Abstract
Ovine leukemia/lymphoma resulting from bovine leukemia virus infection of sheep offers a large animal model for studying mechanisms underlying leukemogenesis. Silencing of viral information including Tax, the major contributor to the oncogenic potential of the virus, is critical if not mandatory for tumor progression. In this study, we have identified epigenetic mechanisms that govern the complete suppression of viral expression, using a lymphoma-derived B-cell clone carrying a silent provirus. Silencing was not relieved by injection of the malignant B cells into sheep. However, exogenous expression of Tax or treatment with either the DNA methyltransferase inhibitor 5′azacytidine or the histone deacetylase (HDAC) inhibitor trichostatin A rescued viral expression, as demonstrated by in vivo infectivity trials. Comparing silent and reactivated provirus, we found mechanistic connections between chromatin conformation and tumor-associated transcriptional repression. Silencing is associated with DNA methylation and decreased accessibility of promoter sequences. HDAC1 and the transcriptional corepressor mSin3A are associated with the inactive but not the reactivated promoter. Silencing correlates with a repressed chromatin structure marked by histone H3 and H4 hypoacetylation, a loss of methylation at H3 lysine 4, and an increase of H3 lysine 9 methylation. These observations point to the critical role of epigenetic mechanisms in tumor-specific virus/oncogene silencing, a potential strategy to evade immune response and favor the propagation of the transformed cell.
Virus-host interactions are important in determining the outcome of infection. In the case of oncogenic viruses, growing evidence suggests that the subtle balance between viral gene expression and efficient host immune responses plays a decisive role in tumor onset and progression. Bovine leukemia virus (BLV) infection of sheep offers a tractable large animal model of disease associated with human T-lymphotropic virus type 1 (HTLV-1) and insight into mechanisms underlying both B-cell and T-cell leukemogenesis. Studying gene expression control mechanisms in BLV-infected leukemic sheep should provide important clues to the significance of gene silencing on the propensity of tumor cells to evade immune control and progress to acute malignancy.
BLV is a complex retrovirus that naturally infects cattle, provoking a chronic disease that culminates in the development of B-lymphoid tumors in a small proportion of infected individuals after a long latency period (5). All experimentally infected sheep, in contrast, consistently develop B-cell leukemia or lymphoma after a shorter latency (9, 29). The preleukemic phase of infection includes the expansion of BLV-infected surface immunoglobulin M-positive (sIgM+) B cells with proviral insertions at multiple sites, whereas a unique integration site represents the molecular signature of the malignant B-cell clone found in each individual after the onset of overt leukemia/lymphoma. BLV shares a number of structural and functional similarities with HTLV-1. Unlike simple retroviruses, which induce tumors by expressing viral products or by proviral insertional mutagenesis, complex oncoretroviruses such as HTLV-1 and BLV exert their oncogenic potential using poorly understood mechanisms which involve Tax, the viral transactivator/oncoprotein (16, 19, 42). Although Tax is an essential contributor to the oncogenic potential of both viruses, mainly through the transcriptional modification of host genes (19, 32, 42), there is compelling evidence that expression of Tax is not sufficient for transformation. Furthermore, the presence of mutations in tumor-associated proviral sequences, including that of tax, suggests that neither virus nor Tax expression is required for the maintenance of the transformed phenotype (18, 45-47).
BLV and HTLV-1 infections are both characterized by low or undetectable viral expression in vivo, but peripheral blood mononuclear cells (PBMCs) isolated from an infected individual during the premalignant phase spontaneously express viral proteins in vitro (13, 36). In contrast, in the B-cell tumors isolated from BLV-infected sheep and the cell lines derived from these tumors, we consistently observed a silent provirus (46-48). A current hypothesis suggests that the complete suppression of viral expression in malignant B cells might be a strategy to circumvent effective immune attack. Sheep infected by BLV mount a strong immune response to viral antigens, and active killing of infected cells might play a decisive role in limiting BLV gene expression during the latency period that precedes tumor onset but seems unable to prevent—or perhaps paradoxically favors—the development of a malignant clone harboring a silent provirus.
The first evidence suggesting the presence of a silent provirus in transformed B-cell clones came from the observation that proviral sequences are frequently altered by genetic changes, including mutation and deletion (45, 47). A study of sheep infected with recombinant BLV proviruses conclusively demonstrated that a mutation in tax, generating a transactivation-deficient provirus, accounts for tumor-associated silencing in at least a proportion of leukemic animals (46). Silencing is not restricted to genetically modified proviruses but was also observed in the case of structurally intact proviruses, suggesting that epigenetic mechanisms might be involved (43, 47; M. Merimi, P. Klener, Y. Cleuter, C. Bagnis, A. Burny, P. Martiat and A. Van den Broeke, submitted for publication). A primary molecular mechanism underlying epigenetics is the alteration of the chromatin structure by covalent DNA modifications, covalent histone modifications, and nucleosome reorganization. Remodeling of the chromatin structure regulates DNA methylation, replication, recombination, and repair as well as gene expression.
Here, in an effort to further understand BLV transcriptional regulation mechanisms contributing to the complete suppression of viral expression in transformed B cells, we investigated potential mechanistic connections between chromatin structure and virus silencing, using L267, a lymphoma-derived B-cell line, as a model. L267 is a clonal population of transformed B cells that contain a single copy of silent BLV provirus. Provirus expression is completely suppressed in the L267 lymphoma B cells, as conclusively demonstrated by their incapacity to generate infection in the in vivo sheep model. In contrast, polyclonally integrated provirus in nontransformed PBMCs isolated from the same animal was found to be transcriptionally active. Both the tumor- and blood-derived proviruses displayed identical wild-type sequences, strongly suggesting that silencing of provirus expression in L267 is not associated with genetic changes but results from epigenetic mechanisms. In this study, we show that silencing in L267 is associated with epigenetic changes leading to a compacted chromatin structure. We demonstrated the impact of DNA methylation, a reduced accessibility of transcription factors to the BLV promoter region and histone modifications, including changes in acetylation and methylation levels characteristic of a repressive histone code.
MATERIALS AND METHODS
Animals and tissues.
Sheep used in this study were housed at the Centre de Recherches Vétérinaires et Agrochimiques (Brussels, Belgium). Animals were injected intradermally with 2 × 107 cells collected from different BLV-positive B-cell populations as previously described (46). Blood was collected from each sheep at weekly intervals for 2 years or until the development of leukemia. PBMCs were isolated using standard Ficoll-Hypaque separation. Lymphoid tumors were collected and handled as described previously (46). Anti-p24 serum antibody titers were determined with a competitive enzyme-linked immunosorbent assay (ELISA) (35). Sheep S2011, S2013, and S2014 were injected with the silent L267 lymphoma-derived B-cell line and did not develop leukemia. Sheep S2233, S2311, and S2350 were inoculated with reactivated L267 cells transduced with the pLTaxSN retroviral vector (L267LTaxSN), resulting in the development of leukemia and lymphoma at 30 and 36 months after injection for S2233 and S2311, respectively. Sheep M2350 died of unrelated causes 8 months after infection. Sheep S5578 was infected with L267 cells treated with a combination of trichostatin A (TSA) and 5′azacytidine and developed leukemia and lymphoma 20 months after infection.
Cells, cell lines, and BLV proviral sequences.
