Abstract
In adult T-cell leukemia (ATL) cells, a defective human T-cell leukemia virus type 1 (HTLV-1) provirus lacking the 5′ long terminal repeat (LTR), designated type 2 defective provirus, is frequently observed. To investigate the mechanism underlying the generation of the defective provirus, we sequenced HTLV-1 provirus integration sites from cases of ATL. In HTLV-1 proviruses retaining both LTRs, 6-bp repeat sequences were adjacent to the 5′ and 3′ LTRs. In 8 of 12 cases with type 2 defective provirus, 6-bp repeats were identified at both ends. In five of these cases, a short repeat was bound to CA dinucleotides of the pol and env genes at the 5′ end, suggesting that these type 2 defective proviruses were formed before integration. In four cases lacking the 6-bp repeat, short (6- to 26-bp) deletions in the host genome were identified, indicating that these defective proviruses were generated after integration. Quantification indicated frequencies of type 2 defective provirus of less than 3.9% for two carriers, which are much lower than those seen for ATL cases (27.8%). In type 2 defective proviruses, the second exons of the tax, rex, and p30 genes were frequently deleted, leaving Tax unable to activate NF-κB and CREB pathways. The HTLV-1 bZIP factor gene, located on the minus strand, is expressed in ATL cells with this defective provirus, and its coding sequences are intact, suggesting its significance in oncogenesis.
Human T-cell leukemia virus type 1 (HTLV-1) is the causative virus of a neoplastic disease, adult T-cell leukemia (ATL), and inflammatory diseases, including HTLV-1-associated myelopathy/tropical spastic paraparesis and HTLV-1-associated uveitis (18, 32). After a long latent period, about 5% of carriers develop ATL (3). These observations suggest that several factors, including the host immune system, genetic background, and viral genes, influence ATL onset.
Retroviruses induce cancers by several different mechanisms (37), including viral oncogenes (51), aberrant transcription of oncogenes by insertion of provirus (24), and actions of virus-encoded genes (14, 55). HTLV-1 is a complex retrovirus, which encodes regulatory and accessory genes in the pX region by four open reading frames. Among those, tax is thought to play a central role in both immortalization and oncogenesis by pleiotropic activity (15, 21, 55). Tax activates the transcription of cellular genes, via nuclear factor κB (NF-κB), cyclic AMP response element-binding protein (CREB), and activator protein-1 pathways, and can inhibit functions of cellular proteins, including p53 and MAD1. In addition, the plus strand of the pX region encodes accessory genes p12, p13, and p30 in the different open reading frames (39). The p12 protein interacts with calreticulin and calnexin (11) and enhances nuclear factor of activated T cells (NFAT)-dependent gene expression (2, 10, 28). The p30 protein binds to CREB binding protein/p300 and modulates the transcription of viral and cellular genes (33, 56). These accessory genes are required for infectivity and for the persistence of HTLV-1 infection (1, 49). Tax and accessory gene products therefore increase the number of HTLV-1-infected cells by promoting proliferation and inhibiting apoptosis. On the other hand, Tax expression makes infected cells a major target of cytotoxic T lymphocytes (CTLs) in vivo, thereby decreasing their number (6). CTLs against p12 and p13 have been also reported in HTLV-1 carriers (43). Thus, proliferation of HTLV-1-infected cells is determined by a balance in activities between viral genes and the host immune system. On the other hand, HTLV-1 has redundant mechanisms to suppress Tax expression. Rex suppresses the transport of doubly spliced viral RNA (25) encoding Tax and Rex. p30 binds to tax gene transcripts and inhibits their transport into the cytoplasm (38). In addition, HTLV-1 bZIP factor (HBZ) suppresses viral gene transcription through the 5′ long terminal repeat (LTR) by binding to c-Jun, a transcription factor critical for tax expression (7).
Tax expression in primary leukemic cells is absent in about 60% of ATL cases. Three mechanisms to suppress or abolish Tax expression have been identified (31). Genetic changes in tax are seen in about 10% of ATL cases (17, 52). DNA methylation of the 5′ LTR also silences tax transcription (29, 52, 54). In addition, the 5′ LTR of HTLV-1, which functions as a promoter/enhancer, is frequently deleted in ATL cases, and such deletion results in loss of viral gene transcription unless the defective provirus traps a cellular promoter. This type of provirus is designated the type 2 defective provirus, and its frequency is higher in aggressive forms of ATL than in chronic or smoldering ATL (53). However, it remains unclear when deletion occurs in the course of disease progression. Although the 5′ LTR is frequently methylated or deleted, the 3′ LTR remains intact in ATL cells. HBZ is transcribed from the minus strand of the provirus using the 3′ LTR as a promoter. We have reported that HBZ is expressed in ATL cells and promotes ATL cell proliferation (44). HBZ suppresses tax gene transcription through the 5′ LTR, simultaneously supporting the growth of ATL cells.
After infection, retroviruses synthesize a double-stranded proviral DNA by using reverse transcriptase (RT) with tRNA as a primer. Double-stranded viral DNA is then integrated by viral integrase and cellular factors (5, 20). During this process, integrase generates short repetitive sequences of 4 to 6 bp adjacent to both LTRs. The length of these repeats is specific to each retrovirus. For example, human immunodeficiency virus type 1 forms a 5-bp short repeat at the ends of both LTRs, and murine leukemia virus generates 4-bp repeats. Identification of these short repeats indicates that integration is mediated by viral integrase.
In this study, we determined the integration sites and neighboring genomic sequences of the HTLV-1 provirus and found that 6-bp repeat sequences adjacent to both ends of the type 2 defective provirus were retained in 8 of 12 cases. This finding indicates that provirus lacking a 5′ LTR can be integrated by the viral integrase and that deletion of the 5′ LTR occurs before integration. In the remaining four cases, the short repeat was absent and host genomic sequences were deleted, indicating that these defective proviruses formed after integration. In type 2 defective proviruses, HBZ sequences were conserved, and HBZ transcripts were detected in ATL cells with type 2 defective provirus, suggesting that they are functional in such ATL cells.
MATERIALS AND METHODS
Samples and cell lines.
Clinical samples were collected from 79 ATL patients after informed consent was obtained. Clinical diagnosis of ATL subtypes was performed according to criteria reported previously (47). Approval for this study was obtained from the institutional review board of Kyoto University. Genomic DNA was extracted from peripheral blood mononuclear cells or lymph node cells using standard phenol-chloroform methods. The ATL cell lines TL-Om1 and ATL-55T were used in this study.
