Abstract
Autologous neutralizing antibodies (NAb) against human immunodeficiency virus type 1 generate viral escape variants; however, the mechanisms of escape are not clearly defined. In a previous study, we determined the susceptibilities of 48 donor and 25 recipient envelope (Env) glycoproteins from five subtype C heterosexual transmission pairs to NAb in donor plasma by using a virus pseudotyping assay, thereby providing an ideal setting to probe the determinants of susceptibility to neutralization. In the present study, acquisition of length in the Env gp120 hypervariable domains was shown to correlate with resistance to NAb in donor plasma (P = 0.01; Kendall's tau test) but not in heterologous plasma. Sequence divergence in the gp120 V1-to-V4 region also correlated with resistance to donor (P = 0.0002) and heterologous (P = 0.001) NAb. A mutual information analysis suggested possible associations of nine amino acid positions in V1 to V4 with NAb resistance to the donor's antibodies, and five of these were located within an 18-residue amphipathic helix (α2) located on the gp120 outer domain. High nonsynonymous-to-synonymous substitution (dN/dS) ratios, indicative of positive selection, were also found at these five positions in subtype C sequences in the database. Nevertheless, exchange of the entire α2 helix between resistant donor Envs and sensitive recipient Envs did not alter the NAb phenotype. The combined mutual information and dN/dS analyses suggest that unique mutational patterns in α2 and insertions in the V1-to-V4 region are associated with NAb resistance during subtype C infection but that the selected positions within the α2 helix must be linked to still other changes in Env to confer antibody escape. These findings suggest that subtype C viruses utilize mutations in the α2 helix for efficient viral replication and immune avoidance.
Circulating human immunodeficiency virus type 1 (HIV-1) strains belong to one of nine subtypes (A, B, C, D, F, G, H, J, and K) and at least 28 circulating recombinant forms (52; see http://www.hiv.lanl.gov/content/hiv-db/CRFs/CRFs.html for a current listing). Because subtype B infections predominate in developed regions of the world, the majority of studies characterizing the structure of gp120, as well as the interplay between HIV-1 and neutralizing antibodies (NAb), have been performed using samples collected from subtype B-infected subjects. More information is therefore needed about the serology of non-subtype B infections because of their pervasiveness in the AIDS pandemic (52, 57) and the urgent need for a vaccine that can induce cross-reactive immune responses. Subtype C viruses, which circulate predominantly in sub-Saharan Africa and India, are often refractory to neutralization by monoclonal antibodies raised from subtype B-infected subjects (4, 7, 27). Consequently, little is known about the mechanisms of autologous neutralization and escape in subtype C infection.
The HIV-1 genome encodes two glycoproteins; the transmembrane subunit gp41 anchors the Env complex in the viral membrane and mediates fusion with the host cell membrane, while the surface subunit gp120 facilitates interactions with receptor molecules (30). HIV-1 gp120 is heavily glycosylated and contains five “hypervariable” domains (V1 to V5) that tolerate sequence heterogeneity (47). The V1V2 and V4 domains, in particular, can also accommodate dramatic insertions, deletions, and variation in the pattern of N-linked glycosylation (N-Gly) sites. The V1V2 domain has been shown to influence sensitivity to NAb in studies of subtype B viruses (6, 13, 26, 33, 35, 36, 49-51, 54, 59, 63, 70, 71, 74) and, more recently, subtype C viruses (66). The V1V2 domain is thought to regulate neutralization sensitivity by masking conserved neutralization targets (39, 40, 44, 59, 66, 76) and can also present type-specific neutralization epitopes (20, 21, 23, 28, 32, 53, 58, 73). The mechanism by which V4 could influence neutralization is less clear, although this region could modulate glycan packing on the outer surface of the Env trimer (74).
There is evidence that distinct selective pressures are active against Env during infections with different subtypes (14, 22). In an analysis of subtype C sequences from the HIV Databases (http://www.hiv.lanl.gov), a high density of positions with nonsynonymous-to-synonymous substitution (dN/dS) ratios of >1 was located in the third conserved region (C3) of gp120, which is downstream in the linear sequence from the third hypervariable domain (V3) (22). High dN/dS ratios are indicative of strong diversifying selection (77). In contrast, high dN/dS ratios in subtype B sequences were concentrated within the boundaries of V3, which, in contrast, is relatively conserved in subtype C viruses (14, 22, 38). These findings suggest that the nature of NAb targets in the V3/C3 region differs between subtypes B and C (7, 24, 25, 46, 48, 74). Furthermore, we recently demonstrated that an amphipathic helix (α2) encoded in C3 exhibits mutational and structural differences in subtypes B and C (22a). Thus, the distributions of amino acid residues under selective pressure in the V3/C3 region are dramatically different in subtypes B and C, but the relationships between mutational patterns, viral subtypes, and sensitivity to autologous NAb have not been elucidated.
We previously demonstrated that Envs from chronically infected (donor) transmission partners exhibited a range of resistance against contemporaneous donor NAb in a pseudovirus assay, but the Envs from their newly infected recipient partners were uniformly sensitive to NAb in this plasma sample (18). Using sequence and NAb data generated in the previous study from 48 donor and 25 recipient Envs, we applied two statistical approaches to probe the determinants of neutralization sensitivity to donor plasma in the present study and used a biological assay to evaluate specific contributions. The results suggest that acquisition of length in the hypervariable domains and mutation of the α2 helix domain track with NAb resistance, but they demonstrate that the positions within the α2 helix must be linked to still other domains in Env that confer antibody escape. These findings suggest that the Env trimers of subtype C viruses utilize mutations in the α2 helix to escape NAb. This could be a pathway unique to subtype C viruses and is therefore important for development of a vaccine that induces potent and cross-reactive NAb responses.
MATERIALS AND METHODS
Sequence data.
Env sequences encompassing the V1-to-V4 region of gp120 (HXB2 residues 136 to 418) were generated from seven subtype C heterosexual transmission pairs and one subtype G heterosexual transmission pair enrolled in a discordant couple cohort in Lusaka, Zambia (18, 72). Evaluation of sensitivity to neutralization by plasma from the donor partner was performed previously for five of the subtype C transmission pairs (18, 72). Details of the cohort and sample collection have also been described previously (18, 72). Sequences are available from GenBank under accession numbers AY423908 to AY424198.
Determination of BL.