L267 is a transformed lymphoma-derived B-cell line established from sheep S267, a BLV-infected sheep injected with naked proviral DNA of an infectious BLV variant described previously (50; M. Merimi et al., submitted for publication). This provirus (pBLVX3C) is isogenic to the wild-type infectious provirus 344 used for in vivo infection of sheep (38, 39, 50-52), except for the longer X3 region that originates from another infectious variant present in the FLK cell line (40). The 383-bp X3 insertion contains a SacI restriction site which is absent from provirus 344 and accounts for the presence of a SacI site in the L267 proviral DNA (Fig. 1A). Sequencing of the provirus integrated in the L267 cell line indicated that it was similar to the sequence of the cloned naked BLVX3C proviral DNA used for injection of sheep S267 and thus structurally capable of transcriptional activity (M. Merimi et al., submitted for publication). YR2 is a transformed B-cell line established from PBMCs isolated from a leukemic sheep (17, 47). YR2LTaxSN results from the transduction of native YR2 cells with the pLTaxSN retroviral vector expressing the tax cDNA following cocultivation with the PG13LTaxSN producer cell line and G418 selection of transduced neomycin-resistant cells (46). L267LTaxSN was generated using a similar strategy for tax delivery into the parental L267 cell line. L267, YR2, L267LTaxSN, and YR2LTaxSN were maintained in OPTIMEM medium (Invitrogen) supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, 2 mM glutamine, nonessential amino acids, and 100 μg/ml kanamycin as previously described (48). All cell lines were cultivated at 37°C in a 5% CO2 humidified atmosphere. L267 cells were treated either for 22 h with 150 nM, 250 nM, 350 nM, 400 nM, 450 nM, or 500 nM TSA (Sigma) or for 48 h with 500 nM, 1,000 nM, or 1,500 nM 5′azacytidine (Sigma) or with 500 nM or 1,000 nM 5′azacytidine for 28 h prior to treatment with the addition of 500 nM TSA for 20 h.
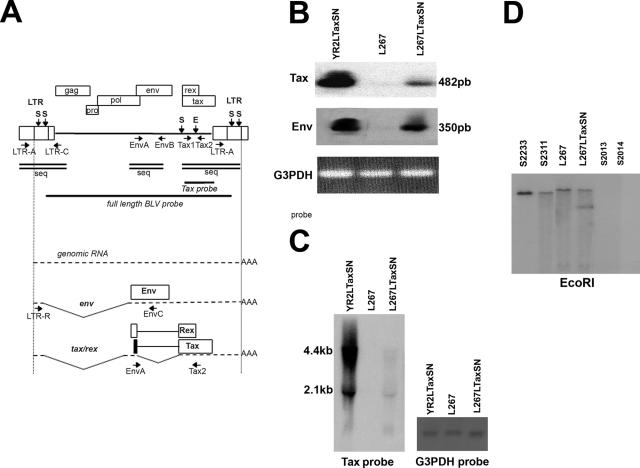
FIG. 1.
Delivery of exogenous Tax leads to viral expression in transformed L267 B cells. (A) Diagram of the BLV provirus and major transcripts. The two LTRs and the genes gag, pro, pol, env, tax, and rex are represented. Vertical arrows indicate restriction sites SacI (S) and EcoRI (E) in the L267 provirus. The positions and directions of the PCR primers are indicated on the provirus map. Horizontal bars indicate regions that were used as probes. Double lines represent the sequenced regions. The genomic env and transcripts tax/rex are represented below. Alternatively spliced RNAs are not shown. The translation products of the singly and doubly spliced transcripts and the positions of the RT-PCR primers are indicated. (B and C) Detection of BLV-specific transcriptional activity in total RNA isolated from L267, L267LTaxSN, and YR2LTaxSN cells. (B) RT-PCR analysis using two sets of splice-specific primers, LTR-R/EnvC and EnvA/Tax2, for detection of the singly and doubly spliced BLV transcripts, respectively. Amplified products (350 bp and 482 bp, respectively) were detected by Southern blot hybridization using a 32P-labeled BLV full-length proviral DNA probe. G3PDH amplification was detected by ethidium bromide staining. (C) Northern blotting analysis of total RNA isolated from the same samples using a 32P-labeled Tax probe. G3PDH hybridization was used as a control for RNA input. Sizes of mRNAs are indicated. (D) Southern blot hybridization of EcoRI-digested DNA extracted from PBMCs isolated from L267LTaxSN-injected leukemic sheep (S2233 and S2311) 30 and 39 months postinoculation, respectively, from PBMCs from sheep injected with the silent L267 cells (S2013 and 2014) collected 24 months postinoculation, and from L267 and L267LTaxSN cell cultures. The BLV full-length proviral DNA probe was used for hybridization. EcoRI-cleaved DNA generates two virus-host junction fragments for each integrated L267 provirus. Shown here in each lane is the fragment containing the unique 5′ flanking genomic region, a hallmark of monoclonal provirus integration characteristic of leukemic cells. The additional smaller band in L267LTaxSN corresponds to the LTaxSN vector-derived EcoRI fragment detected with the BLV probe.
PCR and RT-PCR analysis.
Genomic DNA was prepared and analyzed by PCR as previously described. Primers (and nucleotide positions according to Sagata [40]) were as follows: Tax1 (nucleotides [nt] 7321 to 7340), 5′-GATGCCTGGTGCCCCCTCTG-3′; Tax2 (nt 7604 to 7623), 5′-ACCGTCGCTAGAGGCCGAGG-3′; EnvA (nt 4766 to 4788), 5′-TCCTGGCTACTAACCCCCCCGT-3′; EnvB (nt 5756 to 5777), 5′-TCCAGTGAGCCCCACTGACAGG-3′; LTR-A (nt 1 to 22), 5′-TGTATGAAAGATCATGCAGGCC-3′; and LTR-C (nt 577 to 599), 5′-GCCGCCGAGGGGGTGGGTCCAGA-3′. For reverse transcription (RT)-PCR experiments, total RNA was extracted using Tripure reagent according to the manufacturer's protocol (Roche). RNA (1 μg) was reverse transcribed and amplified using a Titan RT-PCR system according to the protocol supplied by the manufacturer (Roche). Primers EnvA/Tax2 and LTR-R/EnvC (LTR-R [nt 251 to 270], 5′-GTCCTGAGCTCTCTTGCTCC-3′; EnvC [nt 4921 to 4942], 5′-CCTAGGGACAGGGAGCATCTCC-3′, respectively) were used for the detection of the 2.1-kb doubly spliced tax/rex and 4.4-kb singly spliced env mRNAs, respectively, as previously described (46). The positions of the primers are shown in Fig. 1A.
Southern and Northern blotting analysis.
Genomic DNA or PCR-amplified products were analyzed by Southern blotting as previously described (46, 47). The nylon-bound EcoRI-digested DNAs were hybridized with a 32P-labeled BLV full-length proviral DNA probe. Northern blotting was performed using 10 μg of total RNA and hybridization with a 32P-labeled Tax or a glyceraldehyde-3-phosphate dehydrogenase (G3PDH) probe (46). Positions of the BLV probes are shown in Fig. 1A.
Nuclease digestion of purified nuclei.
Exponentially growing cells were harvested and washed twice with ice-cold phosphate-buffered saline. Cells were suspended at 20 × 106 cells/ml in buffer A (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.3 M sucrose) and incubated on ice for 5 min. An equal volume of buffer A containing 0.2% NP-40 was added, and the cells were incubated for another 5 min with intermittent mixing. Nuclei were pelleted, suspended at 108 nuclei/ml in buffer A, and digested using DNase I at concentrations of 0, 50, 60, 70, 80, and 95 units/ml or 0, 80, 85, and 90 units/ml for 10 min at 4°C. Digestion was stopped by adding 1 volume of 2× proteinase K buffer (100 mM Tris [pH 7.5], 200 mM NaCl, 2 mM EDTA, 1% sodium dodecyl sulfate). Samples were treated for 1 h at 55°C and proteinase K was added (20 mg/ml), and the digestion was allowed to continue overnight at 37°C. RNase was added at a final concentration of 50 μg/ml for 1 h at 37°C. DNA was extracted using phenol, followed by chloroform/isoamyl alcohol (24:1) and precipitation with ethanol. Resuspended DNA was digested with BamHI and analyzed by Southern blot hybridization using a probe spanning a region downstream of the 5′ long terminal repeat (LTR) (nt 1422 to 2038, according to Sagata et al. [40]). The amount of hybridized sequence was quantified by conventional autoradiography.
EMSA.
