Abstract
Human coronavirus NL63 (HCoV-NL63), a common human respiratory pathogen, is associated with both upper and lower respiratory tract disease in children and adults. Currently, no antiviral drugs are available to treat CoV infections; thus, potential drug targets need to be identified and characterized. Here, we identify HCoV-NL63 replicase gene products and characterize two viral papain-like proteases (PLPs), PLP1 and PLP2, which process the viral replicase polyprotein. We generated polyclonal antisera directed against two of the predicted replicase nonstructural proteins (nsp3 and nsp4) and detected replicase proteins from HCoV-NL63-infected LLC-MK2 cells by immunofluorescence, immunoprecipitation, and Western blot assays. We found that HCoV-NL63 replicase products can be detected at 24 h postinfection and that these proteins accumulate in perinuclear sites, consistent with membrane-associated replication complexes. To determine which viral proteases are responsible for processing these products, we generated constructs representing the amino-terminal end of the HCoV-NL63 replicase gene and established protease cis-cleavage assays. We found that PLP1 processes cleavage site 1 to release nsp1, whereas PLP2 is responsible for processing both cleavage sites 2 and 3 to release nsp2 and nsp3. We expressed and purified PLP2 and used a peptide-based assay to identify the cleavage sites recognized by this enzyme. Furthermore, by using K48-linked hexa-ubiquitin substrate and ubiquitin-vinylsulfone inhibitor specific for deubiquitinating enzymes (DUBs), we confirmed that, like severe acute respiratory syndrome (SARS) CoV PLpro, HCoV-NL63 PLP2 has DUB activity. The identification of the replicase products and characterization of HCoV-NL63 PLP DUB activity will facilitate comparative studies of CoV proteases and aid in the development of novel antiviral reagents directed against human pathogens such as HCoV-NL63 and SARS-CoV.
To date, five human coronaviruses (HCoV) have been identified as human pathogens. HCoV-229E and HCoV-OC43 were identified in the mid-1960s (1, 39) and were shown to account for 5 to 30% of common-cold-like respiratory diseases (26). The CoV that caused the epidemic of severe acute respiratory syndrome (SARS) in 2002 and 2003 was initially identified by the characteristic “crown-like” projections on the virus particles visualized by electron microscopy (15, 32, 41). Recent studies have shown that a virus similar to SARS-CoV, termed bat-SARS-CoV, is endemic in bats in southern China (33, 34). Sequence analysis indicates that bat-SARS-CoV was likely the source of the epidemic and that the virus evolved, likely during passage through animal intermediates, such as the civet cat, to allow for efficient human-to-human transmission (35, 55). Thus, the potential exists for reemergence of this virus from the animal reservoir. Post-SARS, researchers identified two additional HCoV, HCoV-NL63 (17, 62) and CoV-HKU1 (66), from patients suffering from respiratory infections and pneumonia.
HCoV-NL63 was first isolated from the respiratory tract of a 7-month-old child suffering from bronchiolitis and conjunctivitis in The Netherlands (62) and was later found to be associated with croup in young children (63). Epidemiological studies from several countries revealed that HCoV-NL63 is detected in 0.3 to 10% of clinical specimens from both infants and adults suffering from respiratory tract illnesses (2, 5, 6, 11, 12, 16, 27, 58, 61). Thus, HCoV-NL63 is a common human respiratory pathogen with worldwide distribution that is associated with a substantial proportion of both upper and lower respiratory tract disease. Experimental treatments, such as the use of small interfering RNAs and fusion blockers, have been shown to inhibit replication of HCoV-NL63 in tissue culture cells (44). However, to date there are no approved antiviral reagents for the treatment of any HCoV infection, including HCoV-NL63 and the more severe pathogen SARS-CoV. Therefore, identification of potential targets for the development of anti-CoV drugs is a high priority.
