Abstract
Cells harboring infectious, but transcriptionally latent, human immunodeficiency virus type 1 (HIV-1) proviruses currently pose an insurmountable barrier to viral eradication in infected patients. To better understand the molecular basis for HIV-1 latency, we used the J-Lat model of postintegration HIV-1 latency to assess the kinetic relationship between the induction of NF-κB and the activation of latent HIV-1 gene expression. Chromatin immunoprecipitation analyses revealed an oscillating pattern of RelA recruitment to the HIV-1 long terminal repeat (LTR) during continuous tumor necrosis factor alpha (TNF-α) stimulation. RNA polymerase II (Pol II) recruitment to the HIV-1 LTR closely mirrored RelA binding. Transient stimulation of cells with TNF-α for 15 min induced only a single round of RelA and RNA Pol II binding and failed to induce robust expression of latent HIV-1. Efficient formation of elongated HIV-1 transcripts required sustained induction by NF-κB, which promoted de novo synthesis of Tat. Cyclin-dependent kinase 9 (CDK9) and serine-2-phosphorylated RNA Pol II were rapidly recruited to the HIV-1 LTR after NF-κB induction; however, these elongating polymerase complexes were progressively dephosphorylated in the absence of Tat. Okadaic acid promoted sustained serine-2 phosphorylation of the C-terminal domain of RNA Pol II and stimulated efficient transcriptional elongation and HIV-1 expression in the absence of Tat. These findings underscore important differences between NF-κB and Tat stimulation of RNA Pol II elongation. While NF-κB binding to the HIV-1 LTR induces serial waves of efficient RNA Pol II initiation, elongation is impaired by the action of an okadaic acid-sensitive phosphatase that dephosphorylates the C-terminal domain of RNA Pol II. Conversely, the action of this phosphatase is overcome in the presence of Tat, promoting very efficient RNA Pol II elongation.
Therapeutic efforts to eradicate human immunodeficiency virus type 1 (HIV-1) from infected patients with highly effective combinations of antiviral drugs have been thwarted, in part, by a highly durable and drug-insensitive pool of latently infected memory CD4+ T cells. Latent HIV-1 infection is characterized by integration of the provirus into the host DNA, followed by temporary silencing of viral gene expression. Replication of these latent proviruses can be activated by various cues, including specific antigens and select cytokines (see reference 3 for review). As a consequence of these properties, latently infected cells are unaffected by existing antiretroviral therapies and retain the potential to reseed systemic infection upon activation. The observed half-life of the latent pool of HIV-1 is at least 45 months, and assuming an initial pool of 105 infected cells, eradicating these cells would take over 60 years of continuous treatment (13, 34). If curative therapies for HIV-1 infection are ever to become a reality, new strategies for rapidly eliminating or permanently silencing this pool of persistently infected and drug-insensitive virus must be developed (32). The identification of these strategies will be facilitated by a deeper understanding of the transcriptional events underlying HIV-1 latency.
The principal feature of HIV-1 latency is a failure of viral gene expression, chiefly as a consequence of uninitiated or aborted transcription. Expression of the integrated provirus is regulated by the promoter and enhancer elements in the 5′ long terminal repeat (LTR) (see reference 33 for review). Elongation of HIV-1 mRNA transcripts is strongly dependent on the viral transactivating protein Tat, which binds to a highly structured region of HIV-1 RNA, the trans-acting response element, located at the 5′ end of all initiated transcripts (22). Tat recruits the cellular P-TEFb kinase complex, composed of cyclin T1 and cyclin-dependent kinase 9 (CDK9) (44), to the HIV-1 promoter, which in turn induces serine-2 (S2) phosphorylation of proximal RNA polymerase II (Pol II) complexes (30). This modification promotes efficient transcriptional elongation. Mutations within Tat or the trans-acting response element are responsible for latent proviruses found in U1 and ACH-2 cell lines, respectively (10, 11). Remarkably, infectious virus can be induced from both of these cell lines by a range of NF-κB-inducing agents, indicating that this cellular complex can mimic many of the effects of Tat, albeit with lower efficiency.
The prototypical NF-κB heterodimer composed of RelA and p50 binds to two κB enhancer sites in the HIV-1 LTR located immediately upstream of the transcriptional initiation site (4, 31, 41). Induction of NF-κB is associated with both enhanced transcriptional initiation and elongation of HIV-1 mRNA transcripts (28, 39). RelA contains two strong transcriptional activation domains which mediate recruitment of a number of positively acting transcription factors, including P-TEFb (2). RelA-driven recruitment of P-TEFb is thought to underlie initial elongation of HIV-1 mRNA transcripts, permitting synthesis of Tat (2). Our recent studies further demonstrate that RelA contributes to transcriptional initiation by displacing repressive p50-HDAC1 complexes bound to κB sites of the HIV-1 LTR under basal conditions (41).
In the absence of activating stimuli, NF-κB is predominantly localized within the cytoplasmic compartment and prevented from binding DNA by association with inhibitory IκB proteins (8, 14, 18, 42). Stimulus-coupled activation of NF-κB involves increased activity of the IKK signalsome complex, which promotes phosphorylation of IκBα, a signal for polyubiquitylation and proteasome-mediated degradation of the inhibitor (9, 27, 35, 43). Liberated NF-κB complexes rapidly translocate into the nucleus, bind cognate DNA enhancers, and induce gene expression. Among NF-κB's many cellular targets is the IκBα gene. Thus, NF-κB activation stimulates de novo synthesis of its own inhibitor, leading to auto-regulation of NF-κB activity (35). In the presence of continuous stimulation of cells, NF-κB induction occurs in a dampened oscillatory manner involving steadily decreasing waves of NF-κB moving into the nucleus (7, 29, 38).
These features of NF-κB activation and the apparent functional homology of RelA and Tat as recruiters of P-TEFb prompted us to further investigate the role of NF-κB in the activation of latent HIV-1 gene expression. Owing to the rarity of latently infected cells in HIV-1-infected patients and the absence of specific surface markers for purification of these cells, we employed the J-Lat model of HIV-1 latency (21). J-Lats are Jurkat-derived T-cell clones infected with a single copy of a full-length HIV-1 provirus engineered to express green fluorescent protein (GFP) in lieu of Nef. J-Lat cells produce essentially no GFP under basal conditions, reflecting an absence of detectable RNA Pol II binding to the latent promoter. However, the transcriptional quiescence of HIV-1 in these cells can be reversed by treatment with tumor necrosis factor alpha (TNF-α), which promotes RNA Pol II binding (41). These findings implicate a deficiency in transcriptional initiation as a primary feature of the HIV-1 latency observed in many J-Lat clones.
In the current study, we compared and contrasted changes in RNA Pol II elongation in latently infected cells when NF-κB-inducing stimuli were administered transiently or continuously and when Tat expression was permitted or inhibited.
MATERIALS AND METHODS
Cell lines and culture conditions.
J-Lat 6.3 cells were maintained in RPMI supplemented with 10% fetal calf serum, penicillin, streptomycin, and l-glutamine. For stimulation, cells were treated with 20 ng/ml TNF-α (R&D Systems) or 30 nM okadaic acid (OA; Biomol) alone or in combination. For inhibition experiments, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; Sigma) was used at 10 ng/ml, and cycloheximide (Sigma) was used at 10 μg/ml. To produce NF-κB-DsRed2 reporter cells, Jurkat or various J-Lat cells were transduced with a lentiviral vector, PPT-κB-DsRed2 (12). Briefly, this construct was cotransfected with the vesicular stomatitis virus G protein envelope into HEK 293T cells, and virus-containing supernatants were harvested and concentrated with Centricon filtration units (Millipore). Parental Jurkat or J-Lat 6.3, 8.4, or 15.2 cells were infected with concentrated virus stocks incubated for 72 h, and DsRed2-negative cells were sorted by fluorescence-activated cell sorting. After negative sorting, cells were treated with TNF-α and incubated for 24 h, and cells with TNF-α-inducible DsRed2 expression were selected by flow cytometry and designated JκRed, J6.3κRed, J8.4κRed, and J15.2κRed.
Chromatin immunoprecipitation.
J-Lat 6.3 cells were adjusted to 1 × 106 cells/ml and incubated in medium with or without TNF-α (20 ng/ml) for various times. For pulse-stimulation experiments, cells were treated with TNF-α for 15 min, washed three times in complete culture medium, and then returned to culture. Chromatin immunoprecipitation (ChIP) assays were performed as described elsewhere (40). Antibodies employed were RelA (sc-109), RNA Pol II (sc-899), and CDK9 (H-169) (all three from Santa Cruz Biotechnology) and ser-2-RNA polymerase II (H5) and ser-5-RNA Pol II (H19) (Covance).
