Abstract
Here, we describe a cell-based in vivo assay that probes the specific interaction between nucleocapsid (NC) protein and Psi (Ψ) RNA, the human immunodeficiency virus (HIV) packaging signal. The results demonstrate for the first time a specific NC-Ψ interaction within living cells. The specificity and applicability of the assay were confirmed by mutational studies of NC and deletion-mapping analyses of Ψ-RNA as well as by testing the in vivo NC-binding effects of NC-aptamer RNAs identified previously in vitro. This assay system would facilitate further detailed studies of the NC-Ψ interaction in vivo and the screening of various anti-HIV molecules targeting NC and the specific interaction.
The human immunodeficiency virus (HIV) nucleocapsid (NC) protein derived from the Gag polyprotein precursor is involved in multiple steps of the viral life cycle (4, 16) such as viral replication (26), including the strand transfer reactions during reverse transcription (2, 32), migration of the preintegration complex (8, 17, 20), integration (9), and, most importantly, packaging and assembly of the viral genomic RNA (15, 30). A number of genetic mutation studies of HIV have suggested that viral genomic RNA packaging is mediated by a specific interaction between the NC domain of the Gag and Psi (Ψ) RNA sequence, a so-called packaging signal, located in the HIV 5′ long terminal repeat region (1, 6, 18, 24, 25).
However, studies of NC function with respect to the interaction with Ψ RNA have thus far relied on either mutational studies using a proviral DNA clone (1, 18, 25) in vivo or in vitro analyses such as nitrocellulose filter binding (10, 13), or gel mobility assays (7, 12, 29) using purified recombinant Gag or NC protein. Although those assays revealed important roles for NC, they have limitations in that (i) the direct binding of NC to Ψ RNA in vivo was not fully addressed in the former assays, (ii) the in vivo situation could hardly be reflected in the latter experiments, and, more importantly, (iii) none of them is suitable for specifically screening antiviral agents against NC, which is now regarded as a highly promising target for new anti-HIV drugs (14) and which might overcome the current HIV drug resistance problems, as it is conserved in all HIVs and retroviruses (11). Thus, the development of an easy, specific, and functional assay that can probe the NC-Ψ RNA interaction in vivo is needed for multiple reasons. Moreover, Bacharach and Goff recently described a cell-based genetic assay using a yeast three-hybrid system that could probe the interaction between HIV Gag and Ψ RNA (3). However, they were able to detect the interaction between HIV Gag and Ψ RNA but were unable to detect the specific interaction between NC and Ψ RNA in cells. That result raises the as-yet-unsolved question of whether NC by itself can bind the Ψ RNA in vivo specifically or whether other portions of Gag are necessary for the specific interaction in vivo.
To this end, we set out to examine the specific interaction between NC and Ψ RNA in vivo by developing a cell-based assay. The assay is based on the idea that if NC binds Ψ RNA specifically, this could hinder the translation of reporter gene transcripts containing the Ψ RNA sequence upstream of the translation initiation codon. We thus prepared pMV1psi-LacZ, in which a lacZ open reading frame is immediately fused to the Ψ sequence derived from ARV-2/SF2 and placed downstream of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible trc promoter and operator. The reporter plasmids employed in this study have a tetracycline resistance gene and a p15A replication origin derived from pACYC184 (New England BioLabs). For efficient NC expression, we employed vector pJC1, which harbors an ampicillin resistance gene, a pMB1 replication origin (a relative of the ColE1 origin), and lacIq as described previously (31).
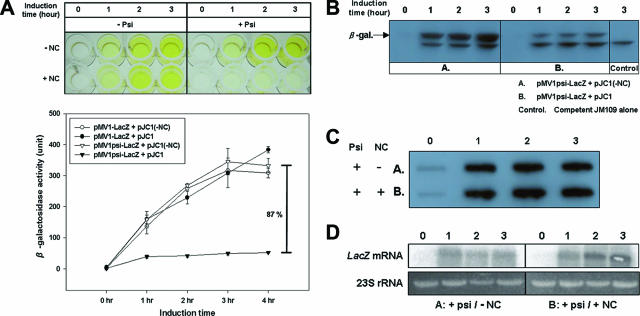
Transformants harboring pMV1psi-LacZ and pJC1 resulted in only about 10% of the β-galactosidase activity of the transformants not containing the NC vector. In contrast, the pMV1-LacZ control vector lacking the Ψ sequence showed nearly the same 400 units of β-galactosidase activity in either the presence or the absence of the NC vector (Fig. 1A). Western blot analysis revealed a good correlation between β-galactosidase activity measured by an ONPG (2-nitrophenyl-β-d-galactopyranoside) assay and the amount of β-galactosidase protein in the transformants (Fig. 1B). Also, slot and Northern blots showed that the level of lacZ mRNA was not influenced by the presence of NC (Fig. 1A and D). It appears rather that NC stabilizes the lacZ transcript containing the Ψ sequence more. Thus, these results together demonstrate that the specific interaction of NC with the Ψ sequence inhibits the translation of reporter transcripts containing an upstream Ψ RNA sequence.
