Abstract
Geminiviruses have circular single-stranded DNA genomes and are important pathogens in tropical and subtropical regions, but their population diversity and variability are poorly understood. Here, we have investigated variations accumulating in Tomato yellow leaf curl China virus (TYLCCNV), a geminivirus in the genus Begomovirus of the family Geminiviridae. The population variation was analyzed in a naturally infected tomato (Solanum lycopersicom) plant and in Nicotiana benthamiana and tomato plants experimentally infected with a swarm of TYLCCNV DNA clones to provide an identical sequence for initiation of infection. Our results demonstrate that the population of TYLCCNV in a naturally infected tomato plant was genetically heterogeneous and that rapid mutation occurred in the populations amplified from N. benthamiana and tomato plants that had been infected with cloned DNA. This feature of the population of TYLCCNV in these plants consisted of the consensus sequence and a pool of mutants that are not identical but are closely related to the consensus sequence, and it coincides with the quasispecies concept described for many RNA viruses. The mutation frequency was circa 10−4 in N. benthamiana and tomato at 60 days postinoculation, a value comparable to that reported for plant RNA viruses. The quasispecies-like nature of the TYLCCNV populations suggested that TYLCCNV is capable of rapid evolution and adaptation in response to changing agricultural practices.
Virus-resistant crops, characterized by their resistance to a particular strain of a virus, offer one of the most cost-effective strategies for the management of plant viral diseases. However, the dynamic nature of virus populations permits the evolution of new strains that can adapt to increasingly changing agricultural practices. In particular, such variability should enable viruses with quasispecies pools to overcome crop resistance and subsequently result in epidemics of viral diseases (22). Thus, the effective use of virus-resistant cultivars and management of viral diseases requires a better understanding of the genetic structures and evolutionary trajectories of virus populations.
Previous research on plant viral population structures and evolution has mainly focused on plant RNA viruses (reviewed in references 9, 12, and 27-29). The populations of plant RNA viruses are intrinsically genetically heterogeneous, and such populations are designated as quasispecies. The quasispecies concept suggests that the population of a virus, even in a single replicating population, is essentially a collection of variants varying around the consensus sequence (6, 7).
Members of the genus Begomovirus of the family Geminiviridae, which have circular single-stranded DNA genomes, are transmitted by the whitefly Bemisia tabaci Genn. in a semipersistent circulating manner (26). Due to the worldwide increases in the population and distribution of the insect vector and global movement of plant materials, begomovirus-induced diseases have become a major constraint on crop production in tropical and subtropical regions (26, 35). Cassava mosaic disease and cotton leaf curl disease, which have caused severe crop losses in African countries and Pakistan, are typical recently emerging begomovirus diseases (reviewed in references 20, 26, and 35). Although the impacts of recombination and pseudorecombination on the evolution of begomoviruses and epidemics of begomovirus-induced diseases have been extensively documented (10, 25, 30-32, 43-44), the effects of mutation on begomovirus evolution have thus far not been documented extensively. Unlike RNA viruses, begomoviruses replicate their genomes inside the nucleus by using the host replication machinery (reviewed in references 14 and 16). Thus, these viruses were assumed to have higher replication fidelity and lower rates of mutation accumulation than RNA viruses (6, 26). However, the large number of species (>100) (8) and the continued reports of new species (3), as well as the high degree of genetic diversity within the species (13, 23, 24, 31, 38), suggest that begomoviruses have a high mutation rate and that they generate highly diverse populations in a short time. Isnard et al. (18) analyzed the genetic diversity of a single isolate population of Maize streak virus (MSV), a virus species in the genus Mastrevirus of the family Geminiviridae, and demonstrated that the population of MSV had a quasispecies structure (18). However, in contrast to plant RNA viruses, the available information about genetic variability is scant, and essentially no quantitative studies have analyzed the genetic structures and variability of begomovirus populations under controlled conditions.
In this study, we investigated the genetic structure and population variability of the Tomato yellow leaf curl China virus (TYLCCNV) populations in a naturally infected Solanum lycopersicom plant and in Nicotiana benthamiana and S. lycopersicom plants inoculated with a cloned TYLCCNV derivative. Our results show that the structure of the TYLCCNV population is quasispecies-like and that rapid accumulation of variation during infection of both plants generated diversity levels comparable to those reported for plant RNA viruses.
MATERIALS AND METHODS
Viral sample source.
Naturally infected tomato plants with symptoms of TYLCCNV infection were collected from Yunnan Province, China, in 2003 (45), and the presence of the virus was later confirmed by PCR and enzyme-linked immunosorbent assay as described previously (4). The samples were stored at −80°C until they were used.
Plant, virus, and viral inoculation.
