Abstract
Human papillomavirus (HPV) infections of the squamous epithelium are associated with high-level expression of the E1^E4 protein during the productive phase of infection. However, the precise mechanisms of how E1^E4 contributes to the replication cycle of the virus are poorly understood. Here, we show that the serine-arginine (SR)-specific protein kinase SRPK1 is a novel binding partner of HPV type 1 (HPV1) E1^E4. We map critical residues within an arginine-rich domain of HPV1 E1^E4, and in a region known to facilitate E1^E4 oligomerization, that are requisite for SRPK1 binding. In vitro kinase assays show that SRPK1 binding is associated with phosphorylation of an HPV1 E1^E4 polypeptide and modulates autophosphorylation of the kinase. We show that SRPK1 is sequestered into E4 inclusion bodies in terminally differentiated cells within HPV1 warts and that colocalization between E1^E4 and SRPK1 is not dependent on additional HPV1 factors. Moreover, we also identify SRPK1 binding of E1^E4 proteins of HPV16 and HPV18. Our findings indicate that SRPK1 binding is a conserved function of E1^E4 proteins of diverse virus types. SRPK1 influences important biochemical processes within the cell, including nuclear organization and RNA metabolism. While phosphorylation of HPV1 E4 by SRPK1 may directly influence HPV1 E4 function during the infectious cycle, the modulation and sequestration of SRPK1 by E1^E4 may affect the ability of SRPK1 to phosphorylate its cellular targets, thereby facilitating the productive phase of the HPV replication cycle.
Human papillomaviruses (HPVs) comprise a family of small double-stranded DNA viruses that have a tropism for human epithelial cells at skin and mucosal surfaces. To date, based on genome sequence, over 100 HPV types have been described, and these virus types vary in both their anatomical tropism and their potential to induce malignant transformation (10). The HPV replication cycle is intimately linked with the differentiation of the host epithelium (21). To initiate an infection, the virus requires access, typically via a cut or abrasion, to basal cells where the virus maintains itself at a low copy number. Subsequently, infected cells undergo differentiation and migrate to the epithelial surface, which initiates the productive phase of virus infection, signified by genome amplification, the synthesis of structural proteins, and assembly of infectious progeny. These events are orchestrated by both transcriptional and posttranscriptional mechanisms and consequently ensure that structural proteins are only synthesized in terminally differentiated cells (61).
High-level expression of the E4 protein accompanies the productive phase of the HPV life cycle (40). E4 is translated from spliced E1^E4 transcripts and encodes the first 5 amino acids from the N terminus of the E1 protein fused to the E4 coding sequence. The E4 open reading frame (ORF) is the most divergent ORF within the HPV family. While there is sequence homology between E1^E4 proteins, this is generally restricted to virus types with similar pathology, and it is predominantly limited to sequences at the amino and carboxy termini of the proteins (43, 45). In natural infections, E4 is expressed as a phosphoprotein (2, 4, 20) that assembles into oligomeric complexes (2, 12) and is further modified by removal of residues from the amino terminus (15, 43).
The role played by E4 in the virus life cycle is uncertain. Loss of full-length E1^E4 expression in experimental systems that recapitulate the infectious cycle of HPV type 16 (HPV16), HPV18, HPV31, and cottontail rabbit papillomavirus correlates with a defective life cycle (35, 39, 57, 57a). Collectively, these studies determined that full-length E1^E4 is necessary for efficient viral genome amplification and up-regulation of late viral gene expression, which are features of the productive phase of the virus life cycle. In one system, based upon HPV16, replication of the viral genome in basal-like cells was also compromised in the absence of full-length E1^E4 (35). Thus, these studies implicate E1^E4 as an important regulator of both early and late stages of the virus life cycle. However, E1^E4 function may not be necessary for productive replication of all papillomavirus types, since impairment of E1^E4 synthesis from HPV11 genomes did not limit genome amplification (17).
Several biological functions have been ascribed to E1^E4, and these observations include the association with keratin intermediate filaments, and in some cases, perturbation of these networks (14, 44); the reorganization of promyelocytic leukemia protein from intranuclear speckles (ND10 bodies) (46); the association with mitochondria (42); and the disruption of normal cell division (9, 28, 34). As the precise mechanisms of how E1^E4 proteins exert these diverse range of effects are not yet fully understood (8, 29, 56), we adopted a proteomics-based approach in an effort to identify HPV E1^E4-associated proteins. Here we show that the serine-arginine (SR)-specific kinase SRPK1 is a novel binding partner of E1^E4 proteins from highly divergent HPV1, -16, and -18. SRPK1 belongs to a family of serine protein kinases that phosphorylate arginine-serine (RS)-rich domains within a subgroup of the small non-RNP particles termed SR proteins. SR proteins have multiple and diverse roles in mRNA metabolism (18), including the control of pre-mRNA splicing through splice site selection, the regulation of the export of spliced mRNA from the nucleus, the stabilization of cytoplasmic transcripts, and promotion of mRNA translation by ribosomes (18, 24, 25, 50). Phosphorylation within the RS domain differentially affects SR protein activity, probably as a result of modulating protein-protein interactions and/or protein-mRNA association (32, 49, 58). The release of SR proteins from distinct storage sites within the nucleus and the movement of SR proteins that shuttle between the nucleus and cytoplasm is under the control of SRPK1 activity (5, 30). SRPK1 also phosphorylates the lamin B receptor (LBR), an integral membrane protein of the nuclear lamina (38). SRPK1 phosphorylation promotes LBR binding to chromatin to facilitate nuclear envelope organization during the cell cycle (54). SRPK1 activity is therefore linked to posttranscriptional mechanisms of control of gene expression and nuclear organization. We provide evidence that SRPK1 binding leads to phosphorylation of an HPV1 E4 species and indicate that this association modulates the ability of SRPK1 to undergo autophosphorylation in vitro. Furthermore, we show that SRPK1 is sequestered to E4-containing structures in HPV1-infected keratinocytes.
MATERIALS AND METHODS
Cell culture and transfections.
293T cells were grown in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with glutamine and 10% fetal calf serum (Invitrogen). The simian virus 40 large T-immortalized keratinocyte cell line SV-JD (44), was grown in Joklick's media (Invitrogen) supplemented with glutamine with 10% fetal calf serum.
For transfection experiments, 293T and SV-JD cells were grown so as to be 80% confluent at the time of transfection. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen) by following the manufacturer's instructions. Following overnight incubation, the cells were subsequently grown in relevant complete media.
Bacterial expression of recombinant proteins.
The HPV1, -16, and -18 E1^E4 cDNAs (44) were separately cloned into pGEX-3X (Amersham, Little Chalfont, United Kingdom) so as to facilitate the expression of glutathione S-transferase (GST) E1^E4 fusion proteins. The HPV1 E1^E4 deletion mutants Δ44-48 and Δ49-53 were amplified with the primer pair 5′-GCGCGAATTCTTACACAGACCACGGGTGGATC-3′ and 5′-GCGCGGATCCGCAGATAATAAAGCTCCCCAAG-3′ combined with pcDNA templates that contained previously described deletions (43). The amplified sequence was cloned into appropriately prepared pGEX-2T. Escherichia coli strain BL21 (Stratagene, La Jolla, CA) containing the different pGEX plasmids was grown in LB media containing 50 μg/ml ampicillin and 2% glucose at 37°C. Overnight cultures of 10 ml were diluted into 200 ml of LB media and incubated with shaking for 1 h. The cultures were transferred to a 25°C incubator and induced for 2 h by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were pelleted at 4°C and resuspended in 4 ml of bacterial lysis (BL) buffer: phosphate-buffered saline, 1% Triton X-100 (vol/vol), and Complete protease inhibitors (Roche Applied Science, Indianapolis, IN). The bacteria were sonicated using a Microson ultrasonic cell disrupter (Misonix, Inc., NY) for three periods of about 30 s, and the lysate was then clarified by high-speed centrifugation at 16,100 g. The soluble recombinant GST-E1^E4 protein was immobilized on glutathione S-agarose (Sigma-Aldrich, St. Louis, MO) prior to washing in BL buffer. Soluble GST proteins were prepared by subsequently eluting the GST protein into BL buffer containing 50 mM reduced glutathione. Soluble proteins were then dialysed overnight in 25 mM Tris, pH 7.5, 200 mM NaCl, 10 mM MgCl2, and Complete protease inhibitors.