Nuclear extracts were prepared from ovine B cells, and electrophoretic mobility shift assays (EMSA) were performed as described previously (42). Briefly, nuclear extract (15 μg of protein) was incubated at room temperature for 20 min in the absence of probe and specific competitor DNA in a 20-μl reaction mixture containing 1 μg of poly(dI-dC) (Amersham Biosciences), 1 mM dithiothreitol, 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, and 5% (vol/vol) glycerol. 32P-labeled Tax-responsive element 1 (TxRE1), TxRE2, TxRE3, cyclic AMP-responsive element-binding (CREB) consensus, or octamer-binding protein (OCT) probe (15,000 cpm) was then added to the mixture with or without a molar excess of unlabeled specific DNA competitor, and the mixture was incubated for 30 min at room temperature. Samples were subjected to electrophoresis on a 6% polyacrylamide gel in 1× TGE buffer (25 mM Tris-acetate [pH 8.3], 190 mM glycine, 1 mM EDTA). For supershift assays, polyclonal antibodies specific for CREB-1 (catalog no. sc-271) and activating transcription factor 1 (ATF1) (catalog no. sc-243; Santa Cruz Biochemicals) were added to the binding mixture (2 μg/reaction) for an additional 30-min incubation at room temperature before electrophoresis.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed essentially by using the Upstate Biotechnology Inc. recommendations with minor modifications. Formaldehyde cross-linking reactions (1% final formaldehyde concentration) from 0.8 × 107 L267 cells, mock treated or treated with 5′azacytidine (1 μM) for 48 h, followed with TSA (500 nM) for 20 h, were quenched with 125 mM glycine after a 10-min incubation at 37°C. Chromatin was sheared by sonication to a mean average DNA length of 500 bp. Following centrifugation, chromatin was precleared using protein A-agarose containing salmon sperm DNA and bovine serum albumin (Upstate Biotechnology). Precleared chromatin (2 ml) was incubated overnight at 4°C with 5 μg of antibody recognizing acetyl-histone H4 (catalog no. 06-866), acetyl-histone H3 (catalog no. 06-866), methyl lysine 9-histone H3 (catalog no. 06-866), methyl lysine 4-histone H3 (catalog no. 06-866), histone deacetylase 1 (HDAC1) (catalog no. 06-866), mSin3A (catalog no. 06-866), or rabbit immunoglobulin G control antibody (catalog no. 12-370) (all antibodies from Upstate Biotechnology), followed by protein A-agarose immunoprecipitation. Eluted protein-DNA cross-links were reversed by heating at 65°C overnight, and 10% of the recovered DNA was used for radioactive PCR amplification (30 cycles) with a primer set amplifying the BLV promoter region encompassing the TxREs from nt 40 to 239, 5′-CCGTAAACCAGACAGAGACGTCAG-3′, and nt 217 to 239, 5′-CACGAGGGTCTCAGGAGAAGAAC-3′. PCR products were analyzed by polyacrylamide gel electrophoresis, and bands were visualized by autoradiography.
RESULTS
In silent L267 lymphoma B cells, viral expression can be restored upon delivery of exogenous Tax.
To unravel the epigenetic mechanisms that account for transformation-associated silencing, we used a B-cell line isolated from sheep S267, called L267 hereafter, as a model. Provirus expression is completely suppressed in L267 lymphoma B cells, as conclusively demonstrated by their incapacity to generate infection in the in vivo sheep model, and the tumor-derived L267 provirus displays a wild-type sequence that is identical to the infectious provirus used for sheep S267 inoculation (detailed in Materials and Methods).
In a previous study, we demonstrated that it was possible to rescue virus expression from a tax-defective provirus integrated into leukemic B cells (YR2) through recombination-mediated mechanisms using a retroviral vector, LTaxSN, for the transfer of exogenous wild-type Tax (YR2LTaxSN) (46). Here, although there was no genetic defect in the endogenous proviral tax sequences in the lymphoma-derived L267 B cells, we delivered LTaxSN to determine whether expressing Tax in trans could restore the transcriptional activity of the integrated provirus. Using RT-PCR analysis of total RNA isolated from the parental and transduced cell lines with primers specific for the BLV tax and env mRNA, respectively, we found that both the singly and doubly spliced viral mRNAs were transcribed in the L267LTaxSN cells (Fig. 1A and B). This unexpected finding suggested that exogenous Tax was capable of restoring viral expression from the silent integrated provirus, though the level of transcription was obviously lower than that of YR2LTaxSN. Using Northern blotting analysis, we confirmed the presence of the viral transcripts detected by RT-PCR. Both the 4.4-kb singly spliced RNA encoding the envelope proteins and the 2.1-kb doubly spliced RNA encoding both Tax and Rex were identified (Fig. 1C). The weak transcriptional activity suggested that Tax-mediated virus reactivation in the L267LTaxSN cells was much less efficient than in the YR2LTaxSN cell line, despite the presence of similar levels of vector-derived Tax in both cell populations (data not shown).
To provide a conclusive proof for virus expression in L267LTaxSN, two naïve sheep were injected with 107 transduced L267LTaxSN tumor B cells (sheep S2233 and S2311). While injection of the parental silent L267 cells did not lead to infection (sheep S2013 and S2014), animals receiving L267LTaxSN cells were shown to be productively infected with the rescued L267 provirus, and both these animals developed leukemia and lymphoid tumors characterized by monoclonal provirus integration at 30 (S2233) and 36 (S2311) months after inoculation, respectively (Fig. 1D). This observation suggested that despite the weak expression of BLV in L267LTaxSN cells, it was sufficient for providing all the elements required for generating an infectious and oncogenic provirus. Proviral sequences including the complete 5′ LTR, the 3′ region from the tax open reading frame to the end of the 3′ LTR R region, and the splice donor- and acceptor-containing regions for the spliced mRNAs encompassing the end of POL and beginning of ENV (Fig. 1A) were examined in biological samples isolated from these sheep. We found that these sequences were identical to those previously identified in both the infectious BLVX3C provirus used for sheep S267 inoculation and the tumor provirus present in the parental L267 cell line described in Materials and Methods (data not shown). These data suggest that the S2233 and S3211 proviruses are structurally capable of transcription. The tax sequence found in S2233 and S2311 matched the parental L267 tax sequence, indicating that reactivated provirus in L267LTaxSN cells used for inoculation did not result from recombination events with the tax cDNA introduced by the LTaxSN vector that belongs to a different variant (46). In conclusion, our data indicate that viral expression in a transformed B cell carrying a silent but potentially functional and genetically unaltered provirus can be restored when Tax is present and active, suggesting that epigenetic mechanisms in which Tax intervenes, such as chromatin remodeling, might participate in tumor-associated BLV silencing.
Treatment with both the HDAC inhibitor TSA and the DNA methyltransferase (DNMT) inhibitor 5′azacytidine results in activation of the silent BLV provirus in L267-transformed B cells.
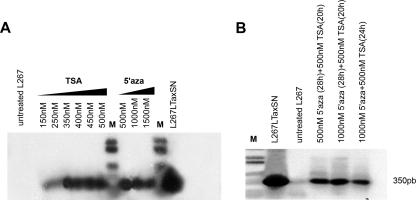
The BLV provirus does not integrate at specific sites in the B-cell genome, although higher-order DNA-chromatin structures might affect the accessibility of proviral DNA to transcriptional factors required for provirus expression (26, 34). The HDAC inhibitors such as TSA or the DNMT inhibitors such as 5′azacytidine are known for their capacity to induce virus expression through mechanisms that involve chromatin remodeling. It has been shown that TSA enhances the level of spontaneous expression resulting from the culture of nontransformed circulating BLV-infected B cells from cattle and sheep (6, 31). However, leukemic or transformed B cells do not spontaneously release virus upon culture, and none of the previous studies addressed the issue of whether a silent provirus integrated in a terminally transformed B-cell clone might be susceptible to reactivation through TSA or 5′azacytidine. To further investigate the molecular mechanisms associated with provirus silencing in tumors, we analyzed the effects of the treatment of L267 B cells with both TSA and 5′azacytidine. First, L267 cells were treated for 22 h with increasing concentrations of either TSA, ranging from 150 nM to 500 nM, or for 48 h with 5′azacytidine concentrations ranging from 500 nM to 1,500 nM, or a combination of both. Using RT-PCR to analyze provirus transcription, we found that TSA and 5′azacytidine individually induced the presence of BLV transcripts and that in addition, treatment with a combination of both drugs was also effective (Fig. 2A and B). Furthermore, the treatment of transduced L267LTaxSN cell lines, characterized by a basic expression level resulting from the presence of LTaxSN, with a combination of 5′azacytidine (1 μM) and TSA (500 nM) did clearly enhance the level of transcription (data not shown). In this case, however, because the drugs might have been activated through the LTaxSN retroviral vector promoter driving Tax expression, it is difficult to draw conclusions as to their effect on the BLV proviral promoter itself. Altogether, our results demonstrate that both 5′azacytidine and TSA are capable of restoring BLV transcription from latency, suggesting that chromatin conformation might play a critical role in establishing and maintaining the silent phenotype of the L267 provirus. Similarly, chromatin-related mechanisms might provide a clue for the unexplained low-level virus reactivation observed when exogenous Tax is expressed in L267LTaxSN cells. These observations support the hypothesis that at least two mechanisms, DNA methylation and histone deacetylation, might contribute to tumor-associated BLV silencing.
