3, 9, 28, 40). Regulated genes are involved in the IFN response, in inflammation, in chemoattraction, and in antigen processing. Furthermore, genes associated with Th1 responses predominate over those associated with Th2 responses. Gene expression in the lung-draining lymph nodes on day 3 was not strongly altered by RSV infection.
A striking feature of the transcriptional response is that only a small subset of genes is down-regulated upon RSV infection and that the level of down-regulation of these genes is not strong (between 1.5- and 2-fold). In contrast, Zhang et al. have shown that infection of human lung epithelial cells in vitro with RSV also down-regulates a large proportion of genes, especially at later time points in infection, indicating that RSV can inhibit gene expression (43). However, later time points in in vitro infection are more likely to result in cell death. In addition, we have to take into account that in our model, we look at the transcription profile of a whole lung, composed of far more than just epithelial cells. Differences between in vivo and in vitro findings are also exemplified by different kinetics of the responses in the in vitro infection model, where there is a gradual increase in gene expression over time (43). Since we used seven biological replicates per group (one tissue per array), our data warrant the conclusion that RSV infection does not clearly down-regulate a large set of genes at early time points postinoculation.
A very limited amount of data is available on in vivo responses to respiratory viruses, and this is, to our knowledge, the first study in which in vivo lung responses to infection with RSV have been studied. This makes comparison with other data difficult. Kash et al. have studied the in vivo lung response to influenza virus infection and showed that infectious virus results in the down-regulation of a large proportion of differentially expressed genes (20). Another interesting difference between our RSV data and those found for influenza virus is the difference in kinetics: responses to influenza virus are stronger on day 3 than on day 1. Although differences in experimental setup and replication kinetics of the viruses have to be taken into account, these data show that RSV alters transcriptional responses differently from influenza virus and suggest that viral load correlates with influenza virus-induced gene expression but not with RSV-induced gene expression. This is consistent with the findings of Zhang et al., who show that ribavirin treatment, which results in reduced viral replication, alters the expression of only a subset of RSV-regulated genes in an in vitro infection model (42). Based on these findings, they postulated that gene expression in response to RSV infection is not dependent on high levels of viral replication.
IFN response in the lung.
RSV has been shown to be a very potent inhibitor of type I IFN expression and responses in a manner that depends on the viral NS1 gene (31, 34, 36). RSV is believed to accomplish this by various mechanisms, including interference with IFN regulatory factor 3 (IRF3) (37) and STAT2 (31, 32). Consistent with this, the type I IFNs, IFN-α and IFN-β, were not up-regulated in the lungs of our RSV-infected mice. Surprisingly, however, a large set of genes that is involved in IFN signaling or that is under IFN-α/β control was strongly up-regulated after RSV infection. In fact, the genes belonging to this category displayed the strongest regulation upon infection (Table 3). How can we explain the apparent discrepancy of such a strong IFN response in the absence of IFN-α/β? One explanation for the absence of type I IFN expression is that the type I IFNs are induced very early after exposure to pathogens (reviewed in reference 18). Therefore, we may have missed a possible increased transcription. However, a more likely explanation is that there is an alternative pathway for the activation of IFN-regulated genes. Indeed, such a pathway has been described previously (reviewed in reference 21). When single-stranded or double-stranded RNA is recognized by TLR7, TLR3, or RIG-I, the transcription factors IRF3, -5 and -7 are activated. Activated IRF3, -5, and -7 can subsequently induce the transcription of type I IFNs but can also directly activate the transcription of genes encoding an IFN-stimulated response element. Thus, although the genes encoding IFN-stimulated response elements can be regulated by type I IFNs in a STAT1- and STAT2-dependent manner, they can also be activated in the absence of type I IFNs themselves. Consistent with this, Zhang et al. have shown that infection of human epithelial cells with RSV results in far greater up-regulation of genes that encode an IFN-stimulated response element than that of the IFN-β gene itself (42). IRF3 is constitutively expressed, and upon activation by single-stranded or double-stranded RNA, it is activated by phosphorylation. IRF7 is activated in a similar way but is also up-regulated at the transcriptional level upon infection with a virus (reviewed in reference 21). Our data are consistent with this model since in the lungs of our infected mice, IRF7 is highly (6.72- and 4.95-fold, respectively) up-regulated on days 1 and 3 after RSV infection and IRF5 is 1.7-fold up-regulated on day 1 after infection, whereas IRF3 expression is unaltered. Although transcription profiles do not give a complete picture of the IRF pathway in the lungs of RSV-infected cells, the type I IFN response in the lungs of RSV-infected mice is much larger than we anticipated, indicating that IFN-α/β production is low upon infection but that the expression of genes regulated by IFN is very high. Interestingly, this subset of genes includes the Prkr and Oas1 genes, involved in the antiviral response (reviewed in reference 29), suggesting that, despite RSV's ability to modulate the IFN response, the antiviral response is still activated upon infection. These findings are consistent with a postulated role for innate IFNs in determining the nature and severity of RSV disease, as postulated by Johnson et al. (19).