S. lycopersicom cv. Hongbaoshi and N. benthamiana plants were grown in an insect-free room with a constant temperature of 25°C and supplementary lighting for 16 h per day. Infectious clones of TYLCCNV isolate Y10 (pBinPLUS-Y10-1.7A) and its associated DNAβ (pBinPLUS-2β) were constructed previously (4). Plants at the six- or seven-leaf stage were inoculated by injecting the stems with a mixture of Agrobacterium tumefaciens strain EHA105 cultures carrying pBinPLUS-1.7A and pBinPLUS-2β as described previously (4). One newly emerging leaf with typical symptoms was taken from each plant at 60 days postinoculation (p.i.) and 120 days p.i. for population analysis.
Total DNA extraction and PCR amplification.
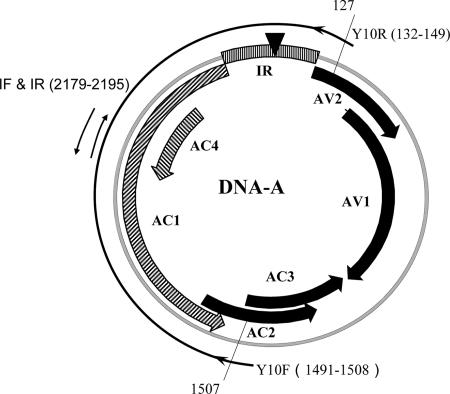
Total DNA was extracted from systemically infected leaves as described previously (42). The primers Y10F (5′-GCATCCACAAGGTAGGTC-3′) and Y10R (5′-CGATGAGCAGAGGATCCC-3′), corresponding to nucleotides (nt) 1491 to 1508 and nt 149 to 132 of TYLCCNV (AJ319675), respectively, were used to amplify a 1,396-bp fragment covering the intergenic region (IR) and the AC1 coding region (Fig. 1). The PCR mixture (50 μl) contained 1.5 to 3.0 μg of the total DNA extract, 0.5 μl Pfu DNA polymerase (5 U/μl; Biobasic Inc., Ontario, Canada), 0.6 μl 1.5 mM MgCl2, 5 μl 2 mM deoxynucleoside triphosphate, and 10 μl of a 5 mM primer mixture of Y10F and Y10R. PCRs were performed on a Minicycler (MJ Research, Inc., MA). After being preheated at 94°C for 3 min, the reaction mixtures were cycled 20 times at 94°C for 45 s, 50°C for 45 s, and 72°C for 1.5 min, followed by an extension at 72°C for 10 min.
FIG. 1.
Genome organization of TYLCCNV DNA-A. The region between the arrows of the outer line was analyzed in this study. The primers Y10F and Y10R were used to amplify the region, and the primers IF and IR were used in sequencing. The numbers represent the nucleotide positions.
Cloning and sequencing.
The PCR products generated from the viral DNA were A tailed, cloned into pGEM-T-Easy vector (Promega, Madison, WI), and transformed into Escherichia coli strain DH5α according to the manufacturer's instructions. About 20 clones were randomly picked from each viral population. The plasmid DNA was extracted from each clone, quantified, and sequenced using the Taq DyeDeoxy terminator cycle-sequencing kit (Applied Biosystems, CA). Sequencing primers included the M13 forward and reverse primers and a pair of primers, IF (5′-GATTGCCTCGGCATATG-3′, identical to nt 2179 to 2195) and IR (5′-CATATGCCGAGGCAATC-3′, complementary to nt 2179 to 2195), located in the middle of the amplified fragment (Fig. 1). About 20 nt at both the 5′ and 3′ ends of the fragment were excluded from sequence analysis. To minimize the potential variation resulting from PCR errors, we took the following measures: high-fidelity polymerase with proofreading capability was used in amplifications of viral DNA of progeny populations; a maximum amount of template DNA from which the target fragment could still be effectively amplified was used to avoid the bias that might result from a small subset of viruses; the PCR parameters were the same, and all components of PCR were from the same lot; and only 20 cycles for each PCR were carried out. In addition, the level of PCR error was determined by sequencing 15 clones generated from a PCR product amplified using the DNA of the infectious clone as a template. Only one of theses clones was shown to bear a single mutation. Thus, the PCR-associated mutation frequency was estimated to be 4.9 × 10−5 mutations per nt (Table 1).
TABLE 1.
