Abstract
The central DNA flap is an important component of lentiviral vectors, but its significance in the context of wild-type human immunodeficiency virus (HIV) is currently unclear. To address this issue, we have compared the in vitro infection kinetics of NL4-3 with those of a flap-deficient mutant and evaluated the in vivo growth characteristics of these viruses by using the SCID-hu mouse model of HIV infection. Flap-deficient virus was only modestly attenuated in vitro, as assessed by single-round and spreading infection assays, and exhibited levels of replication and pathogenesis close to those of the wild-type in vivo. Hence, an intact central flap is not essential for HIV replication.
All lentiviruses, including human immunodeficiency virus (HIV), have the capacity to productively infect nondividing cells (20, 22, 28, 39, 42). However, the specific mechanisms responsible for translocating the HIV preintegration complex through an interphasic nuclear membrane have yet to be fully elucidated (reviewed in references 10 and 43). One component of the HIV preintegration complex with a suggested role in nuclear import is a triple-stranded DNA structure termed the “central DNA flap” (44). The central flap is formed during reverse transcription, when approximately 99 nucleotides between the central polypurine tract (cPPT) and the central termination sequence are displaced during plus-strand DNA synthesis (13-15, 44). The addition of sequences encompassing the central flap to lentiviral vectors usually enhances their infectivity by approximately 2- to 10-fold or more (19, 31, 36, 41). Nevertheless, the level of enhancement bestowed by the central flap may differ substantially between that of lentiviral vectors and that of wild-type (Wt) HIV. This issue has been investigated by several groups, leading to conflicting reports in the literature regarding the requirement for the central DNA flap in viral replication. Some reports asserted that the central DNA flap was either essential (44) or very important (5, 16) for productive HIV infection, but others suggest a much more minor role in virus replication (29, 18). Hence, while the central flap is a highly conserved genetic element, its importance during Wt HIV infection remains uncertain. Furthermore, the contribution of the central flap to HIV replication and pathogenesis within an in vivo system has not previously been described.
To address these issues, we first constructed a flap-deficient (F−) mutant of Wt HIV, NL4-3 (1), by modifying 10 nucleotides in the cPPT and central U box (23) by using site-directed mutagenesis (Fig. 1A) as previously described (18, 29, 44). In order to accurately quantify HIV expression in single-round infection assays, a new NL4-3-based reporter virus was also constructed (Fig. 1B). This virus expresses the enhanced green fluorescent protein (EGFP) (12, 38) fused to firefly luciferase (17) (EGFPLuc) in place of HIV Env but maintains the integrity of all other HIV open reading frames and important cis-acting RNA elements. The proviral plasmid was generated as previously described for NL4-3-GFP (37), except that the primers 5′-ATTGGGTACCTAAGGCCGCGCTACCGGTCGCCACCATG-3′ and 5′-GTCCGTGCTAGCTTACAGCTCGTCCTTCACGGCGATCTTTCCGCCCTT-3′ were used to amplify the EGFPLuc coding sequence by PCR using pEGFPLuc (Clontech) as the template. The cPPT-D modifications were introduced into this denv(Wt) vector to produce a corresponding denv(F−) reporter virus genome. Resultant plasmids were each cotransfected into 293FT cells (Invitrogen) with a vesicular stomatitis virus G glycoprotein-expressing vector to generate pseudotype viruses (11) for use in single-round infection assays. All subsequent in vitro infections were independently performed at least twice.
FIG. 1.
Genetic structure and initial characterization of Wt and F− viruses. (A) Location of the cPPT and central termination sequence (CTS) within HIV-1 NL4-3. The 10 (cPPT-D) nucleotide changes underscored in box F− were introduced into Wt NL4-3 by site-directed mutagenesis. These changes have been previously described as sufficient to abrogate central flap formation in NL4-3 and several other isolates of HIV-1 (18, 29, 44). Mutant (F−) and Wt replication-competent viruses were generated by transient transfection of 293FT cells. (B) Schematic diagram of denv(Wt), a new NL4-3-based reporter virus that expresses the enhanced green fluorescent protein fused to firefly luciferase (EGFPLuc) in place of HIV Env. An endoplasmic reticulum retention sequence (KDEL) (33) is present at the 3′ end of the EGFPLuc coding sequence. The cPPT-D mutation was introduced into this vector to provide a corresponding denv(F−) reporter virus genome. (C) 293FT cells were infected with denv(Wt) or denv(F−) viruses and visualized for GFP expression by fluorescent microscopy (magnification, ×20) at 3 days postinfection. (D) 293FT cells were infected with denv(Wt) and denv(F−) viruses, and quadruplicate wells were harvested daily for a luciferase assay. Mean values ± standard deviations of quadruplicate infections are shown.
