Abstract
Baculovirus DNA binding protein (DBP) binds preferentially single-stranded DNA in vitro and colocalizes with viral DNA replication sites. Here, its putative role as viral replication factor has been addressed by RNA interference. Silencing of DBP in Autographa californica multiple nucleopolyhedrovirus-infected cells increased expression of LEF-3, LEF-4, and P35. In contrast, expression of the structural genes coding for P39 and polyhedrin was suppressed while expression of genes coding for P10 and GP64 was unaffected. In the absence of DBP, viral DNA replication sites were formed, indicating replication of viral DNA. Electron microscopy studies, however, revealed a loss of formation of polyhedra and virus envelopment, suggesting that the primary role of DBP is viral formation rather than viral DNA replication.
The baculovirus DNA binding protein (DBP) has been identified in cells infected with Bombyx mori nucleopolyhedrovirus (BmNPV) as a single-stranded DNA binding protein that is able to unwind partial DNA duplexes in an in vitro system (17). Both the BmNPV dbp and the homologous gene of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) with 96% nucleotide sequence identity colocalize with DNA replication sites (15, 20). Thus, DBP has been suggested to play a role in viral DNA replication.
To address the function of DBP, we performed RNA silencing experiments with AcMNPV-infected Spodoptera frugiperda IPLB-SF21 cells. Previous studies demonstrated the efficiency of RNA interference to knock down baculoviral genes and study their impact during the viral infection cycle (4, 11, 16, 22). To silence the dbp gene, specific double-stranded RNA (dsRNA) was prepared using the T7 RiboMax Express RNAi system (Promega). The cloned cDNA of the AcMNPV dbp gene served as a DNA template to generate dsRNA corresponding to 609 bp at the 5′ terminus of dbp (15) (primer 1a, 5′-GGGTAATACGACTCACTATAGGATGGCAACTAAA CGCAAG-3′; primer 1b, 5′-GGGCACACGTTTGGTTCCAT-3′; primer 2a, 5′-GGGTAATACGACTCACTATAGGGCACACGTTTGGTTCCAT-3′; and primer 2b, 5′-GGATGGCAACTAAACGCAAG-3′). As a control, we used dsRNA corresponding to a 600-bp fragment of the green fluorescent protein (GFP) gene as described recently (11). S. frugiperda cells were transfected with 20 μg of DBP dsRNA or 5 μg of GFP dsRNA as calcium phosphate precipitates (BD BaculoGold) followed by infection with AcMNPV (10 PFU/cell).
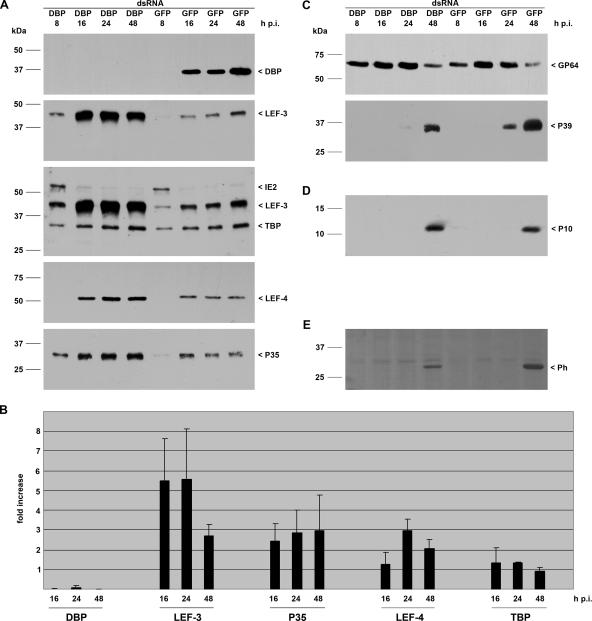
Immunoblot analysis was used to analyze the efficiency of RNA silencing. Detergent-based nuclear and cytoplasmic extracts were prepared at 8, 16, 24, and 48 h postinfection (p.i.) as described previously (19). The polyclonal antiserum directed against BmNPV DBP (20) recognized a polypeptide of about 34 kDa from 16 through 48 h p.i. in nuclear extracts of the cells transfected with control GFP dsRNA. In contrast, no DBP expression was visible upon transfection of DBP dsRNA, indicating efficient silencing of dbp (Fig. 1A).