Variation in progeny populations of TYLCCNV derived from the infection of identical sequence
| Plant | Days p.i. | % Mutated clones (no. mutated/total no. of clones sequenced) | No. of mutations/no. of bases sequenced | Mutation frequency |
|---|---|---|---|---|
| N. benthamiana | ||||
| 1 | 60 | 29 (7/24)a | 10/(1,358 × 24) | 3.1 × 10−4 |
| 2 | 60 | 33 (6/18)b | 8/(1,358 × 18) | 3.2 × 10−4 |
| 3 | 60 | 39 (7/18)c | 10/(1,358 × 18) | 4.1 × 10−4 |
| 1 | 120 | 50 (9/18)d | 13/(1,358 × 18) | 5.3 × 10−4 |
| Tomato | ||||
| 1 | 60 | 35 (7/20) | 11/(1,358 × 20) | 4.1 × 10−4 |
| Field sample (YN-33)e | 29 (4/14)f | 5/(1,358 × 14) | 2.6 × 10−4 | |
| PCR control | 6.7 (1/15) | 1/(1,358 × 15) | 4.9 × 10−5 |
Three identical mutants, each with the same two mutations at C127T and G2312A, were counted as a single mutant (C127T/G2312A).
Mutant C127T/G2312A was detected twice and was counted as a single mutant.
Mutants C127T/G2312A and G2718T were each detected twice and were counted as two mutants.
Three identical mutants (G2718T) were detected and were scored as a single mutant.
TYLCCNV-infected tomato was collected from Yunnan Province, China.
Only the variation within subpopulation II was calculated.
Determination of variability.
Each clone (amplified sequence) from each progeny population was compared with the parental sequence that initiated the infection. Each progeny clone with one or more bases different from the parental sequence was recorded as a mutated clone, and each change was counted as a mutation. Identical mutants detected in the same population were counted as a single unique mutant. The variability of the viral population was indicated by the percentage of mutated clones and the mutation frequency, which was calculated as the total number of mutations observed in all clones of each population divided by the total number of bases sequenced in the population. Testing for statistical significance of the mean of the mutation frequency was conducted by the nonparametric method of Kruskal-Wallis (40).
RESULTS
Rapid generation of genetic heterogeneity in TYLCCNV progeny populations.
The variability of the progeny populations of TYLCCNV in systemically infected N. benthamiana was evaluated at 60 days p.i. by amplifying a region of the TYLCCNV genome (Fig. 1) and sequencing 18 to 24 randomly selected viral clones. The percentages of mutated clones were 29, 33, and 39%, and the mutation frequencies were 3.1 × 10−4, 3.2 × 10−4, and 4.1 × 10−4, respectively, for the three populations in the experimental N. benthamiana plants (Table 1). Statistical tests, using the Kruskal-Wallis method, revealed that the variations for each progeny population of TYLCCNV in N. benthamiana were significantly higher than that generated from the PCR background. Therefore, approximately 9 out of 10 mutations detected in the progeny populations from infected N. benthamiana occurred during TYLCCNV genome replication. The levels of variation among the three progeny populations of TYLCCNV in N. benthamiana plants were not significantly different, suggesting that the population diversity analysis was reliable.
Analysis of population variation in N. benthamiana at 120 days p.i. indicated that the heterogeneous character of the TYLCCNV population did not change over the time course of infection and that the level of diversity increased at 120 days p.i., although the significance of the increase was not supported statistically (Table 1) (Kruskal-Wallis test; P > 0.05).
To determine that the mutation rates were not unusual features of TYLCCNV infection in N. benthamiana, we analyzed the heterogeneity of the TYLCCNV progeny population in an infected tomato plant at 60 days p.i. The results clearly showed that the TYLCCNV progeny population was genetically heterogeneous in the infected tomato plant, and the level of diversity, as indicated by the proportion of mutated clones (35%) and the mutation frequency (4.1 × 10−4), was not significantly different from that observed in N. benthamiana at 60 days p.i. (Table 1) (Kruskal-Wallis test; P > 0.05).
Genetic structure and variability of the TYLCCNV population in a naturally infected plant.
The genetic structure and variability of a TYLCCNV population in a naturally infected tomato (YN-33) was also analyzed. Based on the alignment analysis of 16 sequences of randomly amplified viral clones, we found that the TYLCCNV population contained two distinct clusters, designated as subpopulation I and subpopulation II. Subpopulation I consisted of 2 clones with identical sequences, whereas subpopulation II was composed of 14 clones. Ten of these clones had identical sequences (designated the consensus sequence), and the other four clones harbored five point mutations compared to the consensus sequence. The sequence identity between the consensus sequences of the two subpopulations was 92%, suggesting that they belonged to the same species based on current International Committee on Taxonomy of Viruses guidelines, which set a demarcation value of 89% nucleotide sequence identity between begomovirus DNA-A components (8). Because of the obvious divergence present between subpopulation I and subpopulation II, we assumed that the two subpopulations coexisting in the same plant tissue was the consequence of a mixed infection by two genetically distinct parental viruses. If we assume that the variations within subpopulation II resulted from the accumulation of mutations during the infection, the percentage of mutated clones and the mutation frequency in subpopulation II were 29% and 2.6 × 10−4, respectively. This value is not significantly different from that presented in the TYLCCNV progeny population of experimentally inoculated tomato plants at 60 days p.i. (Table 1) (Kruskal-Wallis test; P > 0.05). Interestingly, the TYLCCNV isolate Y10 (TYLCCNV-[Y10]) and subpopulation I had identical sequences, except for 1 nt difference (62A for TYLCCNV-[Y10] and 62G for subpopulation I), whereas subpopulation II was more divergent from TYLCCNV-[Y10].