Determination of proviral subtypes by PCR and Southern blotting.
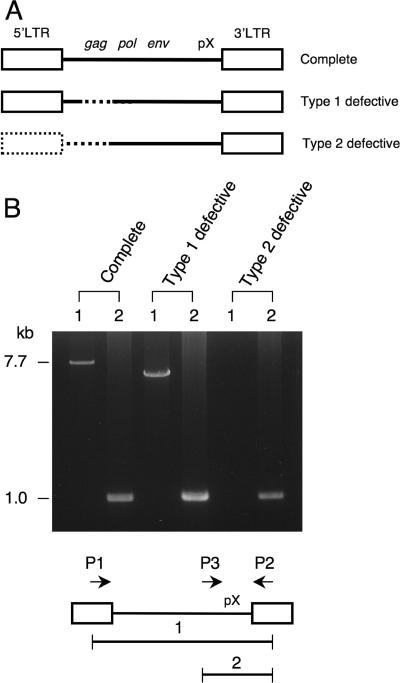
Subtypes of HTLV-1 provirus were determined as reported previously (53). In brief, the whole HTLV-1 provirus was amplified by long PCR using primer 1 (5′-GTTCCACCCCTTTCCCTTTCATTCACGACTGACTGC-3′) and primer 2 (5′-GGCTCTAAGCCCCCGGGGGATATTTGGGGCTCATGG-3′). PCR conditions were as follows: denaturation at 94°C for 30 s and annealing and extension at 64°C for 10 min; 30 cycles were run. The pX region of the provirus was amplified by primer 2 and primer 3 (5′-GGCGACTGGTGCCCCATCTCTGGGGGACTATGTTCG-3′). As a control, we used the genomic DNAs from HTLV-1 carriers and confirmed that there was no band with this condition. When the whole provirus was amplified, the complete provirus (7.7 kb) could be detected using primers 1 and 2, while type 1 defective provirus generated a smaller band. In type 2 defective provirus, no band was amplified. PCR products (1.0 kb) derived from the pX region were detected in all cases by primers 2 and 3. Southern blotting was performed as described previously (53).
Inverse PCR.
Genomic regions flanking the 3′ LTR of the HTLV-1 provirus were amplified by inverse PCR as described previously (13). Conditions for long PCR were as follows: 1 cycle of 98°C for 2 min, 5 cycles of 98°C for 30 s and 64°C for 10 min, and 35 cycles of 94°C for 30 s, 64°C for 10 min, and 72°C for 15 min. Primers were as follows: long-IPCR-F (5′-TGCCTGACCCTGCTTGCTCAACTCTACGTCTTTG-3′) and long-IPCR-R (5′-AGTCTGGGCCCTGACCTTTTCAGACTTCTGTTTC-3′), and pX-3′ F1 (5′-TGGCACGCCTATGATTTCCG-3′) and pX-R1-Ban III (5′-GGGGGTTGTATGAGTGATTGG-3′).
Sequencing of genomic regions adjacent to integration sites and proviruses.
Inverse-PCR products from ATL samples were used as templates for direct sequencing with a primer located at the end of the 3′ LTR (5′-GTTCCACCCCTTTCCCTTTCATTCACGACTGACTGC-3′). After integration sites were determined, the 5′ ends of the integration sites were amplified by PCR using primers based on 5′ human genomic regions at integration sites and based on internal provirus regions. Internal regions of type 2 defective provirus were also amplified by PCR using primers based on gag, pol, env, and the 3′ LTR, and their sequences were determined by direct sequencing. Sequencing was performed using the Big Dye Terminator (version 3.1) cycle sequencing kit and an ABI310 autosequencer (both from Applied Biosystems, Foster City, CA).
RT-PCR.
Spliced HBZ and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts were detected by RT-PCR using a PC-808 thermal cycler (Astec, Fukuoka, Japan) under the following conditions: 1 cycle of 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 57.5°C for 30 s, and 72°C for 30 s. The primers used were as follows: 5′-TAAACTTACCTAGACGGCGG-3′ and 5′-CTGCCGATCACGATGCGTTT-3′ for the HBZ gene and 5′-GCAGGGGGGAGCCAAAAGGG-3′ and 5′-TGCCAGCCCCAGCGTCAAAG-3′ for GAPDH. As a positive control for HBZ gene transcription, we used mRNA derived from an ATL cell line, ATL-55T.
Quantification of type 2 defective provirus in carriers.
First, genomic DNA from a single ATL sample with one type 2 defective provirus was mixed in various proportions (5, 10, 20, 40, and 100%) with DNA from an ATL cell line harboring a complete provirus (TL-Om1). Then we used real-time PCR to quantify provirus loads at three different regions of the provirus: (i) 5′ LTR-gag (deleted only in type 2 defective provirus), (ii) gag (deleted in both type 1 and type 2 defective proviruses), and (iii) pX (conserved in all proviruses). Primer pairs were 5′LTR-SDS-F (5′-AAGTACCGGCGACTCCGTTG-3′) and gag-SDS-R (5′-AGCGCTACGGGAAAAGATTTG); gag-F (5′-GAGTGCCAAAGACCCTTCCT-3′) and gag-R (5′-GTCAAGAGCTATGTTGAGGCG-3′); and tax-exon3-F (5′-GAAGACTGTTTGCCCACCACC) and tax-exon3-R (5′-TGAGGGTTGAGTGGAACGGA). Probes were the 5′ LTR probe (5′-CGTCCGGGATACGAGCGCCCCTT-3′), the gag probe (5′-CAAGGCCTGGAGGAGCCTTACCACG-3′), and the pX probe (5′-CACCCGTCACGCTAACAGCCTGGCAA-3′). Real-time PCR was performed using an ABI Prism 7700 sequence detection system, Taqman Universal PCR master mix, and genomic DNAs (200 ng). PCR amplification consisted of 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. All samples were analyzed in triplicate. Provirus load is shown as the percentage of mononuclear cells that are infected, assuming that one infected cell contains one copy of HTLV-1 provirus.
Following quantification of provirus loads at 5′ LTR-gag and pX, regression lines were drawn using the least-squares method to determine the frequency of type 2 defective provirus. The frequency of defective provirus (type 1 plus type 2) was calculated from provirus loads at the gag and pX regions. We examined the genomic DNAs of two carriers with high proviral loads (64.2% for carrier 1 and 47.2% for carrier 2).
3′-RACE.