FIG. 1.
Effect of dbp silencing on viral gene expression. S. frugiperda cells were transfected with either DBP dsRNA or GFP dsRNA. Cells were subsequently infected with AcMNPV (10 PFU/cell) at 20 h posttransfection, and detergent-based nuclear and cytoplasmic extracts were prepared from infected cells 8, 16, 24, and 48 h p.i. Proteins were resolved on sodium dodecyl sulfate-10% polyacrylamide gels and transferred to nitrocellulose (A, C, and E) or resolved on sodium dodecyl sulfate-15% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Amersham) (D). (A) DBP was stained with rabbit anti-DBP antiserum (20); early gene expression was analyzed with rabbit antisera raised against LEF-3 (3), IE2 (13), LEF-4 (11), or P35 (9); and expression of the cellular TBP was determined with rabbit anti-Sf/TBP antiserum (clone 3890; I. Quadt, unpublished results). (B) Three independent experiments were quantified by ImageJ software, and the increase (fold) in signal intensity in cells with suppressed DBP compared to the level in control cells is given. (C) Structural gene expression was analyzed with anti-GP64 (AcV5) (10) or anti-P39 (P10C6) (25) mouse monoclonal antibodies. (D) Expression of the very late protein P10 was detectable with rabbit anti-P10 serum (23). (E) Polyhedrin was viewed by Ponceau staining. Expression of DBP, LEF-3, IE2, TBP, LEF-4, P39, P10, and polyhedrin was analyzed on samples prepared from nuclear protein extracts, and P35 and GP64 expression was detected in cytosolic extracts that were prepared as described previously (19). Protein size markers are shown on the left, and the identities of the viral proteins are indicated on the right.
To study the effect of DBP suppression on viral gene expression, we analyzed expression of early genes. The immediate-early protein IE2 is detectable from 2 until 12 to 24 h p.i. in S. frugiperda cells (13). Here, we observed IE2 in cells transfected with either GFP dsRNA or DBP dsRNA at 8 h p.i. declining at 16 h p.i. (Fig. 1A). While no effect on the IE2 level was found, the loss of DBP correlated with increased expression of LEF-3, LEF-4, and P35 (Fig. 1A). Quantitation revealed up to fivefold more LEF-3 in the absence of DBP as compared to LEF-3 levels in cells transfected with GFP dsRNA. The effect was most prominent at 16 and 24 h p.i. and decreased at 48 h p.i. The relative increase of p35 and LEF-4 was lower than that of LEF-3. P35 was approximately threefold higher at all times tested, while LEF-4 was highest at 24 h p.i.
To determine whether the effect was specific for viral proteins, we also monitored the cellular TATA-binding protein (TBP). TBP levels increase late in infection, and TBP colocalizes with viral DNA replication centers (21). The loss of DBP, however, influenced neither the increased TBP level during late phases of infection nor the expression level compared to those in cells transfected with GFP dsRNA (Fig. 1A and B).
LEF-3, like DBP, is a single-stranded DNA binding protein (3, 7, 14, 17). In vitro assays revealed multiple functions of LEF-3, suggesting a role in viral replication, repair, and recombination of the viral genome (18). LEF-3, but not DBP, is essential for DNA replication in transient assays (12). The two other factors influenced by the loss of DBP are not directly involved in viral DNA replication. LEF-4, a component of the viral RNA polymerase, is essential for late gene expression as well as for production of viral progeny (6, 11). P35 acts as an apoptotic suppressor and is required to prevent apoptosis in AcMNPV-infected S. frugiperda cells (1, 2, 8). Whether the increased levels of LEF-3, LEF-4, and P35 are a direct effect of DBP silencing has still to be shown. In addition, we cannot exclude effects on further viral genes, since availability of antibodies limits the analysis. Overexpression studies might shed light on the impact of increased LEF-3, LEF-4, and P35 on infection.