We established the functionality of these new reporter viruses by infecting 293FT cells. 293FT cells (105) were infected with denv(Wt) or denv(F−) viruses (6.25 pg of p24/infection) and visualized for GFP expression at 3 days postinfection. GFP expression was absent from mock-infected cells, but abundant GFP was observed for cells infected with each reporter virus (Fig. 1C). A Luc activity time course was also conducted, and increasing levels of Luc activity were detected in infected wells (Fig. 1D).
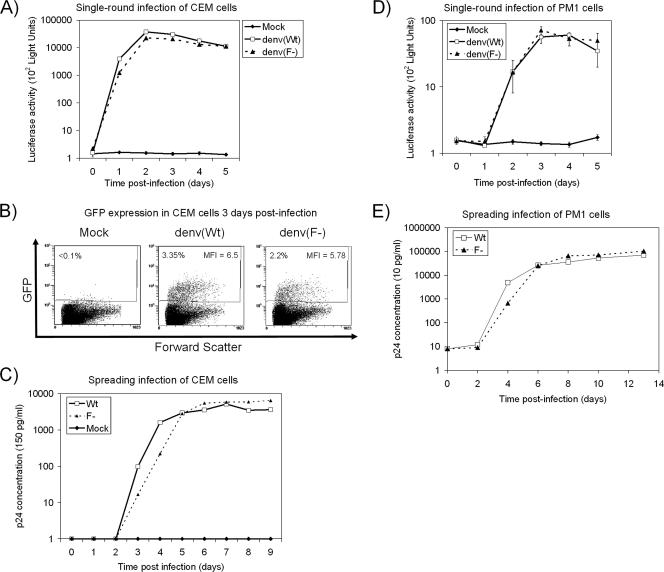
We then tested the Wt and F− viruses with single-round and spreading infection assays using transformed CD4+ T-cell lines. CEM cells (5 × 105) were infected with denv(Wt) or denv(F−) pseudotype viruses (50 ng of p24/infection), and cells were harvested at various times for quantification of Luc activity. Very similar expression kinetics were evident for the two viruses (Fig. 2A). When GFP expression was assessed by flow cytometry at 3 days postinfection, a slightly higher percentage of GFP-positive (GFP+) cells was present in denv(Wt) than in denv(F−) infections (3.35% versus 2.2%, respectively), but the mean fluorescent intensity of GFP+ cells was comparable for each virus (Fig. 2B). Spreading infection characteristics for Wt and F− viruses in CEM cells were also established. CEM cells (105) were infected with Wt or F− replication-competent viruses (10 ng of p24/infection), and aliquots of supernatant were assayed for p24 concentration (Fig. 2C). The F− virus showed slightly delayed replication kinetics (approximately 1 day slower than those of Wt) but was not severely debilitated. Single-round and spreading infection assays performed in the same way using PM1 (30) target cells yielded similar results (Fig. 2D and E).
FIG. 2.
Expression and replication kinetics of Wt and F− viruses in transformed CD4+ T-cell lines. (A) CEM cells were either mock infected or infected with equivalent amounts (50 ng of p24) of denv(Wt) or denv(F−) pseudotype viruses. Infected cells were then assessed for luciferase activity in quadruplicate at the indicated times (error bars representing standard deviations are present at all data points but in most cases are too small to be visible). (B) CEM cells infected as described in the legend to panel A were harvested at 3 days postinfection, and GFP expression was quantified using flow cytometry. The percentage of GFP+ cells in each infection and the mean fluorescent intensity (MFI) of GFP+ cells are indicated on scatter plots. (C) CEM cells were either mock infected or infected with Wt or F− replication-competent viruses. After infections, the cells were washed, and aliquots of cell-free supernatant were removed at the indicated times for quantitation of p24 by enzyme-linked immunosorbent assay. (D) PM1 cells were mock infected or infected with either denv(Wt) or denv(F−) pseudotype viruses in the same manner as described for CEM cells in the legend to panel A, and quadruplicate wells were harvested at the indicated times (mean values ± standard deviations are shown). (E) PM1 cells were infected with replication-competent viruses as described for CEM cells in the legend to panel C and assayed for p24 production.