A maximum likelihood (ML) tree was constructed previously for each transmission pair (18), with gapped regions (including sequences in the first, second, and fourth hypervariable domains [V1V2 and V4]) stripped out of the analysis. Modeltest was used to identify the optimal evolutionary model (60), and for each tree, the model selected included base frequencies, a general reversible model for base substitution, and differences in rate variation at different sites assigned according to a gamma distribution with four categories of rates and invariant sites. A further comparison was made with a REV model with ML assigned rate variation at different sites, but the additional parameters were not justified in a likelihood ratio test for these data, and thus the Modeltest model was used. The specific value for each of the model parameters was estimated for each of the eight trees separately. PAUP was then used to construct a likelihood tree for each transmission pair. BranchLength.pl (B. Korber [www.santafe.edu/∼btk/science-paper/bette.html]; a web-based version of this code is now available at http://hcv.lanl.gov/content/hcv-db/BRANCHLEN/branchlength.html) was used to calculate the branch length (BL) to the patient's ancestral node for each donor and recipient sequence.
Correlation analysis.
A nonparametric Kendall tau rank correlation test was applied to previously described paired sequence and neutralization data (18) to determine whether the 50% inhibitory concentration (IC50) of plasma was correlated with the following different parameters: BL to the patient's ancestral node on the ML tree, amino acid length of the V1-to-V4 region, and the number of N-Gly sites in V1-to-V4. P values of <0.05 were considered statistically interesting for hypothesis forming as a foundation for further experimental exploration. The correlation tests were performed with the donor plasma IC50 and pooled plasma IC50 against each sequence attribute.
Mutual information analysis.
Shannon entropy is a measure of uncertainty used in communication theory (64). It can be applied to other problems where character states are considered, such as sequence analysis (37). For example, for position i in a protein alignment, the Shannon entropy function H(i) is defined by the probability of different amino acids, s, in that position, as follows: H(i) = −S·P(si)·log P(si). This is a measure of the uncertainty in the column of data; if only one amino acid appears in a column, it has zero entropy. Maximal entropy occurs when the frequencies of all of the characters are equal. Here we considered j a character state assigned according to the level of neutralization sensitivity associated with a V1-to-V4 sequence (a, IC50 of <50; b, IC50 of 50 to 249; c, IC50 of 250 to 999; d, IC50 of 1,000 to 4,999; and e, IC50 of >5,000). We asked whether j covaries with amino acid patterns found in any given position in an alignment. Given the joint probability distribution P(si,sj), H(i,j) = −S·P(si,sj)·log P(si,sj) and the mutual information distribution M(i,j) = H(i) + H(j) − H(i,j) (37), we measured how much the data covaries, given that the mutual information is 0 when either i and j are completely independent or there is no variation. To test the significance of these results, 10,000 randomizations of neutralization phenotype assignments to viral sequences were performed, and the number of times the mutual information for the random data was greater than or equal to the original data was tallied and divided by 10,000 to estimate the P value. This statistical approach is based on the assumption that the data are drawn from a random pool; however, the sequence data have strong underlying phylogenetic correlations (2). To partially compensate for this, the randomization of neutralization assignments to sequences was done within donor-recipient pairs; however, the associations identified must therefore still be considered with this limitation in mind and thus viewed in a hypothesis-forming framework to guide experiments and for comparison with other results, such as the dN/dS ratios. Hypervariable loops were omitted from this analysis because we could not exclude the possibility that alignment artifacts could occur in the association patterns.
Codon-specific dN/dS ratios.
Previously described data regarding codon-specific dN/dS ratios were adapted (22). Nonsynonymous mutations become fixed with greater or lesser probability than synonymous mutations depending on whether a site is subject to positive or purifying selection, and thus the dN/dS ratio can be used to characterize specific codons and regions of proteins evolving under positive selection (77). In viruses, regions under strong positive selection occur where immune escape confers a selective advantage to new variants that arise within a host. Accordingly, codon-based models of molecular evolution can be used in conjunction with sequence data and ML parameter estimation to identify sites of potential immunological interest. The dN/dS ratio is indicative of the selection pressure at the protein level, as follows: a dN/dS ratio of <1 is indicative of purifying selection and amino acid conservation because of structural and functional constraints, and a dN/dS ratio of >1 is indicative of diversifying, positive selection where amino acid substitutions confer an advantage. In this study, we asked whether or not the precise positions under positive selective pressure may be lineage specific for HIV-1, comparing subtype B and subtype C, as representatives of both lineages are under consideration as vaccine candidates in trial populations where the HIV-1 C clade dominates. The program CODEML (S8 [http://abacus.gene.ucl.ac.uk/software/paml.html]) was used to determine the regional distributions of dN/dS ratios in the V3 loop and flanking regions for two sets of 25 HIV-1 sequences, one for subtype B and the second for subtype C. The subtype C sequences used did not include those from the Zambian transmission pairs analyzed for neutralization sensitivity here. For each subtype, a three-rate prior with flexible locations and weights was fitted to the data. Bayes's formula was then used to calculate the posterior distribution of the rate ratio at each codon. The posterior mean was taken as a site-specific estimate of the dN/dS ratio.
Three-dimensional mapping.
The core structure used for mapping corresponds to the X-ray structure of CD4-bound YU2 gp120 (43) (PDB code 1RZK). Variable loops V1V2 and V3 were modeled for clarity as described previously (5). Discussions on an unliganded gp120 core structure were based on the fully glycosylated simian immunodeficiency virus (SIV) structure (12) (PDB code 2BF1). Signature positions were mapped onto this structure based on the alignment of sequences. Modeling calculations were performed using a modified version of AMBER (56). Three-dimensional images were generated using VMD (29).
Molecular dynamics simulations.
Comparative all-atom molecular dynamics simulations were carried out on the modeled gp120 to capture the local differences in the V3-C3-V4 regions of gp120 between B and C subtypes. The following three sets of simulations were carried out under identical conditions: (i) gp120 from YU2 (subtype B), (ii) YU2 gp120 with the α2 helix consensus sequence from subtype C, and (iii) gp120 with the subtype C consensus sequence. In all cases, the gp120 protein was solvated in ∼16,000 water molecules. The starting structure for subtype C gp120 was based on the YU2 model described above. The subtype C consensus was homology modeled from the subtype B structure, using MODELLER software (69). All molecular dynamics simulations were performed using the force field described by Cornell et al. (15) and the AMBER 6.0 suite of programs (9). A rectangular box of TIP3P water molecules (31) was added to solvate the complex, with the final system containing ∼55,000 atoms within a box dimension of ∼79 by 79 by 89 Å. Periodic boundary conditions were applied, and the particle mesh Ewald approach (16) was used to accurately treat electrostatic interactions. Both systems were minimized and equilibrated with the same protocol. The temperature was maintained at 300 K, using the Berendsen temperature algorithm (3). A 2-fs time step was used, and all bonds involving hydrogen atoms were constrained using the SHAKE algorithm (68). Simulations were carried out for approximately 4 ns, and the last 2 ns were considered for analysis. Additional details of the simulations and the results are provided in a separate publication (22a). The relative solvation between subtypes was obtained by calculating the number of water molecules in the first solvent shell surrounding a secondary structure (e.g., the α2 helix) or a specific residue. The contact distances between C-alpha carbons were used to identify interactions between regions or differences between the clades. A 10-Å cutoff between C-alpha carbons was used as a measure for contact.