The consensus sequence is stable, but the population composition is varied.
Comparison of the mutant spectra for TYLCCNV progeny populations in three N. benthamiana plants and one tomato plant indicated that the parental sequence from the infectious clone dominated in the populations at 60 days p.i. Thus, the population still represented the consensus sequence, and this status did not change during the time course of infection (Table 2). However, the mutant spectra for the populations at 60 days p.i. varied in each host plant, except for a few mutants that recurred in all populations. Most mutants present at 60 days p.i. were not detected at 120 days p.i. in the same plant; however, some new mutants appeared at 120 days p.i. (Table 2). This result indicates that the consensus sequence of TYLCCNV is stable but that the quasispecies composition varies in a manner similar to that of plant RNA virus populations (33-34).
TABLE 2.
Compositions of TYLCCNV progeny populations in TYLCCNV-infected N. benthamiana
| Populationa | Mutation
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Clone | Nucleotides
|
Genomic region | Amino acids (AC1/AC4)
|
|||||
| Original | Variant | Position | Original | Variant | Position | |||
| 60-1 | 2 | G | T | 27 | IR | NAb | NA | NA |
| 3 | G | A | 2266 | AC1/AC4 | D/T | D/M | 109/57 | |
| G | T | 2373 | AC1/AC4 | R/N | R/K | 74/21 | ||
| 4 | C | T | 127 | IR | NA | NA | NA | |
| 7 | G | T | 2718 | IR | NA | NA | NA | |
| 8, 11, 19 | C | T | 127 | IR | NA | NA | NA | |
| G | A | 2312 | AC1/AC4 | A/L | V/L | 94/42 | ||
| 12 | T | C | 1986 | AC1 | N | S | 203 | |
| G | C | 2526 | AC1 | L | V | 23 | ||
| 17 | G | T | 1972 | AC1 | A | A | 207 | |
| 60-2 | 1 | T | C | 2173 | AC1/AC4 | S/Q | S/R | 140/88 |
| 2, 6 | C | T | 127 | IR | NA | NA | NA | |
| G | A | 2312 | AC1/AC4 | A/L | V/L | 94/42 | ||
| 3 | G | T | 2718 | IR | NA | NA | NA | |
| 4 | G | A | 1807 | AC1 | D | D | 262 | |
| 5 | A | T | 1632 | AC1 | S | T | 321 | |
| 16 | C | T | 2025 | AC1 | D | N | 190 | |
| G | A | 2312 | AC1/AC4 | A/L | V/L | 94/42 | ||
| 60-3 | 4 | G | A | 1687 | AC1 | L | L | 302 |
| 6 | C | G | 2459 | AC1 | R | T | 45 | |
| 10 | C | T | 2268 | AC1/AC4 | D/G | N/G | 109/56 | |
| 11 | C | T | 2419 | AC1/AC4 | L/C | L/Y | 58/6 | |
| 12 | C | T | 127 | IR | NA | NA | NA | |
| C | T | 1706 | IR | NA | NA | NA | ||
| G | A | 2312 | AC1/AC4 | A/L | V/L | 94/42 | ||
| 13, 16 | C | T | 127 | IR | NA | NA | NA | |
| G | A | 2312 | AC1/AC4 | A/L | V/L | 94/42 | ||
| 15, 18 | G | T | 2718 | IR | NA | NA | NA | |
| 120-1 | 3 | G | A | 2589 | AC1 | P | S | 2 |
| 6 | G | A | 1575 | AC1 | L | F | 340 | |
| 7, 9, 13 | G | T | 2718 | IR | NA | NA | NA | |
| 10 | C | T | 1917 | AC1 | G | R | 226 | |
| 12 | G | T | 59 | IR | NA | NA | NA | |
| G | T | 88 | IR | NA | NA | NA | ||
| A | G | 2468 | AC1 | L | S | 42 | ||
| 14 | C | T | 127 | IR | NA | NA | NA | |
| G | A | 2312 | AC1/AC4 | A/L | V/L | 94/42 | ||
| 15 | G | A | 2535 | AC1 | H | Y | 20 | |
| 16 | G | T | 1972 | AC1 | A | A | 207 | |
| 18 | A | C | 1645 | AC1 | D | E | 316 | |
| G | T | 1650 | AC1 | L | I | 315 | ||
Plants were inoculated by agroinjection of an infectious clone with identical sequence. The progeny populations were analyzed at 60 days p.i. for three replicate plants and at 120 days p.i. for one replicate plant.