FIG. 2.
Treatment of silent L267 transformed B cells with TSA and 5′azacytidine (5′aza) leads to the activation of BLV expression. (A) RT-PCR analysis of BLV transcripts in total RNA isolated from L267 cells treated for 22 h with TSA concentrations of 150 nM, 250 nM, 350 nM, 400 nM, 450 nM, and 500 nM or for 48 h with 5′azacytidine concentrations of 500 nM, 1,000 nM, and 1,500 nM. (B) RT-PCR analysis of BLV transcripts in total RNA isolated from L267 cells treated either for 28 h with 5′azacytidine at 500 nM prior to the addition of TSA at 500 nM for 20 h or for 28 h with 5′azacytidine at 1 μM prior to the addition of TSA at 500 nM for 20 h or L267 cells treated simultaneously for 24 h with either 5′azacytidine at 1 μM or TSA at 500 nM. Amplification was performed using a set of singly spliced (env)-specific primers, LTR-R/EnvC (described in the legend to Fig. 1A). RNAs isolated from L267 and L267LTaxSN cells were used as controls. Amplification products were hybridized with a 32P-labeled BLV full-length proviral DNA probe. M, molecular weight markers.
TSA- and 5′azacytidine-treated L267 transformed B cells release virus that possesses in vivo infectious and oncogenic potential.
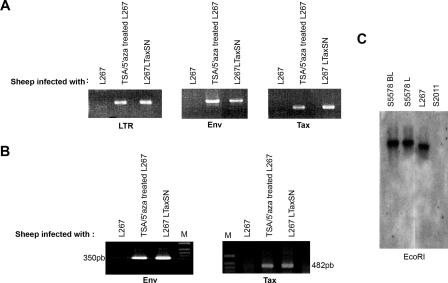
To verify whether the activation of viral expression from the silent L267 tumor provirus was not limited to the production of env and tax transcripts, we examined the infectious potential of the putative rescued BLV in vivo. Sheep were intradermally injected with 2 × 107 5′azacytidine/TSA-treated L267 cells. In parallel, control animals were injected with an equivalent number of either parental L267 lymphoma cells (sheep S2011) or L267LTaxSN cells (sheep S2350). Blood samples were monitored weekly for the detection of BLV-specific seroconversion. Whereas specific antibodies were consistently absent in the control L267-injected animals, both the TSA/5′azacytidine-treated and the L267LTaxSN-infected sheep displayed antibodies to p24 (data not shown). Blood samples were collected 90 days after inoculation and analyzed for the presence of proviral sequences using PCR and different sets of BLV-specific primers. BLV sequences were detected in genomic DNA isolated from the blood of sheep S5578 injected with 5′azacytidine/TSA-treated L267 cells and in that isolated from S2350 inoculated with L267LTaxSN (Fig. 3A). As expected, analysis of BLV expression by RT-PCR in cultured PBMCs isolated from these animals demonstrated the presence of env and tax BLV-specific transcripts (Fig. 3B), suggesting that provirus integrated in PBMCs of 5′azacytidine/TSA-treated L267-injected animals in the aleukemic stage was capable of viral expression upon culture. Sheep S5578 developed B-cell leukemia and lymphoid tumors 20 months after inoculation. Southern blotting analysis of EcoRI-digested DNA isolated from both the leukemic cells and the lymphoid tumors demonstrated the presence of integrated provirus (Fig. 3C). These findings clearly demonstrate the efficient in vivo transfer of BLV genetic information from the TSA/5′azacytidine-treated L267 cells, suggesting that these drugs have the capacity to overcome the strict silencing of a potentially competent provirus and to release its full infectious and oncogenic potential. Altogether, our in vitro and in vivo results using L267 as a model confirm the hypothesis that besides genetic alterations such as provirus mutation, epigenetic chromatin structure-dependent mechanisms play a significant role in the progression and selection of silent transformed B-cell clones.
FIG. 3.
TSA/5′azacytidine treatment of L267 cells results in the rescue of an in vivo infectious and oncogenic virus. (A and B) PCR and RT-PCR analysis of BLV-specific sequences in PBMCs isolated from sheep injected with L267 cells, TSA/5′azacytidine (TSA/5′aza)-treated L267 cells, and L267LTaxSN cells 90 days postinoculation. (A) Proviral DNA was amplified using the LTR-, tax-, and env-specific primers. (B) RT-PCR using two sets of splice-specific primers, LTR-R/EnvC and EnvA/Tax2, for the detection of the singly (env) and doubly (tax) spliced BLV transcripts, respectively. (C) Southern blotting analysis of EcoRI-restricted genomic DNA extracted from PBMCs and lymphoid tumors collected from TSA/5′azacytidine-treated L267-injected sheep S5578 20 months after inoculation (S5578 BL and S5578 L, respectively), PBMCs from sheep injected with the silent L267 parental cell line (S2011), and DNA extracted from the parental L267 cell line. A 32P-labeled BLV full-length proviral DNA probe was used for hybridization in gels shown in panels A, B, and C. Positions of primers are depicted in Fig. 1A.
Rescue from silencing correlates with enhanced DNase I sensitivity in the promoter chromatin region of the L267 lymphoma B-cell integrated provirus.
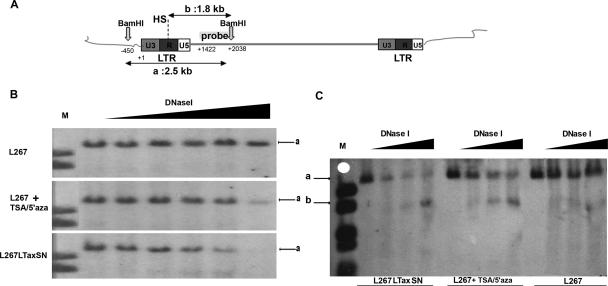
The local chromatin conformation has an impact on the expression of integrated proviral sequences. To determine whether proviral silencing in the L267 transformed B cells was correlated with conformational differences in the chromatin packaging of the proviral LTR region, DNase I treatment hypersensitivity assays were conducted. Intact nuclei isolated from TSA/5′azacytidine-treated L267 and L267LTaxSN cells were treated with increasing amounts of DNase I. DNA was extracted and subsequently digested with the restriction endonuclease BamHI to generate a 2.5-kb fragment encompassing the proviral 5′ LTR (Fig. 4A). After transfer of the digested DNA, filters were hybridized with a probe spanning a 607-bp region recognizing proviral sequences localized downstream from the 5′ LTR to the BamHI site at position 2038 of the provirus. In untreated L267 cells, we identified a single 2.5-kb fragment (Fig. 4B, upper panel, labeled “a”), indicating that the LTR region was protected from DNase I digestion, therefore suggesting a condensed chromatin structure. In contrast, we observed a decrease in the 2.5-kb signal intensity (Fig. 4B and C, labeled “a”) simultaneously with the detection and gradual increase of a band of approximately 1.8 kb (Fig. 4C, labeled “b”) when increasing amounts of DNase I were utilized in TSA/5′azacytidine-treated L267 and L267LTaxSN cells, suggesting that the hypersensitivity of the LTR region to DNase I was associated with the induction of provirus expression. A single hypersensitive site (HS) was detected in both of these virus-expressing cell populations, indicating an open chromatin domain that correlates with the release from silencing. The size of the band resulting from DNase I digestion of the reactivated provirus suggests that the HS is localized within the R region of the 5′ LTR. Altogether, our observations indicate that a compacted close chromatin domain is associated with the silencing of the L267 integrated provirus and that chromatin reorganization results in transcriptional activation.
FIG. 4.