IFN-γ-regulated genes.
RSV infection also enhanced the expression of a set of genes, categorized in the IFN cluster, which are under IFN-γ control. IFN-γ did not reach a level of twofold up-regulation. However, it was 1.7-fold increased, which explains the induction of IFN-γ-regulated genes. The source of early IFN-γ production in the murine model could be either NK cells or CD8 T cells, although the latter cells are probably activated at a later stage in the infection process. Among the up-regulated genes in the lung-draining lymph nodes were the granzyme A and granzyme B genes (Table 5). When less stringent criteria (>1.5-fold up-regulation, P < 0.001) were used, the granzyme K and Klrc2 genes were also up-regulated. All of these genes are expressed by activated NK cells. The first three genes are also expressed by CD8 T cells. However, markers such as CD8 and MHC molecules are not up-regulated in the lymph nodes. Therefore, the observed expression profile is probably associated with NK cell activation in the bronchial lymph nodes, and these cells are the most likely source of early IFN-γ production in infected mice and account for the expression of IFN-γ-regulated genes. This also fits well with the reported observation that NK cell influx into the lung peaks on day 4 after infection (17).
Other genes under the control of IFN-γ include those encoding immunoproteasome subunits, which replace conventional proteasome components in response to IFN-γ stimulation. In addition, up-regulation of immunoproteasome subunits indicates that shortly after infection with RSV, the antigen-processing machinery necessary for the induction of CD8 responses is activated. Consistent with findings in other studies, primary RSV infection induces more genes associated with Th1 responses than with Th2 responses (9, 17).
Chemokines regulated by RSV infection.
Another important category of genes that is regulated upon RSV infection is that of the genes encoding chemokines, indicating that the expression of chemokines is an important early host response during RSV infection and probably initiates cellular influx that is observed at later time points in infection. This is consistent with earlier studies in which RSV infection of lung epithelial cells also resulted in a response dominated by chemokine expression (43) and with studies that analyzed chemokine expression at the protein level in the lung (10, 28). Also, chemokines are transiently expressed in our model and this is consistent with the observations of Miller et al., who showed that Ccl2, Ccl3, and Cxcl10 show a protein peak on day 1 after infection (28) and are not detectable on day 3. Ccl5 displays a similar profile but is expressed at all time points. Our data show a similar pattern for Ccl2, Cxcl10, Ccl3, and Ccl5 expression, although the last two do not reach a change of twofold. Miller et al. also observed a second peak on day 8 after infection (28), a time point which we did not study.
Overall, primary RSV infection induces more Th1- than Th2-associated genes. Specifically looking at chemokines, our data show that genes encoding the IFN-γ-regulated chemokines Cxcl10 (IP10) and Cxcl11 (I-TAC) are up-regulated 9.8- and 8.7-fold, respectively, whereas two Th2-associated chemokines, Ccl11 (eotaxin) and Ccl17 (TARC), were only marginally up-regulated (1.7- and 1.2-fold, respectively). Other chemokines associated with Th2 responses, such as Ccl18 and Ccl22, were not regulated by RSV infection. Taken together, these data indicate that, consistent with the overall response to primary infection, the chemokine response is Th1 skewed.