The RNA genome of HCoV-NL63 is 27,553 nucleotides in length. The 5′ two-thirds of the genomic RNA contains two open reading frames (ORF1a and ORF1b) that are joined by a ribosomal frameshift sequence and encode the replicase polyproteins. Four viral structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), and one accessory ORF protein, termed ORF3, are encoded downstream of the replicase (45, 62). Analysis of susceptibility to infection showed that HCoV-NL63 uses angiotensin-converting enzyme 2 (ACE2), the SARS-CoV receptor, as the cellular receptor to infect host cells (24, 25). Once a CoV enters the cell, the genomic RNA is released into the cytoplasm and translated to generate two polyproteins, pp1a and pp1ab (70). In most CoV, pp1a contains three viral proteases: two papain-like proteases (PLP1 and PLP2) and a picornavirus 3C-like protease (3CLpro). SARS-CoV, infectious bronchitis virus of chickens, and some bat CoV are distinct in that they have only one PLP, PLpro (33, 34, 38, 60). Analysis of proteolytic processing of several CoV has revealed that the replicase polyproteins pp1a and pp1ab are processed cotranslationally and posttranslationally by the viral proteases to generate as many as 16 nonstructural proteins (nsp's), which subsequently assemble with the endoplasmic reticulum membranes to form the replicase complex, which participates in subgenomic RNA synthesis (19, 20, 42, 54). Proteolytic processing of the CoV polyprotein by proteases is essential for correct localization and function of the replicase proteins and ongoing viral RNA synthesis and virus replication (31). Thus, CoV proteases are attractive targets for the development of antiviral drugs for reduction of viral replication and pathogenicity.
Here, we identify processed products nsp3 and nsp4 of the HCoV-NL63 replicase polyprotein and describe cis-cleavage assays that characterize the protease activity required to generate these products. The replicase products can be detected at 24 h postinfection (hpi) and accumulate in perinuclear sites in infected cells. We found that PLP1 processes cleavage site 1 to release nsp1, whereas PLP2 is responsible for processing cleavage sites 2 and 3 to release nsp2 and nsp3. The catalytic residues in PLP1 and PLP2 and the putative cleavage sites between replicase products were also characterized by site-directed mutagenesis of predicted critical residues. Furthermore, we confirmed that, like SARS-CoV PLpro (46), HCoV-NL63 PLP2 has deubiquitinating enzyme (DUB) activity. We found that purified PLP2 hydrolyzed K48-linked hexa-ubiquitin (K48-Ub6) to produce monoubiquitin, and PLP2-Ub adducts were detected using the Ub-vinylsulfone (Ub-VS) inhibitor, which is specific for DUBs. This identification of replicase products and characterization of HCoV-NL63 PLP and DUB activity will facilitate comparative studies of CoV protease activity and aid in the development of novel antiviral reagents directed against HCoV, such as HCoV-NL63 and SARS-CoV.
MATERIALS AND METHODS
Virus and cells.
The HCoV-NL63 (P8) and LLC-MK2 cells were kindly provided by Lia van der Hoek (University of Amsterdam, The Netherlands). LLC-MK2 cells were cultured in a mixture of Hanks minimal essential medium (MEM) and Earle's MEM (two parts Hanks MEM [Invitrogen] and one part Earle's MEM [Invitrogen]), supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) (Invitrogen, Grand Island, NY). The LLC-MK2 cells were cultured in a 37°C incubator with 5.0% CO2. After addition of HCoV-NL63, the LLC-MK2 cells were transferred to a 33°C incubator with 5.0% CO2, as previously described (62). The cells were incubated for 5 to 6 days, after which supernatant containing infectious virus particles was harvested. The initial P8 stock was passaged four times to generate the P12 stock used in these studies.
The 50% tissue culture infectious doses (TCID50s) of the P12 HCoV-NL63 stock were determined by monitoring for cytopathic effects of serial dilutions of the virus applied to LLC-MK2 cells. Six days postinfection, the supernatant was removed, and the cells were washed two times with phosphate-buffered saline (PBS) and stained with 2 ml of crystal violet working solution for 1 h. The crystal violet working solution was made from a stock solution (10% formaldehyde, 1.3 g crystal violet dissolved in 50 ml methanol, in a final volume of 1 liter with distilled water) diluted 1:1 in PBS. After the staining, the cells were washed twice with PBS and allowed to air dry. The wells were inspected for virus-induced cytopathic effects, and the TCID50 for the P12 stock was calculated to be 0.2/ml (47).
Generation of HCoV-NL63 antireplicase sera.