FIG. 1.
Translation inhibition caused by the HIV type 1 (HIV-1) NC-Ψ interaction. (A) Visualization and quantitative analysis of the ONPG assay for translation inhibition due to the interaction of HIV-1 NC and Ψ RNA. JM109 was doubly transformed with the indicated individual vectors shown in the insert, and their lacZ reporter gene activities were determined by ONPG assay. (B) Western blot analysis of the expression of β-galactosidase. A and B indicate JM109 cells cotransformed with pMV1psi-LacZ containing Ψ sequence and pJC1(-NC) or pJC1, respectively. Control indicates nontransformed JM109 cells. The band in the control originates from the endogenous inactive partial lacZα peptide of JM109 (lacZΔM15) cells; it served as an internal control for the amounts of cell lysates used in the analysis. The cotransformants and JM109 were harvested at the indicated times after IPTG induction. Cell lysates were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel and subjected to Western blot analysis. (C and D) Slot blot (C) and Northern blot (D) analyses. Total RNA was prepared from cotransformants A and B as described above. Ten micrograms of DNase I-treated total cellular RNA was loaded onto each lane and subjected to slot blot and Northern analysis.
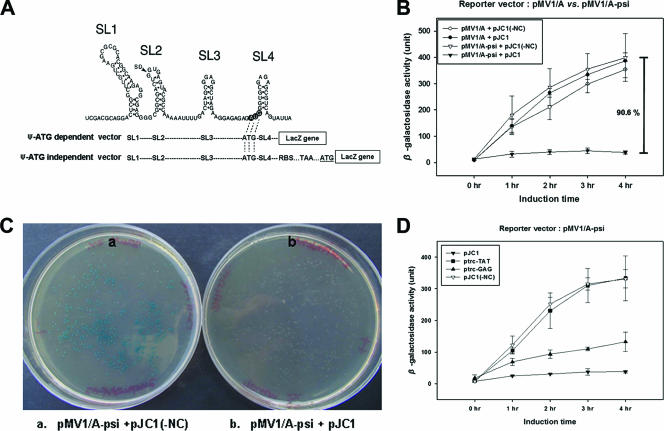
The pMV1psi-LacZ reporter system, designated the “Ψ-ATG-dependent vector,” employed thus far, however, has limitations for further manipulations because of the dependence of lacZ translation on the AUG in the SL4 of the Ψ sequence as shown in Fig. 2A. Thus, we also set out to develop another lacZ reporter vector system independent of the AUG codon. Plasmids pMV1/A-psi and pMV1/AS-psi, in which the ATG of the Ψ sequence is in frame with a stop codon (TAA) placed between the newly engineered ribosome binding site and the ATG of lacZ, were generated and designated the “Ψ-ATG-dependent vector” (Fig. 2A). Using the resulting vectors, we found again that β-galactosidase activity was repressed only in cotransformants harboring the NC expression vector and the pMV1/A-psi vector. In the absence of either NC or Ψ sequence, a high level of β-galactosidase activity was observed (Fig. 2B). pMV1/AS-psi gave the same results as pMV1/A-psi (data not shown). The inhibition of β-galactosidase expression due to the specific NC-Ψ interaction could also be demonstrated nicely on X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates as shown in Fig. 2C. Inhibition was equally evident using the Ψ sequence from HIV isolate NL4-3 (data not shown) and with Gag (albeit a little less efficiently than with NC) but not with Tat, another RNA binding protein in HIV (5) (Fig. 2D), indicating that inhibition is specific to NC or Gag and the Ψ sequence.
FIG. 2.