In conclusion, our observations of the primary RSV infection model show a rapid activation of innate responses, as exemplified by pronounced IFN responses and chemokine activation. IFN-γ-mediated responses are also activated and appear more prolonged. Taken together, these data are suggestive of a proinflammatory response with Th1 characteristics. In addition, the antigen-processing machinery necessary for CD8 cell activation is induced. It is, however, striking that this response is so transient. On day 3 after infection, only a small set of 18 genes is activated and genes that are up-regulated at both time points are almost all expressed at higher levels on day 1 than on day 3. Although we did not measure viral replication in these mice, previous data from our laboratory and data published by others show that viral replication in the BALB/c model peaks between days 4 and 6 and that on day 8 after primary infection no virus is detectable (1, 3, 9, 28, 40). We did not determine transcription profiles at later time points in infection because influx of cells, which appears around and after day 4, would make the interpretation of transcription data very complex, an unfortunate limitation of the microarray technique. Our data, however, strongly suggest that the events necessary for viral clearance are initiated very early in this model for acute self-limiting infection and that this acute response fades rapidly but is sufficient for viral clearance. Genes involved in the IFN response, in antigen processing, in inflammation, and in chemoattraction are highly up-regulated in RSV infection. The importance of these processes and pathways in RSV infection and disease in children can now be studied in human association studies, and this will contribute to our understanding of RSV infection and disease.
Supplementary Material
Acknowledgments
We are grateful for the expert histotechnical assistance of F. M. de Vlugt-van den Koedijk. We thank C. Siezen for critically reading the manuscript. Finally, we thank all of the biotechnicians at our animal facility for performing the animal experiments.
Footnotes
Published ahead of print on 21 March 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderson, J. J., J. Norden, D. Saunders, G. L. Toms, and R. Scott. 1990. Analysis of the local and systemic immune responses induced in BALB/c mice by experimental respiratory syncytial virus infection. J. Gen. Virol. 71:1561-1570. [DOI] [PubMed] [Google Scholar]
- 2.Barends, M., M. van Oosten, C. G. De Rond, J. A. Dormans, A. D. Osterhaus, H. J. Neijens, and T. G. Kimman. 2004. Timing of infection and prior immunization with respiratory syncytial virus (RSV) in RSV-enhanced allergic inflammation. J. Infect. Dis. 189:1866-1872. [DOI] [PubMed] [Google Scholar]
- 3.Boelen, A., A. Andeweg, J. Kwakkel, W. Lokhorst, T. Bestebroer, J. Dormans, and T. Kimman. 2000. Both immunisation with a formalin-inactivated respiratory syncytial virus (RSV) vaccine and a mock antigen vaccine induce severe lung pathology and a Th2 cytokine profile in RSV-challenged mice. Vaccine 19:982-991. [DOI] [PubMed] [Google Scholar]
- 4.Boelen, A., J. Kwakkel, M. Barends, L. de Rond, J. Dormans, and T. Kimman. 2002. Effect of lack of interleukin-4, interleukin-12, interleukin-18, or the interferon-gamma receptor on virus replication, cytokine response, and lung pathology during respiratory syncytial virus infection in mice. J. Med. Virol. 66:552-560. [DOI] [PubMed] [Google Scholar]
- 5.Choi, E. H., H. J. Lee, T. Yoo, and S. J. Chanock. 2002. A common haplotype of interleukin-4 gene IL4 is associated with severe respiratory syncytial virus disease in Korean children. J. Infect. Dis. 186:1207-1211. [DOI] [PubMed] [Google Scholar]