Polyhistidine-tagged SRPK1 was expressed in E. coli strain BL21 containing the plasmid pRSET B-SRPK1 (59) (a kind gift of Bai-Gong Yue). E. coli was manipulated as described above; however, the IPTG induction was at 37°C and extended to 4 h. Subsequently E. coli was lysed using the method described above, and the cleared lysate was then purified on Ni-nitrilotriacetic acid-agarose (QIAGEN, Inc., CA) by mixing at 4°C for 1 h. The beads were washed six times with BL buffer containing 20 mM imidazole (Sigma). His-SRPK1 was eluted off the Ni-nitrilotriacetic acid-agarose by resuspending the beads in BL buffer containing 250 mM imidazole, and the soluble protein was dialyzed overnight in the dialysis buffer described above.
Generation of N-Flag SRPK1.
The Expand (Roche) high-fidelity, proofreading polymerase mix was employed in a PCR to amplify the SRPK1 ORF in a reaction containing IMAGE clone no. 4824261 DNA (MRC Geneservice, Cambridge, United Kingdom) together with the primer pair 5′-GCGCGAATTCCATGGAGCGGAAAGTGCTTGCGCTC-3′ and 5′-GCGCGGATCCTTAGGAGTTAAGCCAAGGGTGCCG-3′. The purified PCR product was subsequently digested with EcoRI and BamHI and ligated into appropriately prepared pCMV-Flag4 (Sigma-Aldrich) to form pCMVFlag4-SRPK1 that encodes an N-terminal Flag tag fused to SRPK1 (Flag-SRPK1).
Coprecipitations.
In proteomic experiments, coprecipitations were performed using 108 SV-JD cells per sample. Cells were lysed in 1 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, and Complete protease inhibitors (NP-40 lysis buffer), and the lysate was cleared by centrifugation at 16,100 × g. Coprecipitations were performed by mixing the cleared SV-JD lysate with GST fusion proteins immobilized to glutathione S-agarose at 4°C for 2 h and were subsequently washed six times with NP-40 lysis buffer. The samples were finally resuspended in 2× Laemmli loading buffer, boiled for 5 min, and then separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on 5 to 15% gradient acrylamide gels. Silver staining was performed using the Silver Stain plus kit (Bio-Rad, Hercules, CA), and after development, specific bands were excised and digested with trypsin (Roche) using the protocol described by Shevchenko et al. (52). Peptides were eluted in 50% acetonitrile-5% formic acid, dried, resuspended, and analyzed on a LCQ DECA XP PLUS (Thermo Electron Corp., Waltham, MA.) using liquid chromatography-tandem mass spectrometry (LC MS/MS). The data were searched using Turbo Sequest (Thermo Electron Corp.).
For coprecipitation of endogenous SRPK1, 5 × 106 SV-JD cells were lysed and coprecipitations performed as described above. Coprecipitation of Flag-tagged SRPK1 involved the preparation of cell lysate from 5 × 106 293T cells transfected with pCMVFlag4-SRPK1, and coprecipitations were performed as above. To coprecipitate His-SRPK1, a bacterial lysate containing His-SRPK1 and supplemented with 100 μg/ml RNase A (Sigma) was prepared, as described above, and mixed with GST fusion proteins immobilized to glutathione S-agarose. After 2 h of mixing, the coprecipitated complexes were washed six times in BL buffer. Samples were resolved by SDS-PAGE, transferred onto nitrocellulose, and analyzed by Western blotting.
Generation of HPV1 E1^E4 mutants.
Previously described deletion mutants of HPV1 E1^E4 (43) were cloned into expression plasmid pcDNA3.1 (Invitrogen). Point mutations within the HPV1 E1^E4 coding sequence were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). HPV1 E1^E4 template was used in conjunction with the primer pairs listed in Table 1. Bidirectional sequencing using a 3100 Genetic Analyzer (ABI Prism) was used to verify the sequences of mutated E1^E4 DNAs.
TABLE 1.
Primers used for alanine-scanning mutagenesis
| Substitution | Primer | Sequence (5′-3′) |
|---|---|---|
| G44A | G44AF | CCCAGGACAGGGCGAGGCCTCGCAGGTCC |
| G44AR | GGACCTGCGAGGCCTCGCCCTGTCCTGGG | |
| R45A | R45AF | CTCCCAGGACAGGGGGGCCCCTCGCAGGTCCGAC |
| R45AR | GTCGGACCTGCGAGGGGCCCCCCTGTCCTGGGAG | |
| P46A | P46AF | CAGGACAGGGGGAGAGCTCGCAGGTCCGACAAAG |
| P46AR | CTTTGTCGGACCTGCGAGCTCTCCCCCTGTCCTG | |
| R47A | R47AF | CAGGACAGGGGGAGGCCTGCCAGGTCCGACAAAGAC |
| R47AR | GTCTTTGTCGGACCTGGCAGGCCTCCCCCTGTCCTG | |
| R48A | R48AF | GGGGGAGACCTCGAGCGTCCGACAAAGACAGCAG |
| R48AR | CTGCTGTCTTTGTCGGACGCTCGAGGTCTCCCCC | |
| Q109A | Q109AF | GGGACATTCTTGCAGACTTAGACGACTTCTGC |
| Q109AR | GTCGTCTAAGTCTGCAAGAATGTCCCTTTTGAG | |
| D110A | D110AF | GACATTCTTCAGGCCTTAGACGACTTCTGCAGG |
| D110AR | GAAGTCGTCTAAGGCCTGAAGAATGTCCCTTTTG | |
| L111A | L111AF | ATTCTTCAAGACGCAGACGACTTCTGCAGGAAG |
| L111AR | GCAGAAGTCGTCTGCGTCTTGAAGAATGTCCCT | |
| D112A | D112AF | ATTCTTCAAGATCTAGCCGACTTCTGCAGGAAGC |
| D112AR | CCTGCAGAAGTCGGCTAGATCTTGAAGAATGTCCC | |
| D113A | D113AF | CAAGACTTAGACGCGTTCTGCAGGAAGCTTGGG |
| D113AR | CTTCCTGCAGAACGCGTCTAAGTCTTGAAGAATG | |
| F114A | F114AF | GACTTAGACGACGCTTGCAGGAAGCTTGGGATC |
| F114AR | AAGCTTCCTGCAAGCGTCGTCTAAGTCTTGAAG |
Immunoprecipitations.
293T cells cotransfected with pcDNA-HPV1 E4 plasmids and pCMVFlag4-SRPK1 or an HPV1 E1-Flag expression vector (a kind gift of Saleem Khan) were harvested 24 h after transfection and lysed on ice in NP-40 lysis buffer containing the Complete cocktail of protease inhibitors. Fifteen minutes later, the lysate was cleared by centrifugation at 16,100 × g. An aliquot of the lysate was removed for subsequent analysis, and the remainder of the lysate was mixed with 3 μg of rabbit anti-FLAG antibody (Sigma-Aldrich) and 50 μl of a 50% slurry of protein A-Sepharose (Cancer Research UK). Immunoprecipitations were incubated at 4°C for 2 h prior to being washed five times in NP-40 lysis buffer. Samples were resolved by SDS-PAGE, transferred onto nitrocellulose, and analyzed by Western blotting.
Western blotting.
Resolved proteins were transferred electrophoretically onto Bio-Trace NT nitrocellulose membranes (Pall Life Sciences, VWR, Poole, Dorset, United Kingdom) and blocked in 2% dried skim milk in phosphate-buffered saline. The mouse anti-HPV1 E1^E4 monoclonal antibody 4.37 (15) was used at a dilution of 1/250, mouse anti-Flag (Sigma-Aldrich) was used at a dilution of 1/2,500, and the goat anti-mouse immunoglobulin G (IgG) conjugated to horseradish peroxidase antibody was used at a dilution of 1/3,000 (Sigma-Aldrich). Blots were developed using chemiluminescence (ECL; Amersham Pharmacia).
Immunofluorescence microscopy.
Frozen sections (4 μm) of HPV1 warts were fixed in 4% paraformaldehyde for 8 min and permeabilized for 10 min in 0.2% Triton X-100. Costaining of sections using the anti-E4 monoclonal antibody 4.37 and an anti-SRPK1 mouse monoclonal antibody purchased from BD Biosciences was performed in a humidified chamber, overnight at 4°C. Appropriate combinations of mouse IgG subclass-specific Alexa 488 or −594 conjugates (Molecular Probes, Inc.) were applied to the sections for 1 h at room temperature. Nuclei were visualized by using 4′,6′-diamidino-2-phenylindole (DAPI) prior to mounting in ProFade (Molecular Probes, Inc.). Images were acquired using a Zeiss LSM510 laser-scanning confocal microscope.