To detect specific HIV-1 LTR and β-actin unique short DNA regions, PCR primers were as described in reference 41. HIV+1000 5′HIV1000 (5′-TATTGCACCAGGCCAGATGA-3′) and 3′HIV1200 (5′-GTCTCTAAAGGGTTCCTTTG-3′). HIV+3000 was amplified with 5′HIV3000 (5′-ACTGGCAGAAAACAGAGAGA-3′) and 3′HIV3200 (5′-TATTTTTTGCACTGCCTCTG-3′). HIV+5000 DNA was amplified with 5′HIV5000 (5′-CATCATGGATCCACTCGACAGAGGAGAG-3′) and HIV3′5200 (5′-ATGATGAAGCTTGAGTCTGACTGTTCTG-3′). HIV+7000 was amplified with 5′HIV7000 (5′-AGTGACACAATCACCCTCCC-3′) and 3′HIV7200 (5′-ATAATTCACTTCTCCAATTG-3′). Amplification was performed with Taq polymerase (QIAGEN) for 35 to 40 cycles, and products were analyzed on 2.5% agarose gels. Images were acquired using an EagleEye II digital imaging system (Stratagene). All ChIP extracts were assessed for enrichment in β-tubulin DNA as a control for nonspecific DNA binding. Specific enrichment in target DNA sequences by a specified antibody was interpreted as DNA binding by the antibody target. ChIP extracts were quantitated with the QuantiTect SYBR Green PCR kit (QIAGEN). Fluorescence profiles were collected on an ABI 7700 real-time thermal cycler and analyzed with SDS v1.91 (Applied Biosystems).
RNA extraction and analysis of initiated and elongated HIV-1 transcripts.
J-Lat 6.3 cells (1 × 106 cells/ml) were either not treated or pretreated with OA (100 nM) for 1 h at 37°C and were either not stimulated or stimulated with TNF-α (20 ng/ml) for 30 min. RNA was extracted from 5 × 106 cells via RNAWiz (Ambion), and transcripts were quantitated using the QuantiTect SYBR Green reverse transcription-PCR (RT-PCR) kit (QIAGEN). To quantitate viral transcripts, serial dilutions of a quantitated RNA stock of full-length HIV-1 genome were employed as a reference standard (gift from R. Grant, Gladstone Institute of Virology and Immunology). PCR primers for detection of initiated and elongated HIV-1 and β-actin transcripts were as described previously (41). Fluorescence profiles were collected on an ABI 7700 real-time thermocycler and analyzed with SDS v1.91 (Applied Biosystems). The absence of nonspecific bands in RT-PCR products was confirmed by electrophoresis of samples on 2% agarose gels and visualization with ethidium bromide and UV light.
Immunoblotting analysis.
For analysis of protein expression, J-Lat 6.3 cells were collected by centrifugation and lysed on ice in egg lysis buffer (50 mM HEPES, pH 7, 250 mM NaCl, 1% Nonidet P-40, 5 mM EDTA) for 20 min, and lysates were clarified by microcentrifugation. Nuclear and cytoplasmic extracts were prepared as described previously (1). The protein concentration was quantitated using the Bradford protein assay (Bio-Rad), and 10 μg of each sample was added to an equal volume of 2× Laemmli buffer and heated to 95°C for 5 min. Samples were separated on 10% acrylamide Tris-HCl-buffered sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Bio-Rad), transferred to polyvinylidene difluoride membranes, and immunoblotted with antibodies specific for Sp1, RelA, or IκBα (Santa Cruz) or α-tubulin or HIV-1 Tat (Advanced Bioscience Laboratories). For detection of HIV-1 Tat, samples were concentrated by immunoprecipitation with antibodies recognizing HIV-1 Tat (116P; Covance) before immunoblotting.
Transfection, nucleofection, and flow cytometric detection of transfected cells.
For DNA transfection by electroporation, 1 × 107 J-Lat 6.3 cells were resuspended in 400 μl of culture medium with 5 μg of FLAG-Tat, T7-RelA, or empty pCMV4 expression vector and 1 μg pMACS H-2Kk marker DNA and transferred to a 0.4-cm gap electroporation cuvette (Bio-Rad). Cells were electroporated with a Bio-Rad Gene Pulser II set to 0.975 μF and 250 V and returned to culture for 16 h before further manipulation. To identify transfected cells, cells were stained with biotin-anti-H-2Kk antibodies, washed, and counterstained with streptavidin-allophyocyanin (Pharmingen). For nucleofection, J-Lat cells were resuspended at a concentration of 5 × 107 cells/ml in 100 μl of reagent R (Amaxa) with 5 μg of high-performance liquid chromatography-purified, annealed RNA oligonucleotides siTat S (5′-CUGCUUGUACCAAUUGCUAdTdT-3′), AS (5′-UAGCAAUUGGUACAAGCAGdTdT-3′), siTatMM S (5′-CUGCUUGUCACAAUUGCUAdTdT-3v), and AS (5′-UAGCAAUUGUGACAAGCAGdTdT-3′) (Ambion). Nucleofections were performed using program O28 with an Amaxa nucleofector. Small interfering RNA (siRNA)-nucleofected cells did not require marking, as 95% of cells were routinely nucleofected with siRNA. Cells were analyzed with a FACSCalibur flow cytometer (Becton Dickinson) and FlowJo software (Treesoft).
RESULTS
Continuous stimulation of NF-κB drives oscillating but synchronous association of RelA and RNA polymerase II with the latent HIV-1 LTR.
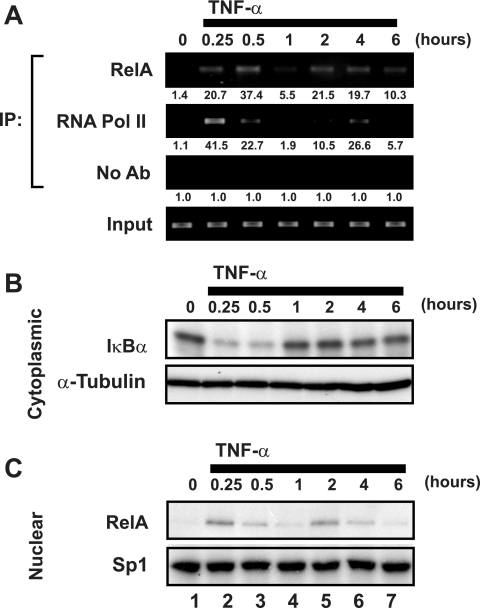
Our prior studies of the latent HIV-1 LTR revealed an absence of RNA polymerase II binding under unstimulated conditions but effective recruitment following TNF-α activation of the J-Lat cells. To further characterize the events underlying activation of the latent HIV-1 LTR, we sought to compare the kinetics of RelA and RNA Pol II recruitment after TNF-α stimulation of these cells. ChIP assays were employed to study the interaction of these factors with the HIV-1 LTR in J-Lat cells in vivo. Sheared formaldehyde-cross-linked chromatin extracts prepared from J-Lat cells untreated or treated with 20 ng/ml TNF-α continuously for up to 6 h were immunoprecipitated with antibodies specific for either RelA or RNA Pol II. The abundance of HIV-1 LTR DNA in immunoprecipitates was assessed by ethidium bromide visualization of PCR products amplified with primers specific for the HIV-1 LTR. Consistent with prior results (41), neither Rel A nor RNA Pol II binding to HIV-1 LTR DNA was detected in untreated cells (Fig. 1A). However, both RelA and RNA Pol II binding were detected 15 min after stimulation with TNF-α, and continued binding was observed after 30 min. ChIP analyses performed after 1 h of stimulation revealed a synchronous diminution of RNA Pol II and RelA binding. In the continuous presence of TNF-α, a second wave of RelA and RNA Pol II binding was observed at 2 h, followed by a less pronounced decline at 6 h. Importantly, levels of input HIV-1 LTR DNA were similar at each time point, and analysis of nonspecific DNA sequences supported specificity in the ChIP procedure (Fig. 1A and data not shown).
FIG. 1.
TNF-α stimulation induces an oscillating pattern of RelA and RNA Pol II binding to the latent HIV-1 LTR. (A) TNF-α stimulation leads to synchronous but oscillating recruitment of RelA and RNA Pol II to the latent HIV-1 LTR. Fixed chromatin extracts from J-Lat 6.3 cells either untreated or treated with 20 ng/ml TNF-α for various times were immunoprecipitated with antibodies specific for RelA, RNA Pol II, or no antibody as a control. Immunoprecipitates were assessed for enrichment in HIV-1 LTR DNA by UV visualization of PCR products in an ethidium bromide-stained gel. Data are representative of three independent experiments. Note the synchronicity of RelA and RNA Pol II recruitment to the HIV-1 LTR, as well as the nadir in binding 1 h after TNF-α stimulation. Quantitation of enrichment (fold increase) above the “no-antibody” control is indicated below each sample. (B) TNF-α-induced recruitment of RelA and RNA Pol II to the latent HIV-1 LTR coincides with IκBα degradation. Cytoplasmic extracts of samples treated as for panel A were prepared, and IκBα levels were assessed by immunoblotting. Cytoplasmic levels of α-tubulin were assessed to confirm equivalent loading of samples. (C) TNF-α induces bimodal nuclear enrichment of RelA. Nuclear extracts of samples treated as for panel A were prepared and analyzed for RelA enrichment by immunoblotting. Nuclear Sp1 levels were assessed to confirm equivalent loading. Note the largely overlapping patterns of nuclear enrichment of RelA and its recruitment to the latent HIV-1 LTR as assessed by ChIP (A).