Construction of chimeric Envs.
PCR amplification and cloning of the donor and recipient Envs from pairs 55 and 135 have been described previously (18). The env genes are in the cytomegalovirus-driven expression vector pCR3.1 (Invitrogen), which is used to generate viral pseudotypes. Each of the donor α2 helix domains shown in Fig. 3A was inserted into the corresponding recipient Env by using a strategy similar to that described previously (66). A DNA duplex corresponding to each donor α2 helix was synthesized as a double-stranded oligonucleotide by IDT Technologies (Coralville, IA) and gel purified to ensure that each fragment was of the correct length. These fragments were each blunt end ligated to an ∼8-kb PCR amplicon corresponding to the recipient Env, excluding the α2 helix and including the pCR3.1 vector sequences. The single-stranded DNA sequences corresponding to the 135 F and 55 M donor α2 helices (HXB2 nucleotides [nt] 7227 to 7279) were as follows: for pair 135, F42a (5′-AAAAAGCTATGGAATAACACTTTAGACGGGATAAAGGAAAAATTAGCAAGATAC-3′), F48a (5′-AAACAAAATTGGACTAAAACTTTAGAAGAGGTAAAGACAGAATTAAGAAAATAC-3′), and F67a (5′-AGACAGAAATGGAATAGCACTTTAGACGGGATAAAGGAAAAATTAGCAAGATAC-3′); and for pair 55, M7a (5′-GAACAAAAATGGAGTACAACTTTAAAAAGGGTAGAGAAAAAATTAAAAGAGCAC-3′), M4.1 (5′-GGAAAAGAATGGAATACAACTTTAACAAGGGTAAAGGAAAGATTAAAAAAGCAC-3′), and M2.13 (5′-AGAAAAAAATGGAATATAACTTTAGAAAGGGTAAAGGAAAGATTAAAAGAGCAC-3′).
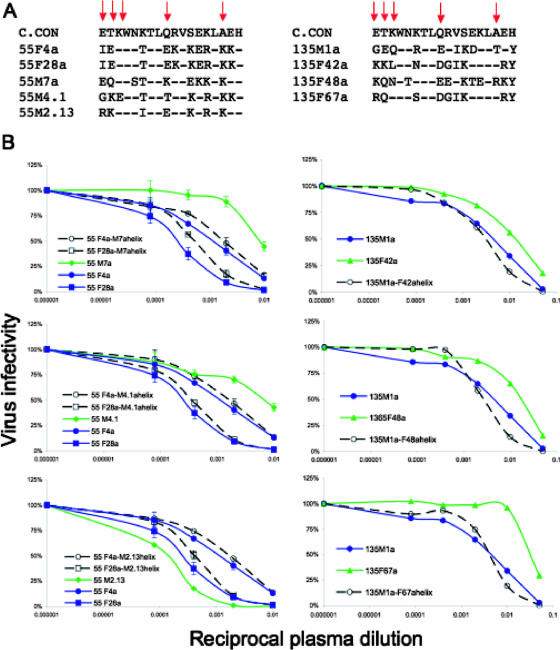
FIG. 3.
Neutralization sensitivities of donor-recipient α2 helix chimeras. (A) An alignment of the predicted amino acid sequences for the α2 helix region is shown for Envs from two transmission pairs (55 [male to female] and 135 [female to male]). The consensus subtype C sequence is shown at the top of each set of sequences. Red arrows show NAb signature residues 335, 336, 337, 343, and 350. The recipient sequences are shown below the consensus (55 F4a, 55 F28a, and 135 M1a). Two recipient Envs with different degrees of neutralization sensitivity were evaluated for pair 55. (B) The neutralization sensitivity of the parental and chimeric Env pseudotypes was evaluated using contemporaneous plasma from donor 55 M or 135 F. The percent virus infectivity relative to the control (lacking donor plasma) is plotted along the vertical axis, and the reciprocal plasma dilution of donor plasma (from 1:20 or 1:100 to 1:62,500) is plotted along the horizontal axis on a log10 scale. Each panel contains infectivity curves for the parental donor (green lines) and recipient (blue lines) Envs and the α2 helix-chimeric recipient Envs (dashed black lines). Infectivity curves for pair 55 and 135 chimeras are shown on the left and right, respectively.
For PCR amplification of the recipient Env (minus the helix) and pCR3.1 vector sequences, 5′-phosphorylated primers were synthesized and gel purified by IDT Technologies. Primer sequences for amplification of the recipient Env backbone (plus vector sequences) and their HXB2 locations were as follows: for pair 135, forward primer 5′-P-CTTCCCTGATAAAATAATAAAATTT-3′ (HXB2 nt 7280 to 7304) and reverse primer 5′-P-ACTAATGTTACAATGTGCTTGTCT-3′ (HXB2 nt 7203 to 7226), and for pair 55, forward primer 5′-P-TTCCCTAATAAAATAATAAAATTT-3′ (HXB2 nt 7281 to 7304) and reverse primer 5′-P-ACTAATGTTACAATGTGCTTGTCT-3′ (HXB2 nt 7203 to 7226).