SV-JD cells were grown on glass slides and cotransfected with plasmids that express HPV1 E1^E4 and Flag-SRPK1, using the transfection protocol described above. The cells were fixed with 4% paraformaldehyde and permeabilized in acetone (−20°C) as previously described (46). Cells were subsequently stained for Flag-SRPK1 with a rabbit anti-Flag antibody (Sigma-Aldrich) and HPV1 E1^E4 with monoclonal antibody 4.37. Immune complexes were detected using the appropriate species-specific IgG-Alexa conjugates, and nuclei were counterstained with DAPI.
In vitro kinase assays.
In vitro kinase reactions were performed following the previously described method (38). Briefly, reaction mixtures were buffered in 25 mM Tris, pH 7.5, 200 mM NaCl, 10 mM MgCl2, and 20 μM ATP and contained approximately 200 ng His-SRPK1, 10 μCi [γ-32P]ATP, and between 1 and 25 μg of substrate protein. The reaction mixtures were incubated at 30°C for 20 min, and then reactions were stopped by the addition of 2× Laemmli loading buffer and resolved by SDS-PAGE. Gels were stained using Bio-Safe (Bio-Rad) Coomassie stain, dried down, and exposed to autoradiography film. Band intensities were separately quantitated using STORM imaging on a Storm860 (GE Healthcare, Waukesha, WI) combined with ImageQuant 5.0 software (GE Healthcare).
RESULTS
HPV E1^E4 proteins interact with the SR kinase SRPK1.
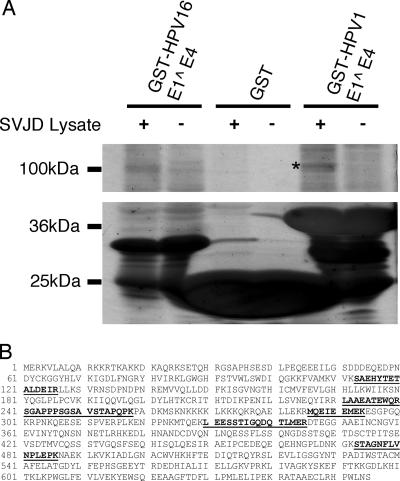
We adopted a proteomics-based approach with the aim of identifying novel HPV E1^E4-associating proteins. Thus, the E1^E4 proteins derived from the cutaneous type 1 virus (HPV1) as well as the anogenital type 16 virus (HPV16) were expressed in bacteria as N-terminally tagged GST fusion proteins and purified on glutathione S-agarose. Immobilized GST and GST-HPV E1^E4 proteins were used to coprecipitate cellular factors from lysates prepared from the simian virus 40 large T-immortalized keratinocyte cell line SV-JD, a line previously used by us to successfully study HPV E1^E4 functions (43-46). The coprecipitations were subsequently resolved by SDS-PAGE separation prior to silver stain analysis. These experiments demonstrated the presence of several bands that were derived from the SV-JD lysate and were only present in the GST-HPV E1^E4 coprecipitations. One prominent band that had an approximate molecular mass of 100 kDa was present in both the HPV1 E1^E4 and HPV16 E1^E4 coprecipitations (Fig. 1A). These bands, along with the respective region from the control lane, were excised from the gel and digested with trypsin prior to elution and analysis of peptides by LC MS/MS (22). The outcome of these analyses indicated the presence of SRPK1, an SR-specific kinase, in the GST-HPV1 E1^E4 coprecipitation, while not being detected in the GST-HPV16 E1^E4 or GST coprecipitations. The peptides that were sequenced from the mass spectrometry analysis, in addition to the full-length SRPK1 sequence, are shown in Fig. 1B.
FIG. 1.
GST-HPV1 E1^E4 coprecipitates SRPK1. (A) Silver-stained 5 to 15% SDS-polyacrylamide gel of GST coprecipitations from lysates prepared from SV-JD keratinocytes showing GST, GST-HPV E1^E4 fusion proteins (lower panel), and associated cellular factors (upper panel). +, lysate present; −, lysate absent. Mass spectrometry analysis by LC MS/MS of trypsin-digested peptides prepared from silver-stained bands identified SRPK1 peptides present in a prominent band (indicated by an asterisk) within the GST-HPV1 E1^E4 coprecipitate. (B) The full-length amino acid sequence of SRPK1 (gene identifier 47419936). Underlined in boldface type are the peptides sequenced from the GST-HPV1 E1^E4 coprecipitation.
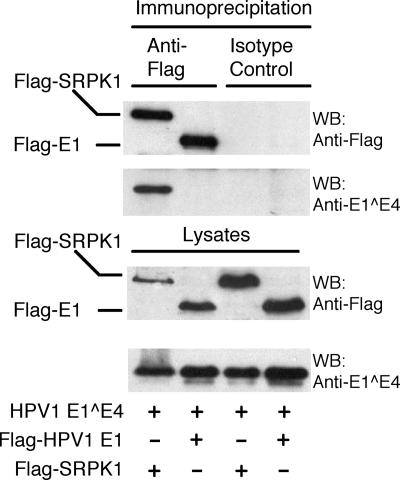
Having identified multiple peptides from SRPK1 in the GST-HPV1 E1^E4 coprecipitation, subsequent efforts were made to validate this interaction in intact cells. Initially, an N-terminally Flag-tagged SRPK1 fusion protein (Flag-SRPK1) was coexpressed in 293T cells along with HPV1 E1^E4. Immunoprecipitation of Flag-SRPK1 with an anti-Flag antibody demonstrated that HPV1 E1^E4 was complexed with Flag-SRPK1 (Fig. 2). The specificity of the coassociation of HPV1 E1^E4 with Flag-SRPK1 was underscored by coexpressing HPV1 E1-Flag with HPV1 E1^E4. Subsequent immunoprecipitation showed that although the E1-Flag protein was efficiently immunoprecipitated by an anti-Flag antibody, no E1^E4 could be detected in the E1-Flag complex (Fig. 2).
FIG. 2.
HPV1 E1^E4 associates with Flag-SRPK1 in human keratinocytes. Western analysis of anti-Flag and isotype control immunoprecipitations from lysates prepared from 293T cells coexpressing Flag-SRPK1 and HPV1 E1^E4 or Flag-HPV1 E1 and HPV1 E1^E4. HPV1 E1^E4 was present in immunoprecipitates of Flag-SRPK1 complexes but not in those containing Flag-HPV1 E1, confirming the specificity of the SRPK1-HPV1 E1^E4 association. WB, Western blot; +, present; −, absent.
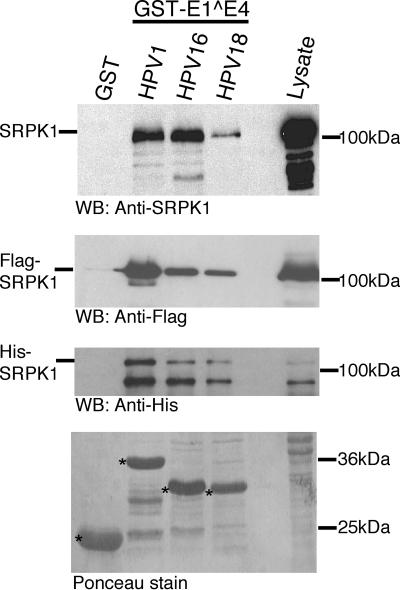
To further validate the HPV1 E1^E4 interaction with SRPK1 and to also examine the capacity of other HPV E1^E4 proteins to associate with SRPK1, a series of coprecipitation studies were initiated. GST E1^E4 fusion proteins of HPV1, -16, and -18 were used to coprecipitate endogenous SRPK1 derived from lysates prepared from SV-JD cells. In addition to GST-HPV1 E1^E4 associating with SRPK1, the GST fusions encoding E1^E4 proteins from the high-risk anogenital types HPV16 and HPV18 also coprecipitated SRPK1 (Fig. 3, top panel). Also, all of the GST-HPV E1^E4 proteins coprecipitated Flag-SRPK1 from lysates prepared from transiently transfected 293T cells (Fig. 3, upper middle panel).
FIG. 3.
The E1^E4 interaction with SRPK1 is conserved between different HPV types. SRPK1 binding to E1^E4 proteins of various HPV types was investigated by using GST-E1^E4 fusion proteins of HPV1, HPV16, and HPV18 to coprecipitate endogenous SRPK1 derived from SV-JD lysates (upper panel), Flag-SRPK1 expressed in 293T cells (middle upper panel), and His-SRPK1 expressed in bacteria (lower middle panel). A Ponceau-stained nitrocellulose membrane used for the SRPK1 coprecipitation demonstrates that the relative amounts of GST and GST fusion proteins (indicated by asterisks) used in these experiments is approximately equal (lower panel). In all cases, SRPK1 forms complexes specifically with all of the different GST-E1^E4 proteins but not GST alone. In the lower middle panel, the upper band migrating with an apparent molecular mass of over 120 kDa represents the full-length His-SRPK1, while the smaller band is likely to be a breakdown product of His-SRPK1. WB, Western blot.