Enhanced DNase I sensitivity correlates with proviral expression. DNase I hypersensitivity mapping is shown. (A) The diagram depicts the localization of the probe, the position, and the length of the fragments generated after DNase I treatment and BamHI digestion. HS, hypersensitive site. (B and C) DNase I hypersensitivity analysis of the BLV LTR region. Cells were treated with 5′azacytidine for 28 h followed by TSA (TSA/5′aza) for 18 h. Nuclei were collected and digested with 0, 50, 60, 70, 80, and 95 units/ml of DNase I (B) and 0, 80, 85, and 90 units/ml DNase I (C) for 10 min at 4°C. Genomic DNA was digested with BamHI and analyzed by Southern blot hybridization using a 607-kb probe spanning provirus sequences downstream of the 5′ LTR (nt 1422 to 2038). Arrow “a” indicates the fragment of approximately 2.5 kb resulting from BamHI digestion and protected from DNase I digestion. Arrow “b” indicates the fragment of approximately 1.8 kb resulting from BamHI restriction and DNase I digestion at the HS.
TSA/5′azacytidine treatment of L267 transformed B cells increases CREB-ATF DNA promoter binding activity in vitro.
It is widely accepted that members of the CREB-ATF complex play an important role in the transcriptional activation of the BLV promoter (1, 2). Tax interacts with cellular transcription factors, including CREB-ATF, to transactivate virus transcription through the TxRE 21-bp repeats localized within the U3 region of the proviral LTR (2). It was previously shown that TSA treatment of BLV-infected aleukemic PBMCs increases the CREB-ATF binding activity (6). In addition, CREB is known for its capacity to recruit histone acetyltransferase (HAT) complexes such as CBP-P300 (27). It is not known, however, whether similar mechanisms might contribute to BLV transcriptional regulation in leukemic B cells. We therefore examined whether TSA/5′azacytidine treatment of L267 cells and reactivation of the silent tumor-associated provirus resulted in altered CREB-ATF binding activity. EMSA were performed using oligonucleotide probes corresponding to either wild-type TxREs or TxREs harboring perfect CRE consensus sequences (6). The nuclear extracts prepared from untreated L267 cells, from L267 cells treated with 5′azacytidine for 24 h followed by TSA treatment for 20 h, and from L267LTaxSN cells were incubated with these different probes. We identified two major retarded protein-DNA complexes, designated C1 and C2 (Fig. 5, upper panel). These complexes correspond to the specific binding of members of the CREB-ATF family, as shown by supershift assays using antibodies directed against CREB-1 and ATF-1 (Fig. 5, lower panel) (1, 6). Whereas binding to the Oct-1 consensus sequence occurred at similar levels for all tested cells, we observed increased binding activity of the CREB-ATF proteins compared to that of the wild-type as well as the perfect CRE consensus versions of the TxRE in both the L267 cells treated with 5′azacytidine/TSA and the L267LTaxSN cells (Fig. 5, upper panel). Our findings indicate that the reactivation of viral expression from a silent integrated provirus in transformed B cells is mediated through the recruitment of CREB-ATF proteins to the TxREs, whether TSA/5′azacytidine or Tax is utilized for restoring viral activity.
FIG. 5.
Release from proviral silencing correlates with enhanced CREB-ATF DNA binding activity in vitro. (Upper panels) Nuclear extracts prepared from L267 cells, TSA/5′azacytidine (TSA/5′aza)-treated L267 cells, and L267LTaxSN cells were incubated with the 32P-end-labeled 21-bp oligonucleotide probes TxRE1, TxRE2, TxRE3, CREB consensus, and OCT (control) and analyzed by EMSA as described in Materials and Methods. The two major retarded DNA-protein complexes C1 and C2 are indicated by arrows. (Lower panels) Supershift assays showing incubation of binding reaction mixtures of L267LTaxSN nuclear extracts with the TxRE1, TxRE2 or TxRE3 probe in the absence of antibody (−) or in the presence of anti-ATF1 or anti-CREB1 antibody. The supershifted complexes are indicated by arrows.
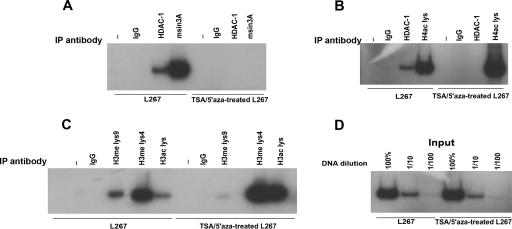
Treatment of L267 cells with TSA and 5′azacytidine decreases the association of HDAC1 and mSin3A with BLV promoter sequences and alters the histone code.
To identify the factors involved in the chromatin conformation associated with L267 provirus silencing in vivo, we sought to determine the effects of both the HDAC inhibitor TSA and the DNMT inhibitor 5′azacytidine on the association of HDAC1 and mSin3a with the BLV promoter sequences. ChIP assays using HDAC-1- and mSin3A-specific antibodies showed that while HDAC1 and mSin3A were associated with the BLV promoter in L267 cells characterized by a silent integrated provirus, TSA/5′azacytidine treatment resulted in the loss of this association (Fig. 6A). To determine whether reactivation of the L267 integrated provirus resulting from TSA/5′azacytidine treatment might be linked to the local hyperacetylation of histone H4 in the vicinity of BLV regulatory sequences, we next performed ChIP assays using antibodies specific for acetylated histone H4 (recognizing histone H4 acetylated at K5, K8, K12, and K16). Acetylated H4 was found to be associated with the BLV promoter in L267 cells, and TSA/5′azacytidine treatment increased the level of this association (Fig. 6B). Furthermore, both methylation and acetylation were examined for histone H3, based on independent studies suggesting a correlation between DNA methylation and histone H3 methylation at lysine 4 (H3-K4) and lysine 9 (H3-K9) in gene repression and heterochromatin organization. The impact of acetylation was assessed using antibodies recognizing both K9- and K14-acetylated histone H3, whereas the extent of methylation was examined using antibodies directed to either K4-dimethylated or K9-dimethylated histone H3. First, we found a robust increase in the ratio of acetylated H3 in TSA/5′azacytidine-treated L267 compared to that of the L267 untreated cells (Fig. 6C), confirming the role of histone hypoacetylation in BLV silencing. Furthermore, although histone H3-K4 was methylated to some extent in L267 cells, the activated promoter in the TSA/5′azacytidine-treated L267 cells appeared to be enriched in methylated H3-K4. H3-K9 methylation, in contrast, was associated with the silent provirus present in the L267 cells, and the level of H3-K9 methylation was significantly decreased in the TSA/5′azacytidine-treated L267 cells. These results are consistent with data that link regional H3-K9 methylation with repressed chromatin and H3-K4 methylation with active chromatin. Taken together, our data suggest that the activation of silent viral genes in malignant BLV-infected B cells involves chromatin remodeling at the promoter sequences where the heterochromatic repressive histone code is reversed and replaced with a euchromatic imprint.
FIG. 6.
ChIP assays reveal a repressive histone code at the silent BLV provirus in L267 lymphoma B cells. The formaldehyde cross-linked chromatin isolated from L267 or TSA/5′azacytidine (TSA/5′aza)-treated L267 cells was incubated with antibodies that detect HDAC1, mSin3A (A and B), H4ac lys (B), H3ac lys, H3me lys9, and H4me lys4 (C) or rabbit immunoglobulin G (IgG) as a control antibody. The immune complexes were pulled down, and the purified DNA was analyzed by radioactive PCR using primers spanning the BLV promoter region containing the three TxREs, from nt 40 to 239. Aliquots of chromatin taken before immunoprecipitation were used as input (D). Different dilutions (100%, 1/10, and 1/100) of input genomic DNA were used for PCR analysis. Chromatins eluted from immunoprecipitations lacking antibody were used as “no antibody” controls and are marked with a −. Data are shown for one representative experiment from three independent assays.