Translation inhibition assay with the Ψ-ATG-independent reporter vector. (A) Schematic representations of the sequence organizations upstream of the lacZ reporter gene in the Ψ-ATG-dependent and Ψ-ATG-independent vector systems. The possible secondary structure of the HIV-1NL4-3 Ψ sequence is shown at the top. The location of the HIV-1 splicing donor site is marked with an arrowhead, and the original translation start codon (AUG) of the HIV-1 Gag polyprotein is shown by open lettering in dark circles. Ψ-ATG-dependent vector refers to lacZ translation initiation starting from the AUG start codon located in the SL4 region of the Ψ sequence. The Ψ-ATG-independent vector initiates the translation of lacZ at its own AUG (underlined) downstream of Ψ RNA (see the text for more details). Dotted lines indicate the same sequence as that in the Ψ RNA sequence. (B) Effect of NC on Ψ-ATG-independent expression of lacZ. (C) Visualization of NC-Ψ-mediated lacZ translation inhibition on agar. The effect of the interaction between NC and the Ψ sequence on β-galactosidase expression in JM109 was monitored using agar plates containing 2% X-gal and 70 μM IPTG. At the left and right are JM109 transformants containing pMV1/A-psi without NC (a) and with NC (b), respectively. (D) Effect of Gag polyprotein and Tat protein on Ψ-ATG-independent expression of lacZ. The expression vectors used are indicated in the insert. All experiments were performed three times.
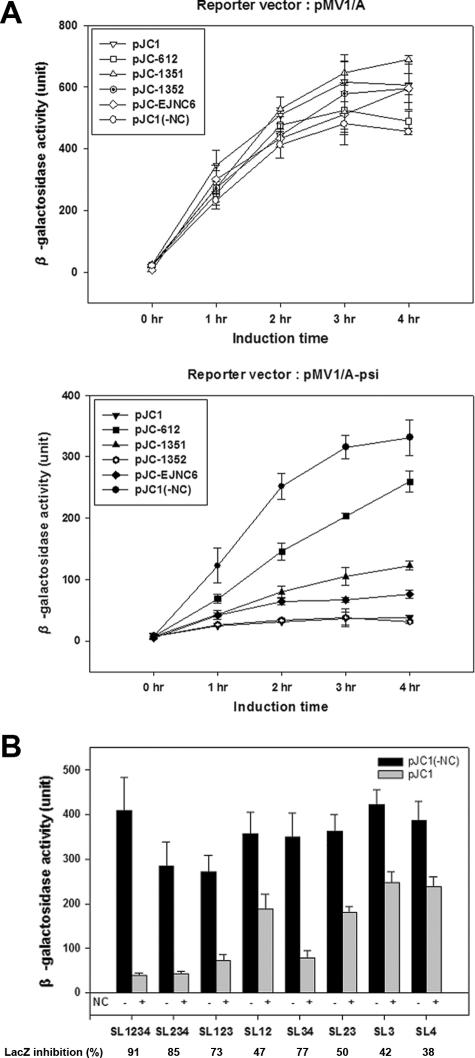
To examine the effect of the NC zinc finger mutations on the interaction with Ψ RNA and thus validate the specificity of our assay further, we employed three kinds of NC zinc finger mutants (a gift of R. Gorelick at NCI) with cysteine-to-serine replacements such as SSHS/CCHC, CCHC/SSHS, and SSHS/SSHS in place of the two original CCHC/CCHC zinc fingers as described previously (19). In the SSHS/SSHS mutant (pJC-DB612), reporter activity was inhibited by about 20 to 25% compared to that without NC protein, as seen in the lower panel of Fig. 3A. While the second zinc finger mutant (pJC-DB1352) still interacted with the Ψ sequence as wild-type NC (over 90% inhibition), the first zinc finger mutant (pJC-DB1351) gave only about 60 to 65% repression, indicating that the first zinc finger is more important than the second zinc finger. This was confirmed further when we tested another NC mutant (pJC-EJNC6) expressing an NC of 37 amino acids instead of 55 amino acids (wild type). It generated 75% inhibition of reporter gene activity, which is stronger than the first zinc finger mutant but a little less than the second zinc finger mutant (Fig. 3A, lower panel). This result demonstrates for the first time that the NC having only the first zinc finger is still capable of specifically interacting with the Ψ sequence. None of these proteins repressed any β-galactosidase activity when the reporter vector did not have the Ψ sequence, as shown in Fig. 3A (upper panel). These results are in good agreement with previous studies where the two zinc finger sequences were shown to be functionally nonequivalent in binding to Ψ and in viral packaging (13, 18, 21, 27, 28). Next, we also examined the contributions of the four stem-loops (SL) in the Ψ sequence to the binding of NC. A series of combinations of the four stem-loops in the Ψ sequence were cloned into pMV1/A and examined for interactions with NC. The results, as shown in Fig. 3B, demonstrated that (i) only the full-length Ψ sequence had the highest binding affinity, indicating that SL1, SL2, SL3, and SL4 each contribute to some extent to NC binding; (ii) SL123 or SL234, containing three stem-loops, had more activity than any of the two stem-loop structures except SL34, and SL234 had a little more activity than SL123, emphasizing the contribution of SL4; (iii) SL34 was more active than SL12 or SL23; and (iv) SL3 and SL4 alone each had substantial binding activity, pointing to the importance of SL3 and SL4. These mapping results suggest that although maximal binding requires the full sequence of Ψ, SL3 and SL4 are the major determinants of the NC-Ψ interaction in vivo. The fact that we observed differential translation inhibition activities by NC with the various SL regions of the Ψ sequence tested points to the usefulness of our assay in terms of its specificity and sensitivity and also suggests that this assay system could be used effectively to further dissect the residues of Ψ RNA in vivo that are critical for NC binding.