Two regions (R3, amino acids [aa] 899 to 998, and R4, aa 2830 to 2939) were selected and were used successfully for the development of antireplicase antisera (see Fig. 1). The HCoV-NL63 RNA was extracted from virus-infected LLC-MK2 cells at 96 hpi using RNeasy mini kits (QIAGEN) according to the manufacturer's instructions. The designated regions were generated by reverse transcription-PCR (RT-PCR) with an Advantage cDNA PCR kit (Clontech Laboratories, Inc.) from HCoV-NL63 RNA by using primers listed in Table 1, according to the manufacturer's instructions. The PCR products were digested with the appropriate restriction enzymes, ligated in frame with glutathione sulfur transferase (GST) in the pGEX-5x-1 vector (Amersham Pharmacia Biotech), and transformed into Escherichia coli. The GST-R3 and GST-R4 fusion proteins were induced, purified, and injected into rabbits for the generation of polyclonal antibodies as previously described (22, 28).
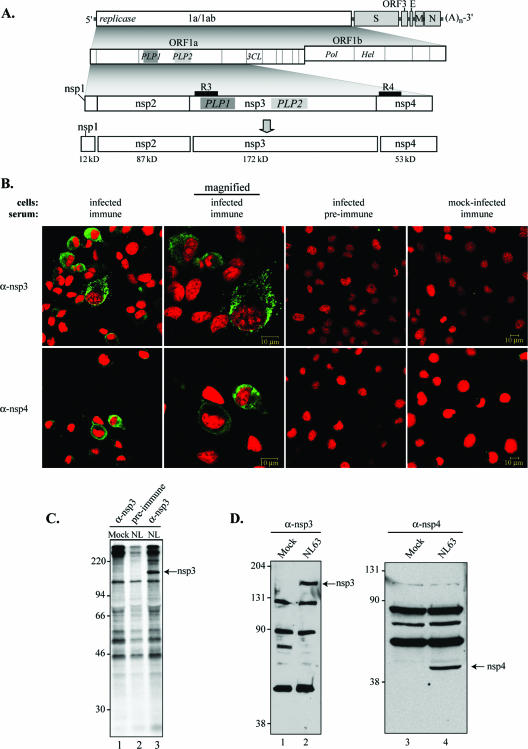
FIG. 1.
Detection of replicase products from HCoV-NL63-infected cells. (A) Schematic diagram illustrating HCoV-NL63 ORFs, the predicted processing of replicase polyprotein to nsp's, and regions used to generate anti-R3 (R3) and anti-R4 (R4) sera. The PLP domain (PLP1 and PLP2) and picornavirus 3C-like protease domain (3CLpro) are indicated. (B) Indirect immunofluorescence assay for HCoV-NL63 nsp3 and nsp4. LLC-MK2 cells were infected with HCoV-NL63, fixed, permeabilized at 24 hpi, and stained with anti-R3 or anti-R4 sera, anti-rabbit Ig Alexa-Fluor-488 secondary antibody, and Sytox orange to stain nuclei as described in Materials and Methods. (C) Detection of nsp3 by immunoprecipitation. HCoV-NL63 (NL)- or mock (M)-infected cells were radiolabeled with [35S]methionine for 24 h from 72 to 96 hpi. Cells were harvested, and the cell lysates were subjected to immunoprecipitation with anti-R3 sera or preimmune sera. Products were analyzed by 10% SDS-PAGE and subjected to autoradiography. (D) Detection of nsp3 and nsp4 by a Western blot assay with anti-R3 and anti-R4 antibodies. LLC-MK2 cells infected with HCoV-NL63 were lysed at 96 hpi, and whole-cell lysates were separated by SDS-PAGE. Immunoblotting was performed using anti-R3 and anti-R4 antibodies. Molecular mass markers (in kDa) are shown on the left of each gel.
TABLE 1.