The dynamic pattern of RelA association with the latent HIV-1 LTR after TNF-α stimulation resembled oscillatory patterns of NF-κB recruitment recently described for various NF-κB target genes (7, 29, 38). This oscillation is attributed to NF-κB-dependent induction of IκBα gene expression, which inhibits DNA binding and promotes nuclear export of RelA. Continued stimulation of the upstream IκB kinases induces secondary rounds of IκBα phosphorylation and degradation, leading to delayed waves of nuclear accumulation of NF-κB. To examine whether this process might underlie the pattern of RelA recruitment to the latent HIV-1 LTR, the abundance of IκBα and nuclear RelA after TNF-α stimulation was assessed by immunoblot analysis. After TNF-α stimulation, IκBα abundance was rapidly ablated; however, resynthesis of the inhibitory protein was apparent after 1 h. The level of IκBα expression after 2 h of TNF-α treatment was slightly depressed relative to untreated samples (Fig. 1B). Nuclear abundance of RelA was inversely correlated to IκBα levels, with a peak in nuclear RelA occurring after 15 min of TNF-α stimulation, a nadir at 1 h, and a second peak at 2 h of stimulation (Fig. 1C). This kinetic pattern of nuclear RelA expression was strikingly similar to the ChIP analyses of RelA and RNA Pol II binding to HIV-1 LTR DNA, suggesting that the oscillating nuclear abundance of RelA likely underlies the temporally dependent recruitment of Rel A and RNA Pol II observed in ChIP analyses.
Transient activation of NF-κB induces a single round of RelA and RNA polymerase II binding to the latent HIV-1 LTR.
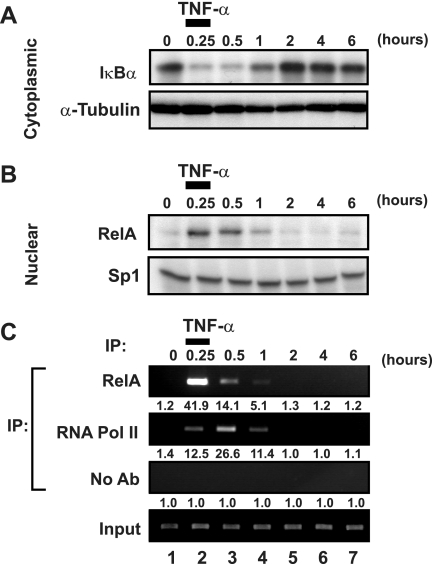
We next investigated whether a transient pulse of TNF-α could be employed to induce a single cycle of RelA activity. To test this possibility, J-Lat 6.3 cells were stimulated with TNF-α for 15 min, washed extensively, and returned to culture for various times. Immunoblot analysis of cytoplasmic IκBα expression demonstrated that the pulsed administration of TNF-α induced rapid degradation of the NF-κB inhibitor, with overall levels falling to levels similar to those observed with continuous TNF-α stimulation (Fig. 2A). Resynthesis of IκBα was apparent 1 h after TNF-α pulse treatment, and in contrast to continuous stimulation, expression levels thereafter appeared to remain slightly higher than in lysates of untreated cells. Correspondingly, early nuclear accumulation of RelA was observed in TNF-α-pulsed samples with kinetics similar to those in samples treated continuously with TNF-α (Fig. 2B). Significantly, TNF-α pulse-stimulation did not induce a second wave of nuclear RelA after 2 h of chase. Taken together, these results indicate that delayed waves of nuclear RelA are not induced by transient TNF-α stimulation.
FIG. 2.
Transient induction of NF-κB induces unimodal recruitment of RNA polymerase II to the latent HIV-1 LTR. (A) Pulsed administration of TNF-α induces IκBα degradation. J-Lat 6.3 cells were stimulated with 20 ng/ml TNF-α or left untreated for 15 min, washed twice in medium, and returned to culture for various times. Cytoplasmic extracts were prepared, and IκBα levels were assessed by immunoblotting. Cytoplasmic α-tubulin levels were assessed to confirm equivalent loading of samples. Note the similarity in depletion of IκBα in transiently and continuously TNF-α treated samples (Fig. 1B). (B) Pulsed administration of TNF-α induces transient activation of NF-κB. Nuclear extracts of samples treated as for panel A were prepared and analyzed for recruitment of RelA by immunoblotting. Nuclear Sp1 levels were assessed to confirm equivalent loading. (C) Pulsed TNF-α administration induces a unimodal pattern of RelA and RNA Pol II recruitment to the latent HIV-1 LTR. Fixed chromatin extracts of samples treated as for panel A were prepared and subjected to immunoprecipitation with antibodies specific to RelA or RNA Pol II or without antibody, as a nonspecific control. Immunoprecipitates were assessed for enrichment in HIV-1 LTR DNA by UV visualization of PCR products in a gel stained with ethidium bromide. Data are representative of three separate experiments. Note the absence of a second wave of RNA Pol II recruitment to the latent HIV-1 LTR. Quantitation of enrichment (fold increase) above the no-antibody control is indicated below each sample.
To examine the effects of transient induction of NF-κB on transcriptional regulation of latent HIV-1, ChIP analyses of RelA and RNA Pol II binding to HIV-1 LTR DNA were conducted in TNF-α-pulsed samples. Consistent with the predicted transient induction of nuclear NF-κB expression, TNF-α pulse treatment induced a single round of RelA recruitment to HIV-1 LTR DNA, with a peak in binding occurring at 15 min (Fig. 2C). Pulse-stimulation with TNF-α also induced only a single round of RNA Pol II binding to the HIV-1 LTR DNA, which appeared to peak at 30 min. Our earlier studies showed that either RelA recruitment or TSA stimulation results in increased acetylation of histones surrounding the HIV-1 LTR (41). Thus, changes in chromatin structure likely contribute to the increased binding of RNA Pol II.
Transient induction of NF-κB fails to activate latent HIV-1 proviral gene expression.
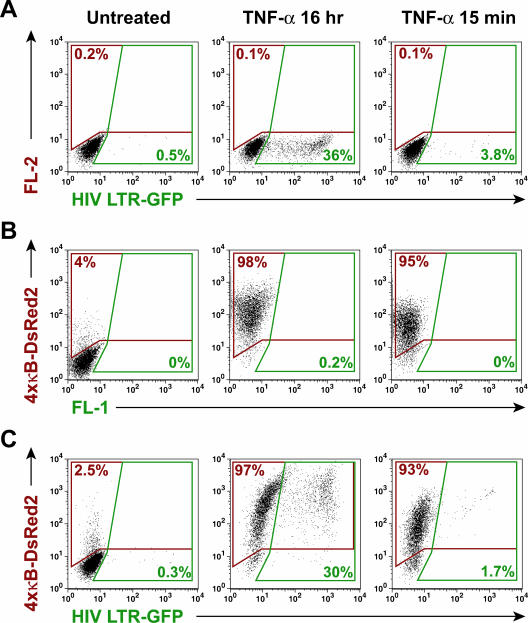
The transient nature of RNA Pol II recruitment to HIV-1 LTR DNA after TNF-α pulse-stimulation suggested that these stimulation conditions might not induce exit of HIV-1 proviruses from latency in J-Lat cells. To test this possibility, J-Lat 6.3 cells were either transiently or continuously stimulated with TNF-α and cultured for 16 h, and HIV-1-dependent expression of the viral GFP reporter (inserted in lieu of Nef) was assessed by flow cytometry. Expression of GFP in unstimulated J-Lat cells was observed in less than 0.5% of cells (Fig. 3A). Continuous stimulation with TNF-α induced robust GFP expression in 36% of the J-Lat cells. In contrast, TNF-α pulse-stimulation induced GFP expression in only 3.8% of cells. Similar results were observed in J-Lat clones 8.4 and 15.2, indicating that this effect can be generalized beyond a single J-Lat clone (data not shown). To test the effectiveness of TNF-α pulse-stimulation in the activation of a κB-dependent integrated gene, induction of DsRed2 expression was assessed in an independent set of Jurkat cells stably transduced with an integrated 4xκ B-DsRed2 reporter vector but lacking HIV-GFP proviruses (JκRed cells). In the absence of stimulation, DsRed2 expression in JκRed cells was largely suppressed, although a low level of constitutive reporter activity was detected (Fig. 3B). After 16 h of continuous TNF-α stimulation, 98% of the JκRed cells expressed DsRed2, indicating a nearly uniform induction of κB-specific gene expression in these cells. Similarly, and in striking contrast to expression of HIV-1-driven GFP, 95% of JκRed cells pulsed for 15 min with TNF-α exhibited DsRed2 expression 16 h after induction.
FIG. 3.
Transient induction of NF-κB is sufficient to induce robust general κB-dependent, but not latent HIV-1, gene expression. (A) Transient TNF-α administration induces poor expression of latent HIV-1. J-Lat 6.3 cells were left untreated or stimulated with 20 ng/ml TNF-α continuously or for 15 min, followed by washing and continued culture. HIV-LTR-dependent expression of GFP was assessed by flow cytometry. Note the overall lack of GFP expression in samples transiently treated with TNF-α. (B) Transient induction of NF-κB is sufficient to stimulate general κB-dependent gene expression. JκRed cells were treated as for panel A, and κB-dependent expression of DsRed2 was assessed by flow cytometry. Note the strong induction of κB-dependent gene expression by transient TNF-α stimulation. (C) Transient NF-κB induction induces robust expression of κB-dependent genes, but not latent HIV-1, in J-Lat 6.3 cells. J6.3κRed cells were treated as for panel A, and HIV-1 LTR-dependent expression of GFP and κB-dependent expression of DsRed2 were assessed by flow cytometry. Note the strong induction of κB-dependent gene expression and relative absence of HIV-1 gene expression induced by transient TNF-α stimulation.