To determine the 3′ ends of the HBZ transcripts, rapid amplification of cDNA ends (RACE) was performed using the SMART RACE cDNA amplification kit (BD Biosciences Clontech), according to the manufacturer's protocol. First-strand cDNAs were synthesized from 1 μg of total RNA with RT and were used for 3′-RACE PCR. For nested 3′-RACE amplifications, primers specific for HBZ (5′-CTAGGTTAGGGCAGGGGGGCTGTAGGGC-3′ and 5′-GGGTCCACGAACAAACTGGCTGGGCAGG-3′) were used. The sequence of the PCR product was determined by direct sequencing.
Mapping of HTLV-1 integration sites and expression of trapped genes.
The BLAT program was used to map identified integration sites in the human genome (UCSC Human Genome Project Working Draft, May 2006 freeze). Sequences were judged authentic only if they showed 95% or higher identity to genomic sequence over the high-quality sequence region and matched only one genomic locus with 95% or greater identity. Gene expression was evaluated by the GeneCards database of the Crown Human Genome Center at the Weizmann Institute of Science (http://www.genecards.org/background.shtml).
RESULTS
Genomic sequences of HTLV-1 integration sites.
Three subtypes of HTLV-1 provirus are seen in ATL cells: the complete type, the type 1 defective type, and the type 2 defective type, as shown in Fig. 1A. Type 1 defective provirus retains the 5′ and 3′ LTRs but lacks internal sequences such as the gag, pol, or env region, whereas type 2 defective provirus lacks the 5′ LTR in addition to internal sequences. We determined the subtypes of HTLV-1 provirus in 79 ATL cases by long PCR (Fig. 1B) and by Southern blotting as reported previously (53). Among samples evaluated, type 2 defective proviruses were found in 27.8% of the cases, while the frequency of type 1 defective proviruses was 11.4% (Table 1).
FIG. 1.
Three subtypes of HTLV-1 provirus in ATL cells. (A) Schemas of HTLV-1 proviruses in ATL. Dotted lines represent deleted regions of HTLV-1 provirus. (B) Representative results of PCR in three subtypes of HTLV-1 proviruses. Whole HTLV-1 provirus was amplified using LTR primers (primers 1 and 2 [P1 and P2] as described in Materials and Methods). The 7.7-kb band (band 1) was detected in the case with complete provirus. In type 1 defective provirus, the smaller band was amplified by primers 1 and 2. In type 2 defective provirus, no band was detected. On the other hand, the 1.0-kb band (band 2) derived from pX-3′ LTR was amplified in all cases.
TABLE 1.
Subtypes of HTLV-I provirus in ATL cases
| Type of provirusa | No. (%) of cases | No. of cases with the following clinical type of ATL:
|
||
|---|---|---|---|---|
| Acute | Chronic | Lymphoma | ||
| Complete | 43 (54.4) | 31 | 8 | 4 |
| Defective | ||||
| Type 1 | 9 (11.4) | 4 | 4 | 1 |
| Type 2 | 22 (27.8) | 18 | 2 | 2 |
| Multiple types | 5 (6.3) | 4 | 1 | 0 |
| Total | 79 (100) | 57 | 15 | 7 |
Subtypes of HTLV-I provirus in ATL have been determined by PCR and Southern blotting as reported previously (54). Multiple types have more than 2 copies of provirus per cell.
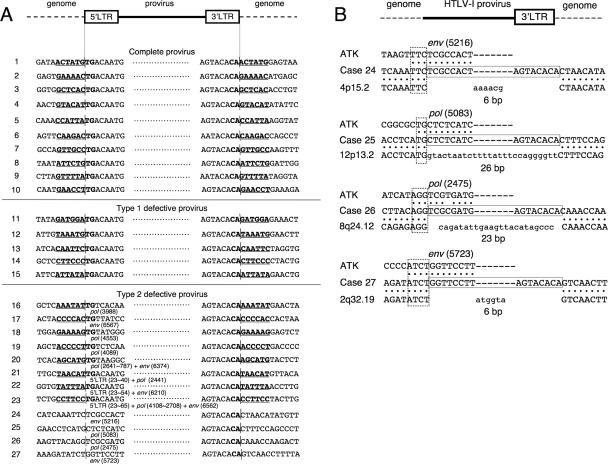
Genomic regions adjacent to HTLV-1 integration sites were amplified by inverse PCR as described in Materials and Methods, and their sequences were determined. Six-base-pair repeats and CA dinucleotides at the ends of both LTRs were conserved in all cases with complete or type 1 defective provirus (cases 1 to 15) (Fig. 2A). These results indicate that these proviruses were likely integrated by the retrovirus integrase.
FIG. 2.
Genomic sequences adjacent to HTLV-1 integration sites in ATL cells. (A) Neighboring genomic regions were amplified by inverse PCR and sequenced. The 6-bp short repeats at the 5′ and 3′ ends of proviruses are underlined. The 6-bp repeats and 2 bp of adjacent LTRs are boldfaced. In type 2 defective proviruses, the position of the deletion in the provirus, which is adjacent to the human genomic sequence, is shown using the numbering system of Seiki et al. (46) (GenBank accession no. J02029). Vertical bars show boundaries between genomic sequences and the provirus. (B) Type 2 defective proviruses lacking the 6-bp repeat. In each case, the sequences from the HTLV-1 provirus (top) (ATK-1 [46]), the integrated provirus (center), and the host genome (bottom) are compared. Proviral sequences are boxed by solid lines. Deleted genomic sequences are lowercased. The chromosomal locations of integration sites are shown. At the 5′ breakpoints, overlapped regions (2 to 4 bp) between the provirus and the genome are boxed by dotted lines.
Among 22 cases with type 2 defective provirus, we identified genomic sequences at integration sites in 12 cases, due to limited availability of DNA samples. Among those 12 cases, 6-bp repeats adjacent to the provirus sequence were observed in 8 cases (cases 16 to 23) (Fig. 2A). These cases could be divided into two groups. In the first, the 5′ LTR was completely deleted and TG dinucleotides in the pol and env regions were directly ligated to a 6-bp repeat of the host genome (cases 16 to 20), indicating that these proviruses were integrated by viral integrase. In these cases, the viral integrase likely recognized a TG sequence in the pol or env region, indicating that the integrase can recognize only a TG dinucleotide for activity. In the second group, a short fragment of the 5′ LTR (18 to 43 bp) remained, but most of the LTR was deleted, in addition to the gag and pol regions (cases 21 to 23). It is not clear whether these regions were deleted before or after integration. In two cases, inverted provirus sequences were observed. In case 20, an inverted gag and pol sequence was ligated to the 6-bp repeat at the 5′ end, and a TG dinucleotide was retained. In case 23, pol sequences were inverted and adjacent to the short U3 sequence.