Primers used for amplification or mutagenesis of HCoV-NL63 sequences
| Purpose | Primer | Oligonucleotide sequencea | Nucleotide no.b | Polarity or mutation |
|---|---|---|---|---|
| Generation of GST-R3 fusion protein | R3-1 | 5′ CCGGAA TTC GGT AAA ATA TCT TTT TCT GAT GAT G 3′ (EcoRI) | 2981-3005 | Forward |
| R3-2 | 5′ CCGCTC GAG ACC ATT ACT ATC ATT ACT AAT AGG 3′ (XhoI) | 3257-3280 | Reverse | |
| Generation of GST-R4 fusion protein | R4-1 | 5′ CCGGAA TTC GAG CTT TTA CCT AAT GTT TTT AAG 3′ (EcoRI) | 8774-8797 | Forward |
| R4-2 | 5′ CCG CTC GAG TTG TAA GGT GGA ATT GTA GCT AAT AG 3′ (XhoI) | 9078-9103 | Reverse | |
| Expression of six-His-tagged PLP2 in E. coli | ZC1 | 5′ CGC GGA TCCGGT TGT AGA GAG TAA TGT TAT GG 3′ | 4979-5000 | Forward |
| ZC2 | 5′ CTA TTG CTC AGC ACC AGT ATC AAG TTT ATC CAT AAC 3′ | 5950-5968 | Reverse | |
| Cloning of pNL-1 | ZC3 | 5′ CCG GAA TTC ACC ATG GCT GTT GCA AGT GAT TCG 3′ (EcoRI) | 311-328 | Forward |
| ZC4 | 5′ AAG CTC GAGCGA CAG TCG TTA ACA TCC ATA ACA TTA C 3′ (XhoI) | 4989-5014 | Reverse | |
| Amplification of PLP2 for cloning into pNL-1 to make pNL-2 | ZC7 | 5′ C CGC TCG AGT TTT AAG AAT GAT AAT GTA GTT TTG 3′ (XhoI) | 5015-5038 | Forward |
| ZC8 | 5′ CG CGG GCC CTC ACT TTT AGA ACA AGC TAC ACA GTC 3′ (ApaI) | 6689-6712 | Reverse | |
| Amplification of nsp123 to make pNL3 by addition of PLP2 to C terminus | ZC5 | 5′ CCG GAA TTC ACC ATG GCT GTT GCA AGT GAT TCG 3′ (EcoRI) | 311-328 | Forward |
| ZC6 | 5′ AA CT CGA GCG TTG CAA CTG TAC AAG TGT GG 3′ (XhoI) | 3483-3502 | Reverse | |
| Cloning of 5′ Flag | ZC9 | 5′ CGG GGT ACC ATG GAC TAC AAA GAC CAT GAC GGT G 3′ (KpnI) | Forward | |
| ZC10 | 5′ CCG GAA TTC CAT ATT CAT ATT CAT CAT CAT GGC CGC AAG CTT GTC ATC GTC ATC 3′ (EcoRI) | Reverse | ||
| Cloning of pNL-4 | ZC11 | 5′ CCG GAA TTC ACC ATG GAT ATT TCA CAC CTT GTA AAC TGT G 3′ (EcoRI) | 4100-4124 | Forward |
| ZC12 | 5′ AAG CTC GAG CGT TGT AAG GTG GAA TTG TAG CTA ATA G 3′ (XhoI) | 9078-9103 | Reverse | |
| Cloning of PLP2 | ZC13 | 5′ C CGG AAT TC ACC ATG GTT GTA GAG AGT AAT GTT ATG 3′ (EcoRI) | 4979-4999 | Forward |
| ZC14 | 5′ CC GCT CGA GCG ACC AGT ATC AAG TTT ATC C 3′ (XhoI) | 5950-5968 | Reverse | |
| Cloning of pNL-5 | ZC15 | 5′ C CGC TCG AGT GCT AGA CTT AAA CGT GTA CCA CTT C 3′ (XhoI) | 6713-6737 | Forward |
| ZC16 | 5′ CG CGG GCC CTC TTG TAA GGT GGA ATT GTA GCT AAT AG 3′ (ApaI) | 9078-9103 | Reverse | |
| Cloning of pNL-6 | ZC17 | 5′ C CGC TCG AGT ACA ACT AAA GCA AAG GGT TTG AC 3′ (XhoI) | 7574-7596 | Forward |