NA, not applicable.
Distribution and types of mutations.
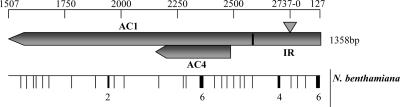
To determine whether the mutations were equally distributed along the sequenced region of the genome, the mutations in TYLCCNV populations from N. benthamiana at 60 days p.i. and 120 days p.i. were pooled, and their distribution is shown in Fig. 2. In general, the mutations occurred throughout the region regardless of the coding capacity. However, the mutation frequencies for the IR and AC1 coding regions were determined to be 6.5 × 10−4 and 3.2 × 10−4, respectively, if we assume that multiple occurrences of mutations at the same position in the same population are one mutation and that those in different populations are independent mutations and if those in close proximity are considered to be independent mutations. These results suggest that variability in the IR is about twice that of the AC1 coding region. In addition, open reading frame (ORF) AC1 completely overlaps ORF AC4, and the mutation frequencies within overlapping AC1-AC4 regions and the left nonoverlapping region were 4.8 × 10−4 and 2.6 × 10−4, respectively. Surprisingly, this suggests that there is a higher selective constraint on the nonoverlapping region than on the overlapping region (Fig. 2).
FIG. 2.
Distribution of mutations detected in progeny populations derived from a TYLCCNV clone. The region sequenced extends from base 1507, the final nucleotide in the AC1 reading frame, to base 127, the final nucleotide of the IR. Sites of mutations are indicated by vertical lines below the map. A line with a number indicates a position where more than one mutation occurred. The actual nucleotide positions of the mutations are shown in Table 2.
All mutations observed in the TYLCCNV progeny populations of N. benthamiana plants were substitutions. No insertion or deletion mutations were detected. The transitions of G to A (12 out of 41) and C to T (11 out of 41) were dominant for viral populations in N. benthamiana plants, whereas the reverse transitions (A to G and T to C) occurred rarely (Table 2). The transversion of G to T occurred commonly (11 out of 41), whereas the transversion of A to C and the reverse transversions of T to G and C to A were not detected (Table 2).
DISCUSSION
The genetic structure and population variability of TYLCCNV was evaluated by sequencing viral-progeny populations derived from infection with a cloned infectious DNA to provide identical parental sequences in N. benthamiana and tomato plants, and the progeny sequences were compared to those arising from a natural population of TYLCCNV. Our results clearly show that (i) TYLCCNV generated variants rapidly upon infection of both host plants to generate a heterogeneous population and (ii) the consensus sequence and diversity level in the population were stable as the infection progressed, whereas the mutant spectra of the population varied. These results demonstrate that the TYLCCNV population is a quasispecies, similar to those described for many RNA virus populations (1, 6-7, 27, 33-34). The levels of genetic variation of TYLCCNV observed in clonal progeny populations in N. benthamiana and tomato were 3.1 × 10−4 to 5.3 × 10−4, respectively, which is comparable to the variation reported for plant RNA viruses (33-34). The presence of high intrapopulation diversity in begomoviruses has been reported previously for naturally infected wild and cultivated hosts (23, 31). However, the population structure and diversity level of begomoviruses under controlled conditions have not been investigated. Hence, the present study is the first report to fill this gap, and our results verify the suggestion that populations of geminiviruses may occur as quasispecies (27, 35).
The high level of population diversity and the uniform distribution of mutations over the region of the genome analyzed in this study suggest that TYLCCNV has high mutation rates during viral-genome replication in vivo. Indeed, similar observations have also been reported for MSV, a geminivirus in the genus Mastrevirus (18). In that case, the observed mutation frequencies ranged from 3.8 × 10−4 to 10.5 × 10−4, depending on the host species, the homogeneity of inocula, and the periods of time the populations were maintained in the hosts (18). The population variation of TYLCCNV estimated in this study is also comparable to that of MSV.