In four cases of type 2 defective provirus (cases 24 to 27 in Fig. 2A), there was no short repeat sequence adjacent to the provirus, and loss of host genomic sequence (6 to 26 bp) was also seen at the integration site (Fig. 2B). In addition, 2- to 4-bp overlaps were identified at breakpoints, features that are characteristic of illegitimate recombination (50). In addition, the 3′ end of the 3′ LTR retained CA sequences in all cases. These observations suggest that this provirus is first integrated by the viral integrase, and then proviral sequences, including the 5′ LTR and host genomic sequences, are deleted.
Structure of regulatory and accessory genes in type 2 defective provirus.
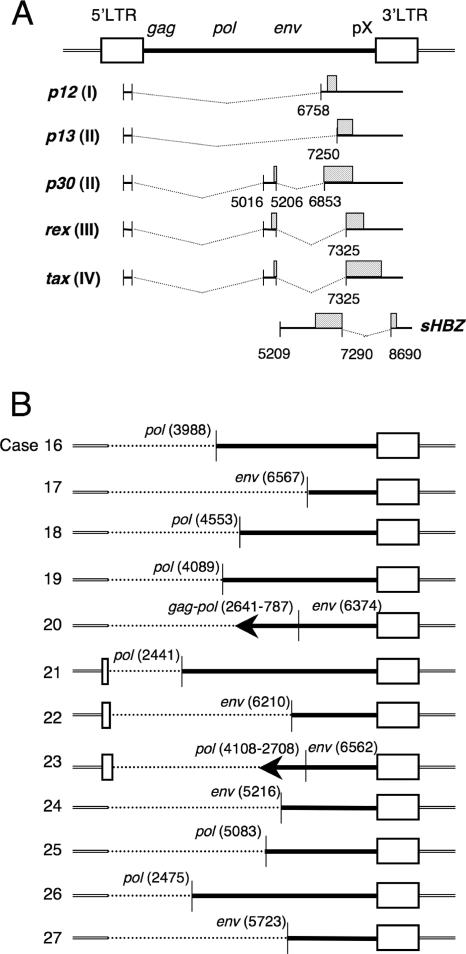
We determined the entire sequence of type 2 defective proviruses in 12 ATL cases by PCR and sequencing (Fig. 3). Although type 2 defective provirus lacks a viral promoter/enhancer (5′ LTR), such a provirus might trap a cellular promoter. The integration sites, trapped cellular genes, and prospective transcription of viral genes are summarized in Table 2. Among five proviruses integrated in a transcriptional unit, the direction of viral transcription matched that of cellular genes in all cases. By use of the GeneCards database (http://www.genecards.org/background.shtml), four of these genes (DONSON, PCDH9, ALPK1, and CLEC7A) were found to be ubiquitously expressed, and the fifth gene, XYLT1, was transcribed in the heart, spleen, and brain. In three of these cases (cases 16, 18, and 21), the second exon of the tax, p30, and rex genes was retained, indicating that these genes could be transcribed in these ATL cells. Among type 2 defective proviruses, the second exon of the tax, rex, and p30 genes was deleted in seven cases (cases 17, 20, 22, 23, 24, 25, and 27 [Fig. 3]). Without this exon, Tax protein lacks NF-κB- and CREB-activating activities (48). Taken together, these data indicate that wild-type Tax proteins cannot be produced in nine cases due to either deletion of an exon or lack of a promoter, while in three cases type 2 defective provirus can generate Tax, Rex, p12, p13, and p30 proteins by using cellular promoters.
FIG. 3.
Schematic diagram of type 2 defective proviruses. (A) The splicing patterns of the p12, p13, p30, rex, tax, and HBZ genes are indicated. Hatched boxes represent coding regions. The number of open reading frames (I, II, III, or IV) in the pX region is given in parentheses. Numbering is according to the work of Seiki et al. (46). (B) For cases with type 2 defective proviruses, dotted lines represent deleted portions, and arrows (cases 20 and 23) indicate inverted regions of proviruses. Positions of defective proviruses correspond to those of the wild-type virus diagramed in panel A. Type 2 defective provirus lacks the second exon of tax in 7 of 12 cases (cases 17, 20, 22, 23, 24, 25, and 27). In cases 21, 22, and 23, parts of the 5′ LTR remain.
TABLE 2.
Predicted transcription of regulatory and accessory genes in type 2 defective proviruses
| Case no. | Coding gene | Transcriptional direction | Integration site | Predicted transcriptiona of the following gene:
|
Clinical subtype | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| p12 | p13 | p30 | tax | rex | HBZ | |||||
| 16 | XYLT1 | Sense | 16p12 | + | + | + | + | + | + | Acute |
| 17 | None | 7q21 | − | − | − | − | − | + | Acute | |
| 18 | DONSON | Sense | 21q22 | + | + | + | + | + | + | Acute |
| 19 | None | 4p15 | − | − | − | − | − | + | Acute | |
| 20 | None | 1q12 | − | − | − | − | − | + | Lymphoma | |
| 21 | PCDH9 | Sense | 13q21 | + | + | + | + | + | + | Acute |
| 22 | None | 5q23 | − | − | − | − | − | + | Acute | |
| 23 | ALPK1 | Sense | 4q25 | + | + | − | − | − | + | Chronic |
| 24 | None | 4p15 | − | − | − | − | − | + | Acute | |
| 25 | CLEC7A | Sense | 12p13 | + | + | − | − | − | + | Lymphoma |
| 26 | None | 8q24 | − | − | − | − | − | + | Acute | |
| 27 | None | 2q32 | − | − | − | − | − | + | Acute | |
The transcription of regulatory (tax and rex) and accessory genes (p12, p13, p30, and HBZ) is predicted from data on integration sites and sequences. +, possible transcription; −, transcription is not expected by loss of promoters or exons. In five cases, cellular gene promoters are trapped (cases 16, 18, 21, 23, and 25). Transcriptional units near integration sites were analyzed by the BLAT program (UCSC Human Genome Project Working Draft, May 2006 freeze).
HBZ coding sequences in type 2 defective proviruses.