PCR amplification conditions for the recipient Env backbones were as follows: 1 cycle of 95°C for 3 min; 35 cycles of 95°C for 1 min, 62°C for 30 s, and 72°C for 10 min; 1 cycle of 72°C for 15 min; and storage at 4°C. Pfu Turbo DNA polymerase (Stratagene) was used to generate a blunt-ended PCR amplicon, which was gel purified from an agarose gel by using a QIAquick gel extraction kit (QIAGEN) prior to ligation. The 25-μl PCR mix contained 50 ng of each primer, 10 ng of the plasmid template, 2.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, and 1× reaction buffer. Each synthesized α2 helix DNA fragment was ligated to the purified recipient env backbone by using T4 DNA ligase (5 U/μl; Roche) at 4°C overnight. A portion of the ligation reaction mix was transformed into maximum efficiency XL2-Blue supercompetent cells (1 × 109 CFU/μg DNA; Stratagene). The entire transformation mix was plated onto LB-ampicillin agar plates, generally resulting in ∼50 colonies per ligation reaction. These colonies were screened by PCR to identify colonies in which the fragments ligated together in the correct orientation, using forward primer EnvA and a reverse primer that anneals to the 3′ end of the helix. The colony screening primers were as follows: forward, 5′-CAGCACAGTACAATGTACACATGGAA-3′ (HXB2 nt 6950 to 6975); and reverse, 5′-TTCCTTTATCCCGTCTAAAGT-3′ (HXB2 nt 7254 to 7274) for 135 F42a or 135 F67a α2 helix screening, 5′-TGTCTTTACCTCTTCTAAAGT-3′ (HXB2 nt 7254 to 7274) for 135 F48a α2 helix screening, 5′-TTTTAATCTTTCCTTTACCCT-3′ (HXB2 nt 7254 to 7274) for 55 M4.1 or 55 M2.13 α2 helix screening, or 5′-TTTTAATTTTTTCTCTACCCT-3′ (HXB2 nt 7254 to 7274) for 55 M7a α2 helix screening. Colonies that were positive by PCR screening were inoculated into LB-ampicillin overnight cultures, and the plasmids were prepared using a QIAprep Spin miniprep kit (QIAGEN). Plasmids were then screened by (i) restriction digestion, (ii) biological function assay (described below), and (iii) nucleotide sequencing of the α2 helix and flanking regions to confirm that they contained the correct sequences.
Neutralization assay.
All neutralization assays were performed using a pseudotyped virion assay as described previously (17, 18, 48, 66, 74). In this assay, the NAb activity of plasmas collected contemporaneously with the Envs from donors 135 F and 55 M was evaluated against virions pseudotyped with recipient Envs containing chimeric α2 helix regions in parallel with the parental donor and recipient Envs. For the present study, to evaluate the neutralization sensitivities of the donor, recipient, and chimeric Envs together, it was necessary to conserve limited amounts of donor plasma. Therefore, the highest plasma dilution tested was 1:100 for donor 55 M and 1:20 for donor 135 F.
RESULTS
Sequence divergence that occurs during chronic infection is correlated with NAb resistance.
In our previous study of heterosexual transmission, phylogenetic trees of gp120 V1-to-V4 sequences from donor-recipient pairs were created using ML to investigate the phylogenetic relationships between donor and recipient sequences (18). In the present study, we calculated the BL from each patient's ancestral node for 48 donor and 27 recipient Env sequences from five transmission pairs for which neutralization sensitivity had also been evaluated (18). Informative sites in regions lacking insertions and deletions drove the BL, since most sequences from the V1V2 and V4 hypervariable domains were excluded from the tree. To investigate the relationship between sequence evolution outside the hypervariable domains and neutralization resistance, Kendall's tau rank correlation test was applied to determine whether BL was significantly correlated with sensitivity to NAb in the infecting partner's (donor) plasma or in a heterologous plasma pool from subtype C-infected subjects in Zambia (Fig. 1A and B). BLs for sequences from the chronically infected donors ranged from 0.002 to 0.138 but were significantly shorter for the newly infected recipients (ranging from 0.000 to 0.005; P = 0.001). When the recipient and donor BLs from their respective ancestral nodes were considered together, as the BL increased, so did resistance to NAb in plasma from the linked donor (Fig. 1A) (P = 0.0002) and the heterologous pool (Fig. 1B) (P = 0.001). These correlations held even when the donor Envs were considered without the recipient Envs for both autologous (P = 0.038) and heterologous (P = 0.01) antibodies (data not shown). Thus, sequence diversification in the donor Envs is driven at least in part by autologous NAb, and these sequence adaptations also provide some protection against heterologous NAb.
FIG. 1.
Correlation between sequence adaptation and neutralization sensitivity. Each correlation plot (A to F) contains data points generated from NAb (at the IC50)-Env combinations. Viruses were pseudotyped with either a donor (green) or recipient (blue) Env protein, and neutralization was performed with the linked donor plasma (A, C, and E) or a heterologous subtype C plasma pool (B, D, and F). Data for 48 donor and 27 recipient Envs from five previously described heterosexual transmission pairs in Zambia are shown (18). The P value for correlation between each set of parameters was calculated using Kendall's tau rank correlation test. P values of <0.05 are considered significant. Correlation plots show reciprocal NAb IC50 titers (A to F), BLs from the patients' ancestral nodes on ML trees (A and B), amino acid lengths for V1 to V4 (C and D), and the numbers of N-Gly sites in V1 to V4 (NXS or NXT, where X is any amino acid except proline) (E and F). A reciprocal NAb IC50 titer of 0.1 corresponds to a 1:10 dilution of plasma, a titer of 0.01 corresponds to a 1:100 dilution, and a titer of 0.001 corresponds to a 1:1,000 dilution. As previously described (18), reciprocal NAb IC50 titers were rounded to the nearest two decimal places to facilitate statistical analysis; NAb-sensitive Envs with reciprocal titers of <0.005 were therefore plotted at 0.
Acquisition of length in hypervariable domains is correlated with autologous NAb resistance in donor Envs.
Length polymorphisms in the hypervariable domains V1V2 and V4 were reflected in the donor sequences, where the V1-to-V4 region ranged from 264 to 314 amino acids in length (Fig. 1C and D). The variation in the recipient sequences was less broad, with lengths between 264 and 282 amino acids. Longer hypervariable domains (i.e., increased V1-to-V4 length) were significantly associated with resistance to NAb in linked donor plasma (P = 0.01) (Fig. 1C). This association was recently confirmed for a subset of these Envs, using chimeric recipient Env pseudoviruses in which the native V1V2 domain was replaced with “long” V1V2 domains derived from matched donor Envs (66). In the referenced study, five of seven long donor V1V2 domains tested conferred NAb resistance to the chimeric recipient Env. A significant correlation was not reached between V1-to-V4 length and neutralization by heterologous pooled plasma (Fig. 1D), but there was a trend in this direction (P = 0.08). Thus, hypervariable domain insertions could have the strongest effects on masking strain-specific epitopes, but some protection against cross-neutralization could also be afforded.
Accumulation of N-Gly sites is not correlated with NAb resistance.
Tandem repeat sequences inserted within the longer V1V2 and V4 domains often encoded N-Gly sites (66; data not shown), and in four of the five donors, the number of N-Gly sites was correlated with V1-to-V4 length, using a Spearman rank correlation test (data not shown). Nevertheless, the number of N-Gly sites was not significantly correlated with neutralization by either type of plasma (Fig. 1E and F). This finding could reflect a lack of statistical power for the number of samples analyzed, given the relatively small window of N-Gly sites (17 to 23 sites for all but one Env). Alternatively, the relative positions and packing of the glycans could be more important than net gains or losses for neutralization escape (74).