DISCUSSION
Suppression of Tax and virus expression is associated with the malignant phenotype in BLV-transformed cells. A current hypothesis suggests that silencing of provirus expression, a late but critical step for tumor onset and progression, might be a strategy to escape host immune responses. We have previously shown that deletions in the BLV provirus or mutations into functionally important domains of the Tax protein are responsible for transformation-associated virus silencing in ovine B-cell tumors (17, 46, 47). Our rationale for initiating this study was to identify the mechanisms that lead to the complete suppression of viral expression in malignant lymphoid tumors carrying a nondefective provirus. The study presented here suggests that besides genetic changes, epigenetic mechanisms are contributing to tumor-associated BLV silencing. The frequency with which those mechanisms might occur has not been evaluated and would require further investigation. We examined the contribution of epigenetic changes in silencing by using a transformed B-cell clone, L267, isolated from a lymphomatous BLV-infected sheep and carrying a single full-length, nondefective, silent provirus. We found that virus expression was rescued by either the expression of exogenous Tax (L267LTaxSN) or the treatment of L267 B cells with TSA, an HDAC1 inhibitor, or 5′ azacytidine, a DNMT inhibitor, or a combination of both drugs. We examined epigenetic differences between the silent and reactivated single-copy provirus, using the L267 cells, the L267 cells treated with TSA/5′azacytidine, and the L267LTaxSN B-cell populations as a model system for comparison. We demonstrated that silencing was associated with promoter DNA methylation, DNase I protection, and decreased CREB-ATF binding activity. Furthermore, HDAC1 and mSin3A, a corepressor generally found in association with HDAC1 and DNMTs, are associated with the inactive promoter, while they are excluded after TSA/5′azacytidine treatment. Concurrently, both histone H3 and H4 lysines are extensively acetylated at the active promoter. Histone H3 lysine 9, an epigenetic tag characteristic of heterochromatin and DNA sequences that are constitutively methylated in normal cells (21), is methylated in silent L267 cells. It loses its methyl group(s) as a result of TSA/5′azacytidine treatment, while H3 lysine 4 exhibits an increase in methylation.
The compactness of the 5′ LTR chromatin is so tight that it cannot be relieved by ex vivo culture of fresh L267 lymphoma cells or of the cell line derived from this tumor (M. Merimi et al., submitted for publication). Highly sensitive techniques such as RT-PCR or in vivo infection trials with naïve sheep show no evidence of transcription of the provirus in the lymphoma cells submitted to these treatments. In contrast, transformed cells treated with either 5′azacytidine or TSA or both drugs produce viral RNA in cell culture and readily infect naïve sheep in infectivity trials, suggesting evidence that favors the release of infectious virus particles by the injected cells. Our data, therefore, suggest that both DNA methylation and histone deacetylation cooperate to abrogate BLV expression in sheep lymphoma cells transformed by the virus.
Our findings suggesting a correlation of DNA methylation with the silent phenotype in the transformed L267 B cells are in contrast with those of earlier studies indicating the absence of or minimal CpG methylation of the U3/R region of the BLV LTR in infected cattle and sheep at various clinical stages (44). In the case of sheep, however, the analysis was limited to cells collected before tumor onset, while our work aimed at identifying transformation-associated mechanisms through the study of a clonal transformed B-cell population with a single integrated provirus. DNA methylation of CpG dinucleotides, which represent the target for covalent modification of DNA, is one of the major mechanisms in the epigenetic regulation of genes. Protrusion of the methyl group into the major groove of the DNA gives two major effects: displacement of transcription factors, such as CREB-ATF, that normally bind to the DNA and attraction of methyl-binding proteins, such as MeCP2, which are functionally associated with gene silencing and chromatin compaction, probably via interactions with complexes containing corepressor molecules such as HDACs and mSin3A that modify the tails of histone proteins. On the other hand, histone tails can be modified by several posttranslational mechanisms including acetylation, methylation, phosphorylation, and ubiquitination to create potential combinations that have been referred to as histone codes in which regulatory information is concealed.
Interestingly, ectopic expression of Tax via the LTaxSN vector relieves the repressive impact acting on the provirus, as shown by viral RNA expression in transduced cells and by virus release in the naïve host as shown by infectivity trials. Retroviral transactivators, such as Tax of HTLV-1 (10, 25) or Tat of human immunodeficiency virus type 1 (HIV-1) (3, 49), have been shown previously to have a relaxing impact on epigenetic silencers of integrated proviruses. Our data corroborate these observations. The mechanisms governing Tax-associated activation might, in part, result from the interaction of the LTaxSN-derived Tax with transcriptional repressors such as HDACs, known to play a crucial role in maintaining the balance between the acetylated and deacetylated states of lysine residues in the histone tails (14, 37). Studies by Lemasson et al. suggest that transcriptional regulation at the HTLV-1 promoter sequences is mediated through the mutually exclusive binding of Tax and HDACs (23-25). HDAC1 directly interacts with Tax both in vivo and in vitro, and levels of acetylated H3 and H4 in the vicinity of the HTLV-1 LTR were enhanced in the presence of Tax, consistent with the association of CBP with the LTR (28). It is well documented that Tax interacts with HATs such as p300, CBP, and PCAF (4, 11). Furthermore, p300 is capable of activating Tax-mediated HTLV-1 chromatin transcription through targeted acetylation of nucleosomal histones (11, 27). The potential interaction of Tax with HDAC1 in L267LTaxSN cells might thus be part of the transcriptional activation pathway. Tax can decrease HDAC1 binding to the template DNA by inhibiting the binding and/or by dissociating bound HDAC1.
The stimulatory effect of TSA on virus expression correlates positively with the positive effect of Tax and points to an important role for HDACs in provirus silencing. Previous data from our group and others showed an increase in BLV expression after TSA treatment of cultured PBMCs (6, 31), but tumor cells with a completely silent provirus had not been examined so far. Interestingly, sheep injected with TSA- and 5′azacytidine-treated L267 cells were infected and developed tumors, conclusively demonstrating that the induction of silent provirus was not limited to mRNA expression but also leads to the production of infectious and oncogenic virus.
Comparing L267 cells, TSA/5′azacytidine-treated L267 cells, and L267LTaxSN cells, we identified a single DNase I hypersensitive site located in the R region of the LTR. In the parental L267 cell line, the LTR region was resistant to DNase I digestion, suggesting the presence of a nucleosome at the R region, as shown in the case of HIV (8), indicating that chromatin condensation at the LTR region is contributing to the silent phenotype and that activation mediated by TSA/5′azacytidine or Tax is associated with chromatin remodeling. Furthermore, reactivation of the silent integrated provirus after treatment with either TSA/5′azacytidine or Tax resulted in a significant increase in CREB-ATF binding activity to the BLV CREs, emphasizing conclusions from previous studies with nontransformed BLV-infected PBMC or YR2, a B-cell line characterized by a silent-defective Tax-mutated provirus (6, 33).
Using ChIP experiments, we found that HDAC1 and mSin3A were associated with the inactive promoter, whereas enhanced levels of acetylated histones H3 and H4 were present at the BLV LTR when the provirus was reactivated using TSA and 5′azacytidine treatment. Histone acetylation is among the best-characterized amino-terminal modifications and has been shown to correlate with transcriptional stimulation through changes in the chromatin structure. Among other histone tail modifications, histone lysine methylation, through histone methyltransferases (HMTases) that direct the site-specific methylation of, for example, the lysine 4 and lysine 9 in the histone H3 amino terminus, is a central modification for the epigenetic organization of the genome (20, 21). At least five methylable lysines exist in the N terminus of H3 (K4, K9, K27, and K36) and H4 (K20) histones. Fully activated promoters appear to be enriched in methylated H3 lysine 4. H3 lysine 9 methylation, by contrast, has been linked mainly to gene silencing (21). We have examined the methylation status of H3 lysine 9 and lysine 4 in L267 native chromatin, and our findings are consistent with opposite roles in structuring repressive or accessible chromatin domains. Interestingly, a model suggests that histone lysine methylation can direct DNA methylation. H3 lysine 9-methylated chromatin recruits a DNMT for subsequent DNA methylation. Since methylated DNA is recognized by DNA methyl binding proteins such as MeCP2, which in turn can associate with HDACs, DNA methylation could even feed back to facilitate histone methylation of hypoacetylated chromatin, thereby reinforcing the repressive impact of these two distinct methylation layers on the silent integrated L267 provirus.