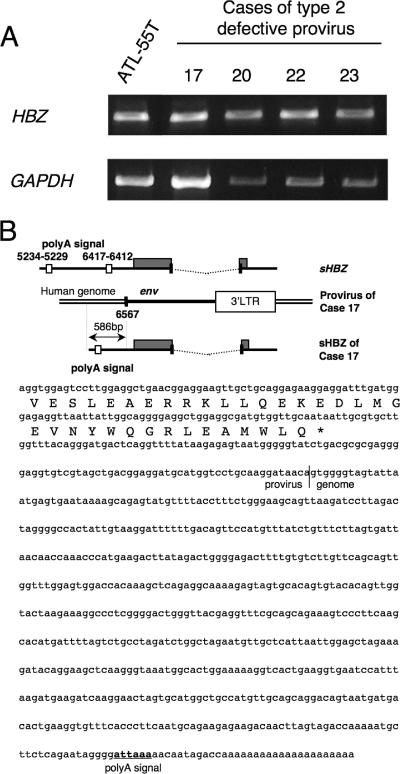
Recently, we reported that HBZ was transcribed in all ATL cases examined and that it supported the growth of ATL cells, suggesting that it is critical for leukemogenesis (44). Examination of sequences of HBZ in type 2 defective proviruses revealed no nonsense mutations, insertions, or deletions, although several missense mutations were detected (Fig. 4 and 5). Two HBZ polyadenylation signal sequences have been identified (8, 35, 44), both deleted in case 17, as shown in Fig. 6B. Next, we examined HBZ transcription in ATL cells with type 2 defective proviruses. In four cases in which RNAs were available for analysis, HBZ was transcribed in all cases, including case 17 (Fig. 6A). We determined the 3′end of the HBZ transcript in case 17 by 3′-RACE and found that the HBZ gene trapped a cellular polyadenylation signal (Fig. 6B). In case 17, the proviral sequence recombined with a human genomic sequence (7q21) in the env region (nucleotide 6567 according to the numbering of Seiki et al. [46]). The polyadenylation signal (attaaa [underlined in Fig. 6B]) in the host genome sequences was detected upstream of the poly(A) tail.
FIG. 4.
Nucleotide sequences of the HBZ gene in ATL cells with type 2 defective provirus. The coding sequences of the HBZ gene in type 2 defective proviruses were amplified, and their sequences were determined. Case numbers are given on the left. The nucleotide sequence of the HBZ gene in ATK-1 is shown as a control.
FIG. 5.
Amino acid sequences of HBZ protein in ATL cells with type 2 defective provirus. The predicted amino acid sequences (206 amino acids) of HBZ protein are shown. Case numbers are given on the left. The ATK-1 sequence is used as a control.
FIG. 6.
HBZ transcription in ATL cells with type 2 defective provirus. (A) HBZ transcription in ATL cases with type 2 defective proviruses was analyzed by RT-PCR. GAPDH transcripts served as internal controls. RNA from ATL-55T cells served as a positive control for HBZ transcription. (B) 3′-RACE was undertaken to determine the polyadenylation signal in case 17. In that case, previously reported HBZ polyadenylation signals were deleted. 3′-RACE identified a polyadenylation signal (attaaa) (underlined) in the host genomic sequence. sHBZ, spliced form of the HBZ gene.
Frequencies of defective proviruses in HTLV-1 carriers.
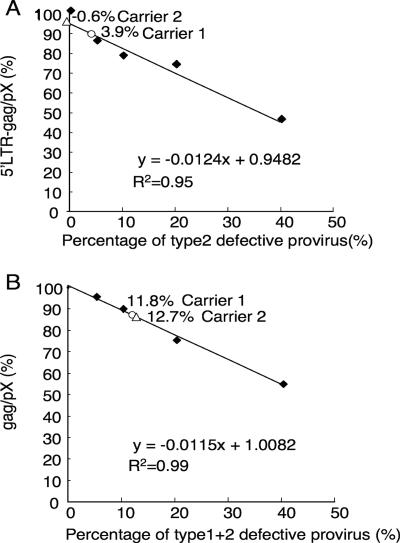
As shown in Table 1, the frequencies of type 1 and type 2 defective proviruses in ATL cases were 11.4% and 27.8%, respectively. However, their frequencies in the carrier state were unknown. To estimate those frequencies, we quantified provirus loads at three different regions of the provirus, including the region conserved in all types of provirus (pX), the region deleted in both types of defective provirus (gag), and the region deleted only in type 2 defective provirus (5′ LTR-gag). We determined that part of the gag region (nucleotides 1591 to 1681 according to the numbering of Seiki et al. [46]) was deleted in 95% of defective proviruses (data not shown). The 5′ LTR-gag region was absent only in type 2 defective provirus, whereas gag nucleotides 1591 to 1681 were deleted in both type 1 and type 2 proviruses. By contrast, the pX region was present in all proviruses. As controls, we mixed various proportions of genomic DNA derived from ATL cases exhibiting type 2 defective provirus with DNA from an ATL cell line, TL-Om1, carrying the complete provirus, in various proportions, i.e., 0, 5, 10, 20, and 40%. Based on data for provirus loads from these samples, regression lines were drawn using the least-squares method (Fig. 7). Evaluation of provirus loads at the 5′ LTR-gag and pX regions enabled us to estimate the frequency of type 2 defective provirus (Fig. 7A). Quantification of the gag and pX regions indicated the frequencies of total defective (type 1 plus type 2) provirus (Fig. 7B). We measured provirus loads at three provirus regions using DNA samples from two carriers with high provirus loads (64.2% in carrier 1, and 47.2% in carrier 2) and then calculated the frequencies of type 2 and total defective proviruses (Fig. 7A and B). Due to the limitations of this assay, we could not determine the frequencies of defective provirus in carriers with moderate or low provirus loads. The results showed that the frequencies of type 2 defective provirus in the two carriers were 3.9 and −0.6%. Likewise, the frequency of total defective provirus was 11.8% in carrier 1 and 12.7% in carrier 2. These results suggest that the frequency of type 2 defective provirus in the carrier state is much lower than that in ATL cases, while the frequencies of type 1 defective provirus in ATL cases and the carrier state seem to be similar.
FIG. 7.
Quantification of defective proviruses in carriers. To estimate the frequencies of type 1 and 2 defective proviruses in carriers, we quantified HTLV-1 provirus at three different provirus regions: 5′ LTR-gag (deleted in type 2 defective provirus), gag (deleted in both type 1 and 2 defective proviruses), and pX (conserved in all proviruses). Genomic DNA from an ATL patient with type 2 defective provirus was mixed with genomic DNA from an ATL cell line with a complete provirus (TL-Om1) in various proportions. These DNAs were used to quantify provirus, and regression lines were drawn using the least-squares method (solid diamond). The frequencies of type 2 and total defective proviruses in carriers were calculated from quantitative data at the 5′ LTR-gag and pX (A) or gag and pX (B) regions, respectively. The frequencies of defective proviruses in carrier 1 (open circle) and carrier 2 (open triangle) were analyzed. R2, coefficient of determination.