FIG. 3.
(A) Effect of mutations of NC protein on the translation inhibition assay. pJC-DB1351, pJC-DB1352, pJC-DB612, and pJC-EJNC6 are described in the text. Data from ONPG assays of wild-type and mutant NC proteins with pMV1/A, a control reporter vector not containing Ψ, and with vector pMV1/A-psi are shown in upper and lower panels, respectively. (B) Interaction strengths of different regions of the Ψ stem-loop sequence with NC using our cell-based assay. Shown are β-galactosidase activities of individual reporter vectors examined in the absence (black bar) and presence (gray bar) of NC. The percentages of lacZ inhibition that indicate β-galactosidase activity obtained with NC compared to that without NC are also shown. All experiments were performed three times.
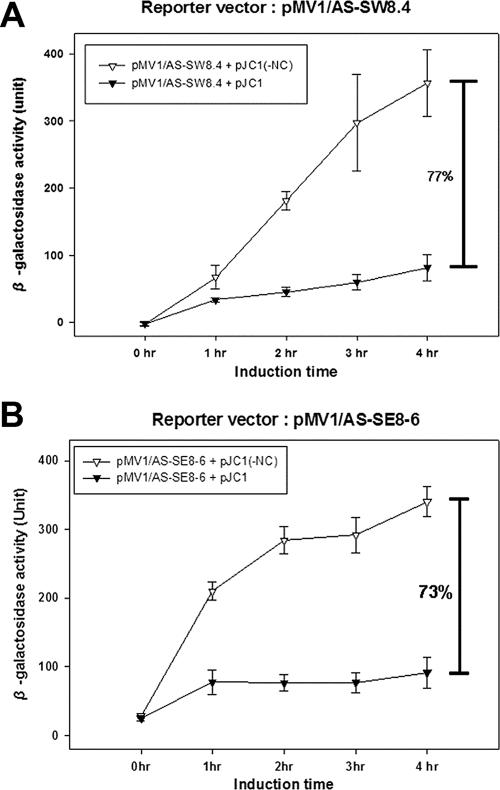
Finally, we examined the applicability of our assay by testing whether the aptamers identified in vitro could also interact with NC in this cell-based assay. We inserted some of the systematic evolution of ligands by exponential enrichment (SELEX) RNA aptamers, isolated previously by Lochrie et al. (23) and in our laboratory (22), in place of the Ψ sequence in pMV1/AS and measured lacZ expression in the presence and absence of NC (Fig. 4). In each case, we observed an inhibition of translation, which is indicative of a specific interaction between NC and the aptamers. This suggests that the assay may also be used to screen and identify high-affinity NC binding RNA molecules in vivo without going through the rather laborious complex in vitro selection method that has been used thus far.
FIG. 4.
Effect of various SELEX RNAs on NC binding in the cell-based assay. The SELEX RNA aptamers described in the text were cloned into pMV1/AS, resulting in (A) pMV1/AS-SW8.4 and (B) pMV1/AS-SE8-6. They were tested for translation inhibition of lacZ in the presence or absence of NC. All these experiment were performed three times.