To exclude the possibility of confounding mutations and to confirm that similar transient inducibility of κB-dependent gene expression is possible within J-Lat cells, populations of J-Lat 6.3 cells were stably transduced with the κB-DsRed2 reporter and then pulsed or continuously stimulated with TNF-α and analyzed by flow cytometry after 16 h of culture. As observed in JκRed cells, J6.3κRed cells exhibited a modest background of DsRed2-expressing cells (Fig. 3C). In cells continuously stimulated with TNF-α, 97% expressed κB-driven DsRed2, and 30% expressed HIV-1 LTR-driven GFP. Similarly, 93% of TNF-α-pulsed J6.3κRed cells exhibited DsRed2 expression, but only 1.7% of these cells expressed HIV-1 LTR-GFP. Similar results were obtained in J8.4κRed and J15.2κRed cells. Thus, failure of robust expression of HIV-1 in response to transient NF-κB induction is not a general feature of κB-dependent transcription but rather appears to reflect properties unique to the latent HIV-1 LTR.
TNF-α induction of efficient RNA Pol II elongation on the HIV-1 LTR requires prolonged stimulation.
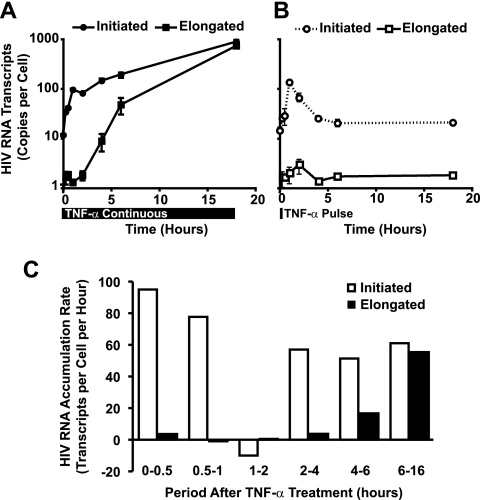
To further explore differences at the RNA level occurring in J-Lat cells after transient or continuous stimulation with TNF-α, the levels of initiated versus elongated HIV-1 mRNA transcripts were assessed by quantitative real-time RT-PCR. Continuous TNF-α treatment induced a rapid 10-fold increase in initiated HIV-1 mRNA transcripts. These plateaued temporarily at 1 h and then increased (Fig. 4A). Conversely, elongated HIV-1 mRNA transcripts were only modestly induced during the first 4 h of TNF-α treatment, with exponential increases occurring thereafter. Extrapolation of the rate of transcript accumulation revealed a similar rate of initiated HIV-1 transcript accumulation in early and late transcription (Fig. 4C). In contrast, elongated HIV-1 transcripts fail to accumulate at an appreciable rate until after 4 h of TNF-α stimulation.
FIG. 4.
Efficient elongation of HIV-1 mRNA transcripts is delayed in TNF-α-activated J-Lat cells. (A) TNF-α treatment of J-Lat cells induces rapid accumulation of initiated, but not elongated, HIV-1 mRNA transcripts. J-Lat 6.3 cells were treated with 20 ng/ml TNF-α for various times, and total RNA was extracted. Initiated and elongated HIV-1 mRNA transcripts were quantitated by real-time RT-PCR. Note the delayed emergence of elongated HIV-1 transcripts relative to the rapid increase in initiated transcripts. (B) Transient induction of NF-κB does not induce accumulation of elongated HIV-1 mRNA transcripts. J-Lat 6.3 cells were treated with 20 ng/ml TNF-α for 15 min, washed twice, and returned to culture for various times. Initiated and elongated HIV-1 mRNA transcript abundance were assessed as for panel A. (C) The kinetics of initiated and elongated HIV-1 transcript formation in TNF-α-induced J-Lat cells are dynamic. The rate of transcript formation in continuously TNF-α-stimulated J-Lat 6.3 cells was determined from the data in panel A. Note that the rate of initiated HIV-1 mRNA transcript formation is relatively constant across time, in contrast to the accelerating rate of elongated transcript formation.
Transient activation of NF-κB with TNF-α pulse-stimulation induced initiated HIV-1 mRNA transcripts with kinetics similar to those observed at early time points in continuously stimulated cells; however, after 1 h, initiated transcript abundance declined rapidly, with a t1/2 of 60 min (Fig. 4B). Elongated HIV-1 mRNA transcripts in TNF-α-pulsed samples were modestly induced in the first hour; however, sustained induction was not observed, and after 2 h the abundance of elongated transcripts declined to less than one copy per cell. These findings indicate that transient induction of NF-κB is sufficient to elicit a brief but temporary increase in transcriptional activity of the latent HIV-1 provirus. Continuous activation of the cells with TNF-α sharply increases the number of initiated HIV-1 transcripts; however, marked increases in HIV-1 elongation do not occur until after 4 h of stimulation.
TNF-α induction of HIV-1 Tat synergizes with active NF-κB to promote efficient transcriptional elongation of latent HIV-1 mRNA.
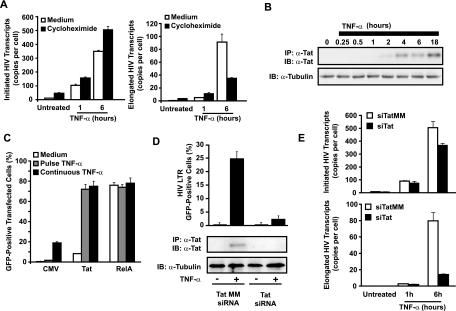
The delayed accumulation of elongated HIV-1 mRNA transcripts relative to initiated transcripts in TNF-α-stimulated J-Lat cells suggested that efficient elongation is dependent on de novo synthesis of an NF-κB-responsive factor. Alternatively, binding of RelA to the HIV-1 LTR might slowly induce chromatin rearrangements required for effective elongation. To distinguish between these possibilities, new protein synthesis in J-Lat 6.3 cells was inhibited with the translational inhibitor cycloheximide, and initiated and elongated HIV-1 transcripts were quantitated after TNF-α stimulation. Cycloheximide strongly impaired resynthesis of IκBα in TNF-α-treated cells, confirming its bioactivity in these assays (data not shown). Early initiated and elongated HIV-1 transcript levels were modestly enhanced in cycloheximide-treated J-Lat cells stimulated with TNF-α continuously for 1 h, indicating that these early transcripts are not dependent on de novo protein synthesis (Fig. 5A). In contrast, accumulation of elongated HIV-1 transcripts in samples treated with TNF-α for 6 h was markedly reduced in the presence of cycloheximide. These findings indicate that the initial NF-κB induction of transcriptional initiation and low-level elongation of HIV-1 mRNA do not require de novo protein synthesis while the high-efficiency elongation occurring after 4 to 6 h is dependent on new protein synthesis in J-Lat cells. The increased levels of transcriptional initiation induced in cycloheximide-treated cells are likely a consequence of the inhibition of IκBα resynthesis, resulting in prolonged NF-κB activity. Levels of β-actin mRNA and cell viability remained stable for the duration of the experiment, suggesting cycloheximide-induced toxicity was not a factor in the observed impairment of elongated HIV-1 mRNA transcript accumulation (data not shown).
FIG. 5.
TNF-α-induced expression of HIV-1 is dependent on de novo synthesis of Tat. (A) TNF-α-induced accumulation of elongated, but not initiated, HIV-1 mRNA transcripts is dependent on de novo protein synthesis. J-Lat 6.3 cells were preincubated with 10 μg/ml cycloheximide for 30 min or left in complete culture medium before a 15-min pulse or continuous stimulation with TNF-α for 1 or 6 h. Total RNA was extracted and initiated, and elongated HIV-1 mRNA transcripts were quantitated by real-time RT-PCR. Note the continued accumulation of initiated HIV-1 transcripts in cycloheximide-treated samples at both 1 and 6 h after TNF-α stimulation, whereas accumulation of elongated transcripts is blunted by cycloheximide at 6, but not 1, h after TNF-α treatment. Data are representative of three independent experiments. (B) Expression of Tat is delayed in TNF-α-stimulated J-Lat cells. Tat was immunoprecipitated from whole-cell lysates of J-Lat 6.3 cells treated with TNF-α for various times, and expression levels were assessed by immunoblotting. Note that efficient elongation of HIV-1 transcripts in Fig. 4A coincides with the kinetics of Tat expression. (C) Ectopic expression of HIV-1 Tat rescues HIV-1 gene expression in response to transient TNF-α stimulus. J-Lat 6.3 cells were cotransfected with control empty CMV, Tat, or RelA expression vectors and a plasmid expressing the cell surface H-2Kk marker to identify transfected cells. Transfected cells were stimulated with TNF-α for 15 min or continuously, and GFP expression was assessed in the H-2Kk-expressing cells. (D) Tat is required for efficient TNF-α-induced expression of latent HIV-1. J-Lat 6.3 cells were nucleofected with siRNA targeting Tat mRNA or a mismatched sequence, and knockdown of Tat expression was confirmed by immunoblotting (bottom panel). siRNA-treated cells were stimulated with TNF-α, and the percentage of cells expressing HIV-GFP was quantitated by flow cytometry (top). (E) Total RNA was extracted from cells treated as for panel D, and initiated (left) or elongated (right) transcripts were quantitated by RT-PCR.