DISCUSSION
A defective provirus lacking the 5′ LTR has been reported in B-cell neoplasms caused by the avian leukosis virus (ALV). In these tumors, defective ALV proviruses were integrated into the 5′ region of the c-myc gene, and the 3′ LTR drove expression of c-myc (16, 36, 41). The 5′ LTR causes transcriptional interference (9) and is deleted in tumors induced by ALV. In contrast to ALV, HTLV-1 provirus integration sites are random in ATL cells (12, 40, 45), indicating that the 3′ LTR does not activate the transcription of specific cellular genes. By contrast, we have reported that the HBZ gene, which is transcribed from the 3′ LTR, is required for the proliferation of ATL cells (44). The observations that HBZ is transcribed in ATL cells with type 2 defective proviruses and that the HBZ sequence is intact suggest that its expression is necessary for leukemogenesis. In particular, the finding that type 2 defective provirus lacking proviral polyadenylation signal sequences trapped a cellular polyadenylation signal suggests a critical role for HBZ in HTLV-1-induced oncogenesis. A recent study reported that HBZ protein is not necessary for in vitro immortalization of T lymphocytes but is critical for infectivity and persistence in vivo (4). Furthermore, this study indicates the significance of the HBZ gene in oncogenesis by HTLV-1, suggesting that persistent infection by HTLV-1 carrying the HBZ gene is essential for oncogenesis.
Retrovirus integrases generate short repeat sequences adjacent to an LTR during integration. It has been reported that human immunodeficiency virus with mutant integrase lacking activity still produces proviruses that are integrated into the host genome but exhibit no adjacent repeats (19), suggesting that cellular recombination and DNA repair machinery likely integrate the provirus. Therefore, the appearance of short repeats adjacent to LTRs indicates that viral integrase functions in integration. Since LTRs are conserved in integrated proviruses, they are thought to be essential for integration. Here we identified three kinds of type 2 defective provirus in ATL patients. In the first, the 5′ LTR was deleted and the internal proviral sequence was flanked by short repeat sequences at the 5′ end of the provirus. In these cases, TG in pol or env sequences was ligated to short repeats, indicating that the viral integrase recognized TG dinucleotides in these internal sequences rather than TG at end of the LTR. In a second type, short 5′ regions of the 5′ LTR U3 (18 to 43 bp) were retained at the 5′ end of the provirus. In these cases, it is unclear whether deletion occurred before or after integration. In a third type, short repeat sequences were not seen at integration sites, and short host genomic sequences were also deleted. In these cases, CA dinucleotides were retained at the 3′ end of the LTR, indicating that the viral integrase recognized these sequences at the integration step and suggesting that deletion of the 5′ LTR occurred after integration. Such deletions may block Tax expression, enabling ATL cells to escape the host immune system.
Quantitative analyses of defective provirus indicate that the frequency of type 2 defective provirus (27.8%) in ATL is much higher than that seen in carriers (less than 3.9%), suggesting that HTLV-1-infected cells with type 2 defective provirus tend to become leukemic. The presence of type 2 defective provirus without short repeats supports the hypothesis that the provirus 5′ LTR is deleted during oncogenesis. Such cells would lose Tax expression and escape host immunosurveillance but still transcribe HBZ. Type 2 defective proviruses with short repeats are thought to be generated before integration. It is likely that such infected cells are selected during leukemogenesis. Thus, two mechanisms generating type 2 defective provirus increase its frequency in ATL cells.
Analyses of type 2 defective provirus can provide important information regarding the minimum components of the HTLV-1 viral sequence necessary for oncogenesis. We have reported that all ATL cases examined express HBZ and that HBZ RNA has growth-promoting activity, while tax expression was observed only in some (44). All type 2 defective proviruses analyzed in this study retained the HBZ coding sequence, and ATL cells with type 2 defective provirus expressed HBZ transcripts. Since viral integrase recognizes CA sequences inside the provirus as shown here, defective proviruses without a 3′ LTR could in theory be generated during integration. However, no such defective provirus was observed in ATL cells, indicating that only proviruses capable of HBZ expression are selected during oncogenesis. With regard to the tax, p30, and rex genes, seven cases analyzed here lacked the second exon (Fig. 3). At least two cases (cases 17 and 20) likely lost the second exon before integration. Without this exon, Tax cannot activate NF-κB and CREB (48), suggesting that these activities of Tax are not essential for oncogenesis. Type 2 defective provirus trapped the cellular promoters in 5 of 12 cases. In three of these cases, regulatory genes (tax and rex) and accessory genes could be transcribed, since they retained the second exon. In the remaining two cases, only p12 and p13 genes could be expressed. Taken together, regulatory genes and most of the accessory genes, which are transcribed from the 5′ LTR, were not transcribed in 9 of 12 cases with type 2 defective proviruses. However, all cases could transcribe the HBZ gene from the 3′ LTR.
Tax has been demonstrated to be oncogenic in transgenic animals (22, 23, 27). In these animals, tumor types depend on the tax promoter, indicating that Tax is oncogenic in different cell types. It has been reported that Tax can induce chromosomal instabilities (26, 30, 42), which along with its other functions, such as NF-κB activation and functional inactivation of p53, should play an important role in oncogenesis. Our previous (44) and present studies suggest that HBZ also functions in oncogenesis. In human papillomavirus-induced tumors, the E6 and E7 viral proteins cooperate in oncogenesis (34). It is likely that the tax and HBZ genes function cooperatively in HTLV-1-induced oncogenesis. Further studies are required to clarify the roles of the tax and HBZ genes in ATL.
Here we have demonstrated that a defective provirus lacking the 5′ LTR was generated before and after provirus integration. Detailed analyses of such defective proviruses also suggested that HBZ activity promotes oncogenesis by HTLV-1.