The different viruses were then compared using primary target cells. First, CD4+ T cells were isolated by negative selection from peripheral blood leukocytes, using immunomagnetic beads (CD4+ T-cell isolation kit II; Miltenyi Biotech). Cells were costimulated for 2 days and then subjected to both single-round (Fig. 3A) and spreading infection (Fig. 3B) assays as described above for CEM cells. The differences between Wt and F− virus infections with these primary target cells were greater than those for transformed T-cell lines, but expression levels were still quite robust for the mutant viruses. A spreading infection assay using 2 × 107 primary fetal thymocyte target cells (prepared as previously described in reference 27) and 100 ng of p24 for each virus showed a similar, modest reduction in replication capacity with the F− virus (Fig. 3C).
FIG. 3.
Expression and replication kinetics of Wt and F− viruses in primary cells. (A) Primary CD4+ T cells were costimulated using anti-CD3 and anti-CD28 antibodies in the presence of 20 U/ml interleukin-2 and then infected with denv(Wt) or denv(F−) pseudotype viruses as described for CEM cells in the Fig. 2A legend. Cells were harvested for luciferase assay at the indicated times (mean values ± standard deviations of quadruplicate wells are shown). (B) Stimulated primary CD4+ T cells were infected with Wt or F− replication-competent viruses as described for CEM cells in the Fig. 2C legend, and aliquots of cell-free supernatant were removed at the indicated times for quantification of p24 by enzyme-linked immunosorbent assay. (C) Primary thymocytes were infected with Wt or F− virus and then washed and resuspended in 4 ml of medium containing 20 U/ml interleukin-2 and 20 ng/ml interleukin-4. Cell-free supernatants were removed at the indicated times for quantitation of p24.
It was possible that the central flap would have had a more profound effect upon HIV spread and pathogenesis in vivo than was evident during in vitro assays. The SCID-hu thymus/liver (Thy/Liv) mouse model (32, 35) of HIV infection is an extremely versatile in vivo system for characterizing HIV replication efficiency and pathogenesis (2, 7, 34). This model has been successfully utilized for comparing different HIV strains (6, 9, 25, 26), as well as accessory gene mutants (3, 4, 24) and viruses harboring mutations in other regions of the HIV genome such as the Rev response element (40). Hence, this model is particularly well suited for testing the in vivo effects of disrupting the HIV central DNA flap.
Thy/Liv implants within SCID-hu mice were mock infected or inoculated with equivalent amounts (10 ng of p24) of Wt or F− viruses, and then wedge biopsies of implants were done at week 3 postinfection, and the same mice were sacrificed and reassessed at week 5 postinfection, as previously described (3, 8). Levels of HIV replication within the tissue samples were determined by real-time PCR using primers designed to detect the R/U5 regions of the viral long terminal repeat (21). A PCR assay for human β-globin sequences was performed in parallel to standardize DNA input (Fig. 4A). Sequencing of several week-5 DNA samples (Fig. 4A and B) from F−-infected implants confirmed that the cPPT-D mutations had not reverted during the infections (data not shown).
FIG. 4.
In vivo replication and pathogenesis of mutant and Wt viruses. Implants of SCID-hu (Thy/Liv) mice were mock infected or inoculated with Wt or F− replication-competent viruses and assessed at week 3 and week 5 postinfection. (A) Real-time PCR assays for HIV DNA and human β-globin DNA were performed using samples obtained at each time point. Values from all samples are shown individually. Gray icons represent those samples where sequencing was performed to verify maintenance of the cPPT-D mutations throughout infections. Black icons represent the samples where detailed flow cytometry scatter plots are shown in panel C. Differences between viral loads of mock-infected and infected samples are significant (P < 0.001) at both time points. The viral loads in tissues infected with Wt virus are significantly higher than those of tissues infected with the F− mutant at week 3 (P < 0.01) but not at week 5 (P > 0.2). (B) CD45+ (human) cells were analyzed for CD4:CD8 profiles by flow cytometry to assess depletion of CD4+ CD8+ cells by the different viruses. Values are expressed as the percentage of CD4+ CD8+ double-positive cells present in each implant. Mock values are significantly higher than those of infected samples at both time points (P < 0.001). Percentage values for the F− virus are significantly higher than those for Wt at both week 3 (P < 0.05) and week 5 (P < 0.02). (C) Scatter plots showing flow cytometry profiles of samples at, or closest to, the median percentage of CD4+ CD8+ cells within each group. All statistical comparisons were performed using the Wilcoxon rank-sum test (two sided). A second, independent experiment using different human donor tissue to generate the SCID-hu (Thy/Liv) mice yielded very similar results to those presented here (data not shown).