The restricted transcriptional elongation apparent at early time points after TNF-α stimulation could reflect limited expression of Tat, which is under control of the viral LTR and regulated by NF-κB. Immunoblot analysis of Tat expression in J-Lat cells stimulated with TNF-α did not detect Tat at 0, 0.25, 0.5, and 1 h after induction. Low levels of Tat were first detected at 2 h, and higher levels were found at 4, 6, and 18 h (Fig. 5B). Of note, this slow accumulation of Tat tracked the observed delay in transcriptional elongation noted in Fig. 4B, suggesting that an absence of Tat may underlie the lack of latent gene expression in TNF-α-pulse-treated cells.
To examine whether expression of Tat alone was sufficient to rescue expression of latent HIV-1 in response to transient NF-κB induction, J-Lat 6.3 cells were transfected with control, Tat, or RelA expression vectors, pulse-stimulated or continuously stimulated with TNF-α, and analyzed by flow cytometry for HIV-directed GFP expression. Ectopic expression of Tat induced HIV-1 LTR-driven GFP expression in 9.8% of cells in the absence of stimulation (Fig. 5C). In contrast, >70% of Tat-transfected cells either pulse-stimulated or continuously stimulated with TNF-α expressed GFP. Of note, mean GFP fluorescence in J-Lat cells transfected with Tat and continuously stimulated with TNF-α was threefold higher than that observed in TNF-α pulse-stimulated cells (data not shown). These findings indicate that the initial wave of NF-κB recruited to the HIV-1 LTR is sufficient to drive effective gene expression in the context of Tat expression and argues against the necessity for a second wave of NF-κB recruitment. Importantly, ectopic expression of RelA strongly drove expression of latent HIV-1 without TNF-α, demonstrating inducibility of transfected cells in the absence of stimulation.
To further confirm a role for Tat as the key NF-κB-induced factor mediating TNF-α-induced HIV-1 gene expression, we examined the effect of Tat “knockdown” with siRNAs specific to the viral transactivator. Tat siRNA, but not a mismatched siRNA control, decreased TNF-α-induced Tat expression to undetectable levels (Fig. 5D). Tat siRNA reduced expression of HIV-GFP in J-Lat cells continuously stimulated with TNF-α from 24.7% to 3.8% but did not affect early HIV-1 transcriptional initiation (Fig. 5E). Taken together with studies of Tat overexpression, these studies demonstrate that expression of Tat is both necessary and sufficient for efficient TNF-α-induced expression of HIV-1 in latently infected cells.
TNF-α induces recruitment of CDK9 to the latent HIV-1 LTR.
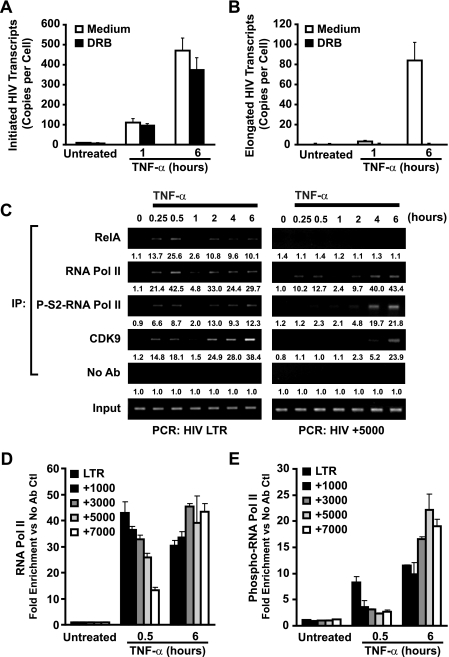
Elongation of HIV-1 mRNA is strongly dependent on phosphorylation of the carboxy-terminal domain (CTD) of RNA Pol II by P-TEFb (19), a factor recruited by Tat to the HIV-1 LTR. Recent studies suggested that RelA is similarly capable of recruiting P-TEFb to κB-responsive genes, such as interleukin-8 (2); however, the role of NF-κB-dependent P-TEFb recruitment to the HIV-1 LTR has not been explored. To determine whether early TNF-α-induced elongation of HIV-1 mRNA transcripts is dependent on P-TEFb or is rather a consequence of “slip-through” elongation of unphosphorylated polymerase complexes, we examined the effect of the P-TEFb inhibitor DRB on elongated HIV-1 mRNA transcript formation in cells stimulated with TNF-α for 1 h. As a positive control, DRB inhibition of delayed Tat-dependent elongation of HIV-1 mRNA was assessed in samples treated with TNF-α for 6 h. Prolonged TNF-α stimulation induced accumulation of ∼500 initiated and ∼80 elongated transcripts per cell (Fig. 6A and B). Addition of DRB did not markedly alter the number of initiated transcripts but did greatly reduce the number of elongated transcripts. Samples treated with TNF-α for 1 h accumulated ∼90 initiated transcripts and ∼4 elongated transcripts per cell. DRB treatment had little effect on the accumulation of initiated HIV-1 mRNA transcripts in samples treated for 1 h, whereas elongated transcript formation was reduced to levels below the limit of detection (Fig. 6A and B). These findings suggest that both early Tat-independent and late Tat-dependent TNF-α-induced transcriptional elongation of HIV-1 mRNA likely depend on active P-TEFb and that P-TEFb is recruited to the HIV-1 LTR quickly after TNF-α stimulation before Tat is synthesized, likely by NF-κB. ChIP analyses of CDK9 binding to the HIV-1 LTR in TNF-α-stimulated J-Lat cells further supported this notion (Fig. 6B). CDK9 binding to HIV LTR DNA was not detected in unstimulated cells; however, abundant enrichment was noted within 15 min of TNF-α stimulation and followed the oscillating pattern observed with RelA and RNA Pol II (Fig. 6C). CDK9 binding to the HIV LTR exhibited kinetics similar to those of RelA recruitment and certainly preceded expression of Tat (Fig. 5C). These findings are consistent with P-TEFb recruitment to the HIV LTR by RelA.
FIG. 6.
NF-κB induces promoter-proximal, but not downstream, recruitment of CDK9 and serine-2 phosphorylation of RNA Pol II. (A) Early and late TNF-α-induced HIV-1 mRNA transcript elongation is DRB sensitive. J-Lat 6.3 cells were either untreated or pretreated with DRB for 30 min and then stimulated with 20 ng/ml TNF-α for 1 or 6 h or left unstimulated. Total RNA was extracted and initiated, or elongated HIV-1 mRNA transcripts were quantitated by real-time RT-PCR (B). (C) TNF-α stimulation induces rapid recruitment of CDK9 and serine-2-phosphorylated RNA Pol II to the HIV-1 LTR. Conversely, recruitment to downstream DNA is delayed. Fixed chromatin extracts prepared from J-Lat 6.3 cells stimulated with 20 ng/ml TNF-α continuously for various times were immunoprecipitated with antibodies specific for serine-2-phosphorylated RNA Pol II and assessed for enrichment in HIV-1 LTR DNA (left) or HIV+5000 DNA (right) by UV visualization of PCR products in a gel stained with ethidium bromide. Data are representative of three independent experiments. Note the presence of serine-2-phosphorylated RNA Pol II on the HIV-1 LTR in samples stimulated for 15 min; this association was markedly decreased when HIV+5000 DNA was analyzed. Quantitation of enrichment (fold increase) above the no-antibody control is indicated below each sample. (D) RNA Pol II is decreasingly associated with downstream regions of HIV-1 DNA in early, but not late, TNF-α-induced transcription of latent HIV-1. Samples stimulated with TNF-α for 30 min and immunoprecipitated with RNA Pol II or phospho-S2-RNA Pol II antibodies were subjected to real-time PCR quantitative analysis for the indicated HIV-1 DNA regions (E).
TNF-α induces rapid association of serine-2 phosphorylation of RNA polymerase II with the HIV-1 LTR but not distal HIV-1 DNA.