Finally, to conclude these studies, we wanted to determine whether we could demonstrate an interaction between SRPK1 and HPV E1^E4 proteins in the absence of any other mammalian factors. Therefore, the GST-HPV E1^E4 proteins were mixed with a lysate prepared from bacteria expressing SRPK1 as a polyhistidine-tagged fusion protein (His-SRPK1). These studies, shown in the lower middle panel of Fig. 3, indicate that His-SRPK1 associated with the GST-HPV E1^E4 fusion proteins while not binding to GST alone. Therefore, given that this interaction was performed in the absence of any other eukaryotic factors, and in the presence of RNase, it is highly likely that SRPK1 is making a direct interaction with the GST-HPV E1^E4 proteins. These experiments provide compelling evidence to indicate that the binding of SRPK1 is a conserved function of E1^E4 proteins derived from divergent HPV types. A Ponceau stain of the nitrocellulose membrane used for the endogenous SRPK1 coprecipitation indicates that comparable amounts of GST fusion proteins were used throughout these studies (Fig. 3, lower panel).
Mapping the domains and key amino acids within HPV1 E1^E4 that mediate the association with SRPK1.
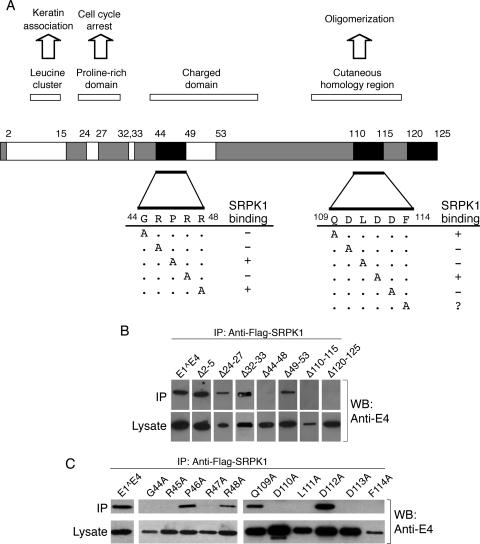
To identify HPV1 E1^E4 sequences involved in the association with SRPK1, a panel of previously described HPV1 E1^E4 deletion mutants (43) were coexpressed in 293T cells along with Flag-SRPK1. Cell lysates were prepared from these transfections, and simultaneously, the lysate was examined for the presence of soluble E1^E4 protein, while the presence of E1^E4 complexed with Flag-SRPK1 was determined by an anti-Flag immunoprecipitation. The summary of these analyses is given in Fig. 4A, with the immunoprecipitation data shown in Fig. 4B. Loss of extreme N-terminal sequences of E1^E4 (amino acids 2 to 15), including the keratin association motif (10LLGLL14) did not perturb the association with SRPK1. Sequences (amino acids 24 to 27) that form part of a proline-rich motif important in a G2 arrest function of a modified form of HPV1 E1^E4 (28) also were not necessary for the association. However, the deletion of residues 44 to 48 that form part of a bipartite arginine-rich motif (32RR33-44GRPRR48) abrogated the association with SRPK1. The arginine dipeptide (32RR33), however, was not necessary for maintenance of the interaction. Neighboring charged residues (49SDKDS53) also did not contribute to the association with the kinase. Sequences at the C terminus of the E1^E4 protein do participate in binding SRPK1. A self-association domain comprising amino acids 95 to 115 lies within this region of the protein (1, 43). SRPK1-containing complexes were not formed upon loss of E1^E4 residues 110 to 115, although it was not possible to evaluate the contribution of other regions of the self-association domain because of inadequate expression of soluble forms of the mutant proteins. Sequences at the extreme C terminus (amino acids 120 to 125) that do not seem necessary for oligomerization (1) are, however, required for the association with SRPK1. Our analysis of E1^E4 sequences necessary for the interaction with SRPK1 has not been exhaustive, and it is possible that other regions of E1^E4 not covered in our analysis may be involved in this association. However, at least two regions, an arginine-rich region (amino acids 44 to 48) and the C-terminal domain (amino acids 110 to 115 and 120 to 125), each abrogated the association with SRPK1 when deleted (Fig. 4A).
FIG. 4.
Mapping the domains within HPV1 E1^E4 that mediate the association with SRPK1. Anti-Flag immunoprecipitations (IP) from lysates prepared from 293T cells coexpressing Flag-SRPK1 and mutant HPV1 E1^E4 proteins were performed to identify key residues in HPV1 E1^E4 necessary for SRPK1 binding. (A) A diagrammatic summary of the data from the immunoprecipitation experiments is shown. The white domains correspond to regions within E1^E4 that are not required for the interaction with Flag-SRPK1, and the black domains identify residues found necessary for the association to occur. Alanine-scanning mutagenesis of residues in regions 44 to 48 and 109 to 114 involved in SRPK1 binding identified individual amino acids that are key participants in the association. +, binding; −, no binding. The relationship of the different domains to known E1^E4 functions and sequence characteristics is also shown. (B) Western blot (WB) analysis of immunoprecipitations between Flag-SRPK1 and E1^E4 proteins containing various deletions. (C) Western blot analysis of immunoprecipitations between alanine point substitutions and Flag-SRPK1. All of these mutants aside from F114A are expressed to a similar or greater level than the wild-type E1^E4. Analysis of the immunoprecipitations indicate that the amino acids G44, R45, R47, D110, L111, and D113 are each required to maintain the association of HPV1 E1^E4 with Flag-SRPK1.
The E1^E4 domains implicated in SRPK1 binding were further analyzed by alanine scanning mutagenesis with the objective of identifying critical amino acids that contribute to this interaction. Since arginine-rich regions constitute a common feature of E1^E4 proteins and residues 110 to 115 form a region within HPV1 E1^E4 that possesses homology to other E1^E4 proteins (43), the mutagenesis studies focused on these two regions. The results of the immunoprecipitations suggested that the point mutations G44A, R45A, and R47A located within the second arginine-rich region, each failed to bind SRPK1 (Fig. 4C). Furthermore, analysis of the alanine point mutations incorporated within the C-terminal domain revealed that D110A, L111A, and D113A each failed to associate with SRPK1 (Fig. 4C). In summary, individual key amino acid residues were identified in each of the two regions implicated by the deletion studies to be important in the association with SRPK1 (Fig. 4A).
SRPK1 accumulates at inclusion bodies formed by the HPV1 E1^E4 protein.
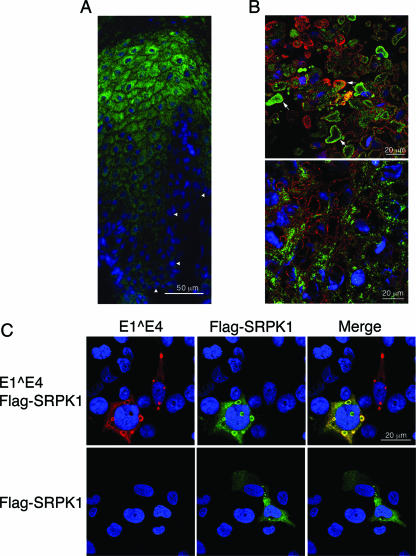
Up-regulation of E1^E4 expression in HPV infections coincides with a switch from the nonproductive phase of the life cycle to the productive phase. In HPV1 infections, the switch occurs immediately as cells move up from the basal layer and is marked by an accumulation of E4 into numerous cytoplasmic inclusion bodies of an undefined nature (2). Nuclear E4 inclusions are observed, and these are found to be associated with the ND10 component promyelocytic leukemia protein (46). To identify the cellular localization of SRPK1 in HPV warts, frozen sections of HPV1 warts were costained with anti-E4 and anti-SRPK1 monoclonal antibodies and examined by confocal microscopy. In areas of the wart showing no evidence of productive infection or E4 expression, SRPK1 was shown to be present in basal and suprabasal cells, with levels increasing as the cells became more differentiated, but was largely absent from the most differentiated cells of the cornified layers (Fig. 5A). SRPK1 staining was predominantly limited to the cytoplasm, an observation consistent with SRPK1 distribution in cervical keratinocytes grown in tissue culture (11). Examination of SRPK1 distribution in productive areas of the wart that expressed E4 revealed that SRPK1 was present in the cytoplasm of keratinocytes in the spinous cell layer but was not associated with E4 inclusions (Fig. 5B, lower panel). However, in infected cells of the granular layer of the wart, SRPK1 was localized to cytoplasmic E4 inclusion bodies (Fig. 5B, upper panel). In general, both E4 and SRPK1 staining of the inclusions is concentrated toward the periphery of the inclusion bodies and probably reflects epitope inaccessibility toward the center of these electron-dense structures (47). Similar productive regions of the wart showed no evidence of nonspecific staining following omission of the E4 and SRPK1 monoclonal antibodies from the staining protocol (data not shown).