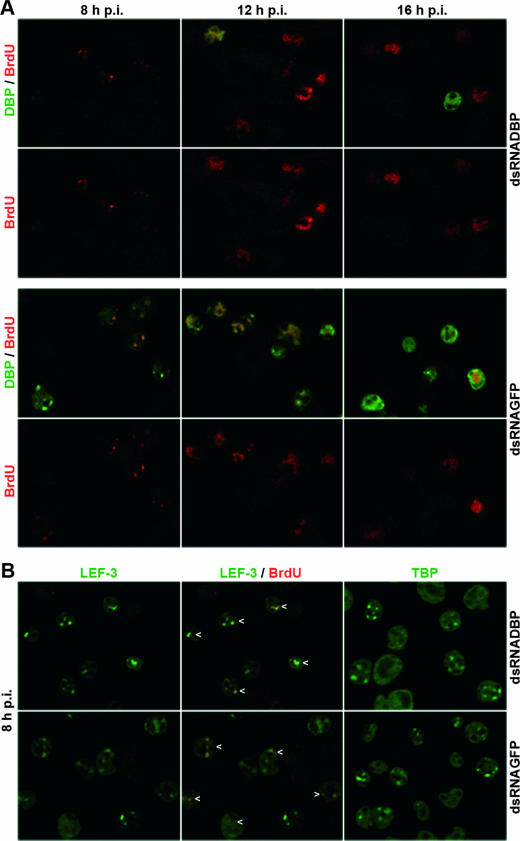
DBP and LEF-3 localize in viral DNA replication centers, and the formation of distinct DBP and LEF-3 sites is thought to be coupled to the initiation of viral DNA replication (15, 20). Hence, we analyzed whether replication sites are still formed in cells with suppressed DBP and increased levels of LEF-3. Nuclear DNA replication sites were visualized after incorporation of 5-bromo-2′-deoxyuridine (BrdU) by staining with an anti-BrdU antibody (15). Indirect immunofluorescence studies of AcMNPV-infected S. frugiperda cells treated with GFP dsRNA confirmed the presence of distinct DNA replication foci at 8 h p.i. The foci enlarged and fused at 12 h p.i., eventually occupying most of the nucleus at 16 h p.i. (Fig. 2A) (15). Confocal images indicated colocalization of DBP and BrdU in early and enlarged replication sites, confirming recent results in AcMNPV-infected TN-368 cells (Fig. 2A) (15). By 16 h p.i., DBP staining still corresponded to replication sites, although it was also observed at the rim of nuclei as described recently (Fig. 2A) (15).
FIG. 2.
Colocalization of DBP, LEF-3, and TBP with viral DNA replication sites upon dbp silencing in AcMNPV-infected S. frugiperda cells. Cells were transfected with either DBP dsRNA or GFP dsRNA, infected with AcMNPV (10 PFU/cells) at about 20 h posttransfection, and analyzed at 8, 12, and 16 h p.i. Cells were fixed in 2% paraformaldehyde and permeabilized with 0.1% Triton X-100; viral DNA synthesis was visualized by adding thymidine analogue BrdU to the cell medium 1 h prior to fixation. (A) Fixed cells were costained with mouse monoclonal anti-BrdU antibody (1:50 diluted) (clone B44; Becton Dickinson) and rabbit anti-DBP antiserum (1:4,000 diluted) (20). (B) Cells fixed at 8 h p.i. were costained with mouse monoclonal anti-BrdU antibody and with rabbit anti-LEF-3 antiserum (1:2,000 diluted) (3) or stained with rabbit anti-Sf/TBP antiserum (1:1,000 diluted) (clone 3890; I. Quadt, unpublished results). Rabbit antisera were visualized with Alexa Fluor 488-conjugated anti-rabbit immunoglobulin G (Molecular Probes) diluted 1:2,000 (green), and mouse monoclonal antibodies were visualized with Alexa Fluor 555-conjugated anti-mouse immunoglobulin G (Molecular Probes) diluted 1:2,000 (red). Specimens were mounted and viewed under a Leica DM RE microscope linked to a Leica SP/2 confocal unit as described previously (15). Images were captured with a 63× objective and a 1.32 NA. Confocal sections and merges of confocal sections are shown. Arrowheads mark colocalization of LEF-3 and BrdU.