| ZC18 | 5′ CG CGG GCC CTC TTG TAA GGT GGA ATT GTA GCT AAT AG 3′ (ApaI) | 9078-9103 | Reverse | |
| Site-directed mutagenesis | ||||
| PLP1 C1062A | ZC19 | 5′ ATA ATG CTT GGA TTA GTA CCA CAC TTG TAC AGT TG 3′ | 3465-3499 | C1062A |
| ZC20 | 5′ TCC AAG CAT TAT TAT CAG ATT GAT CTA AAA CAC G 3′ | 3443-3476 | ||
| PLP2 C1678A | ZC21 | 5′ CCA CCG ACA ATA ATGCTT GGG TTA ATG CAA C 3′ | 5304-5334 | C1678A |
| ZC22 | 5′ GTT GCA TTA ACC CAAGCA TTA TTG TCG GTG G 3′ | 5304-5334 | ||
| PLP2 C1684A | ZC23 | 5′ GTT GGG TTA ATG CAA CTGCTA TAA TTT TAC AGT ATC 3′ | 5319-5354 | C1684A |
| ZC24 | 5′ GAT ACT GTA AAA TTA TAGCAG TTG CAT TAA CCC AAC 3′ | 5319-5354 | ||
| PLP2 H1836A | ZC25 | 5′ GGT TCT TTT GAT AAC GGT GCC TAT GTA GTT TAT GAT GC 3′ | 5774-5811 | H1836A |
| ZC26 | 5′ GCA TCA TAA ACT ACA TAGGCA CCG TTA TCA AAA GAA CC 3′ | 5774-5811 | ||
| CS3 G2461A | ZC27 | 5′ GTA TAG TTG CAA AAC AGGCTG CTG GTT TTA AAC GTA C 3′ | 7650-7686 | G2461A |
| ZC28 | 5′ GTA CGT TTA AAA CCA GCAGCC TGT TTT GCA ACT ATA C 3′ | 7650-7686 | ||
| CS3 A2462N | ZC29 | 5′ GTT GCA AAA CAG GGT AAC GGT TTT AAA CGT ACT 3′ | 7655-7687 | A2462N |
| ZC30 | 5′ AGT ACG TTT AAA ACC GTT ACC CTG TTT TGC AAC 3′ | 7655-7687 | ||
| CS3 LXGG | ZC31 | 5′ TT GCA CTA CAG GGT GGT GGT TTT AAA CGT ACT TAT AA 3′ | 7656-7692 | LXGG |
| ZC32 | 5′ A ACC ACC ACC CTG TAG TGC AAC TAT ACT AGT AG 3′ | 7644-7676 | ||
| CS2 L896A | ZC33 | 5′ ACA AAA GCA GCT GGT GGT AAA ATA TCT TTT TCT GAT G 3′ | 2966-3002 | L896A |
| ZC34 | 5′ ACC AGC TGC TTT TGT AAA AGC AAC AGG CAA TAC ACC 3′ | 2945-2980 | ||
| CS2 A897N | ZC35 | 5′ AAA TTA AAT GGT GGT AAA ATA TCT TTT TCT GAT GAT G 3′ | 2969-3005 | A897N |
| ZC36 | 5′ ACC ACC ATT TAA TTT TGT AAA AGC AAC AGG CAA TAC 3′ | 2948-2983 | ||
| CS2 G898A | ZC37 | 5′ TTA GCT GCT GGT AAA ATA TCT TTT TCT GAT GAT GTT 3′ | 2972-3007 | G898A |
| ZC38 | 5′ TTT ACC AGC AGC TAA TTT TGT AAA AGC AAC AGG CAA 3′ | 2951-2986 | ||
| CS2 G899A | ZC39 | 5′ GCT GGT GCT AAA ATA TCT TTT TCT GAT GAT GTT ATA G 3′ | 2975-3011 | G899A |
| ZC40 | 5′ TAT TTT AGC ACC AGC TAA TTT TGT AAA AGC AAC AGG 3′ | 2954-2989 | ||
| CS1 A109N | ZC41 | 5′ CAT GGT AAC GGA AGT GTG GTT TTT GTG GAT AAG TAC 3′ | 605-640 | A109N |
| ZC42 | 5′ ACT TCC GTT ACC ATG GCC AAA AAC AAC ATC AAA GTC 3′ | 584-619 | ||
| CS1 G110A | ZC43 | 5′ GGT GCA GCA AGT GTG GTT TTT GTG GAT AAG TAC ATG 3′ | 608-643 | G110A |
| ZC44 | 5′ CAC ACT TGC TGC ACC ATG GCC AAA AAC AAC ATC AAA G 3′ | 586-622 |
Underlined nucleotides were added for cloning purposes or mutated sequences. Restriction enzymes are indicated in parentheses.