- 6.Connors, M., N. A. Giese, A. B. Kulkarni, C. Y. Firestone, H. C. Morse III, and B. R. Murphy. 1994. Enhanced pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of interleukin-4 (IL-4) and IL-10. J. Virol. 68:5321-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallas, P. B., N. G. Gottardo, M. J. Firth, A. H. Beesley, K. Hoffmann, P. A. Terry, J. R. Freitas, J. M. Boag, A. J. Cummings, and U. R. Kees. 2005. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR—how well do they correlate? BMC Genomics 6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Swart, R. L., T. Kuiken, H. H. Timmerman, G. van Amerongen, B. G. Van Den Hoogen, H. W. Vos, H. J. Neijens, A. C. Andeweg, and A. D. Osterhaus. 2002. Immunization of macaques with formalin-inactivated respiratory syncytial virus (RSV) induces interleukin-13-associated hypersensitivity to subsequent RSV infection. J. Virol. 76:11561-11569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, B. S., M. D. Perkins, P. F. Wright, and D. T. Karzon. 1988. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 26:153-162. [DOI] [PubMed] [Google Scholar]
- 10.Haeberle, H. A., W. A. Kuziel, H.-J. Dieterich, A. Casola, Z. Gatalica, and R. P. Garofalo. 2001. Inducible expression of inflammatory chemokines in respiratory syncytial virus-infected mice: role of MIP-1α in lung pathology. J. Virol. 75:878-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzmann, A., S. P. Jerkic, K. Ganter, T. Kurz, S. Blattmann, L. Schuchmann, K. Gerhold, R. Berner, and K. A. Deichmann. 2003. Association study of the IL13 variant Arg110Gln in atopic diseases and juvenile idiopathic arthritis. J. Allergy Clin. Immunol. 112:735-739. [DOI] [PubMed] [Google Scholar]
- 12.Hoebee, B., L. Bont, E. Rietveld, M. van Oosten, H. M. Hodemaekers, N. J. Nagelkerke, H. J. Neijens, J. L. Kimpen, and T. G. Kimman. 2004. Influence of promoter variants of interleukin-10, interleukin-9, and tumor necrosis factor-alpha genes on respiratory syncytial virus bronchiolitis. J. Infect. Dis. 189:239-247. [DOI] [PubMed] [Google Scholar]
- 13.Hoebee, B., E. Rietveld, L. Bont, M. van Oosten, H. M. Hodemaekers, N. J. Nagelkerke, H. J. Neijens, J. L. Kimpen, and T. G. Kimman. 2003. Association of severe respiratory syncytial virus bronchiolitis with interleukin-4 and interleukin-4 receptor alpha polymorphisms. J. Infect. Dis. 187:2-11. [DOI] [PubMed] [Google Scholar]
- 14.Hosack, D. A., G. Dennis, Jr., B. T. Sherman, H. C. Lane, and R. A. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hull, J., A. Thomson, and D. Kwiatkowski. 2000. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 55:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussell, T., A. Georgiou, T. E. Sparer, S. Matthews, P. Pala, and P. J. Openshaw. 1998. Host genetic determinants of vaccine-induced eosinophilia during respiratory syncytial virus infection. J. Immunol. 161:6215-6222. [PubMed] [Google Scholar]
- 17.Hussell, T., and P. J. Openshaw. 1998. Intracellular IFN-gamma expression in natural killer cells precedes lung CD8+ T cell recruitment during respiratory syncytial virus infection. J. Gen. Virol. 79:2593-2601. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, T. R., S. E. Mertz, N. Gitiban, S. Hammond, R. Legallo, R. K. Durbin, and J. E. Durbin. 2005. Role for innate IFNs in determining respiratory syncytial virus immunopathology. J. Immunol. 174:7234-7241. [DOI] [PubMed] [Google Scholar]
- 20.Kash, J. C., C. F. Basler, A. García-Sastre, V. Carter, R. Billharz, D. E. Swayne, R. M. Przygodzki, J. K. Taubenberger, M. G. Katze, and T. M. Tumpey. 2004. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J. Virol. 78:9499-9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 22.Kuperman, D. A., C. C. Lewis, P. G. Woodruff, M. W. Rodriguez, Y. H. Yang, G. M. Dolganov, J. V. Fahy, and D. J. Erle. 2005. Dissecting asthma using focused transgenic modeling and functional genomics. J. Allergy Clin. Immunol. 116:305-311. [DOI] [PubMed] [Google Scholar]
- 23.Kurt-Jones, E. A., L. Popova, L. Kwinn, L. M. Haynes, L. P. Jones, R. A. Tripp, E. E. Walsh, M. W. Freeman, D. T. Golenbock, L. J. Anderson, and R. W. Finberg. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398-401. [DOI] [PubMed] [Google Scholar]