In cells treated with DBP dsRNA, the formation of replication foci and their increase in size and intensity exactly paralleled those in the control cells, suggesting that viral DNA replication was relatively normal in the absence of DBP (Fig. 2A). As compared to control cells, DBP was observed in nine-times-fewer infected cells, which most likely represented those cells that were not transfected with DBP dsRNA (data not shown).
To further characterize the DNA replication sites, we analyzed the localization of LEF-3 and TBP, which both colocalize with viral DNA replication sites (15, 21). At 8 h p.i. LEF-3 was found in distinct nuclear dots that colocalized with DNA replication sites in both DBP-suppressed and control cells (Fig. 2B). Quantitation revealed approximately the same number of cells with LEF-3 and BrdU foci in GFP and DBP dsRNA-treated cells (data not shown). In addition, the loss of DBP did not influence the localization of TBP in nuclear dots which have been shown to resemble early viral replication sites (Fig. 2B) (21). Our results further support that viral DNA replication sites are formed in the absence of DBP and irrespectively of increased LEF-3 levels.
To investigate whether silencing of DBP affected structural genes and production of viral progeny, we initially analyzed expression of late and very late genes. Immunoblots revealed no effect on the accumulation of the envelope protein GP64 or on the very late protein P10 in cells with suppressed DBP (Fig. 1C and D). Interestingly, levels of the capsid protein P39 and the very late polyhedrin protein were reduced in DBP-suppressed cells compared to control cells (Fig. 1C and E).
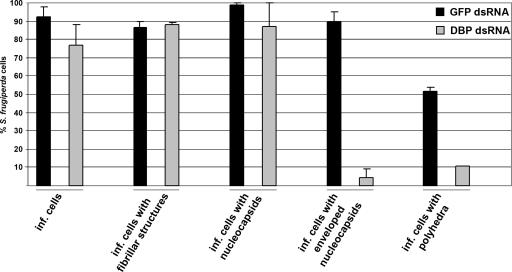
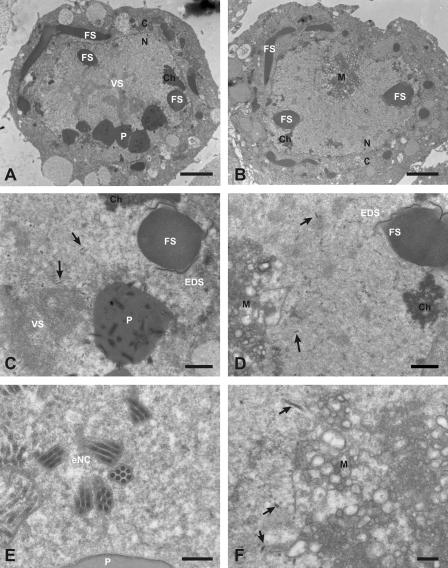
The reduction in capsid and polyhedrin proteins suggested that virus assembly may be affected by the suppression of DBP. Thus, electron microscopy (EM) studies were performed. Cells transfected with either GFP or DBP dsRNAs were infected and fixed with 2% glutaraldehyde-4% paraformaldehyde at 48 h p.i., followed by standard embedding, thin sectioning, and staining for EM analysis as described previously (23). Based on the presence of nucleocapsids and/or fibrillar structures, the numbers of infected cells were comparable in DBP-suppressed and control cells (see Fig. 4). Furthermore, no alterations were observed in the formation of fibrillar structures, which are composed of P10 and are localized in the cytoplasm as well as in the nucleus (Fig. 3A and B). The number of cells with polyhedra, however, was significantly lower in DBP-suppressed cells (Fig. 3A to D and Fig. 4). These findings correlate with the unchanged protein levels of P10 and the reduction of polyhedrin in DBP-suppressed cells (Fig. 1D and E). While nucleocapsids were present in all cells, enveloped virions were only detectable in a few cells, which likely represent those cells that did not take up DBP dsRNA (Fig. 3E and F and Fig. 4). Interestingly, membranous aggregates appeared in DBP-suppressed cells, in addition to alterations of the virogenic stroma, suggesting that silencing of DBP interferes with envelopment of nucleocapsids (Fig. 3C, D, and F).