The observed early recruitment of P-TEFb to the HIV LTR was somewhat surprising in light of the restricted elongation of HIV-1 mRNA observed at early time points. P-TEFb phosphorylates S2 residues within the CTD of RNA Pol II, a modification that promotes transcriptional elongation. We hypothesized that P-TEFb directed to the HIV-1 LTR might have reduced access to RNA Pol II relative to P-TEFb recruited by Tat and would consequently serve as a “weaker” kinase. S2 phosphorylation of the CTD of RNA Pol II bound at proximal or distal sites on the HIV-1 proviral DNA in TNF-α-treated J-Lat 6.3 cells was assessed by ChIP. However, binding of phospho-S2 RNA Pol II to proximal sites in the HIV-1 LTR DNA was readily detected at 15 and 30 min after TNF-α stimulation (Fig. 6B, left panel) and was followed by a decline at 1 h that paralleled the decline in RelA binding. Binding increased at 2, 4, and 6 h in cells continuously stimulated with TNF-α, returning to levels observed in 15- and 30-min samples. The similarity in RNA Pol II phosphorylation at early and late time points suggests that the kinase complex driving phosphorylation of RNA Pol II on the HIV-1 LTR has similar activity when directed by RelA or by Tat. Consequently, the inefficiency of elongation in early TNF-α-induced HIV-1 mRNA transcription is unlikely to be solely the consequence of insufficient P-TEFb kinase activity localized to the LTR or its ability to modify the CTD of RNA Pol II.
To further explore the disparity in transcriptional outcomes in early and late TNF-α-induced transcriptional elongation of HIV-1 mRNA, the binding of RNA Pol II and phospho-S2 RNA Pol II to regions downstream of the HIV-1 LTR was assessed. ChIP analysis of RNA Pol II binding to coding sequences of HIV-1 DNA 5,000 bp downstream (HIV+5000 DNA) of the transcription start site revealed detectable binding of the polymerase 15 and 30 min after TNF-α induction. This binding was followed by a characteristic decrement at 1 h (like RelA) and a second wave of binding at 2, 4, and 6 h (Fig. 6C, right panel). In contrast, binding of phospho-S2-RNA Pol II to HIV+5000 DNA was markedly limited for the first 2 h of TNF-α induction but became readily apparent at later time points where effective Tat production occurred (Fig. 5B). The combined observations of efficient phosphorylation of RNA Pol II at the proximal HIV-1 LTR and the absence of phosphorylation at downstream coding regions following early TNF-α stimulation suggest that RNA Pol II is progressively dephosphorylated during elongation. Further, the restoration of downstream RNA polymerase II phosphorylation during late TNF-α-induced transcription suggests that this dephosphorylation is somehow overcome in the presence of Tat.
To further test this possibility and to exclude potential target site-specific effects, a panel of HIV-1 coding regions composed of +1,000, +3,000, +5,000, and +7,000 bp downstream of the HIV-1 transcriptional initiation site were examined for binding by RNA Pol II and phospho-S2-RNA Pol II. Samples stimulated with TNF-α for 30 min revealed a progressive reduction of RNA Pol II and phospho-S2-RNA Pol II at successively distal coding regions (Fig. 6D and E). Notably, phospho-S2-RNA Pol II binding appeared to be reduced more rapidly within early downstream regions than general RNA Pol II binding, suggesting that RNA Pol II dephosphorylation might precede stalling and dissociation of the holoenzyme. In contrast, samples treated with TNF-α for 6 h (conditions allowing for effective production of Tat) revealed strong association of both general RNA Pol II as well as phospho-S2-RNA Pol II along all regions of the HIV-1 DNA assessed. Thus, the progressive dephosphorylation of the CTD of RNA Pol II at downstream regions observed during early TNF-α stimulation may be responsible for the observed defect in Pol II elongation, and that this defect can be rescued by Tat.
CDK9 associates with downstream coding regions of actively transcribed DNA, suggesting that CDK9 can transit with the elongating RNA Pol II complex. To assess whether CDK9 is associated with downstream HIV-1 transcription complexes, ChIP analysis of CDK9 binding to HIV+5000 DNA in J-Lat 6.3 cells stimulated with TNF-α for various times was performed. The kinetics of CDK9 association with HIV+5000 DNA (Fig. 6C) was quite similar to the patterns observed with phospho-S2-RNA Pol II. Specifically, no appreciable downstream binding was detected until 4 h after TNF-α addition. These findings indicate that the lack of downstream CDK9 binding coupled with the action of an “unopposed” phosphatase could underlie the lack of effective RNA Pol II elongation occurring during the early phases of TNF-α stimulation.
HIV-1 transcriptional elongation is restricted by an OA-sensitive phosphatase.
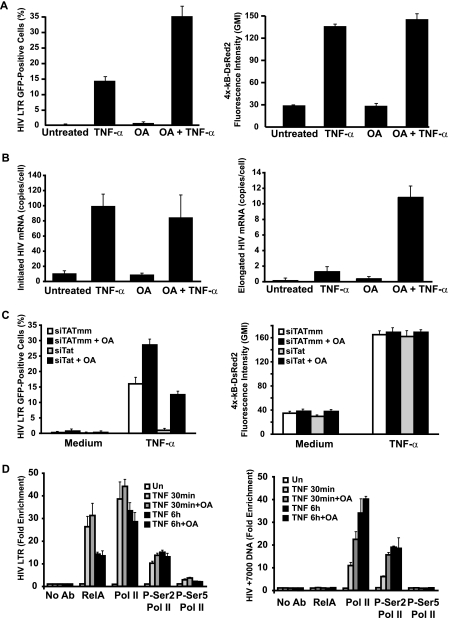
CDK9 and phospho-S2-RNA Pol II were absent from downstream regions of HIV-1 DNA during early TNF-α-induced transcription, despite their effective binding to proximal regions within the HIV-1 LTR. This observation raised the possibility that a cellular phosphatase dephosphorylates the elongating RNA Pol II complex, provided that Tat has not been synthesized. We hypothesized that inhibition of this phosphatase might promote effective elongation during early TNF-α-stimulated transcription. OA is an effective inhibitor of RNA Pol II dephosphorylation by protein phosphatase 1 (36). We tested the effect of OA on TNF-α-induced expression of latent HIV-1 in J6.3κRed cells. OA synergized with TNF-α, inducing HIV-GFP expression in ∼35% of cells, compared to ∼15% and <1% in samples treated with TNF-α or OA alone, respectively (Fig. 7A, left panel). In contrast, no significant difference in κB-DsRed2 expression was detected between samples treated with a combination of TNF-α and OA or with TNF-α alone (Fig. 7A, right panel).
FIG. 7.
OA rescues NF-κB induction of efficient expression of latent HIV. (A) OA synergizes with TNF-α to promote expression of latent HIV. J6.3κRed cells were incubated with or without 30 nM OA for 1 h, and stimulated with 20 ng/ml TNF-α or left unstimulated, and HIV-GFP (left panel) and kB-DsRed2 expression (right) was quantitated 18 h later by flow cytometry. (B) OA promotes early TNF-α-induced transcriptional elongation. Total RNA was extracted from cells treated as for panel A, and initiated (left panel) and elongated (right panel) HIV-1 transcripts were quantitated by real-time RT-PCR. Note that OA does not affect initiated transcript abundance but effectively promotes elongated HIV-1 mRNA abundance. (C) OA promotes TNF-α-induced expression of latent HIV-1 in the absence of Tat. J6.3κRed cells were nucleofected with siRNA directed against Tat mRNA or a mismatch sequence, treated with 30 nM OA or left untreated, and stimulated with TNF-α or left unstimulated, and HIV-GFP (left) or κB-DsRed2 (right) expression was assessed by flow cytometry. Note the rescue of HIV-GFP expression in Tat siRNA-treated cells with OA and the absence of effect on κB-DsRed2 expression. (D) OA promotes downstream association of P-S-RNA Pol II in early transcription. J6.3κRed cells were pretreated for 1 h with 30 nM OA or left untreated and stimulated with TNF-α for 30 min, 1 h, or left untreated, and ChIP assessment for RelA, RNA Pol II, P-S2-Pol II, and P-S5-Pol II was conducted at the HIV-1 LTR and HIV+7000 DNA by quantitative PCR. Note the increase in Pol II and P-S2-Pol II enrichment in HIV+7000 DNA in samples treated for 30 min with TNF with OA versus without OA.
To further confirm these effects of OA on TNF-α-induced expression of latent HIV, initiated and elongated HIV-1 mRNAs were quantitated from J-Lat cells treated with OA and/or TNF-α for 30 min. OA did not significantly enhance the abundance of TNF-α-induced initiated HIV-1 mRNA (Fig. 7B, left panel), suggesting that the inhibitor was not acting by enhancing transcriptional initiation. In contrast, cells treated with OA and TNF-α expressed eightfold more elongated HIV-1 mRNA than cells treated with TNF-α alone (Fig. 7B, right panel). These data suggest that OA enhances latent HIV-1 gene expression by promoting transcriptional elongation.
The augmentation of early TNF-α-induced HIV-1 mRNA elongation by OA suggested that this inhibitor might promote effective expression of latent HIV-1 in the absence of Tat. To assess this possibility, J6.3κRed cells were nucleofected with siRNA targeting Tat mRNA or a mismatched sequence and cells were incubated with or without OA, stimulated with TNF-α or left unstimulated, and assessed for HIV-GFP expression by flow cytometry 18 h later. Expression of HIV-GFP was strongly blunted in siTat-treated cells stimulated with TNF-α alone (Fig. 7C, left panel). In contrast, HIV-GFP expression was enhanced in siTat-treated samples incubated with both OA and TNF-α. Of note, OA induced more expression of HIV-GFP in siTatMM samples than siTat samples. Similar results were obtained in J15.2κRed and J8.4κRed cells. Importantly, OA did not enhance general κB-DsRed2 expression, nor did it promote expression of Tat in siTat-treated cells (Fig. 7C, right panel, and data not shown).