FIG. 5.
SRPK1 distribution is altered in the presence of HPV1 E1^E4. (A and B) Confocal analysis of 4-μm sections of an HPV1-induced wart costained for E4 (red) and SRPK1 (green); nuclei were identified using DAPI (blue). (A) SRPK1 expression in regions of the wart tissue that are E4 negative and do not show evidence of productive HPV1 infection. Arrowheads indicate the basal cell layer. (B) In regions of the wart positive for E4 expression, SRPK1 is contained within E4 inclusions present in cells of the granular layers (upper panel, examples of costained inclusions are indicated by arrows) but not in those formed in cells of the lower (spinous) layers (bottom panel). (C) Confocal analysis of distribution of E4 (red) and SRPK1 (green) in SV-JD cells cotransfected with plasmids that express HPV1 E1^E4 and Flag-SRPK1 or Flag-SRPK1 alone. Nuclei were identified using DAPI (blue).
Since transient overexpression of HPV1 E4 in monolayer cultures of keratinocytes, including SV-JD cells, reproduces the formation of E4 inclusion bodies (46), we used this system to determine whether SRPK1 would localize to the E4 granules in the absence of HPV factors. SV-JD cells were cotransfected with plasmids expressing full-length HPV1 E1^E4 protein and Flag-SRPK1 or the Flag-SRPK1 expression vector alone. Confocal analysis of cells costained for E4 and the Flag epitope revealed localization of SRPK1 to cytoplasmic E4, including E4 associated with inclusion structures (Fig. 5C). Flag-SRPK1 did not accumulate into inclusion bodies in the absence of E4 (Fig. 5C).
In summary, SRPK1 kinase is sequestered to E4 cytoplasmic inclusions in cells of the upper layers of the wart, indicating that this is a late event in the productive cycle of the virus. Studies in transfected cells reveal that this E4 function is not dependent on additional HPV factors.
SRPK1 binding is associated with in vitro phosphorylation of HPV1 E1^E4.
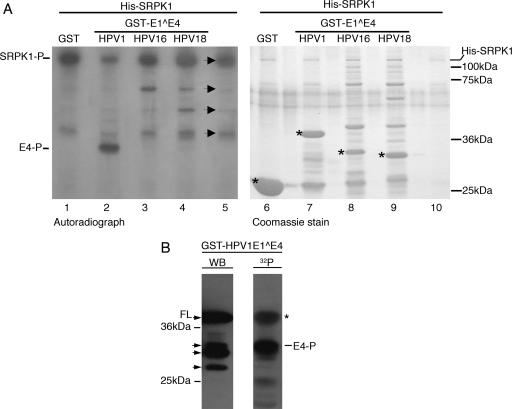
In vitro kinase reactions were used to determine whether SRPK1 phosphorylates HPV E1^E4 proteins. Reactions were performed using His-SRPK1 and GST-HPV E1^E4 fusion proteins which were all generated by expression in bacteria. Figure 6A shows the Coomassie-stained gel and an autoradiograph of a typical SRPK1 kinase experiment. In the Coomassie-stained gel, the location of the full-length substrates, GST (lane 6), GST-HPV1 E1^E4 (lane 7), GST-HPV16 E1^E4 (lane 8), and GST-HPV18 E1^E4 (lane 9) are each marked with an asterisk. Analysis of the autoradiograph indicated that, in the reaction mixture containing only His-SRPK1 (lane 5), four distinct phosphorylated species could be observed. The largest of these, with a molecular mass of approximately 120 Da, corresponds to phosphorylated full-length His-SRPK1, which migrates at this size (see the Coomassie-stained gel), and indicates that SRPK1 undergoes autophosphorylation. The three smaller species correspond to either phosphorylated breakdown products of His-SRPK1 or bacterial contaminants that have subsequently undergone phosphorylation. In reaction mixtures that contained test substrates for SRPK1, no phosphorylated bands could be observed in reaction mixtures containing GST, GST-HPV16 E1^E4, or GST-HPV18 E1^E4 (lanes 1, 3 and 4, respectively) that were not also present within the reaction mixture containing His-SRPK1 alone (lane 5). This suggests that, although SRPK1 interacts with the HPV16 and -18 GST-E1^E4 fusion proteins (Fig. 3), at this point we have no evidence to indicate that SRPK1 is phosphorylating these proteins. In contrast, analysis of the in vitro kinase reaction containing GST-HPV1 E1^E4 identifies a major phosphospecies specific to the GST-HPV1 E1^E4 sample (Fig. 6A, lane 2). However, this species migrates 3 to 4 kDa faster than the full-length GST-E1^E4 fusion protein. Western blot analysis of the kinase reactions identifies a number of products smaller than the full-length GST-HPV1 E1^E4 protein that are recognized by HPV1 E4-specific antibodies, and the phosphospecies corresponds to one of these smaller E4 polypeptides (Fig. 6B). We therefore conclude that the full-length GST-HPV1 E1^E4 protein is not phosphorylated by SRPK1, but a smaller E4-containing species is a target. Further evidence to support the fact that this phosphorylated species is derived from GST-HPV1 E1^E4 comes from in vitro kinase reactions where a GST fusion encoding a naturally processed N-terminal truncation (16 kDa) of the full-length 17-kDa E1^E4 protein was used as a substrate. In these studies, the migration of the major phosphospecies was only about 1 kDa faster than the phosphorylated species present in the reaction mixture containing the full-length GST-E1^E4 protein (data not shown), thus demonstrating that truncation of HPV1 E1^E4 resulted in a concomitant shift in the size of the phosphorylated polypeptide.
FIG. 6.
SRPK1 phosphorylates HPV1 E1^E4 in vitro. (A) SDS-polyacrylamide gel of in vitro kinase reaction mixtures containing His-SRPK1 and GST-E1^E4 proteins of HPV1, HPV16, and HPV18. The amounts of GST-E1^E4 fusion proteins (indicated by asterisks) and His-SRPK1 in the kinase reactions are shown in the Coomassie-stained gel. The autoradiograph demonstrates the presence of phosphorylated protein species. The arrows adjacent to lane 5, which contained only His-SRPK1, indicate the presence of phosphorylated full-length His-SRPK1 (SRPK1-P) as well as smaller phosphopeptide species, which may be breakdown products of SRPK1 or phosphorylated bacterial contaminants. Lanes 1, 3, and 4 show similar patterns of protein phosphorylation, indicating that neither GST, GST-HPV16 E1^E4, nor GST-HPV18 E1^E4 is phosphorylated by SRPK1 under these conditions. In contrast, the GST-HPV1 E1^E4 substrate (lane 2), produces a distinct phosphorylated species (E4-P) which is not present in the reaction mixture containing His-SRPK1 alone (lane 5). (B) An in vitro kinase reaction mixture containing His-SRPK1 and GST-HPV1 E1^E4 proteins was analyzed by Western blotting (WB) with an HPV1 E1^E4-specific monoclonal antibody prior to autoradiography (32P). Major E4 phosphospecies (E4-P) corresponds to one of a series of peptides (identified with arrowheads) smaller than the full-length GST fusion (FL). The faint band (marked with an asterisk) appearing in the autoradiograph is derived from the strong chemiluminescence signal of the full-length E1^E4 fusion protein.
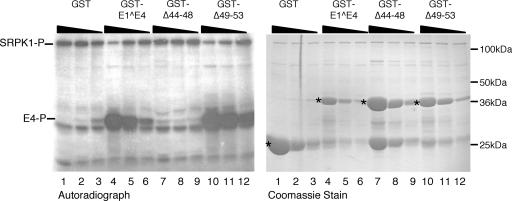
Having demonstrated that SRPK1 was able to phosphorylate GST-HPV1 E1^E4, we then examined the ability of SRPK1 to phosphorylate HPV1 E1^E4 deletion mutants. Thus, the deletion mutants Δ44-48, which is unable to bind SRPK1, and Δ49-53, which retains SRPK1 binding, were expressed as GST fusion proteins and used as substrates in in vitro kinase reactions. In these experiments, the GST substrates were titrated at 1, 5, and 25 μg per kinase reaction (Fig. 7). Both HPV1 E1^E4 (lanes 4 to 6) and the deletion mutant Δ49-53 (lanes 10 to 12) were phosphorylated by His-SRPK1. Again, the major phosphospecies migrated slightly faster that the full-length GST E1^E4 fusion. In contrast, the deletion mutant Δ44-48 was not phosphorylated by His-SRPK1 (lanes 7 to 9). Thus, with respect to HPV1 E1^E4, proteins which interact with SRPK1 appear to be phosphorylated by SRPK1.