Acknowledgments
We thank Elise Lamar for excellent proofreading of the manuscript.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan and by an anticancer project grant from the Ministry of Health, Welfare, and Labor of Japan.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Albrecht, B., N. D. Collins, M. T. Burniston, J. W. Nisbet, L. Ratner, P. L. Green, and M. D. Lairmore. 2000. Human T-lymphotropic virus type 1 open reading frame I p12I is required for efficient viral infectivity in primary lymphocytes. J. Virol. 74:9828-9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, B., and M. D. Lairmore. 2002. Critical role of human T-lymphotropic virus type 1 accessory proteins in viral replication and pathogenesis. Microbiol. Mol. Biol. Rev. 66:396-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arisawa, K., M. Soda, S. Endo, K. Kurokawa, S. Katamine, I. Shimokawa, T. Koba, T. Takahashi, H. Saito, H. Doi, and S. Shirahama. 2000. Evaluation of adult T-cell leukemia/lymphoma incidence and its impact on non-Hodgkin lymphoma incidence in southwestern Japan. Int. J. Cancer 85:319-324. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, J., B. Yamamoto, M. Li, A. J. Phipps, I. Younis, M. D. Lairmore, and P. L. Green. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 107:3976-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asante-Appiah, E., and A. M. Skalka. 1997. Molecular mechanisms in retrovirus DNA integration. Antivir. Res. 36:139-156. [DOI] [PubMed] [Google Scholar]
- 6.Bangham, C. R. 2003. Human T-lymphotropic virus type 1 (HTLV-1): persistence and immune control. Int. J. Hematol. 78:297-303. [DOI] [PubMed] [Google Scholar]
- 7.Basbous, J., C. Arpin, G. Gaudray, M. Piechaczyk, C. Devaux, and J. M. Mesnard. 2003. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 278:43620-43627. [DOI] [PubMed] [Google Scholar]
- 8.Cavanagh, M. H., S. Landry, B. Audet, C. Arpin-Andre, P. Hivin, M. E. Pare, J. Thete, E. Wattel, S. J. Marriott, J. M. Mesnard, and B. Barbeau. 2006. HTLV-I antisense transcripts initiating in the 3′ LTR are alternatively spliced and polyadenylated. Retrovirology 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, B. R., P. T. Lomedico, and G. Ju. 1984. Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature 307:241-245. [DOI] [PubMed] [Google Scholar]
- 10.Ding, W., B. Albrecht, R. E. Kelley, N. Muthusamy, S. J. Kim, R. A. Altschuld, and M. D. Lairmore. 2002. Human T-cell lymphotropic virus type 1 p12I expression increases cytoplasmic calcium to enhance the activation of nuclear factor of activated T cells. J. Virol. 76:10374-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding, W., B. Albrecht, R. Luo, W. Zhang, J. R. Stanley, G. C. Newbound, and M. D. Lairmore. 2001. Endoplasmic reticulum and cis-Golgi localization of human T-lymphotropic virus type 1 p12I: association with calreticulin and calnexin. J. Virol. 75:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi, K., X. Wu, Y. Taniguchi, J. Yasunaga, Y. Satou, A. Okayama, K. Nosaka, and M. Matsuoka. 2005. Preferential selection of human T-cell leukemia virus type I provirus integration sites in leukemic versus carrier states. Blood 106:1048-1053. [DOI] [PubMed] [Google Scholar]
- 13.Etoh, K., S. Tamiya, K. Yamaguchi, A. Okayama, H. Tsubouchi, T. Ideta, N. Mueller, K. Takatsuki, and M. Matsuoka. 1997. Persistent clonal proliferation of human T-lymphotropic virus type I-infected cells in vivo. Cancer Res. 57:4862-4867. [PubMed] [Google Scholar]
- 14.Fan, H., M. Palmarini, and J. C. DeMartini. 2003. Transformation and oncogenesis by jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275:139-177. [DOI] [PubMed] [Google Scholar]
- 15.Franchini, G., R. Fukumoto, and J. R. Fullen. 2003. T-cell control by human T-cell leukemia/lymphoma virus type 1. Int. J. Hematol. 78:280-296. [DOI] [PubMed] [Google Scholar]
- 16.Fung, Y. K., A. M. Fadly, L. B. Crittenden, and H. J. Kung. 1981. On the mechanism of retrovirus-induced avian lymphoid leukosis: deletion and integration of the proviruses. Proc. Natl. Acad. Sci. USA 78:3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa, Y., R. Kubota, M. Tara, S. Izumo, and M. Osame. 2001. Existence of escape mutant in HTLV-I tax during the development of adult T-cell leukemia. Blood 97:987-993. [DOI] [PubMed] [Google Scholar]
- 18.Gallo, R. C. 2002. Human retroviruses after 20 years: a perspective from the past and prospects for their future control. Immunol. Rev. 185:236-265. [DOI] [PubMed] [Google Scholar]
- 19.Gaur, M., and A. D. Leavitt. 1998. Mutations in the human immunodeficiency virus type 1 integrase D,D(35)E motif do not eliminate provirus formation. J. Virol. 72:4678-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff, S. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1939. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 21.Grassmann, R., M. Aboud, and K. T. Jeang. 2005. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene 24:5976-5985. [DOI] [PubMed] [Google Scholar]
- 22.Grossman, W. J., J. T. Kimata, F. H. Wong, M. Zutter, T. J. Ley, and L. Ratner. 1995. Development of leukemia in mice transgenic for the tax gene of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 92:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasegawa, H., H. Sawa, M. J. Lewis, Y. Orba, N. Sheehy, Y. Yamamoto, T. Ichinohe, Y. Tsunetsugu-Yokota, H. Katano, H. Takahashi, J. Matsuda, T. Sata, T. Kurata, K. Nagashima, and W. W. Hall. 2006. Thymus-derived leukemia-lymphoma in mice transgenic for the Tax gene of human T-lymphotropic virus type I. Nat. Med. 12:466-472. [DOI] [PubMed] [Google Scholar]
- 24.Hayward, W. S., B. G. Neel, and S. M. Astrin. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475-480. [DOI] [PubMed] [Google Scholar]
- 25.Inoue, J., M. Yoshida, and M. Seiki. 1987. Transcriptional (p40x) and post-transcriptional (p27x-III) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc. Natl. Acad. Sci. USA 84:3653-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, D. Y., F. Spencer, and K. T. Jeang. 1998. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell 93:81-91. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi, K., H. Ikeda, T. Tsuchikawa, T. Tsuji, S. Tanaka, K. Fugo, T. Sugaya, Y. Tanaka, M. Tateno, N. Maruyama, and T. Yoshiki. 2002. A novel animal model of thymic tumour: development of epithelial thymoma in transgenic rats carrying human T lymphocyte virus type I pX gene. Int. J. Exp. Pathol. 83:247-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. J., W. Ding, B. Albrecht, P. L. Green, and M. D. Lairmore. 2003. A conserved calcineurin-binding motif in human T lymphotropic virus type 1 p12I functions to modulate nuclear factor of activated T cell activation. J. Biol. Chem. 278:15550-15557. [DOI] [PubMed] [Google Scholar]