The phenomenon of high population genetic diversity was also observed in a field population of Beet curly top virus, genus Curtovirus, family Geminiviridae (38). In addition, a high level of genetic diversity was reported in TT virus, a small nonenveloped, single-stranded, circular DNA virus of humans (39, 41), and in Canine parvovirus, a small single-stranded DNA virus of animals (36-37). The mutation frequency for a virus is determined by a combination of the intrinsic frequency of misincorporation and the capability for mismatch repair, as well as the extent of specific selection or stochastic drift resulting from genetic bottlenecks imposed on the virus population. The high mutation frequency associated with RNA viruses is presumed to be due in large part to their lack of proofreading capability during replication (6-7, 27). In comparison, geminiviruses replicate using their host DNA replication machinery (14, 16). Theoretically, these viruses should have less population variation. However, there is no information about the nature of plant DNA polymerase or the polymerase factors involved in the replication of geminiviruses. In particular, it is not known whether only a subset of cellular DNA replication and/or mismatch repair machinery is activated for geminivirus replication or whether the cellular environment affects the fidelity of those polymerases. Brough et al. (2) reported that DNA methylation inhibits the replication of tomato gold mosaic virus in tobacco protoplasts (2), implying that geminivirus DNA may not be methylated and that the normal mechanisms for mismatch repair probably do not operate during the tomato gold mosaic virus replication cycle (17). Thus, it is possible that the mechanisms of mismatch repair may function differently during geminivirus DNA and cellular DNA replication and that the lack of postreplication repair may be responsible for higher misincorporation in the geminivirus progeny DNA (27, 31, 35).
Analyses of the composition of the TYLCCNV progeny populations in both N. benthamiana and tomato indicate that the progenitor sequence is still dominant. This implies that the rule of genetic stability is obeyed by the TYLCCNV quasispecies, as is the case for quasispecies of plant RNA viruses (11-12, 33-34). Moreover, the minor sequences (excluding a few presumed mutational hot spots) were inconsistent in the TYLCCNV populations analyzed and varied as infection progressed (Table 2). In addition, the mutations must have occurred during TYLCCNV replication, and the mutation frequency in TYLCCNV should theoretically have increased in the quasispecies population as the infections progressed. However, no statistically significant diversity increase was observed for the TYLCCNV population in N. benthamiana at 120 days p.i. compared to 60 days p.i. (Tables 1 and 2), suggesting that some mechanisms must operate on the TYLCCNV population to reduce particular variants and to maintain the diversity at a certain level. For the plant RNA viruses, predominantly negative selection, bottlenecks arising during movement and transmission, and population differentiation during plant growth and development have been demonstrated to be responsible for maintaining population diversity (5, 9, 15, 19, 21). It is clear that selection must play an important role in TYLCCNV populations, since mutations based on the bias of coding capacity are not evenly distributed throughout the viral genome (Fig. 2). The presence and impact of bottleneck and population allocation on quasispecies diversity of plant DNA virus remains to be explored.
The mutants C127T/G2312A and G2718T were detected in all three N. benthamiana plants analyzed at 60 and 120 days p.i. These nucleotides are most likely the hot spots for misincorporation, based on the fact that the parental sequence still dominated in the various populations analyzed and that the ratio of these mutants in the 120-day-p.i. population did not increase obviously compared to the 60-day-p.i. population. The C-to-T change at 127 is 1 bp upstream of the ATG for AV1, which may impact promoter activity or translation of the AV1 mRNA. The G-to-A change at 2312 results in an atypical residue at position 94 in AC1. The G-to-T change at 2718 alters the sequence of one side the stem of the conserved hairpin. In future studies, we plan to reintroduce these mutations into the TYLCCNV genome to elucidate their functional consequences. Sequence variability in different genomic regions of TYLCCNV could reflect selective constraints or hot spots for misincorporation of those regions. The mutation frequency in the IR is higher than that in the AC1 coding region, suggesting that the IR is subjected to less-selective constraints. This result agrees with previous reports that the IR is a variable region for geminiviruses (32). ORF AC1 overlaps ORF AC4 completely. Theoretically, the overlapping region should be subjected to stricter evolutionary constraints than the nonoverlapping region and should be less variable. However, as indicated by the mutation frequency, the current results show that the AC1-AC4 overlapping region is more variable than the AC1 nonoverlapping region (4.8 × 10−4 versus 2.6 × 10−4). Higher variations in the AC1-AC4 overlapping region were also found in cotton leaf curl virus, and these were thought to be related to the mechanism by which the AC4 gene was generated (by overprinting) and the lack of its universal presence in geminiviruses (31).
With the introduction and spread of the begomovirus whitefly vector (B. tabaci B biotype), begomovirus diseases have posed serious threats to crop production in tropical and subtropical regions. These threats will continue to expand with the increase of global temperatures and the spread of the whitefly. The quasispecies-like nature of geminivirus populations clearly provides a mechanism for their rapid evolution and adaptation to ever-changing environments. Thus, a study of the genetic structure of a geminivirus and the determinants that affect its population evolution will require durable strategies to manipulate geminivirus-induced disease epidemics and to limit the emergence of these viruses.
Acknowledgments
We thank Marilyn J. Roossinck (The Samuel Roberts Noble Foundation, Oklahoma), Stephane Blanc (Biologie et Génétique des Interactions Plante-Parasite, CIRAD-INRA-ENSAM, France), and Andrew O. Jackson (Plant and Microbial Biology Department, University of California—Berkeley) for critical reviews of the manuscript. We also thank Xiaoguang Liu (University of Illinois at Chicago) for his helpful suggestions.