FIG. 7.
In vitro phosphorylation of HPV1 E1^E4 is dependent on SRPK1 binding. In vitro kinase reactions containing His-SRPK1 and GST-HPV1 E1^E4 fusion proteins that bind SRPK1 (E1^E4 and Δ49-53) or are defective for binding (Δ44-48) titrated at 25, 5, and 1 μg. The relative amounts of His-SRPK1 and the different GST proteins (indicated by asterisks) present in the kinase reactions are shown in the Coomassie-stained gel. A major phosphospecies (E4-P) is present in in vitro kinase reaction mixtures containing the wild-type E1^E4 protein (lanes 4 to 6) and the Δ49-53 deletion mutant (lanes 10 to 12) but is not detected in reactions containing the Δ44-48 deletion mutant (lanes 7 to 9) or the GST control protein (lanes 1 to 3). In the titrations of E1^E4 substrates that interact with SRPK1, we observed a dose-dependent reduction in autophosphorylation of the full-length His-SRPK1 polypeptide (SRPK1-P).
Further examination of the autoradiograph of the in vitro kinase reactions shown in Fig. 6 indicated that, in comparison to reactions containing GST (lane 1), HPV16 (lane 3), or HPV18 (lane 4) E1^E4 fusions or no substrate (lane 5), there was a marked decrease in the level of the autophosphorylated form of His-SRPK1 in the presence of the HPV1 E1^E4 fusion protein (lane 2). Titration of increasing amounts of GST-HPV1 E1^E4 protein in the in vitro kinase reactions revealed that HPV1 E1^E4 inhibition of SRPK autophosphorylation was dose dependent (Fig. 7, autoradiograph, lanes 5 to 7). A similar dose-dependent inhibition of SRPK1 autophosphorylation occurred following titration of the deletion mutant Δ49-53 that retains SRPK1 binding (lanes 10 −12). In contrast, GST (lanes 1 to 3) and GST-HPV1 E1^E4 Δ44-48 (lanes 7 to 9), neither of which interact with SRPK1, show no inhibitory effect on the autophosphorylation of SRPK1.
DISCUSSION
We have shown that the E1^E4 protein of HPV1, a cutaneous virus type, interacts with the cellular protein kinase SRPK1. Moreover, our data confirm that E1^E4 proteins of HPV16 and HPV18, viruses that are more frequently associated with infections of the mucosa, also interact with SRPK1, indicating that this association is conserved across different phylogenetic groups of HPV. These studies have also identified key residues, notably G44, R45, R47, D110, L111, and D113, within HPV1 E1^E4 that, when substituted by alanine, abrogate binding. We have previously reported that a C-terminal region encompassing amino acids 95 to 115 encodes sequences that facilitate oligomerization of E1^E4 and, more specifically, deletion of the sequence 110DLD112 abrogated self-association (1). It is therefore plausible that D110, L111, and D113 contribute to self-association and that E1^E4 oligomerization is required for SRPK1 binding. The other critical residues, G44, R45, and R47 lie within an arginine-rich domain that bears resemblance to the arginine-rich sequences present within SRPK1 substrates, including SR proteins. However, it remains to be determined which serine residues in E1^E4 are phosphorylated by SRPK1, although we can confirm that deletion of the sequence 49SDKDS53, which lies within the arginine-rich domain, does not diminish the ability of SRPK1 to bind nor phosphorylate E1^E4 and suggests that other serine residues must be sites of SRPK1 phosphorylation. Close inspection of the major phosphorylated species from the in vitro kinase experiments indicates that it is migrating faster than the major GST-HPV1 E1^E4 product. Loss of a proximal C-terminal sequence may enhance phosphorylation or phosphorylation of E4 is affecting the migration of the E4 polypeptide, which is possible given that the full-length E1^E4 protein migrates anomalously high at 17-kDa, 2.6-kDa higher than its calculated mass of 14.4 kDa. While we have been unable to detect phosphorylation of HPV16 and HPV18 E1^E4 proteins in vitro, we cannot rule out the possibility that these may be substrates of SRPK1 and indeed that the GST component of these proteins may induce steric changes that impairs phosphorylation in these assays. Phosphorylation is likely to be an important determinant of E4 function, and to date, a number of kinases have been implicated in E4 phosphorylation (4, 7, 20). It will therefore be interesting to determine whether E1^E4 phosphorylation by SRPK1 does contribute to a specific E4 function.
The oligomerization domain of HPV1 E4 displays significant homology with the C-terminal sequences of E4 proteins derived from other HPV types that infect cutaneous surfaces. Within this domain, D110, L111, and D113 form part of a conserved motif (D-L-[D/E]-X-[Y/F], where X is a hydrophilic residue) that has been shown to be necessary for oligomerization (1, 43). A similar function has been assigned to the C terminus of HPV16 E4 (56). While sequence alignment identifies homology across this region of HPV16 E4 with E4s of other mucosal types, it is not highly conserved within the cutaneous viruses (43, 45). The lack of homology between the oligomerization domains of cutaneous and mucosal viruses would suggest that D110, L111, and D113 may not be involved in direct binding of SRPK1 but that oligomerization of the protein is perhaps necessary. With regard to the other HPV1 residues necessary for SRPK1 binding (G44, R45, and R47), they are contained within a region of HPV1 E4 rich in basic amino acids (arginine, lysine, and histidine). Similar basic regions are a common feature of E4 proteins (16). In some types they form a bipartite arginine-rich motif, similar to the one present in the type 1 protein. Comparative analysis of the basic regions of HPV1, -16, and -18, show that, although all are rich in arginines, or a mixture of basic amino acids, they are variable in both length and sequence (Fig. 8). Indeed, alignment of this region with HPV63, which is phylogenetically a close relation to HPV1, would suggest that the only conserved feature of this domain is the positive charge (Fig. 8). Thus, further analysis of the association between SRPK1 and the type 16 and 18 E4 proteins will be necessary to reveal whether amino acids in their oligomerization domains and basic regions contribute to their interaction with SRPK1.
FIG. 8.
The sequence of regions of HPV E4 proteins that are rich in basic amino acids. The amino acid sequences of E4 regions of HPV1, -16, -18, and -63 that are particularly rich in basic amino acids are shown with arginine residues identified in boldface type. Note that bipartite arginine-rich motifs are present in HPV1, -18, and -63 but not HPV16. Amino acids within the HPV1 E4 region involved in the interaction with SRPK1 are underlined.
It has been shown that SRPK1 undergoes modulation during the cell cycle, translocating into the nucleus during the G2-to-M transition (11). The importance of this translocation is not known, although it likely permits the kinase access to its substrates, including the SR proteins contained within nuclear speckles (30), and the LBR (36). Other regulatory mechanisms of SRPK1 activity could include autophosphorylation of the kinase, as is apparent for CLK family kinases, which also phosphorylate the RS domains of SR factors, and is found to modulate substrate recognition and lead to changes in the phosphorylation patterns on specific SR proteins (41). It is therefore of interest that, in the in vitro kinase assays, the ability of SRPK1 to undergo autophosphorylation appeared to be inhibited by HPV1 E1^E4, and this inhibition was abrogated by mutations which blocked HPV1 E1^E4 binding of SRPK1. Thus, whether E1^E4 does indeed modulate SRPK1 autophosphorylation and substrate specificity in vivo warrants further investigation.