Thymocytes were stained for a flow cytometry assay, and CD4:CD8 profiles were used to assess the pathogenic potential of each virus, as defined by their ability to deplete CD4+ CD8+ double-positive cells (Fig. 4C). The F− virus replicated with an efficiency level close to that of the Wt (Fig. 4A) and caused depletion of CD4+ CD8+ thymocytes in a manner similar to that of the Wt but with slightly delayed kinetics (Fig. 4B).
The results described here demonstrate that the central flap can modestly enhance HIV replication in vitro and in vivo, which may explain its evolutionarily conserved nature. However, virus bearing a disrupted central flap was quite capable of initiating and maintaining a robust infection in all cell types tested and replicated efficiently in vivo, showing only slightly reduced pathogenesis compared with that of Wt HIV. Hence, the central flap is not essential for HIV replication.
Acknowledgments
We thank Gregory Bristol for excellent technical assistance.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3 proviral plasmid from Malcolm Martin and the PM1 cell line from Paolo Lusso and Marvin Reitz.
This work was supported by NIH grants AI070010 and AI36059 and by the UCLA CFAR.
Footnotes
Published ahead of print on 28 March 2007.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrovandi, G. M., G. Feuer, L. Gao, B. Jamieson, M. Kristeva, I. S. Chen, and J. A. Zack. 1993. The SCID-hu mouse as a model for HIV-1 infection. Nature 363:732-736. [DOI] [PubMed] [Google Scholar]
- 3.Aldrovandi, G. M., L. Gao, G. Bristol, and J. A. Zack. 1998. Regions of human immunodeficiency virus type 1 nef required for function in vivo. J. Virol. 72:7032-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldrovandi, G. M., and J. A. Zack. 1996. Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in SCID-hu mice. J. Virol. 70:1505-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arhel, N., S. Munier, P. Souque, K. Mollier, and P. Charneau. 2006. Nuclear import defect of human immunodeficiency virus type 1 DNA flap mutants is not dependent on the viral strain or target cell type. J. Virol. 80:10262-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz, R. D., A. B. van't Wout, N. A. Kootstra, M. E. Moreno, V. D. Linquist-Stepps, C. Bare, C. A. Stoddart, H. Schuitemaker, and J. M. McCune. 1999. R5 strains of human immunodeficiency virus type 1 from rapid progressors lacking X4 strains do not possess X4-type pathogenicity in human thymus. J. Virol. 73:7817-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonyhadi, M. L., L. Rabin, S. Salimi, D. A. Brown, J. Kosek, J. M. McCune, and H. Kaneshima. 1993. HIV induces thymus depletion in vivo. Nature 363:728-732. [DOI] [PubMed] [Google Scholar]
- 8.Bristol, G. C., L. Y. Gao, and J. A. Zack. 1997. Preparation and maintenance of SCID-hu mice for HIV research. Methods 12:343-347. [DOI] [PubMed] [Google Scholar]
- 9.Brooks, D. G., S. G. Kitchen, C. M. Kitchen, D. D. Scripture-Adams, and J. A. Zack. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7:459-464. [DOI] [PubMed] [Google Scholar]
- 10.Bukrinsky, M. 2004. A hard way to the nucleus. Mol. Med. 10:1-5. [PMC free article] [PubMed] [Google Scholar]
- 11.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 13.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charneau, P., and F. Clavel. 1991. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J. Virol. 65:2415-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 16.De Rijck, J., and Z. Debyser. 2006. The central DNA flap of the human immunodeficiency virus type 1 is important for viral replication. Biochem. Biophys. Res. Commun. 349:1100-1110. [DOI] [PubMed] [Google Scholar]