All nucleotide numbers are based on the NL63 genome sequence (GenBank accession no. NC_005831).
Radioimmunoprecipitation and Western blot assays.
LLC-MK2 cells (∼5 × 106 cells per 60-mm2 dish) were infected with 2 ml of HCoV-NL63 P12 virus (∼0.2 TCID50/ml) and incubated at 33°C. The medium was removed at 72 hpi, the cells were washed once with PBS, and methionine-free Dulbecco's modified Eagle's medium (Invitrogen) with actinomycin (5 μg/ml) was added to the cells for 1 h. Then, 100 μCi of 35S-labeled methionine (MP Biomedicines, Inc.) was added to each dish and incubated at 33°C for 24 h. At 96 hpi, the radiolabel was removed, and the cells were washed three times with PBS. Whole-cell lysates were prepared by the addition of 250 μl of lysis buffer A (4% sodium dodecyl sulfate [SDS], 3% dithiothreitol [DTT], 40% glycerol, 0.065 M Tris-HCl [pH 6.8]). The cells were then scraped together with a rubber policeman, and the lysate was passed through a 25-gauge needle to shear the cellular DNA. The lysates were either used directly for immunoprecipitation or stored at −80°C for further experiments.
To immunoprecipitate radiolabeled protein, 100 μl of the cell lysate (with equal cell equivalents) was diluted in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (0.5% Triton X-100, 0.1% SDS, 300 mM NaCl, 4 mM EDTA, and 50 mM Tris-HCl, pH 7.4). Five microliters of the preimmune rabbit serum or the designated antiserum and 40 μl of protein A-Sepharose beads (Amersham Pharmacia Biotech AB, Uppsala, Sweden) were added to the above-described sample mixture and then rocked overnight at 4°C. The beads were pelleted for 1 min at 12,000 rpm in a bench-top microcentrifuge, the supernatant was removed, and the beads were washed three times with 1 ml of RIPA buffer. The beads were again pelleted, and 50 μl of 2× sample buffer was added to each sample and incubated for 30 min at 37°C. The immunoprecipitated protein was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were then fixed for 30 min in 25% methanol-10% acetic acid in water, treated with Amplify (Amersham Biosciences) for 30 min, dried, and exposed to X-ray film for 1 to 5 days.
For the Western blot assay, HCoV-NL63-infected LLC-MK2 cells were harvested by adding 250 μl of lysis buffer A at 96 hpi. The cell lysate was mixed 1:1 with 2× sample buffer and incubated for 30 min at 37°C. The cell lysate was separated on an SDS-PAGE gel, followed by a transfer to a polyvinylidene fluoride membrane in transfer buffer (3 liters containing 9.09 g Tris base, 43.28 g glycine, and 600 ml methanol) for 2 h at 4°C. The membrane was blocked using 5% dried skim milk in Tris-buffered saline (TBS) (0.9% NaCl, 10 mM Tris-HCl [pH 7.5]) plus 0.1% Tween 20 (TBST) for 2 h at room temperature. The blot was probed with the designated antibody by incubation overnight at 4°C. The membrane was washed in TBST three times for 20 min each. Following the washes, the membrane was incubated with peroxidase-conjugated secondary antibody (Amersham) at a dilution of 1:10,000 for 2 h at room temperature. The membrane was then washed three times with TBST, and bound antibody was detected with Western Lightning Chemiluminescence Reagent Plus (PerkinElmer LAS Inc.).
Immunofluorescence staining.