Individual positions in V1 to V4 are associated with the neutralization phenotype.
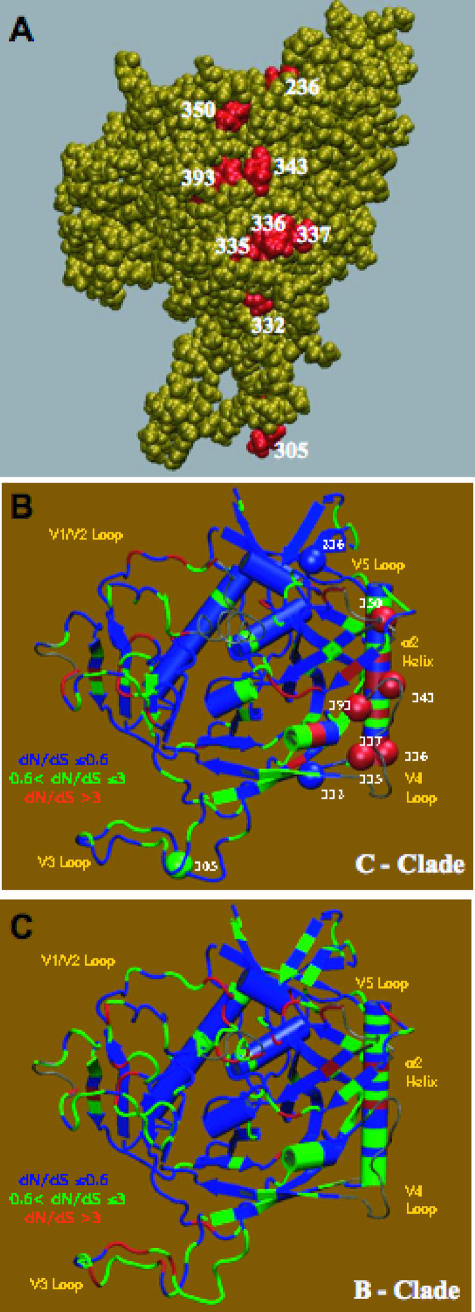
A mutual information analysis based on Shannon entropy was applied to the V1-to-V4 sequence and NAb data for the donor and recipient Envs to search for individual positions in the sequence that were predictive of a neutralization-resistant phenotype. Nine “signature” positions were significantly associated with the NAb phenotype after a Bonferroni correction for multiple tests (uncorrected P value, <0.0001) and were mapped three-dimensionally onto a gp120 model that was based on the X-ray structure of the CD4-bound YU2 gp120 core with modeled variable loops (Fig. 2A). Together, the signature residues form a linear display along the immunologically silent face of gp120 (75) and are proximal to the α2 helix (Fig. 2B). Five of the nine residues are located within the α2 helix, and these five positions overlap those previously demonstrated to be under strong positive selection, with dN/dS ratios of >3, using subtype C sequences in the database (Fig. 2B) (22). The convergence of these two independent analyses supports a role for these sequences in neutralization escape. Also, since the database sequences were drawn from a broader sampling that did not include the Envs studied here, these findings could be generally relevant to subtype C viruses and not limited to those circulating in Zambia. Importantly, the positions of the NAb escape signature residues are not subject to the same degree of positive selection in subtype B sequences, where the highest dN/dS ratios are concentrated in V3 (Fig. 2C). Taken together, these data suggest that substitutions in this region could be driven by the autologous NAb response and that changes in this region, in conjunction with alterations in the hypervariable domains, could contribute to neutralization escape during subtype C infection.
FIG. 2.
Three-dimensional structural illustration of positions associated with NAb escape and positive selection. Locations were mapped onto a model of gp120 based on the X-ray structure of the CD4-bound YU2 gp120 core (43) (PDB code 1RZK). V1V2 and V3 loops were modeled onto the core for completeness. In the orientation shown, the viral and cellular membranes would be located above and below the protein, respectively. (A) Space-filling model showing locations that were significantly associated with the neutralization phenotype in the mutual information analysis (red balls; HXB2 positions 236, 305, 332, 335, 336, 337, 343, 350, and 393). Positions 335, 336, 337, 343, and 350 are located in the α2 helix. (B and C) Cartoon diagrams showing locations under positive selection, as determined by dN/dS ratios for subtype C sequences (B) and subtype B sequences (C) from the Los Alamos Database (22). Red indicates strong positive selection (dN/dS ratio of >3). Residues associated with the neutralization phenotype in the mutual information analysis are shown as spheres in panel B. In the orientation shown in panels B and C, the viral and cellular membranes would be located above and below the protein, respectively. The orientation in panel A roughly corresponds to rotating the structure in panel B about an axis normal to the viral membrane ∼90 degrees counterclockwise.
Exchange of the α2 helix does not directly alter the neutralization phenotype.
The α2 helix is located on the outer domain of gp120, and as an amphipathic helix, the outer face could be partially exposed (Fig. 2A). Based on the mutual information analysis and dN/dS ratios from the database analysis, five positions on the outer surface of this structure appear to be under strong positive selection in subtype C viruses. To investigate whether determinants of neutralization resistance are actually contained within the α2 helix, we selected Envs from transmission pairs 55 (male to female) and 135 (female to male) for further study (18). In pair 55, only 6 of the 18 positions in the helix were completely conserved (Fig. 3A), and these were located on the inner surface facing the gp120 core (22a). Residues in all but one of the five NAb signature positions were heterogeneous in the pair 55 Envs (Fig. 3A). Likewise, for pair 135, the five internal positions of the helix were completely conserved, and all NAb signature residues were heterogeneous (Fig. 3A). Chimeric Envs were created in which the entire helix of a recipient Env was replaced with the corresponding sequences from three different donor Envs. In the case of pair 55, two recipient Envs with different degrees of neutralization sensitivity determined by a charged residue in V2 were used (66). The entire α2 helix domain was exchanged instead of performing targeted site-directed mutagenesis due to the limited quantities of donor plasma available and the extensive sequence heterogeneity in this region (Fig. 3A). Despite the strong association between neutralization sensitivity and the amino acid residues in this region that emerged in the mutual information analysis, exchange of this region resulted in only slight changes in NAb sensitivity between the recipient and chimeric Envs for both transmission pairs (Fig. 3B). The reciprocal constructs were created and evaluated for pair 135 (recipient helix into donor Envs), and again, only minimal changes in neutralization phenotype were observed (data not shown). These results argue that despite the strong selective pressure on the α2 helix and its association with NAb resistance, this structure does not directly modulate resistance to autologous NAb.