FIG. 4.
Quantification of the effects of suppressed DBP. About 50 cells transfected with either GFP dsRNA or DBP dsRNA followed by infection (inf.) were investigated at 48 h p.i. Results are the mean + absolute error from two independent experiments.
FIG. 3.
EM analysis of the effects of suppressed DBP in AcMNPV-infected S. frugiperda cells. Cells were transfected with either DBP dsRNA or GFP dsRNA, infected with AcMNPV (10 PFU/cells), and processed for EM at 48 h p.i. as described previously (23). White letters indicate viral structures (P, polyhedra; FS, fibrillar structures; VS, virogenic stroma; EDS, electron-dense spacer; eNC, enveloped nucleocapsids), black letters indicate cellular structures (C, cytoplasm; N, nucleus; Ch, chromatin; M, membranous aggregate), and arrows indicate nucleocapsids. Cells transfected with GFP dsRNA are shown on the left (A, C, and E), and cells transfected with DBP dsRNA are shown on the right (B, D, and F). (A and B) Overview of infected cells. (C) Detail of the nucleus shown in panel A. (F) Detail of the nucleus shown in panel B. Bars are 2 μm in panels A and B, 0.5 μm in panels C and D, and 0.2 μm in panels E and F. Samples were examined and photographed with a Philips CM12 electron microscope.
In summary, our results support an essential role of DBP for the production of viral progeny. The EM studies demonstrate the loss of polyhedra and enveloped nucleocapsids in DBP-suppressed cells, while fibrillar structures and nucleocapsids were still detectable. The nucleocapsids had a dense core, indicating that they are filled with DNA (5); yet, it is still an open question as to whether nucleocapsids were properly assembled. At this point, we do not know whether the decrease in enveloped nucleocapsids is due to a block at envelopment or to misassembly of the nucleocapsids. In this context, it is interesting that DBP was not detected in budded or in occlusion-derived virions of BmNPV, suggesting that DBP is not associated with mature viral DNA, at least in BmNPV (20). Taken together, the results of our studies suggest that DBP plays an essential role during virus assembly. Although DBP is found in DNA replication sites, its absence does not affect the formation of these sites.
Acknowledgments
We thank Keiju Okano for the antiserum against BmNPV DBP, George F. Rohrmann for anti-LEF-3 antiserum, Linda Guarino for anti-LEF-4 antiserum, Paul D. Friesen for anti-p35 antiserum, Gary W. Blissard for monoclonal antibody gp64 (AcV5), Loy Volkman for monoclonal antibody p39 (P10C6), and Monique van Oers for anti-p10 antiserum.