To assess the mechanistic action of OA, the binding of RelA, RNA Pol II, phospho-S2-RNA Pol II, and phospho-S5-RNA Pol II (a marker of early transcriptional events [24]) to HIV-1 LTR and downstream DNA was assessed in OA-treated and untreated cells stimulated with TNF-α for 30 min or 6 h. OA did not significantly affect binding of RelA, Pol II, or either of its phosphorylated forms to the HIV-1 LTR at either time point (Fig. 7D, left panel). In contrast, binding of Pol II and phospho-S2-Pol II to HIV+7000 DNA was enhanced by OA treatment after 30 min of TNF-α stimulation (Fig. 7D, right panel). No enhancement was observed in samples treated with TNF-α for 6 h, indicating this effect was specific to early transcriptional events. Induction of S5 phosphorylation of the CTD was also observed at the LTR, in agreement with a prior report (23). These findings suggest that an OA-sensitive phosphatase progressively dephosphorylates the CTD of RNA Pol II in cells treated for short periods with TNF-α. The absence of effective recruitment of CDK9 to this elongating RNA Pol II complex creates a situation where the action of this phosphatase is unopposed.
DISCUSSION
To further delineate the role of NF-κB in the activation of integrated but transcriptionally latent HIV-1 proviruses, we performed kinetic analyses of RelA and the RNA polymerase II interaction with the LTR in the J-Lat cellular model of HIV-1 latency. We observed a dynamic bimodal pattern of RNA Pol II recruitment to the LTR that paralleled and was likely driven by the inherent oscillatory nature of nuclear NF-κB. Two recent studies have similarly demonstrated that sustained induction of the NF-κB signaling pathway produces similar oscillations of RelA and RNA Pol II at host gene promoters (7, 38), likely through the induction of NF-κB-dependent expression of IκBα and A20, both potent inhibitors of NF-κB. The pattern of RelA and RNA Pol II recruitment to the HIV-1 LTR is coincident with the pattern of overall nuclear enrichment of RelA in response to a TNF-α stimulus, implying that LTR binding reflects general NF-κB abundance. Indeed, when NF-κB signaling was limited to a single round by administration of TNF-α for a brief period of time, the bimodal pattern of RelA and RNA polymerase II recruitment to the latent HIV-1 LTR was lost.
Transient induction of NF-κB is sufficient to induce the synthesis of many κB-responsive genes, most notably IκBα (35). Consequently, it was unexpected that expression of latent HIV-1 required sustained rounds to NF-κB induction for efficient activation of the virus. To confirm that general κB-responsive gene expression is achieved with transient induction of NF-κB, we constructed new Jurkat- and J-Lat-based cell lines containing integrated κB-DsRed2 reporter plasmids. Analysis of these cell lines showed that transient induction of NF-κB is sufficient for generalized κB-dependent gene expression, but not for expression of latent HIV. These results prompted us to analyze the mechanistic basis for the lack of responsiveness by the HIV-1 LTR.
Analysis of HIV-1 mRNA transcript formation immediately after induction of NF-κB revealed that, while initiated transcripts readily accumulate, transcriptional elongation is impaired. Consistent with this observation, early studies of HIV-1 transcription described NF-κB as a strong inducer of transcriptional initiation (28). However, more recent studies of NF-κB in HIV-1 transcription have suggested a role in RNA Pol II elongation as well as initiation (39). Low-level accumulation of elongated HIV-1 mRNA transcripts is apparent immediately after TNF-α stimulation; however, the rate of accumulation is greatly reduced relative to the rate of initiated transcript synthesis. During sustained stimulation with TNF-α, the rate of elongated transcript formation begins to approximate the rate of initiated transcript formation only at 4 to 6 h after induction. Further, this delayed increase in RNA Pol II elongation is dependent on de novo protein synthesis. These results raised the possibility that the late increase in elongation reflects synthesis of Tat, which through its recruitment of P-TEFb is known to greatly increase elongation. We hypothesized that the initial round of NF-κB recruitment to the HIV-1 LTR, while inefficiently inducing elongation, is sufficient to produce small quantities of Tat. This newly synthesized Tat then synergizes with subsequent rounds of transcriptional initiation induced by NF-κB to promote highly efficient RNA Pol II elongation. Additionally, variations in Tat synthesis may underlie the fractional response to TNF-α observed in J-Lat cells (37). This hypothesis is supported by the finding that ectopic expression of Tat drives robust expression of latent HIV-1 in response to transient induction of NF-κB, suggesting that the absence of this transactivator is key to the initial ineffectiveness of RNA Pol II elongation. These conclusions also agree with a recent computational model of the effects of Tat on HIV-1 gene expression (37).
The ability of NF-κB to induce transcriptional elongation has been ascribed to its ability to recruit P-TEFb to sites of transcription, the same RNA Pol II kinase partner employed by Tat to induce elongation (2). Our studies with the P-TEFb inhibitor DRB suggest that early TNF-α driven, NF-κB-dependent transcriptional elongation involves this kinase complex. ChIP analyses of CDK9, the kinase component of P-TEFb, revealed a pattern of recruitment to the HIV-1 LTR coincident with RelA. Assessment of functional phosphorylation of the CTD of RNA Pol II in phospho-S2 RNA Pol II ChIP analyses confirmed that the polymerase complex is similarly phosphorylated in both early and late TNF-α-stimulated samples at proximal sites on the HIV-1 LTR. In contrast, we observed that the pattern of S2 RNA Pol II phosphorylation at more distal regions of HIV-1 DNA was greatly reduced during NF-κB-dependent versus Tat-dependent Pol II elongation. Similarly, we observed that CDK9 is associated with downstream HIV-1 DNA in late, but not early, NF-κB-driven transcription. These findings raise the possibility that CDK9 transits with the RNA Pol II complex in the context of Tat and reinforces S2 phosphorylation during elongation (Fig. 8). This is in agreement with a recent study demonstrating Tat-induced rephosphorylation of RNA Pol II in vitro (5), as well as several other studies demonstrating CDK9 association with coding regions of actively transcribed host genes (15, 16, 20).
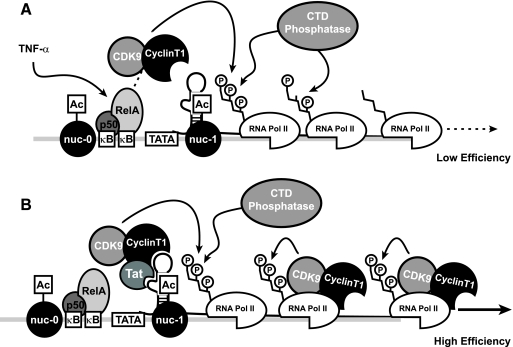
FIG. 8.
Model for events influencing early and late TNF-α-induced HIV-1 transcription. (A) TNF-α stimulation of J-Lat cells induces NF-κB-mediated recruitment of PTEF-b to the HIV LTR, which drives serine-2 phosphorylation of the CTD of coincidentally recruited RNA Pol II. This phosphorylated polymerase is progressively dephosphorylated during elongation, limiting processivity. (B) Early, inefficient elongation produces low levels of HIV-1 Tat, which recruits CDK9 to the HIV-1 LTR in a context capable of transiting with the elongating polymerase. This continued association permits reinforcement of serine-2 phosphorylation of the CTD and prevents transcriptional stalling due to the action of CTD phosphatases.
A recent study of HIV-1 latency reported by Kim et al. (23), published during the preparation of the manuscript, similarly demonstrated a minimally processive RNA Pol II complex in TNF-α-stimulated infected Jurkat T cells in the absence of Tat. Also consistent with our own work, this study demonstrates increased binding of CDK9 to coding regions of HIV-1 DNA in the presence of Tat. However, Kim et al. observed significant RNA Pol II binding to the unstimulated latent HIV-1 LTR, while our ChIP studies consistently do not detect such binding. Additionally, whereas we observe TNF-α-induced binding of CDK9 to the HIV-1 LTR at early time points when Tat is not present, Kim et al. did not observe appreciable binding of CDK9 to the TNF-α-induced HIV-1 LTR in the absence of Tat. These differences may be a function of the experimental systems employed. Of note, the model of latency employed by Kim et al. includes a significant level of active HIV-1 expression in the unstimulated state (10% GFP-positive cells). In contrast, very little basal expression occurs in the J-Lat cell model, as indicated by <1% GFP-positive cells. The relatively high level of GFP under basal conditions reported by Kim et al. could account for the presence of bound RNA Pol II in the absence of stimulation. Finally, differences in the experimental protocol for ChIP assays (absence of micrococcal nuclease in the fragmentation step) might contribute to the differences in the detection of CDK9 binding.