DISCUSSION
Longitudinal studies of humans and animal models have demonstrated that the V1V2 and V4 regions are susceptible to insertions, deletions, and alterations in the pattern of glycosylation during chronic infection of HIV-1-infected individuals and experimentally SIV-infected macaques (8, 10, 11, 13, 34, 55, 61, 62, 67, 71). Acquisition of length in V1V2 and/or V4 in the current study was associated with decreased sensitivity to neutralization by linked donor plasma, but less so with heterologous plasma. Changes in this region may therefore be driven by escape from NAb that target mainly strain-specific epitopes. Using a V1V2 domain exchange approach, we have confirmed that long V1V2 domains from chronically infected donors frequently mask determinants of sensitivity to autologous NAb (66). The effect of V1V2, however, was highly dependent on the Env background. We also observed cases in which donor Envs had acquired NAb resistance to autologous neutralization independently of V1V2, consistent with the presence of neutralization determinants that are not contained within this domain.
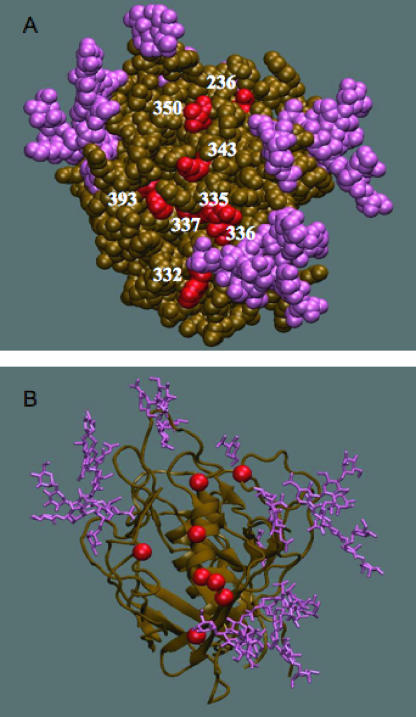
Increasing sequence divergence, as measured by BL on ML trees, showed a strong association with decreased sensitivity to neutralization by donor plasma and a heterologous plasma pool. This result suggests that point mutations in regions outside the hypervariable domains V1V2 and V4, which were excluded from the trees, facilitate escape from antibodies that target some cross-neutralizing epitopes and is consistent with the correlation observed previously between the IC50 titers for neutralization by donor and pooled plasmas (18) and V1V2-independent pathways to NAb resistance (66). Despite the inherent limitations, given that the analysis was based on multiple sequences from five transmission pairs, a striking relationship between signature sites was apparent, in that all nine neutralization “signature” residues identified in the mutual information analysis cluster together on the outer face of the gp120 molecule, nestled among glycans that were resolved on the unliganded SIV gp120 structure (Fig. 4A and B). Variation in one residue identified in the mutual information analysis, i.e., residue 236 (based on HXB2 numbering) in C2, can alter glycosylation at N234, and its effect appeared to differ among the transmission pairs. For example, the presence of a glycan at N234 was associated with neutralization resistance in pair 135, while the loss of the same glycan was associated with resistance in pair 53; this sort of patient-specific gain or loss of a glycan illustrates why there may be no discernible association between the net number of N-Gly sites and the resistance phenotype. Notably, glycosylation at the same position was previously associated with cumulative escape from autologous neutralization in a subtype B-infected subject (74).
FIG. 4.
Three-dimensional structural illustration of positions associated with NAb escape on the unliganded SIV gp120 structure. Locations were mapped onto a model of unliganded gp120 based on the fully glycosylated X-ray structure of SIVmac32H gp120 (12) (PDB code 2BF1). The V1V2 and V3 loops were truncated in the crystallized protein and replaced with a GAG sequence spanning the positions where the loop would have been. N-linked carbohydrate moieties are shown in purple. In the orientation shown, the viral and cellular membranes would be located above and below the protein, respectively. (A) Space-filling model showing locations that were significantly associated with the neutralization phenotype in the mutual information analysis (in red; HXB2 positions 236, 332, 335, 336, 337, 343, 350, and 393). Positions 335, 336, 337, 343, and 350 are located in the α2 helix. Position 305 is missing from the structure. (B) Cartoon diagram showing residues associated with the neutralization phenotype in the mutual information analysis as red spheres.
One potentially complicating factor for the mutual information analysis used here is that the recipient Envs were always NAb sensitive and all NAb-resistant Envs were from the donors (18). It is therefore possible that the associations detected by the mutual information analysis reflect positions in V1 to V4 that track with transmission or establishment of infection. This remains a formal possibility, as the transmission bottleneck would constrict variation at all sites and thus could produce an association with the neutralization phenotype simply due to the overall sensitivity of the recipient Envs. However, the coincidence of high mutual information scores and high levels of positive selection (high dN/dS ratios) at the population level for subtype C viruses for the same sites supports their role in neutralization escape. Moreover, an independent mutual information analysis was performed on 515 V1-to-V4 sequences from an additional 13 transmission pairs from the same cohort (including 502 donor sequences and 13 “consensus” sequences from each recipient) in the absence of NAb data. In this analysis, four residues (two located in V2, one in α2, and one in V4) showed a trend of association with transmission status (P. Hawkins et al., unpublished data), and none of these overlapped with the nine residues shown here to track with NAb resistance. Thus, the positions we have identified in the current study as being associated with neutralization resistance appear to be independent of transmission status, given the sample size under consideration.
In the trimer model of gp120, based on the structure of HXB2 (45), the 18-residue α2 helix is located on the outer surface of gp120. Even though this model is based on CD4-bound gp120, a comparison of liganded and unliganded gp120 structures does not indicate major changes in the outer domain upon CD4 binding (12). Five of the nine NAb signature residues were located within the α2 helix (Fig. 2B), and the same residues were under strong positive selection in subtype C database sequences (22); the coincidence of the two independent results raised the interesting hypothesis that substitutions in this region could be related to antibody escape. Nevertheless, exchange of the α2 helix between donor and recipient Envs did not significantly alter the neutralization sensitivity of the related recipient Env, suggesting that it alone does not contain direct determinants of NAb resistance. It is not surprising that an exchange of the α2 helix failed to produce direct changes in patterns of neutralization resistance, since this region of Env is likely protected from antibody recognition by the glycan shield and thus, in itself, is unlikely to represent a neutralization epitope (74). More likely, this region of the Env glycoprotein is responsible for contributing to the three-dimensional structure of the protomer and the quaternary structure of the trimer that confers neutralization escape by mechanisms in addition to epitope variation (e.g., epitope masking, steric hindrance, conformation energetics, and glycan packing) (42, 74). Molecular dynamics simulation studies provided evidence for an extensive interaction between the α2 helix and a flexible V4 loop in subtype C, with certain residues in the helix associated with restricting V4 loop length (22a). The nonpolar face of the α2 helix is predicted to interact with structural domains β10, β11, β14, and β24, which could help to stabilize the gp120 core structure. It is therefore possible that to see a phenotypic effect of the α2 helix on neutralization sensitivity, other domains must be present. Perhaps the outer face of the α2 helix compensates structurally for the adaptations that are directly involved in NAb escape of subtype C viruses. If so, mutations in the helix would track with a resistant phenotype without physically defining or altering an epitope.