This research was supported by grant KN536/11-1 from the Deutsche Forschungsgemeinschaft and the Köln Fortune Program/Faculty of Medicine, University of Cologne.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Clem, R. J., and L. K. Miller. 1993. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J. Virol. 67:3730-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clem, R. J., M. Fechheimer, and L. K. Miller. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388-1390. [DOI] [PubMed] [Google Scholar]
- 3.Evans, J. T., and G. F. Rohrmann. 1997. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J. Virol. 71:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores-Jasso, C. F., V. J. Valdes, A. Sampieri, V. Valadez-Graham, F. Recillas-Targa, and L. Vaca. 2004. Silencing structural and nonstructural genes in baculovirus by RNA interference. Virus Res. 102:75-84. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, M. J. 1986. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J. Ultrastruct. Mol. Struct. Res. 95:189-195. [Google Scholar]
- 6.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 69:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershberger, P. A., J. A. Dickson, and P. D. Friesen. 1992. Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J. Virol. 66:5525-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershberger P. A., D. J. LaCount, and P. D. Friesen. 1994. The apoptotic suppressor P35 is required early during baculovirus replication and is targeted to the cytosol of infected cells. J. Virol. 68:3467-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125:432-444. [DOI] [PubMed] [Google Scholar]
- 11.Knebel-Mörsdorf, D., I. Quadt, Y. Li, L. Montier, and L. Guarino. 2006. Expression of baculovirus late and very late genes depends on LEF-4, a component of the viral RNA polymerase whose guanyltransferase function is essential. J. Virol. 80:4168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kool, M., C. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krappa, R., R. Roncarati, and D. Knebel-Mörsdorf. 1995. Expression of PE38 and IE2, viral members of the C3HC4 finger family, during baculovirus infection: PE38 and IE2 localize to distinct nuclear regions. J. Virol. 69:5287-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y., A. L. Passarelli, and L. K. Miller. 1993. Identification, sequence, and transcriptional mapping of lef-3, a baculovirus gene involved in late and very late gene expression. J. Virol. 67:5260-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mainz, D., I. Quadt, and D. Knebel-Mörsdorf. 2002. Nuclear IE2 structures are related to viral DNA replication sites during baculovirus infection. J. Virol. 76:5198-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Means, J. C., I. Muro, and R. J. Clem. 2003. Silencing of the baculovirus Op-iap3 gene by RNA interference reveals that it is required for prevention of apoptosis during Orgyia pseudotsugata M nucleopolyhedrovirus infection of Ld652Y cells. J. Virol. 77:4481-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikhailov, V. S., A. L. Mikhailova, M. Iwanaga, S. Gomi, and S. Maeda. 1998. Bombyx mori nucleopolyhedrovirus encodes a DNA-binding protein capable of destabilizing duplex DNA. J. Virol. 72:3107-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikhailov, V. S., K. Okano, and G. F. Rohrmann. 2006. Structural and functional analysis of the baculovirus single-stranded DNA-binding protein LEF-3. Virology 346:469-478. [DOI] [PubMed] [Google Scholar]
- 19.Murges, D., I. Quadt, J. Schroer, and D. Knebel-Mörsdorf. 2001. Dynamic nuclear localization of the baculovirus proteins IE2 and PE38 during the infection cycle: the promyelocytic leukemia protein colocalizes with IE2. Exp. Cell Res. 264:219-232. [DOI] [PubMed] [Google Scholar]
- 20.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quadt, I., D. Mainz, R. Mans, A. Kremer, and D. Knebel-Mörsdorf. 2002. Baculovirus infection raises the level of TATA-binding protein that colocalizes with viral DNA replication sites. J. Virol. 76:11123-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdes, V. J., A. Sampieri, J. Sepulveda, and L. Vaca. 2003. Using double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J. Biol. Chem. 21:19317-19324. [DOI] [PubMed] [Google Scholar]
- 23.van Lent, J. W. M., J. T. Groenen, E. C. Klinge-Roode, G. F. Rohrmann, D. Zuidema, and J. M. Vlak. 1990. Localization of the 34 kDa polyhedron envelope protein in Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. Arch. Virol. 111:103-114. [DOI] [PubMed] [Google Scholar]
- 24.van Oers, M. M., J. T. M. Flipsen, C. B. E. M. Reusken, and J. M. Vlak. 1994. Specificity of baculovirus p10 functions. Virology 200:513-523. [DOI] [PubMed] [Google Scholar]
- 25.Whitt, M. A., and J. S. Manning. 1988. A phosphorylated 34-kDa protein and a subpopulation of polyhedron are thiol-linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology 163:33-42. [DOI] [PubMed] [Google Scholar]