This work was supported by the National Key Basic Research and Development Program (2006CB101903) and the National Natural Foundation of Science of China (30470078 and 30530520).
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Ambrose, S., C. Hernandez, and R. Flores. 1999. Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J. Gen. Virol. 80:2239-2252. [DOI] [PubMed] [Google Scholar]
- 2.Brough, C. L., W. E. Gardiner, N. M. Inamdar, X.-Y. Zhang, M. Ehrlich, and D. M. Bisaro. 1992. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 18:702-712. [DOI] [PubMed] [Google Scholar]
- 3.Bull, S. E., R. W. Briddon, W. S. Sserubombwe, K. Ngugi, P. G. Markham, and J. Stanley. 2006. Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J. Gen. Virol. 87:3053-3065. [DOI] [PubMed] [Google Scholar]
- 4.Cui, X. F., X. R. Tao, Y. Xie, C. M. Fauquet, and X. P. Zhou. 2004. A DNAβ associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78:13966-13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietrich, C., and E. Maiss. 2003. Fluorescent labeling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J. Gen. Virol. 84:2871-2876. [DOI] [PubMed] [Google Scholar]
- 6.Domingo, E., and J. J. Holland. 1994. Mutation rates and rapid evolution of RNA viruses, p. 161-184. In S. S. Morse (ed.), The evolutionary biology of viruses. Raven Press, Ltd., New York, NY.
- 7.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 8.Fauquet, C. M., D. M. Bisaro, R. W. Briddon, J. K. Brown, B. D. Harrison, E. P. Rybicki, D. C. Stenger, and J. Stanley. 2003. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch. Virol. 148:405-421. [DOI] [PubMed] [Google Scholar]
- 9.French, R., and D. C. Stenger. 2003. Evolution of wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu. Rev. Phytopathol. 41:199-214. [DOI] [PubMed] [Google Scholar]
- 10.García-Andres, S., F. Monci, J. Navas-Castillo, and E. Moriones. 2006. Begomovirus genetic diversity in the native plant reservoir Solanum nigrum: Evidence for the presence of a new virus species of recombinant nature. Virology 350:433-442. [DOI] [PubMed] [Google Scholar]
- 11.García-Arenal, F., A. Fraile, and J. M. Malpica. 2003. Variation and evolution of plant virus populations. Int. Microbiol. 6:225-232. [DOI] [PubMed] [Google Scholar]
- 12.García-Arenal, F., A. Fraile, and J. M. Malpica. 2001. Variability and genetic structure of plant virus population. Annu. Rev. Phytopathol. 39:157-186. [DOI] [PubMed] [Google Scholar]
- 13.Gilbertson, R. L., M. R. Rojas, D. R. Russel, and D. P. Maxwell. 1991. Use of the asymmetric polymerase chain reaction and DNA sequencing to determine genetic variability of bean golden mosaic geminivirus in the Dominican Republic. J. Gen. Virol. 72:2843-2848. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez, C. 1999. Geminivirus DNA replication. Cell Mol. Life Sci. 56:313-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, J. S., R. French, T. J. Morris, and D. C. Stenger. 2001. Structure and temporal dynamics of populations within wheat streak mosaic virus isolates. J. Virol. 75:10231-10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 2000. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35:105-140. [PubMed] [Google Scholar]
- 17.Inamdar, N. M., X.-Y. Zhang, C. L. Brough, W. E. Gardiner, D. M. Bisaro, and M. Ehrlich. 1992. Transfection of heteroduplexes containing uracil-guanine or thymine-guanine mispairs into plant cells. Plant Mol. Biol. 20:123-131. [DOI] [PubMed] [Google Scholar]
- 18.Isnard, M., M. Granier, R. Frutos, B. Reynaud, and M. Peterschmitt. 1998. Quasispecies nature of three maize streak virus isolates obtained through different modes of selection from a population used to assess response to infection of maize cultivars. J. Gen. Virol. 79:3091-3099. [DOI] [PubMed] [Google Scholar]
- 19.Jridi, C., J. F. Martin, V. Marie-Jeanne, G. Labonne, and S. Blanc. 2006. Distinct viral populations differentiate and evolve independently in a single perennial host plant. J. Virol. 80:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legg, J. P., and C. M. Fauquet. 2004. Cassava mosaic geminiviruses in Africa. Plant Mol. Biol. 56:585-599. [DOI] [PubMed] [Google Scholar]
- 21.Li, H. Y., and M. J. Roossinck. 2004. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J. Virol. 78:10582-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansoor, S., I. Amin, S. Iram, M. Hussain, Y. Zafar, K. A. Malik, and R. W. Briddon. 2003. Breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. Plant Pathol. 52:784. [Google Scholar]