Epigenetic mechanisms such as DNA methylation are tightly connected to cancer. It is well documented that oncogenesis is promoted by local hypermethylation of tumor suppressor genes (7, 41). Here, local hypermethylation and chromatin condensation result in silencing of viral information, which might lead to immune evasion and facilitate tumor progression. The finding that the provirus in L267 cells is marked by silent chromatin established via the dynamic interplay of multiple epigenetic mechanisms identifies a second distinct mechanism involved in the complete suppression of virus/oncogene Tax expression. In attributing genetic or epigenetic modifications, there is now evidence supporting both pictures, and they are not mutually exclusive. The data presented in this study suggest that there is cross talk between genes that contributes to cell transformation and genes that provoke the epigenetic repression of provirus transcription. The two gene populations might overlap to some extent. Cellular memory of such modifications has to be ensured for the initiation and propagation of cancer cells. It will be important to study the regulation of host gene expression and to characterize the signaling pathways (such as, for example, the Wnt, Hedgehog, Rb, Akt, and Notch pathways) that can be disrupted at the onset of the malignant clone (15, 22).
Overall, these results allow us to approach the mechanistic interplay between B-cell transformation and BLV silencing and point to the possibility of using HDAC and DNMT inhibitors to combat virus-induced tumors (12, 30). In the case of BLV-infected sheep, it is expected that the reversal of virus and Tax epigenetic silencing in response to these inhibitors might elicit effective antitumor immune responses. It remains to be determined whether the immune response resulting from the activation of Tax expression will have the capacity to limit tumor progression and to override the potential oncogenic effects that might result from this rescued expression. More generally, the derepression of silenced genes in tumor cells might lead to progress in the clinical treatment of some cancers, especially in combination with chemotherapeutic drugs.
Acknowledgments
This work was supported by the Fonds National de la Recherche Scientifique (FNRS), the Medic Foundation, the International Brachet Foundation, the Fondation Bekales, and les Amis de l'Institut Bordet (Y.C.) and by Télévie grants to M.M. and M.S.
Sheep S267, from which the cell lines examined in this work were derived, is an animal from a study conducted by L. Willems.
The authors are grateful to J. M. Londes and G. Vandendaele for experimental help.
Footnotes
Published ahead of print on 28 March 2007.
REFERENCES
- 1.Adam, E., P. Kerkhofs, M. Mammerickx, A. Burny, R. Kettman, and L. Willems. 1996. The CREB, ATF-1, and ATF-2 transcription factors from bovine leukemia virus-infected B lymphocytes activate viral expression. J. Virol. 70:1990-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, E., P. Kerkhofs, M. Mammerickx, R. Kettmann, A. Burny, L. Droogmans, and L. Willems. 1994. Involvement of the cyclic AMP-responsive element binding protein in bovine leukemia virus expression in vivo. J. Virol. 68:5845-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agbottah, E., L. Deng, L. O. Dannenberg, A. Pumfery, and F. Kashanchi. 2006. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azran, I., K. T. Jeang, and M. Aboud. 2005. High levels of cytoplasmic HTLV-1 Tax mutant proteins retain a Tax-NF-kappaB-CBP ternary complex in the cytoplasm. Oncogene 24:4521-4530. [DOI] [PubMed] [Google Scholar]
- 5.Burny, A., L. Willems, I. Callebaut, E. Adam, I. Cludts, F. Dequiedt, L. Droogmans, C. Grimonpont, P. Kerkhofs, M. Mammerickx, D. Portetelle, A. Van den Broeke, and R. Kettman. 1994. Bovine leukemia virus: biology and mode of transformation, p. 313-334. In A. C. Minson, J. C. Neil, and M. A. McRae (ed.), Viruses and cancer. Cambridge University Press, Cambridge, United Kingdom.
- 6.Calomme, C., A. Dekoninck, S. Nizet, E. Adam, T. L. Nguyen, A. Van den Broeke, L. Willems, R. Kettmann, A. Burny, and C. Van Lint. 2004. Overlapping CRE and E box motifs in the enhancer sequences of the bovine leukemia virus 5′ long terminal repeat are critical for basal and acetylation-dependent transcriptional activity of the viral promoter: implications for viral latency. J. Virol. 78:13848-13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, P. M., V. Bovenzi, and M. Szyf. 2004. Methylated DNA-binding protein 2 antisense inhibitors suppress tumourigenesis of human cancer cell lines in vitro and in vivo. Carcinogenesis 25:499-507. [DOI] [PubMed] [Google Scholar]
- 8.Demonte, D., V. Quivy, Y. Colette, and C. Van Lint. 2004. Administration of HDAC inhibitors to reactivate HIV-1 expression in latent cellular reservoirs: implications for the development of therapeutic strategies. Biochem. Pharmacol. 68:1231-1238. [DOI] [PubMed] [Google Scholar]
- 9.Djilali, S., and A. L. Parodi. 1989. The BLV-induced leukemia-lymphosarcoma complex in sheep. Vet. Immunol. Immunopathol. 22:233-244. [DOI] [PubMed] [Google Scholar]
- 10.Ego, T., Y. Ariumi, and K. Shimotohno. 2002. The interaction of HTLV-1 Tax with HDAC1 negatively regulates the viral gene expression. Oncogene 21:7241-7246. [DOI] [PubMed] [Google Scholar]
- 11.Georges, S. A., H. A. Giebler, P. A. Cole, K. Luger, P. J. Laybourn, and J. K. Nyborg. 2003. Tax recruitment of CBP/p300, via the KIX domain, reveals a potent requirement for acetyltransferase activity that is chromatin dependent and histone tail independent. Mol. Cell. Biol. 23:3392-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gore, S. D., S. Baylin, E. Sugar, H. Carraway, C. B. Miller, M. Carducci, M. Grever, O. Galm, T. Dauses, J. E. Karp, M. A. Rudek, M. Zhao, B. D. Smith, J. Manning, A. Jiemjit, G. Dover, A. Mays, J. Zwiebel, A. Murgo, L. J. Weng, and J. G. Herman. 2006. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 66:6361-6369. [DOI] [PubMed] [Google Scholar]
- 13.Hanon, E., R. E. Asquith, G. P. Taylor, Y. Tanaka, J. N. Weber, and C. R. Bangham. 2000. High frequency of viral protein expression in human T cell lymphotropic virus type 1-infected peripheral blood mononuclear cells. AIDS Res. Hum. Retroviruses 16:1711-1715. [DOI] [PubMed] [Google Scholar]
- 14.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 15.Hayward, S. D., J. Liu, and M. Fujimuro. 2006. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci. STKE 2006:re4. [DOI] [PubMed] [Google Scholar]
- 16.Jeang, K. T., C. Z. Giam, F. Majone, and M. Aboud. 2004. Life, death, and tax: role of HTLV-I oncoprotein in genetic instability and cellular transformation. J. Biol. Chem. 279:31991-31994. [DOI] [PubMed] [Google Scholar]
- 17.Kettmann, R., Y. Cleuter, D. Gregoire, and A. Burny. 1985. Role of the 3′ long open reading frame region of bovine leukemia virus in the maintenance of cell transformation. J. Virol. 54:899-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kettmann, R., J. Deschamps, Y. Cleuter, D. Couez, A. Burny, and G. Marbaix. 1982. Leukemogenesis by bovine leukemia virus: proviral DNA integration and lack of RNA expression of viral long terminal repeat and 3′ proximate cellular sequences. Proc. Natl. Acad. Sci. USA 79:2465-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klener, P., M. Szynal, Y. Cleuter, M. Merimi, H. Duvillier, F. Lallemand, C. Bagnis, P. Griebel, C. Sotiriou, A. Burny, P. Martiat, and A. Van den Broeke. 2006. Insights into gene expression changes impacting B-cell transformation: cross-species microarray analysis of bovine leukemia virus tax-responsive genes in ovine B cells. J. Virol. 80:1922-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lachner, M., R. O'Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117-2124. [DOI] [PubMed] [Google Scholar]