- 24.Liu, P., M. Jamaluddin, K. Li, R. P. Garofalo, A. Casola, and A. R. Brasier. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 81:1401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNamara, P. S., and R. L. Smyth. 2002. The pathogenesis of respiratory syncytial virus disease in childhood. Br. Med. Bull. 61:13-28. [DOI] [PubMed] [Google Scholar]
- 26.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135-145. [DOI] [PubMed] [Google Scholar]
- 27.Meissner, H. C. 2003. Selected populations at increased risk from respiratory syncytial virus infection. Pediatr. Infect. Dis. J. 22:S40-S44. [DOI] [PubMed] [Google Scholar]
- 28.Miller, A. L., T. L. Bowlin, and N. W. Lukacs. 2004. Respiratory syncytial virus-induced chemokine production: linking viral replication to chemokine production in vitro and in vivo. J. Infect. Dis. 189:1419-1430. [DOI] [PubMed] [Google Scholar]
- 29.Player, M. R., and P. F. Torrence. 1998. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Ther. 78:55-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puthothu, B., M. Krueger, J. Forster, and A. Heinzmann. 2006. Association between severe respiratory syncytial virus infection and IL13/IL4 haplotypes. J. Infect. Dis. 193:438-441. [DOI] [PubMed] [Google Scholar]
- 31.Ramaswamy, M., L. Shi, M. M. Monick, G. W. Hunninghake, and D. C. Look. 2004. Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am. J. Respir. Cell Mol. Biol. 30:893-900. [DOI] [PubMed] [Google Scholar]
- 32.Ramaswamy, M., L. Shi, S. M. Varga, S. Barik, M. A. Behlke, and D. C. Look. 2006. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344:328-339. [DOI] [PubMed] [Google Scholar]
- 33.Rudd, B. D., E. Burstein, C. S. Duckett, X. Li, and N. W. Lukacs. 2005. Differential role for TLR3 in respiratory syncytial virus-induced chemokine expression. J. Virol. 79:3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlender, J., V. Hornung, S. Finke, M. Günthner-Biller, S. Marozin, K. Brzózka, S. Moghim, S. Endres, G. Hartmann, and K.-K. Conzelmann. 2005. Inhibition of Toll-like receptor 7- and 9-mediated alpha/beta interferon production in human plasmacytoid dendritic cells by respiratory syncytial virus and measles virus. J. Virol. 79:5507-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simoes, E. A., and X. Carbonell-Estrany. 2003. Impact of severe disease caused by respiratory syncytial virus in children living in developed countries. Pediatr. Infect. Dis. J. 22:S13-S18. [DOI] [PubMed] [Google Scholar]
- 36.Spann, K. M., K.-C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sparer, T. E., S. Matthews, T. Hussell, A. J. Rae, B. Garcia-Barreno, J. A. Melero, and P. J. Openshaw. 1998. Eliminating a region of respiratory syncytial virus attachment protein allows induction of protective immunity without vaccine-enhanced lung eosinophilia. J. Exp. Med. 187:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tal, G., A. Mandelberg, I. Dalal, K. Cesar, E. Somekh, A. Tal, A. Oron, S. Itskovich, A. Ballin, S. Houri, A. Beigelman, O. Lider, G. Rechavi, and N. Amariglio. 2004. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J. Infect. Dis. 189:2057-2063. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, G., E. J. Stott, M. Hughes, and A. P. Collins. 1984. Respiratory syncytial virus infection in mice. Infect. Immun. 43:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y., M. Jamaluddin, S. Wang, B. Tian, R. P. Garofalo, A. Casola, and A. R. Brasier. 2003. Ribavirin treatment up-regulates antiviral gene expression via the interferon-stimulated response element in respiratory syncytial virus-infected epithelial cells. J. Virol. 77:5933-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Y., B. A. Luxon, A. Casola, R. P. Garofalo, M. Jamaluddin, and A. R. Brasier. 2001. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75:9044-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.