- 23.Ooi, K., S. Ohshita, I. Ishii, and T. Yahara. 1997. Molecular phylogeny of geminivirus infecting wild plants in Japan. J. Plant Res. 110:247-257. [Google Scholar]
- 24.Patil, B. L., S. Rajasubramaniam, C. Bagchi, and I. Dasgupta. 2005. Both Indian cassava mosaic virus and Sri Lankan cassava mosaic virus are found in India and exhibit high variability as assessed by PCR-RFLP. Arch. Virol. 150:389-397. [DOI] [PubMed] [Google Scholar]
- 25.Pita, J. S., V. N. Fondong, A. Sangare, G. W. Otim-Nape, S. Ogwal, and C. M. Fauquet. 2001. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82:655-665. [DOI] [PubMed] [Google Scholar]
- 26.Rojas, M. R., C. Hagen, W. J. Lucas, and R. L. Gibertson. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43:361-394. [DOI] [PubMed] [Google Scholar]
- 27.Roossinck, M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 28.Roossinck, M. J. 2003. Plant RNA virus evolution. Curr. Opinion Microbiol. 6:406-409. [DOI] [PubMed] [Google Scholar]
- 29.Roossinck, M. J., and W. L. Schneider. 2005. Mutant clouds and occupation of sequence space in plant RNA viruses, p. 337-348. In E. Domingo (ed.), Quasispecies. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 30.Rothenstein, D., D. Haible1, I. Dasgupta, N. Dutt, B. L. Patil, and H. Jeske. 2006. Biodiversity and recombination of cassava-infecting begomoviruses from southern India. Arch. Virol. 151:55-69. [DOI] [PubMed] [Google Scholar]
- 31.Sanz, A. I., A. Fraile, J. M. Gallego, J. M. Malpica, and F. Garcia-Arenal. 1999. Genetic variability of natural populations of cotton leaf curl geminivirus, a single-stranded DNA virus. J. Mol. Evol. 49:671-681. [DOI] [PubMed] [Google Scholar]
- 32.Sanz, A. I., A. Fraille, F. Garcia-Arenal, X. Zhou, D. J. Robinson, S. Khalid, T. Butt, and B. D. Harrison. 2000. Multiple infection, recombination and genome relationships among begomovirus isolates found in cotton and other plants in Pakistan. J. Gen. Virol. 81:1839-1849. [DOI] [PubMed] [Google Scholar]
- 33.Schneider, W. L., and M. J. Roossinck. 2000. Evolutionarily related Sindbis like plant viruses maintain different levels of population diversity in a common host. J. Virol. 74:3130-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider, W. L., and M. J. Roossinck. 2001. Genetic diversity in RNA viral quasispecies is controlled by host-virus interactions. J. Virol. 75:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seal, S. E., F. vandenBosch, and M. J. Jeger. 2006. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 25:23-46. [Google Scholar]
- 36.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 102:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shackelton, L. A., and E. C. Holmes. 2006. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J. Virol. 80:3666-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenger, D. C. 1995. Genotypic variability and occurrence of less than genome-length viral DNA forms in a field population of beet curly top geminivirus. Phytopathology 85:1316-1322. [Google Scholar]
- 39.Sugiyama, K., K. Goto, T. Ando, F. Mizutani, K. Terabe, and T. Yokoyama. 2001. Highly diverse TTV population in infants and their mothers. Virus Res. 73:183-188. [DOI] [PubMed] [Google Scholar]
- 40.Tang, Q. Y., and M. G. Feng. 2002. DPS—Data Process System. Science Press, Beijing, China.
- 41.Vasconcelos, H. C., S. A. Gomes, M. Cataldo, and C. Niel. 2003. Prevalence and genetic diversity of TT virus genotype 21 (YONBAN virus) in Brazil. Arch. Virol. 48:517-529. [DOI] [PubMed] [Google Scholar]
- 42.Xie, Y., X. P. Zhou, Z. K. Zhang, and Y. J. Qi. 2002. Tobacco curly shoot virus isolated in Yunnan is a distinct species of Begomovirus. Chin. Sci. Bull. 47:197-200. [Google Scholar]
- 43.Zhou, X., Y. Liu, L. Calvert, C. Munoz, G. W. Otim-Nape, D. J. Robinson, and B. D. Harrison. 1997. Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol. 78:2101-2111. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, X., Y. Liu, D. J. Robinson, and B. D. Harrison. 1998. Four DNA-A variants among Pakistan isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol. 79:915-923. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, X. P., Y. Xie, X. R. Tao, Z. K. Zhang, Z. H. Li, and C. M. Fauquet. 2003. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J. Gen. Virol. 84:237-247. [DOI] [PubMed] [Google Scholar]