- 17.de Wet, J. R., K. V. Wood, M. DeLuca, D. R. Helinski, and S. Subramani. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 20.Gendelman, H. E., O. Narayan, S. Kennedy-Stoskopf, P. G. Kennedy, Z. Ghotbi, J. E. Clements, J. Stanley, and G. Pezeshkpour. 1986. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. J. Virol. 58:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorry, P. R., G. Bristol, J. A. Zack, K. Ritola, R. Swanstrom, C. J. Birch, J. E. Bell, N. Bannert, K. Crawford, H. Wang, D. Schols, E. De Clercq, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, D. D., T. R. Rota, and M. S. Hirsch. 1986. Infection of monocyte/macrophages by human T lymphotropic virus type III. J. Clin. Investig. 77:1712-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilyinskii, P. O., and R. C. Desrosiers. 1998. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 17:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamieson, B. D., G. M. Aldrovandi, V. Planelles, J. B. Jowett, L. Gao, L. M. Bloch, I. S. Chen, and J. A. Zack. 1994. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J. Virol. 68:3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamieson, B. D., S. Pang, G. M. Aldrovandi, J. Zha, and J. A. Zack. 1995. In vivo pathogenic properties of two clonal human immunodeficiency virus type 1 isolates. J. Virol. 69:6259-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneshima, H., L. Su, M. L. Bonyhadi, R. I. Connor, D. D. Ho, and J. M. McCune. 1994. Rapid-high, syncytium-inducing isolates of human immunodeficiency virus type 1 induce cytopathicity in the human thymus of the SCID-hu mouse. J. Virol. 68:8188-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitchen, S. G., C. H. Uittenbogaart, and J. A. Zack. 1997. Mechanism of human immunodeficiency virus type 1 localization in CD4-negative thymocytes: differentiation from a CD4-positive precursor allows productive infection. J. Virol. 71:5713-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limón, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manganini, M., M. Serafini, F. Bambacioni, C. Casati, E. Erba, A. Follenzi, L. Naldini, S. Bernasconi, G. Gaipa, A. Rambaldi, A. Biondi, J. Golay, and M. Introna. 2002. A human immunodeficiency virus type 1 pol gene-derived sequence (cPPT/CTS) increases the efficiency of transduction of human nondividing monocytes and T lymphocytes by lentiviral vectors. Hum. Gene Ther. 13:1793-1807. [DOI] [PubMed] [Google Scholar]
- 32.McCune, J. M., R. Namikawa, H. Kaneshima, L. D. Shultz, M. Lieberman, and I. L. Weissman. 1988. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science 241:1632-1639. [DOI] [PubMed] [Google Scholar]
- 33.Munro, S., and H. R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899-907. [DOI] [PubMed] [Google Scholar]
- 34.Namikawa, R., H. Kaneshima, M. Lieberman, I. L. Weissman, and J. M. McCune. 1988. Infection of the SCID-hu mouse by HIV-1. Science 242:1684-1686. [DOI] [PubMed] [Google Scholar]
- 35.Namikawa, R., K. N. Weilbaecher, H. Kaneshima, E. J. Yee, and J. M. McCune. 1990. Long-term human hematopoiesis in the SCID-hu mouse. J. Exp. Med. 172:1055-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen, T. H., J. Oberholzer, J. Birraux, P. Majno, P. Morel, and D. Trono. 2002. Highly efficient lentiviral vector-mediated transduction of nondividing, fully reimplantable primary hepatocytes. Mol. Ther. 6:199-209. [DOI] [PubMed] [Google Scholar]
- 37.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasher, D. C., V. K. Eckenrode, W. W. Ward, F. G. Prendergast, and M. J. Cormier. 1992. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111:229-233. [DOI] [PubMed] [Google Scholar]
- 39.Sellon, D. C., S. T. Perry, L. Coggins, and F. J. Fuller. 1992. Wild-type equine infectious anemia virus replicates in vivo predominantly in tissue macrophages, not in peripheral blood monocytes. J. Virol. 66:5906-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valentin, A., G. Aldrovandi, A. S. Zolotukhin, S. W. Cole, J. A. Zack, G. N. Pavlakis, and B. K. Felber. 1997. Reduced viral load and lack of CD4 depletion in SCID-hu mice infected with Rev-independent clones of human immunodeficiency virus type 1. J. Virol. 71:9817-9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Maele, B., J. De Rijck, E. De Clercq, and Z. Debyser. 2003. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 77:4685-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita, M., and M. Emerman. 2006. Retroviral infection of nondividing cells: old and new perspectives. Virology 344:88-93. [DOI] [PubMed] [Google Scholar]
- 44.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]