The manipulation of SRPK1 by a virus is not without precedent, indeed the herpes simplex virus type 1 encodes a protein ICP27 which associates with SRPK1, relocating it to the nucleus, resulting in hypophosphorylation of SR proteins and a consequent impairment in spliceosome assembly (51). This action has the effect of inhibiting cellular pre-mRNA splicing and promotes export and expression of herpes simplex virus type 1 transcripts. There are also a number of studies that implicate substrates of SRPK1 in the replication cycle of viruses. These studies range from human immunodeficiency virus type 1 (HIV-1), where virion production is regulated by SR proteins, including ASF/SF2, SC35, and 9G8 (26), while SRp75 acts to upregulate HIV-1 gene expression (19); adenovirus, where SR proteins purified from late infected cells are functionally inactivated (27); and vaccinia, where SR proteins are hypophosphorylated (23). Thus, viruses appear to have adopted mechanisms to act upon SR proteins as a means of modifying host gene expression and affecting efficient viral gene expression. Pertinently with respect to HPV, an SRPK1 substrate, SF2/ASF, is part of a complex that associates with a negative regulatory mRNA element that is central to the posttranscriptional regulation of late gene expression in HPV16, and both expression and phosphorylation of this SR factor increase in response to differentiation of the HPV16-infected cells (33). In fact, numerous cis-acting elements appear to contribute to the control of HPV late gene expression, including HPV16 (48, 60), and similar structures that are present in bovine papillomavirus transcripts are associated with multiple SR proteins (61). Further studies are required to examine the differentiation-specific regulation of other SR proteins and to identify whether E1^E4 proteins, via their interaction with SRPK1, impact upon their regulation. The suggestion that E1^E4 may modify posttranscriptional processing is not novel; indeed, HPV16 E4^E4 has previously been shown to associate with an RNA helicase, an association that was proposed to affect mRNA stability and ribosome biogenesis (13).
In addition to phosphorylating SR proteins, SRPK1 also phosphorylates LBR, an inner nuclear membrane protein which interacts with B-type lamins (53) and chromatin (55). The association of LBR with chromatin is cell cycle dependent and regulated by multiple kinases, including SRPK1 (54), and acts to maintain the organization of the nuclear envelope during cell division (6). Inhibition, modulation, or sequestration of SRPK1 by E1^E4 may inhibit the attachment of LBR to chromatin and result in the loss of nuclear envelope integrity. Since the sequestration of SRPK1 to HPV1 E4 inclusion bodies is restricted to cells of the upper wart regions, and as HPV virion assembly is confined to the nucleus of these cells, destabilization of the nuclear envelope may facilitate egress of virions from the nucleus. Indeed, the human polyomavirus JC virus perturbs the structure of the nuclear envelope by abrogating the interaction between LBR and heterochromatin and thus promoting nuclear egress of progeny JC virus virions (37). This possible E1^E4 function, in addition to its action on the cornified cell envelope (3) and the keratin cytoskeleton (14, 44) and an ability to promote apoptosis (42), may all contribute to ease the passage of newly synthesized virions from the nucleus to the cytoplasm and promote their subsequent shedding from the upper cells of the lesion.
SRPK1 binding by E1^E4 may influence the function of other HPV proteins. Candidate molecules such as the HPV5 E2 protein, which contains a hinge region that possesses RS domain characteristics (31), may be an SRPK1 substrate, and as such, SRPK1 may influence HPV5 E2 function by modulating its ability to activate transcription/replication.
SRPK1 appears to play an important role in the regulation of some fundamental cellular processes, including the organization of components of the nucleus and mRNA processing, and this may explain why it appears to be a strategically important cellular target for a diverse number of viruses. Our study identifies SRPK1 as a novel cellular binding partner of E4 proteins derived from genetically diverse HPV types. With regard to HPV1 E1^E4, we have demonstrated that the ability of SRPK1 to undergo autophosphorylation is impaired, while it is sequestered into E4 inclusions in terminally differentiated cells within wart lesions. Future studies are necessary to dissect the biochemical implications of this finding and to determine whether this interaction has a role in the regulation of aspects of HPV replication. The biochemistry of SRPK1 is poorly understood and the role it plays in the HPV replication cycle is unknown, but novel substrates encoded by HPV and/or the host cell may exist. Indeed this kinase may prove to be a target for therapeutic intervention, as has been proposed for HIV-1, where the use of small-molecule inhibitors of SRPK1 has been demonstrated to suppress HIV-1 replication (19).
Acknowledgments
We thank Saleem Khan (University of Pittsburgh) for his kind gift of the HPV1 E1-Flag expression vector and Bai-Gong Yue (University of Leicester) for the generous gift of the SRPK1-His expression plasmid. We are grateful to Gillian Knight for her help in generating the HPV1 44-48 single-alanine substitutions and to Michele McNally for her excellent technical assistance.
This study was supported by a Cancer Research UK Programme Grant (C427/A3919) to S.R.
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Ashmole, I., P. H. Gallimore, and S. Roberts. 1998. Identification of conserved hydrophobic C-terminal residues of the human papillomavirus type 1 E1E4 protein necessary for E4 oligomerisation in vivo. Virology 240:221-231. [DOI] [PubMed] [Google Scholar]
- 2.Breitburd, F., O. Croissant, and G. Orth. 1987. Expression of human papillomavirus type-1 E4 gene products in warts, p. 115-122. In B. M. Steinberg, J. Brandsma, and L. B. Taichman (ed.), Papillomaviruses, vol. 5. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 3.Brown, D. R., D. Kitchin, B. Qadadri, N. Neptune, T. Batteiger, and A. Ermel. 2006. The human papillomavirus type 11 E1-E4 protein is a transglutaminase 3 substrate and induces abnormalities of the cornified cell envelope. Virology 345:290-298. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, J. T., A. Han, K. H. Fife, and D. R. Brown. 2000. The human papillomavirus type 11 E1E4 protein is phosphorylated in genital epithelium. Virology 268:430-439. [DOI] [PubMed] [Google Scholar]
- 5.Caceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, A., R. Rassadi, and U. Stochaj. 1998. Velcro in the nuclear envelope: LBR and LAPs. FEBS Lett. 441:165-169. [DOI] [PubMed] [Google Scholar]
- 7.Davy, C. E., M. Ayub, D. J. Jackson, P. Das, P. McIntosh, and J. Doorbar. 2006. HPV16 E1-E4 protein is phosphorylated by Cdk2/cyclin A and relocalizes this complex to the cytoplasm. Virology 349:230-244. [DOI] [PubMed] [Google Scholar]
- 8.Davy, C. E., D. J. Jackson, K. Raj, W. L. Peh, S. A. Southern, P. Das, R. Sorathia, P. Laskey, K. Middleton, T. Nakahara, Q. Wang, P. J. Masterson, P. F. Lambert, S. Cuthill, J. B. Millar, and J. Doorbar. 2005. Human papillomavirus type 16 E1 E4-induced G2 arrest is associated with cytoplasmic retention of active Cdk1/cyclin B1 complexes. J. Virol. 79:3998-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davy, C. E., D. J. Jackson, Q. Wang, K. Raj, P. J. Masterson, N. F. Fenner, S. Southern, S. Cuthill, J. B. Millar, and J. Doorbar. 2002. Identification of a G(2) arrest domain in the E1 wedge E4 protein of human papillomavirus type 16. J. Virol. 76:9806-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 324:17-27. [DOI] [PubMed] [Google Scholar]
- 11.Ding, J. H., X. Y. Zhong, J. C. Hagopian, M. M. Cruz, G. Ghosh, J. Feramisco, J. A. Adams, and X. D. Fu. 2006. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol. Biol. Cell 17:876-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doorbar, J., I. Coneron, and P. H. Gallimore. 1989. Sequence divergence yet conserved physical characteristics among the E4 proteins of cutaneous human papillomaviruses. Virology 172:51-62. [DOI] [PubMed] [Google Scholar]
- 13.Doorbar, J., R. C. Elston, S. Napthine, K. Raj, E. Medcalf, D. Jackson, N. Coleman, H. M. Griffin, P. Masterson, S. Stacey, Y. Mengistu, and J. Dunlop. 2000. The E1E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J. Virol. 74:10081-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 15.Doorbar, J., H. S. Evans, I. Coneron, L. V. Crawford, and P. H. Gallimore. 1988. Analysis of HPV-1 E4 gene expression using epitope-defined antibodies. EMBO J. 7:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doorbar, J., and G. Myers. 1996. The E4 protein, p. 58-80. In G. Myers, H. Delius, J. Icenogel, H.-C. Bernard, C. Baker, A. Halpern, and C. Wheeler (ed.), Human papillomaviruses, vol. 3. Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 17.Fang, L., L. R. Budgeon, J. Doorbar, E. R. Briggs, and M. K. Howett. 2006. The human papillomavirus type 11 E1^E4 protein is not essential for viral genome amplification. Virology 351:271-279. [DOI] [PubMed] [Google Scholar]