Sequence and structural studies revealed at least two major differences in the α2 helix between subtypes B and C. Even though the helix of subtype C exhibits high sequence entropy, the amphipathicity of the helix is preserved to a higher degree than that of subtype B (22a). The helix of subtype C is also more solvated than that of subtype B (S. Gnanakaran, unpublished data), with high-sequence-entropy positions occurring at the solvent-exposed face. All five signature residues identified here occupy solvent-exposed positions of the helix, while residues that face toward the core are well conserved (22a). Thus, in subtype C viruses, the α2 helix may act as a flexible interface to maintain the integrity between NAb escape mutations occurring on the outer domain and the more highly conserved gp120 inner core. Alternatively, these mutations could accommodate escape mechanisms occurring in the hypervariable regions. In subtype C, the N-terminal half of the helix interacts extensively with the V4 loop, while the C-terminal half interacts with the V5 domain.
Finally, mutations at some of the signature positions could affect the local stability of the α2 helix. Alpha helices represent a common secondary structural motif in proteins and are stabilized by intramolecular hydrogen bonding between the i and i + 4 residues on parallel turns of the helix. However, the first four and last four residues of an α helix lack this intramolecular hydrogen bonding and are often stabilized by alternative hydrogen bonding partners and hydrophobic interactions. This leads to differences in the propensities of amino acids found in the N-terminal and C-terminal capping motifs. Several systematic studies on the positional frequency of each of the amino acids found in helices in protein structures show statistical deviations from a random distribution (1, 19, 41, 65). For example, asparagine has a higher preference of occurring at the N-terminal cap position, whereas the structurally similar glutamic acid does not. Further studies have established a relationship between capping motifs and the stability of an α helix. Interestingly, five of the nine signature positions identified here have the potential to play a structural role as capping residues of the α2 helix. Four of them coincide with the N-terminal capping positions, and one coincides with a C-terminal capping position in the α2 helix. Furthermore, the first turn of the N-terminal capping sequence is located adjacent to the antiparallel beta strand just downstream from the V3 domain. Thus, any local unfolding of the N-terminal region of the α2 helix could potentially impact the projection of the V3 domain out from the core. Thus, the α2 helix of gp120 appears to be a novel and important source of structural adaptation during subtype C infection, and its structural role in the context of the entire gp120 molecule needs to be explored further.
The present study combines statistical, biological, and molecular dynamics simulation analyses to elucidate the poorly understood pathways to neutralization resistance in subtype C virus infection. NAb are clearly a strong evolutionary force during chronic infection with subtype C HIV-1, driving sequence divergence in structural domains and also dramatic length changes in the hypervariable regions of Env. Moreover, mutations in a purely structural domain (in a region that historically has been considered well conserved) track with NAb resistance, supporting a novel and potentially subtype-specific mechanism for neutralization escape. Further investigations will be required to understand the selective forces directed at the α2 helix and to determine why this region is differentially targeted depending on the viral subtype, as these questions have direct implications for global vaccine design.
Acknowledgments
We gratefully acknowledge the contributions of the staff, lab technicians, participants, and project management group of the Lusaka cohort. We thank Peter Kwong for providing us with the HXB2 trimer coordinates.
This work was supported by NIH grants R01-AI-58706 (C.D.) and R01-AI-51231 (E.H.), by The Bill & Melinda Gates Grand Challenges Program (G.M.S.), and by Yerkes National Primate Research Center base grant RR-00165, awarded by the National Center for Research Resources of the National Institutes of Health.
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Aurora, R., and G. D. Rose. 1998. Helix capping. Protein Sci. 7:21-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Naim, E., and A. S. Lapedes. 1999. Genetic correlations in mutation processes. Phys. Rev. E 59:7000-7007. [DOI] [PubMed] [Google Scholar]
- 3.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684-3690. [Google Scholar]
- 4.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blay, W. M., S. Gnanakaran, B. Foley, N. A. Doria-Rose, B. T. Korber, and N. L. Haigwood. 2006. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J. Virol. 80:999-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouma, P., M. Leavitt, P. F. Zhang, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. J. Virol. 77:8061-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns, D. P., and R. C. Desrosiers. 1994. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr. Top. Microbiol. Immunol. 188:185-219. [DOI] [PubMed] [Google Scholar]
- 9.Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, R. V. Stanton, A. L. Cheng, J. J. Vincent, M. Crowley, V. Tsui, R. J. Radmer, Y. Duan, J. Pitera, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 1999. Amber 6 software. University of California, San Francisco.
- 10.Chackerian, B., W. R. Morton, and J. Overbaugh. 1994. Persistence of simian immunodeficiency virus Mne variants upon transmission. J. Virol. 68:4080-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 13.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choisy, M., C. H. Woelk, J. F. Guegan, and D. L. Robertson. 2004. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J. Virol. 78:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force-field for the simulation of proteins; nucleic acids; and organic molecules. J. Am. Chem. Soc. 117:5179-5197. [Google Scholar]
- 16.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089-10092. [Google Scholar]
- 17.Decker, J. M., F. Bibollet-Ruche, X. P. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 19.Engel, D. E., and W. F. DeGrado. 2004. Amino acid propensities are position-dependent throughout the length of alpha-helices. J. Mol. Biol. 337:1195-1205. [DOI] [PubMed] [Google Scholar]
- 20.Etemad-Moghadam, B., G. B. Karlsson, M. Halloran, Y. Sun, D. Schenten, M. Fernandes, N. L. Letvin, and J. Sodroski. 1998. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J. Virol. 72:8437-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fung, M. S., C. R. Sun, W. L. Gordon, R. S. Liou, T. W. Chang, W. N. Sun, E. S. Daar, and D. D. Ho. 1992. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1 gp120. J. Virol. 66:848-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 22a.Gnanakaran, S., D. Lang, M. Daniels, T. Bhattacharya, C. A. Derdeyn, and B. Korber. 2007. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J. Virol. 81:4886-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorny, M. K., and S. Zolla-Pazner (ed.). 2003. Human monoclonal antibodies that neutralize HIV-1. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM.