We suspect that NF-κB-dependent RNA Pol II elongation is inefficient because of the absence of CDK9 coupled with the unopposed action of one or more phosphatases that progressively dephosphorylate the CTD of RNA Pol II at downstream DNA regions. These changes do not occur with Tat. Whether Tat enhancement of RNA Pol II elongation is solely due to effective recruitment of the CDK9 kinase or also involves active inhibition of the key phosphatases attacking the CTD is currently unclear. We further found a significant restoration of TNF-α-induced expression of latent HIV-1 in the presence of OA, an inhibitor of various phosphatases. These findings suggest that a component of the restricting CTD phosphatase is sensitive to OA. These findings support a model in which NF-κB-dependent RNA Pol II elongation is greatly undermined by progressive dephosphorylation of the CTD. Preliminary siRNA knockdown studies suggest that FCP1, a well-characterized CTD phosphatase (6, 26), is not a key component of this process. Future studies directed to identifying the phosphatases limiting the processivity of RNA Pol II will be of interest.
It is not yet clear whether latency as it exists in vivo is a consequence of low-level viral gene expression or complete suppression. We have chosen to evaluate cells displaying a more complete suppression, as this highly restrictive environment would seem to prove most difficult to activate and purge. These studies were conducted using the J-Lat cellular model of HIV-1 latency, a model that though fulfilling many criteria for HIV-1 latency bears an uncertain relationship to HIV-1 latency occurring in infected patients. Analyses of NF-κB oscillation within quiescent primary T cells activated by various inducers have largely recapitulated the findings observed in the J-Lat system (7, 38). Additionally, latent proviruses contained in J-Lat cells are preferentially integrated into actively transcribed genes, mirroring findings of infected resting CD4+ T cells in HIV-infected patients with effective suppression of viremia (17, 25). Thus, we strongly suspect the biochemical changes we have detected also occur in latently infected cells in vivo.
The finding that sustained induction of NF-κB is required for κB-driven activation of HIV-1 latency has important implications for therapeutic efforts to eliminate this viral reservoir. Potential agonists of latent HIV-1 should be designed that take into account the importance of sustained induction of NF-κB. Alternatively, these findings support the consideration of combined application of soluble and cell-permeable forms of Tat and transient agonists of NF-κB, or perhaps the elective use of specific phosphatase inhibitors. However, given the broad assortment of host genes induced by NF-κB and the sensitivity of many gene products to phosphorylation-dependent regulation, the combination of Tat and transient induction of NF-κB might avoid the toxicity likely to be associated with sustained systemic induction of NF-κB or manipulation of phosphatase activity.
Acknowledgments
The authors thank G. Howard and S. Ordway for editorial assistance and R. Givens for assistance with the preparation of the manuscript.
This work was supported by funds from NIH grant P01 AI058708, the University of California San Francisco—Gladstone Institute of Virology and Immunology Center for AIDS Research (UCSF-GIVI CFAR) (P30 AI27763), and an NIH core equipment grant awarded to the J. David Gladstone Institutes (RR1 892801).
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 3.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 4.Bohnlein, E., J. W. Lowenthal, M. Siekevitz, D. W. Ballard, B. R. Franza, and W. C. Greene. 1988. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell 53:827-836. [DOI] [PubMed] [Google Scholar]
- 5.Bres, V., N. Gomes, L. Pickle, and K. A. Jones. 2005. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 19:1211-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, H., T. K. Kim, H. Mancebo, W. S. Lane, O. Flores, and D. Reinberg. 1999. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13:1540-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covert, M. W., T. H. Leung, J. E. Gaston, and D. Baltimore. 2005. Achieving stability of lipopolysaccharide-induced NF-κB activation. Science 309:1854-1857. [DOI] [PubMed] [Google Scholar]
- 8.Davis, N., S. Ghosh, D. L. Simmons, P. Tempst, H. C. Liou, D. Baltimore, and H. R. Bose, Jr. 1991. Rel-associated pp40: an inhibitor of the Rel family of transcription factors. Science 253:1268-1271. [DOI] [PubMed] [Google Scholar]
- 9.DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature 388:548-554. [DOI] [PubMed] [Google Scholar]
- 10.Emiliani, S., W. Fischle, M. Ott, C. Van Lint, C. A. Amella, and E. Verdin. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emiliani, S., C. Van Lint, W. Fischle, P. Paras, Jr., M. Ott, J. Brady, and E. Verdin. 1996. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. USA 93:6377-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenard, D., W. Yonemoto, C. de Noronha, M. Cavrois, S. A. Williams, and W. C. Greene. 2005. Nef is physically recruited into the immunological synapse and potentiates T cell activation early after TCR engagement. J. Immunol. 175:6050-6057. [DOI] [PubMed] [Google Scholar]
- 13.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 14.Ganchi, P. A., S. C. Sun, W. C. Greene, and D. W. Ballard. 1992. I kappa B/MAD-3 masks the nuclear localization signal of NF-kappa B p65 and requires the transactivation domain to inhibit NF-kappa B p65 DNA binding. Mol. Biol. Cell 3:1339-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud, S., A. Hurlstone, S. Avril, and O. Coqueret. 2004. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene 23:7391-7398. [DOI] [PubMed] [Google Scholar]
- 16.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, X. Liu, T. C. Pierson, J. B. Margolick, R. F. Siliciano, and J. D. Siliciano. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henkel, T., U. Zabel, K. van Zee, J. M. Muller, E. Fanning, and P. A. Baeuerle. 1992. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-kappa B subunit. Cell 68:1121-1133. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y. K., C. F. Bourgeois, R. Pearson, M. Tyagi, M. J. West, J. Wong, S. Y. Wu, C. M. Chiang, and J. Karn. 2006. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 25:3596-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewinski, M. K., D. Bisgrove, P. Shinn, H. Chen, C. Hoffmann, S. Hannenhalli, E. Verdin, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandal, S. S., H. Cho, S. Kim, K. Cabane, and D. Reinberg. 2002. FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol. Cell. Biol. 22:7543-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercurio, F., H. Zhu, B. W. Murray, A. Shevchenko, B. L. Bennett, J. Li, D. B. Young, M. Barbosa, M. Mann, A. Manning, and A. Rao. 1997. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science 278:860-866. [DOI] [PubMed] [Google Scholar]
- 28.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711-713. [DOI] [PubMed] [Google Scholar]
- 29.Nelson, D. E., A. E. Ihekwaba, M. Elliott, J. R. Johnson, C. A. Gibney, B. E. Foreman, G. Nelson, V. See, C. A. Horton, D. G. Spiller, S. W. Edwards, H. P. McDowell, J. F. Unitt, E. Sullivan, R. Grimley, N. Benson, D. Broomhead, D. B. Kell, and M. R. White. 2004. Oscillations in NF-κB signaling control the dynamics of gene expression. Science 306:704-708. [DOI] [PubMed] [Google Scholar]
- 30.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 31.Perkins, N. D., N. L. Edwards, C. S. Duckett, A. B. Agranoff, R. M. Schmid, and G. J. Nabel. 1993. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J. 12:3551-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomerantz, R. J. 2003. Reservoirs, sanctuaries, and residual disease: the hiding spots of HIV-1. HIV Clin. Trials 4:137-143. [DOI] [PubMed] [Google Scholar]
- 33.Rohr, O., C. Marban, D. Aunis, and E. Schaeffer. 2003. Regulation of HIV-1 gene transcription: from lymphocytes to microglial cells. J. Leukoc. Biol. 74:736-749. [DOI] [PubMed] [Google Scholar]
- 34.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727-728. [DOI] [PubMed] [Google Scholar]
- 35.Sun, S. C., P. A. Ganchi, D. W. Ballard, and W. C. Greene. 1993. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science 259:1912-1915. [DOI] [PubMed] [Google Scholar]
- 36.Washington, K., T. Ammosova, M. Beullens, M. Jerebtsova, A. Kumar, M. Bollen, and S. Nekhai. 2002. Protein phosphatase-1 dephosphorylates the C-terminal domain of RNA polymerase-II. J. Biol. Chem. 277:40442-40448. [DOI] [PubMed] [Google Scholar]
- 37.Weinberger, L. S., J. C. Burnett, J. E. Toettcher, A. P. Arkin, and D. V. Schaffer. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169-182. [DOI] [PubMed] [Google Scholar]
- 38.Werner, S. L., D. Barken, and A. Hoffmann. 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309:1857-1861. [DOI] [PubMed] [Google Scholar]
- 39.West, M. J., A. D. Lowe, and J. Karn. 2001. Activation of human immunodeficiency virus transcription in T cells revisited: NF-κB p65 stimulates transcriptional elongation. J. Virol. 75:8524-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, S. A., L. F. Chen, H. Kwon, D. Fenard, D. Bisgrove, E. Verdin, and W. C. Greene. 2004. Prostratin antagonizes HIV latency by activating NF-κB. J. Biol. Chem. 279:42008-42017. [DOI] [PubMed] [Google Scholar]
- 41.Williams, S. A., L. F. Chen, H. Kwon, C. M. Ruiz-Jarabo, E. Verdin, and W. C. Greene. 2006. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zabel, U., T. Henkel, M. S. Silva, and P. A. Baeuerle. 1993. Nuclear uptake control of NF-kappa B by MAD-3, an I kappa B protein present in the nucleus. EMBO J. 12:201-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zandi, E., D. M. Rothwarf, M. Delhase, M. Hayakawa, and M. Karin. 1997. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell 91:243-252. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]