- 29.Koiwa, T., A. Hamano-Usami, T. Ishida, A. Okayama, K. Yamaguchi, S. Kamihira, and T. Watanabe. 2002. 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 76:9389-9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, Y. L., and C. Z. Giam. 2006. Activation of the anaphase promoting complex by HTLV-1 tax leads to senescence. EMBO J. 25:1741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuoka, M. 2005. Human T-cell leukemia virus type I (HTLV-I) infection and the onset of adult T-cell leukemia (ATL). Retrovirology 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuoka, M., and K. T. Jeang. 2005. Human T-cell leukemia virus type I at age 25: a progress report. Cancer Res. 65:4467-4470. [DOI] [PubMed] [Google Scholar]
- 33.Michael, B., A. M. Nair, H. Hiraragi, L. Shen, G. Feuer, K. Boris-Lawrie, and M. D. Lairmore. 2004. Human T lymphotropic virus type-1 p30II alters cellular gene expression to selectively enhance signaling pathways that activate T lymphocytes. Retrovirology 1:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78:11451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murata, K., T. Hayashibara, K. Sugahara, A. Uemura, T. Yamaguchi, H. Harasawa, H. Hasegawa, K. Tsuruda, T. Okazaki, T. Koji, T. Miyanishi, Y. Yamada, and S. Kamihira. 2006. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J. Virol. 80:2495-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neel, B. G., W. S. Hayward, H. L. Robinson, J. Fang, and S. M. Astrin. 1981. Avian leukosis virus-induced tumors have common proviral integration sites and synthesize discrete new RNAs: oncogenesis by promoter insertion. Cell 23:323-334. [DOI] [PubMed] [Google Scholar]
- 37.Nevins, J. 2001. Cell transformation by viruses, p. 245-283. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 38.Nicot, C., M. Dundr, J. M. Johnson, J. R. Fullen, N. Alonzo, R. Fukumoto, G. L. Princler, D. Derse, T. Misteli, and G. Franchini. 2004. HTLV-1-encoded p30II is a post-transcriptional negative regulator of viral replication. Nat. Med. 10:197-201. [DOI] [PubMed] [Google Scholar]
- 39.Nicot, C., R. L. Harrod, V. Ciminale, and G. Franchini. 2005. Human T-cell leukemia/lymphoma virus type 1 nonstructural genes and their functions. Oncogene 24:6026-6034. [DOI] [PubMed] [Google Scholar]
- 40.Ohshima, K., A. Ohgami, M. Matsuoka, K. Etoh, A. Utsunomiya, T. Makino, M. Ishiguro, J. Suzumiya, and M. Kikuchi. 1998. Random integration of HTLV-1 provirus: increasing chromosomal instability. Cancer Lett. 132:203-212. [DOI] [PubMed] [Google Scholar]
- 41.Payne, G. S., S. A. Courtneidge, L. B. Crittenden, A. M. Fadly, J. M. Bishop, and H. E. Varmus. 1981. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell 23:311-322. [DOI] [PubMed] [Google Scholar]
- 42.Peloponese, J. M., Jr., K. Haller, A. Miyazato, and K. T. Jeang. 2005. Abnormal centrosome amplification in cells through the targeting of Ran-binding protein-1 by the human T cell leukemia virus type-1 Tax oncoprotein. Proc. Natl. Acad. Sci. USA 102:18974-18979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pique, C., A. Ureta-Vidal, A. Gessain, B. Chancerel, O. Gout, R. Tamouza, F. Agis, and M. C. Dokhelar. 2000. Evidence for the chronic in vivo production of human T cell leukemia virus type I Rof and Tof proteins from cytotoxic T lymphocytes directed against viral peptides. J. Exp. Med. 191:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satou, Y., J. Yasunaga, M. Yoshida, and M. Matsuoka. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. USA 103:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seiki, M., R. Eddy, T. B. Shows, and M. Yoshida. 1984. Nonspecific integration of the HTLV provirus genome into adult T-cell leukaemia cells. Nature 309:640-642. [DOI] [PubMed] [Google Scholar]
- 46.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimoyama, M. 1991. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984-87). Br. J. Haematol. 79:428-437. [DOI] [PubMed] [Google Scholar]
- 48.Shuh, M., S. A. Hill, and D. Derse. 1999. Defective and wild-type human T-cell leukemia virus type I proviruses: characterization of gene products and trans-interactions between proviruses. Virology 262:442-451. [DOI] [PubMed] [Google Scholar]
- 49.Silverman, L. R., A. J. Phipps, A. Montgomery, L. Ratner, and M. D. Lairmore. 2004. Human T-cell lymphotropic virus type 1 open reading frame II-encoded p30II is required for in vivo replication: evidence of in vivo reversion. J. Virol. 78:3837-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stary, A., and A. Sarasin. 1992. Molecular analysis of DNA junctions produced by illegitimate recombination in human cells. Nucleic Acids Res. 20:4269-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stehelin, D., H. E. Varmus, J. M. Bishop, and P. K. Vogt. 1976. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260:170-173. [DOI] [PubMed] [Google Scholar]
- 52.Takeda, S., M. Maeda, S. Morikawa, Y. Taniguchi, J. Yasunaga, K. Nosaka, Y. Tanaka, and M. Matsuoka. 2004. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer 109:559-567. [DOI] [PubMed] [Google Scholar]
- 53.Tamiya, S., M. Matsuoka, K. Etoh, T. Watanabe, S. Kamihira, K. Yamaguchi, and K. Takatsuki. 1996. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood 88:3065-3073. [PubMed] [Google Scholar]
- 54.Taniguchi, Y., K. Nosaka, J. Yasunaga, M. Maeda, N. Mueller, A. Okayama, and M. Matsuoka. 2005. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 2:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida, M. 2001. Multiple viral strategies of HTLV-1 for dysregulation of cell growth control. Annu. Rev. Immunol. 19:475-496. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, W., J. W. Nisbet, B. Albrecht, W. Ding, F. Kashanchi, J. T. Bartoe, and M. D. Lairmore. 2001. Human T-lymphotropic virus type 1 p30II regulates gene transcription by binding CREB binding protein/p300. J. Virol. 75:9885-9895. [DOI] [PMC free article] [PubMed] [Google Scholar]