- 26.Gram, G. J., A. Hemming, A. Bolmstedt, B. Jansson, S. Olofsson, L. Akerblom, J. O. Nielsen, and J. E. Hansen. 1994. Identification of an N-linked glycan in the V1-loop of HIV-1 gp120 influencing neutralization by anti-V3 antibodies and soluble CD4. Arch. Virol. 139:253-261. [DOI] [PubMed] [Google Scholar]
- 27.Gray, E. S., T. Meyers, G. Gray, D. C. Montefiori, and L. Morris. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 3:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho, D. D., M. S. Fung, Y. Z. Cao, X. L. Li, C. Sun, T. W. Chang, and N. C. Sun. 1991. Another discontinuous epitope on glycoprotein gp120 that is important in human immunodeficiency virus type 1 neutralization is identified by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 88:8949-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—Visual Molecular Dynamics. J. Mol. Graphics 14:33-38. [DOI] [PubMed] [Google Scholar]
- 30.Hunter, E. 1997. Viral entry and receptors, p. 71-121. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 31.Jorgensen, W. L., J. Chandreshkar, J. D. Madura, R. W. Imprey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926-935. [Google Scholar]
- 32.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinsey, N. E., M. G. Anderson, T. J. Unangst, S. V. Joag, O. Narayan, M. C. Zink, and J. E. Clements. 1996. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology 221:14-21. [DOI] [PubMed] [Google Scholar]
- 34.Kodama, T., K. Mori, T. Kawahara, D. J. Ringler, and R. C. Desrosiers. 1993. Analysis of simian immunodeficiency virus sequence variation in tissues of rhesus macaques with simian AIDS. J. Virol. 67:6522-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolchinsky, P., E. Kiprilov, and J. Sodroski. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korber, B. T., R. M. Farber, D. H. Wolpert, and A. S. Lapedes. 1993. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc. Natl. Acad. Sci. USA 90:7176-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korber, B. T., K. MacInnes, R. F. Smith, and G. Myers. 1994. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J. Virol. 68:6730-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krachmarov, C., A. Pinter, W. J. Honnen, M. K. Gorny, P. N. Nyambi, S. Zolla-Pazner, and S. C. Kayman. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J. Virol. 79:780-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krachmarov, C. P., W. J. Honnen, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 80:7127-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar, S., and M. Bansal. 1998. Dissecting alpha-helices: position-specific analysis of alpha-helices in globular proteins. Proteins 31:460-476. [DOI] [PubMed] [Google Scholar]
- 42.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 43.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 44.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LaRosa, G. J., J. P. Davide, K. Weinhold, J. A. Waterbury, A. T. Profy, J. A. Lewis, A. J. Langlois, G. R. Dreesman, R. N. Boswell, P. Shadduck, et al. 1990. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science 249:932-935. [DOI] [PubMed] [Google Scholar]
- 47.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinksy, and B. Korber. 2003. HIV sequence compendium. The Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 48.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Losman, B., A. Bolmstedt, K. Schonning, A. Bjorndal, C. Westin, E. M. Fenyo, and S. Olofsson. 2001. Protection of neutralization epitopes in the V3 loop of oligomeric human immunodeficiency virus type 1 glycoprotein 120 by N-linked oligosaccharides in the V1 region. AIDS Res. Hum. Retrovir. 17:1067-1076. [DOI] [PubMed] [Google Scholar]
- 50.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCutchan, F. 2006. Global epidemiology of HIV. J. Med. Virol. 78:S7-S12. [DOI] [PubMed] [Google Scholar]
- 53.McKeating, J. A., C. Shotton, J. Cordell, S. Graham, P. Balfe, N. Sullivan, M. Charles, M. Page, A. Bolmstedt, S. Olofsson, et al. 1993. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J. Virol. 67:4932-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nabatov, A. A., G. Pollakis, T. Linnemann, A. Kliphius, M. I. Chalaby, and W. A. Paxton. 2004. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 and V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J. Virol. 78:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmer, C., P. Balfe, D. Fox, J. C. May, R. Frederiksson, E. M. Fenyo, and J. A. McKeating. 1996. Functional characterization of the V1V2 region of human immunodeficiency virus type 1. Virology 220:436-449. [DOI] [PubMed] [Google Scholar]
- 56.Pearlman, D. A., D. A. Case, J. W. Caldwell, W. L. Ross, T. E. Cheatham, D. M. Ferguson, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 1995. AMBER 4.1 software. University of California, San Francisco.
- 57.Peeters, M. 2000. Recombinant HIV sequences: their role in the global epidemic, p. 39-54. In F. B. Kuiken, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, J. Sodroski, S. Wolinksy, and B. Korber (ed.), HIV sequence compendium 2000. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 58.Pinter, A., W. J. Honnen, P. D'Agostino, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2005. The C108g epitope in the V2 domain of gp120 functions as a potent neutralization target when introduced into envelope proteins derived from human immunodeficiency virus type 1 primary isolates. J. Virol. 79:6909-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 61.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 73:5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poss, M., A. G. Rodrigo, J. J. Gosink, G. H. Learn, D. de Vange Panteleeff, H. L. Martin, Jr., J. Bwayo, J. K. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pugach, P., S. E. Kuhmann, J. Taylor, A. J. Marozsan, A. Snyder, T. Ketas, S. M. Wolinsky, B. T. Korber, and J. P. Moore. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8-22. [DOI] [PubMed] [Google Scholar]
- 64.Reza, F. 1994. Introduction to information theory. Dover, Mineola, NY.
- 65.Richardson, J. S., and D. C. Richardson. 1988. Amino acid preferences for specific locations at the ends of alpha helices. Science 240:1648-1652. [DOI] [PubMed] [Google Scholar]
- 66.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ryckaert, J. P., G. Ciccotti, and H. C. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327-341. [Google Scholar]
- 69.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 70.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trask, S., C. A. Derdeyn, F. Kasolo, R. Musonda, F. Gao, S. A. Allen, and B. Hahn. 2002. Epidemiological linkage analysis in a heterosexual transmission cohort in Zambia. J. Virol. 76:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Warrier, S. V., A. Pinter, W. J. Honnen, M. Girard, E. Muchmore, and S. A. Tilley. 1994. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J. Virol. 68:4636-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 75.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 76.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, W., J. P. Bielawski, and Z. Yang. 2003. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J. Mol. Evol. 57:212-221. [DOI] [PubMed] [Google Scholar]