- 21.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 22.Lan, K., T. Choudhuri, M. Murakami, D. A. Kuppers, and E. S. Robertson. 2006. Intracellular activated Notch1 is critical for proliferation of Kaposi's sarcoma-associated herpesvirus-associated B-lymphoma cell lines in vitro. J. Virol. 80:6411-6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2002. Transcription factor binding and histone modifications on the integrated proviral promoter in human T-cell leukemia virus-I-infected T-cells. J. Biol. Chem. 277:49459-49465. [DOI] [PubMed] [Google Scholar]
- 24.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2004. Transcription regulatory complexes bind the human T-cell leukemia virus 5′ and 3′ long terminal repeats to control gene expression. Mol. Cell. Biol. 24:6117-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2006. Tax-dependent displacement of nucleosomes during transcriptional activation of human T-cell leukemia virus type 1. J. Biol. Chem. 281:13075-13082. [DOI] [PubMed] [Google Scholar]
- 26.Li, H. P., Y. W. Leu, and Y. S. Chang. 2005. Epigenetic changes in virus-associated human cancers. Cell Res. 15:262-271. [DOI] [PubMed] [Google Scholar]
- 27.Lu, H., C. A. Pise-Masison, T. M. Fletcher, R. L. Schiltz, A. K. Nagaich, M. Radonovich, G. Hager, P. A. Cole, and J. N. Brady. 2002. Acetylation of nucleosomal histones by p300 facilitates transcription from tax-responsive human T-cell leukemia virus type 1 chromatin template. Mol. Cell. Biol. 22:4450-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, H., C. A. Pise-Masison, R. Linton, H. U. Park, R. L. Schiltz, V. Sartorelli, and J. N. Brady. 2004. Tax relieves transcriptional repression by promoting histone deacetylase 1 release from the human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 78:6735-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mammerickx, M., R. Palm, D. Portetelle, and A. Burny. 1988. Experimental transmission of enzootic bovine leukosis to sheep: latency period of the tumoral disease. Leukemia 2:103-107. [PubMed] [Google Scholar]
- 30.Maslak, P., S. Chanel, L. H. Camacho, S. Soignet, P. P. Pandolfi, I. Guernah, R. Warrell, and S. Nimer. 2006. Pilot study of combination transcriptional modulation therapy with sodium phenylbutyrate and 5-azacytidine in patients with acute myeloid leukemia or myelodysplastic syndrome. Leukemia 20:212-217. [DOI] [PubMed] [Google Scholar]
- 31.Merezak, C., M. Reichert, L. C. Van, P. Kerkhofs, D. Portetelle, L. Willems, and R. Kettmann. 2002. Inhibition of histone deacetylases induces bovine leukemia virus expression in vitro and in vivo. J. Virol. 76:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, P. W., H. Iha, Y. Iwanaga, M. Bittner, Y. Chen, Y. Jiang, G. Gooden, J. M. Trent, P. Meltzer, K. T. Jeang, and S. L. Zeichner. 2001. Genome-wide expression changes induced by HTLV-1 Tax: evidence for MLK-3 mixed lineage kinase involvement in Tax-mediated NF-kappaB activation. Oncogene 20:4484-4496. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, T. L., C. Calomme, G. Wijmeersch, S. Nizet, E. Veithen, D. Portetelle, Y. de Launoit, A. Burny, and C. Van Lint. 2004. Deacetylase inhibitors and the viral transactivator TaxBLV synergistically activate bovine leukemia virus gene expression via a cAMP-responsive element- and cAMP-responsive element-binding protein-dependent mechanism. J. Biol. Chem. 279:35025-35036. [DOI] [PubMed] [Google Scholar]
- 34.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 35.Portetelle, D., M. Mammerickx, and A. Burny. 1989. Use of two monoclonal antibodies in an ELISA test for the detection of antibodies to bovine leukaemia virus envelope protein gp51. J. Virol. Methods 23:211-222. [DOI] [PubMed] [Google Scholar]
- 36.Powers, M. A., and K. Radke. 1992. Activation of bovine leukemia virus transcription in lymphocytes from infected sheep: rapid transition through early to late gene expression. J. Virol. 66:4769-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quivy, V., C. Calomme, A. Dekoninck, D. Demonte, F. Bex, I. Lamsoul, C. Vanhulle, A. Burny, and L. C. Van Lint. 2004. Gene activation and gene silencing: a subtle equilibrium. Cloning Stem Cells 6:140-149. [DOI] [PubMed] [Google Scholar]
- 38.Rice, N. R., R. M. Stephens, A. Burny, and R. V. Gilden. 1985. The gag and pol genes of bovine leukemia virus: nucleotide sequence and analysis. Virology 142:357-377. [DOI] [PubMed] [Google Scholar]
- 39.Rice, N. R., R. M. Stephens, D. Couez, J. Deschamps, R. Kettmann, A. Burny, and R. V. Gilden. 1984. The nucleotide sequence of the env gene and post-env region of bovine leukemia virus. Virology 138:82-93. [DOI] [PubMed] [Google Scholar]
- 40.Sagata, N., T. Yasunaga, J. Tsuzuku-Kawamura, K. Ohishi, Y. Ogawa, and Y. Ikawa. 1985. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 82:677-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szyf, M. 2003. Targeting DNA methylation in cancer. Ageing Res. Rev. 2:299-328. [DOI] [PubMed] [Google Scholar]
- 42.Szynal, M., Y. Cleuter, T. Beskorwayne, C. Bagnis, C. Van Lint, P. Kerkhofs, A. Burny, P. Martiat, P. Griebel, and A. Van den Broeke. 2003. Disruption of B-cell homeostatic control mediated by the BLV-Tax oncoprotein: association with the upregulation of Bcl-2 and signaling through NF-kappaB. Oncogene 22:4531-4542. [DOI] [PubMed] [Google Scholar]
- 43.Tajima, S., Y. Ikawa, and Y. Aida. 1998. Complete bovine leukemia virus (BLV) provirus is conserved in BLV-infected cattle throughout the course of B-cell lymphosarcoma development. J. Virol. 72:7569-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tajima, S., M. Tsukamoto, and Y. Aida. 2003. Latency of viral expression in vivo is not related to CpG methylation in the U3 region and part of the R region of the long terminal repeat of bovine leukemia virus. J. Virol. 77:4423-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda, S., M. Maeda, S. Morikawa, Y. Taniguchi, J. Yasunaga, K. Nosaka, Y. Tanaka, and M. Matsuoka. 2004. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer 109:559-567. [DOI] [PubMed] [Google Scholar]
- 46.Van den Broeke, A., C. Bagnis, M. Ciesiolka, Y. Cleuter, H. Gelderblom, P. Kerkhofs, P. Griebel, P. Mannoni, and A. Burny. 1999. In vivo rescue of a silent tax-deficient bovine leukemia virus from a tumor-derived ovine B-cell line by recombination with a retrovirally transduced wild-type tax gene. J. Virol. 73:1054-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van den Broeke, A., Y. Cleuter, G. Chen, D. Portetelle, M. Mammerickx, D. Zagury, M. Fouchard, L. Coulombel, R. Kettmann, and A. Burny. 1988. Even transcriptionally competent proviruses are silent in bovine leukemia virus induced tumor cells. Proc. Natl. Acad. Sci. USA 85:9263-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van den Broeke, A., Y. Cleuter, L. Droogmans, A. Burny, and R. Kettmann. 1997. Isolation and culture of B lymphoblastoid cell lines from bovine leukemia virus-induced tumors, p. 2127-2132. In Y. Lefkovits (ed.), Immunology methods manual: in vitro experimental immunology in sheep. Academic Press, St. Louis, MO.
- 49.Van Lint, C. 2000. Role of chromatin in HIV-1 transcriptional regulation. Adv. Pharmacol. 48:121-160. [DOI] [PubMed] [Google Scholar]
- 50.Willems, L., R. Kettmann, F. Dequiedt, D. Portetelle, V. Voneche, I. Cornil, P. Kerkhofs, A. Burny, and M. Mammerickx. 1993. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 67:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willems, L., D. Portetelle, P. Kerkhofs, G. Chen, A. Burny, M. Mammerickx, and R. Kettmann. 1992. In vivo transfection of bovine leukemia provirus into sheep. Virology 189:775-777. [DOI] [PubMed] [Google Scholar]
- 52.Willems, L., E. Thienpont, P. Kerkhofs, A. Burny, M. Mammerickx, and R. Kettmann. 1993. Bovine leukemia virus, an animal model for the study of intrastrain variability. J. Virol. 67:1086-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]