- 18.Fu, X. D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuhara, T., T. Hosoya, S. Shimizu, K. Sumi, T. Oshiro, Y. Yoshinaka, M. Suzuki, N. Yamamoto, L. A. Herzenberg, L. A. Herzenberg, and M. Hagiwara. 2006. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. USA 103:11329-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grand, R. J., J. Doorbar, K. J. Smith, I. Coneron, and P. H. Gallimore. 1989. Phosphorylation of the human papillomavirus type 1 E4 proteins in vivo and in vitro. Virology 170:201-213. [DOI] [PubMed] [Google Scholar]
- 21.Hebner, C. M., and L. A. Laimins. 2005. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 16:83-97. [DOI] [PubMed] [Google Scholar]
- 22.Hillenkamp, F., M. Karas, R. C. Beavis, and B. T. Chait. 1991. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 63:1193A-1203A. [DOI] [PubMed] [Google Scholar]
- 23.Huang, T. S., C. E. Nilsson, T. Punga, and G. Akusjarvi. 2002. Functional inactivation of the SR family of splicing factors during a vaccinia virus infection. EMBO Rep. 3:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899-905. [DOI] [PubMed] [Google Scholar]
- 25.Huang, Y., and J. A. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 26.Jacquenet, S., D. Decimo, D. Muriaux, and J. L. Darlix. 2005. Dual effect of the SR proteins ASF/SF2, SC35 and 9G8 on HIV-1 RNA splicing and virion production. Retrovirology 2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanopka, A., O. Muhlemann, S. Petersen-Mahrt, C. Estmer, C. Ohrmalm, and G. Akusjarvi. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185-187. [DOI] [PubMed] [Google Scholar]
- 28.Knight, G. L., J. R. Grainger, P. H. Gallimore, and S. Roberts. 2004. Cooperation between different forms of the human papillomavirus type 1 E4 protein to block cell cycle progression and cellular DNA synthesis. J. Virol. 78:13920-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight, G. L., A. S. Turnell, and S. Roberts. 2006. Role for Wee1 in inhibition of G2-to-M transition through the cooperation of distinct human papillomavirus type 1 E4 proteins. J. Virol. 80:7416-7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koizumi, J., Y. Okamoto, H. Onogi, A. Mayeda, A. R. Krainer, and M. Hagiwara. 1999. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs). J. Biol. Chem. 274:11125-11131. [DOI] [PubMed] [Google Scholar]
- 31.Lai, M. C., B. H. Teh, and W. Y. Tarn. 1999. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 274:11832-11841. [DOI] [PubMed] [Google Scholar]
- 32.Lin, S., R. Xiao, P. Sun, X. Xu, and X. D. Fu. 2005. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell 20:413-425. [DOI] [PubMed] [Google Scholar]
- 33.McPhillips, M. G., T. Veerapraditsin, S. A. Cumming, D. Karali, S. G. Milligan, W. Boner, I. M. Morgan, and S. V. Graham. 2004. SF2/ASF binds the human papillomavirus type 16 late RNA control element and is regulated during differentiation of virus-infected epithelial cells. J. Virol. 78:10598-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakahara, T., A. Nishimura, M. Tanaka, T. Ueno, A. Ishimoto, and H. Sakai. 2002. Modulation of the cell division cycle by human papillomavirus type 18 E4. J. Virol. 76:10914-10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakahara, T., W. L. Peh, J. Doorbar, D. Lee, and P. F. Lambert. 2005. Human papillomavirus type 16 E1^E4 contributes to multiple facets of the papillomavirus life cycle. J. Virol. 79:13150-13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolakaki, E., J. Meier, G. Simos, S. D. Georgatos, and T. Giannakouros. 1997. Mitotic phosphorylation of the lamin B receptor by a serine/arginine kinase and p34(cdc2). J. Biol. Chem. 272:6208-6213. [DOI] [PubMed] [Google Scholar]
- 37.Okada, Y., T. Suzuki, Y. Sunden, Y. Orba, S. Kose, N. Imamoto, H. Takahashi, S. Tanaka, W. W. Hall, K. Nagashima, and H. Sawa. 2005. Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: role in nuclear egress of viral particles. EMBO Rep. 6:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papoutsopoulou, S., E. Nikolakaki, and T. Giannakouros. 1999. SRPK1 and LBR protein kinases show identical substrate specificities. Biochem. Biophys. Res. Commun. 255:602-607. [DOI] [PubMed] [Google Scholar]
- 39.Peh, W. L., J. L. Brandsma, N. D. Christensen, N. M. Cladel, X. Wu, and J. Doorbar. 2004. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. J. Virol. 78:2142-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and J. Doorbar. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 76:10401-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad, J., and J. L. Manley. 2003. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol. Cell. Biol. 23:4139-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raj, K., S. Berguerand, S. Southern, J. Doorbar, and P. Beard. 2004. E1^E4 protein of human papillomavirus type 16 associates with mitochondria. J. Virol. 78:7199-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, S., I. Ashmole, L. J. Gibson, S. M. Rookes, G. J. Barton, and P. H. Gallimore. 1994. Mutational analysis of human papillomavirus E4 proteins: identification of structural features important in the formation of cytoplasmic E4/cytokeratin networks in epithelial cells. J. Virol. 68:6432-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts, S., I. Ashmole, G. D. Johnson, J. W. Kreider, and P. H. Gallimore. 1993. Cutaneous and mucosal human papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology 197:176-187. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, S., I. Ashmole, S. M. Rookes, and P. H. Gallimore. 1997. Mutational analysis of the human papillomavirus type 16 E1-E4 protein shows that the C terminus is dispensable for keratin cytoskeleton association but is involved in inducing disruption of the keratin filaments. J. Virol. 71:3554-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, S., M. L. Hillman, G. L. Knight, and P. H. Gallimore. 2003. The ND10 component promyelocytic leukemia protein relocates to human papillomavirus type 1 E4 intranuclear inclusion bodies in cultured keratinocytes and in warts. J. Virol. 77:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogel-Gaillard, C., G. Pehau-Arnaudet, F. Breitburd, and G. Orth. 1993. Cytopathic effect in human papillomavirus type 1-induced inclusion warts: in vitro analysis of the contribution of two forms of the viral E4 protein. J. Investig. Dermatol. 101:843-851. [DOI] [PubMed] [Google Scholar]
- 48.Rush, M., X. Zhao, and S. Schwartz. 2005. A splicing enhancer in the E4 coding region of human papillomavirus type 16 is required for early mRNA splicing and polyadenylation as well as inhibition of premature late gene expression. J. Virol. 79:12002-12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanford, J. R., J. D. Ellis, D. Cazalla, and J. F. Caceres. 2005. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. USA 102:15042-15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanford, J. R., N. K. Gray, K. Beckmann, and J. F. Caceres. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18:755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sciabica, K. S., Q. J. Dai, and R. M. Sandri-Goldin. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 22:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 53.Simos, G., and S. D. Georgatos. 1992. The inner nuclear membrane protein p58 associates in vivo with a p58 kinase and the nuclear lamins. EMBO J. 11:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takano, M., Y. Koyama, H. Ito, S. Hoshino, H. Onogi, M. Hagiwara, K. Furukawa, and T. Horigome. 2004. Regulation of binding of lamin B receptor to chromatin by SR protein kinase and cdc2 kinase in Xenopus egg extracts. J. Biol. Chem. 279:13265-13271. [DOI] [PubMed] [Google Scholar]
- 55.Takano, M., M. Takeuchi, H. Ito, K. Furukawa, K. Sugimoto, S. Omata, and T. Horigome. 2002. The binding of lamin B receptor to chromatin is regulated by phosphorylation in the RS region. Eur. J. Biochem. 269:943-953. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Q., H. Griffin, S. Southern, D. Jackson, A. Martin, P. McIntosh, C. Davy, P. J. Masterson, P. A. Walker, P. Laskey, M. B. Omary, and J. Doorbar. 2004. Functional analysis of the human papillomavirus type 16 E1^E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization. J. Virol. 78:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, R., F. Fehrmann, and L. A. Laimins. 2005. Role of the E1^E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J. Virol. 79:6732-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.Wilson, R., G. B. Ryan, G. L. Knight, L. A. Laimins, and S. Roberts. 13 February 2007, posting date. The full-length E1^E4 protein of human papillomavirus type 18 modulates differentiation-dependent viral DNA amplification and late gene expression. Virology. doi: 10.1016/j.virol.2007.01.005. [DOI] [PubMed]
- 58.Xiao, S. H., and J. L. Manley. 1998. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 17:6359-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yue, B. G., P. Ajuh, G. Akusjarvi, A. I. Lamond, and J. P. Kreivi. 2000. Functional coexpression of serine protein kinase SRPK1 and its substrate ASF/SF2 in Escherichia coli. Nucleic Acids Res. 28:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, X., M. Rush, and S. Schwartz. 2004. Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene. J. Virol. 78:10888-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng, Z. M., and C. C. Baker. 2006. Papillomavirus genome structure, expression, and posttranscriptional regulation. Front Biosci. 11:2286-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]