In conclusion, we have developed a cell-based in vivo assay that probes the specific interaction between NC and Ψ RNA, demonstrating for the first time the ability of the specific interaction of NC by itself with Ψ within living cells. This assay provides an easy and efficient means of studying the NC-Ψ interaction in detail in vivo and may also facilitate the screening and identification of antiviral chemicals and bioactive molecules against NC and the NC-Ψ interaction.
Acknowledgments
We are grateful to Robert Gorelick at NCI-Frederick Cancer Research Center for providing the NC zinc finger mutants and to Tristram Parslow at Emory University for SW8.4 aptamer RNA. We thank Hee-Yeon Cho and Hyukjun Nam for their technical assistance in the early stage of this work.
This work was supported by a National Research Laboratory grant (M10500000148-06J0000-14810) from the Korean Ministry of Science and Technology and a research fund (R01-2005-000-11258) from the Korea Science and Engineering Foundation.
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Aldovini, A., and R. A. Young. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J. L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacharach, E., and S. P. Goff. 1998. Binding of the human immunodeficiency virus type 1 Gag protein to the viral RNA encapsidation signal in the yeast three-hybrid system. J. Virol. 72:6944-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bampi, C., S. Jacquenet, D. Lener, D. Decimo, and J. Darlix. 2004. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Int. J. Biochem. Cell Biol. 36:1668-1686. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz, R. D., A. Ohagen, S. Hoglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz, R. D., J. Luban, and S. P. Goff. 1993. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 67:7190-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever, J., C. Sassetti, and T. G. Parslow. 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 69:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covey, S. N. 1986. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 14:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damgaard, C. K., H. Dyhr-Mikkelsen, and J. Kjems. 1998. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 26:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein psi RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 15.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643-655. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 17.Gally, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 18.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, J., T. Wu, J. Anderson, B. F. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. G. Levin. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 74:8980-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hameau, L., J. Jeusset, S. Lafosse, D. Coulaud, E. Delain, T. Unge, T. Restle, E. Le Cam, and G. Mirambeau. 2001. Human immunodeficiency virus type 1 central DNA flap: dynamic terminal product of plus-strand displacement DNA synthesis catalyzed by reverse transcriptase assisted by nucleocapsid protein. J. Virol. 75:3301-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, M., A. Maynard, J. A. Turpin, L. Graham, G. M. Janini, D. G. Covell, and W. G. Rice. 1998. Anti-HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc finger proteins. J. Med. Chem. 41:1371-1381. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. J., M. Y. Kim, J. H. Lee, J. C. You, and S. Jeong. 2002. Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 291:925-931. [DOI] [PubMed] [Google Scholar]
- 23.Lochrie, M. A., S. Waugh, D. G. Pratt, Jr., J. Clever, T. G. Parslow, and B. Polisky. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 Gag polyprotein. Nucleic Acids Res. 25:2902-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride, M. S., and A. T. Panganiban. 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 71:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meric, C., and S. P. Goff. 1989. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His boxes of the nucleocapsid protein. J. Virol. 63:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rein, A., L. E. Henderson, and J. G. Levin. 1998. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 27.Rice, W. G. 1997. Azodicarbonamide inhibits HIV-1 replication by targeting the nucleocapsid protein. Nat. Med. 3:341-345. [DOI] [PubMed] [Google Scholar]
- 28.Rice, W. G., J. G. Supko, L. Malspeis, R. W. Buckheit, Jr., D. Clanton, M. Bu, L. Graham, C. A. Schaeffer, J. A. Turpin, J. Domagala, R. Gogliotti, J. P. Bader, S. M. Halliday, L. Coren, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1995. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science 270:1194-1197. [DOI] [PubMed] [Google Scholar]
- 29.Sakaguchi, K., N. Zambrano, E. T. Baldwin, B. A. Shapiro, J. W. Erickson, J. G. Omichinski, G. M. Clore, A. M. Gronenborn, and E. Appella. 1993. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein (RNA binding/dimerization). Proc. Natl. Acad. Sci. USA 90:5219-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scarlata, S., and C. Carter. 2003. Role of HIV-1 Gag domains in viral assembly. Biochim. Biophys. Acta Biomembr. 1614:62-72. [DOI] [PubMed] [Google Scholar]
- 31.You, J. C., and C. S. McHenry. 1993. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J. Biol. Chem. 268:16519-16527. [PubMed] [Google Scholar]
- 32.You, J. C., and C. S. McHenry. 1994. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 269:31491-31495. [PubMed] [Google Scholar]