LLC-MK2 cells (∼4 × 104 cells per chamber) were plated onto an eight-chamber culture slide (Nalge Nunc International, Rochester, NY). When the cells were approximately 80 to 90% confluent, they were infected with 200 μl HCoV-NL63 P12 virus stock. At 24 hpi, the cells were fixed, permeabilized, and stained with the indicated antibody. For fixation and permeabilization of the cells, the medium was removed, and the slides were washed twice with PBS, fixed for 30 min with 3.7% paraformaldehyde, washed three times with PBS containing 10 mM glycine, and permeabilized for 10 min in PBS containing 0.1% Triton X-100. To block nonspecific binding, the fixed cells were treated with blocking solution (5% fetal bovine serum in PBS) for 30 min at room temperature. After removal of the blocking solution, 100 μl of anti-R3 or anti-R4 serum (1:2,000 dilution in blocking solution) was added to each well and incubated for 2 h at room temperature in a humidified chamber. The cells were then washed three times for 1 min with blocking solution. Anti-rabbit immunoglobulin (Ig; heavy and light chain) Alexa Fluor-488 secondary antibody (Molecular Probes) was diluted 1:1,000 in blocking solution, and 100 μl was added per well and incubated in the dark for 1 h at room temperature in a humidified chamber. To detect nuclear DNA, cells were stained with Sytox orange (Molecular Probes) at a 1:80,000 dilution for 10 min at room temperature. The slides were washed four times with PBS and mounted with DAKO fluorescent mounting medium (DAKO Corporation, Carpinteria, CA). Immunofluorescence staining was visualized using a Zeiss LSM-510 confocal microscope at the Loyola University Medical Center Core Imaging Facility.
Generating HCoV-NL63 PLP constructs and site-directed mutants.
Constructs expressing the HCoV-NL63 replicase-coding regions were generated using specific primers (Table 1) to RT-PCR amplify the designated regions from RNA isolated from HCoV-NL63-infected cells. RT-PCRs were performed using an Advantage cDNA PCR kit (Clontech Laboratories, Inc.) according to the manufacturer's protocol. The amplified products were then digested with the appropriate restriction enzymes and ligated into the corresponding sites in the pcDNA3.1/V5-HisB expression vector (Stratagene, La Jolla, CA). For some constructs, a 5′ Flag tag sequence was generated by PCR and cloned into pcDNA3.1/V5-HisB, and the replicase-coding regions were then cloned into the vector downstream of 5′ Flag to obtain the amino-terminal Flag-tagged construct. The ligated DNA was transformed into XL1-Blue competent cells (Stratagene) according to the manufacturer's instructions, with the exception that bacteria were grown at 25°C. To generate specific mutations in the catalytic domain or substrate cleavage sites, mutagenic primers (Table 1) were incorporated into newly synthesized DNA by using the QuikChange site-directed mutagenesis protocol (Stratagene, La Jolla, CA) as previously described (4, 22). A number of the mutagenic primers were designed according to modified methods (69). All introduced mutations were confirmed by DNA sequencing.
For purposes of overexpressing and purifying protein, the DNA region encoding the core domain of HCoV-NL63 PLP2, polyprotein residues 1565 to 1894, was PCR amplified using primers listed in Table 1 and cloned into pET15b (Novagen) between the BamHI and Bpu1102I sites. Site-directed mutagenesis was used to generate a PLP2-C1678A catalytic site mutant using primers listed in Table 1. The final clones were verified by DNA sequencing and designated pET-NL63-PLP2(aa 1565 to 1894) and pET-NL63-PLP2-C1678A.
HCoV-NL63 PLP cis-cleavage assays.
HCoV-NL63 replicase-coding regions cloned into pcDNA3.1/V5-HisB (Invitrogen) under T7 promoter control were transfected into HeLa-mouse hepatitis virus receptor (MHVR) cells infected with recombinant vaccinia virus expressing bacteriophage T7 polymerase, as previously described (4, 22, 28). Briefly, newly synthesized proteins were metabolically labeled with 50 μCi/ml Tran 35S label (MP Biomedicines, Inc.) from 4.5 to 9.5 hpi. Cells were washed three times in PBS, and cell lysates were prepared by scraping the cells in 250 μl lysis buffer A. The cell lysate (50 to 100 μl) was diluted in 1.0 ml RIPA buffer and subjected to immunoprecipitation with anti-V5 antibody (Invitrogen, Carlsbad, CA) or anti-Flag M2 antibody (Sigma) and protein-A Sepharose beads (Amersham Biosciences, Piscataway, NJ). The immunoprecipitated products were separated by electrophoresis on a 7.5 to 10% polyacrylamide gel containing 0.1% SDS. Eight to 16% Criterion gels (Bio-Rad) were used for analysis of anti-Flag M2 immunoprecipitated products. Following electrophoresis, the gels were fixed for 30 min in 25% methanol-10% acetic acid, enhanced for 30 min with Amplify (Amersham Biosciences), dried, and then exposed to Kodak X-ray film.
Purification of HCoV-NL63 PLP2.
One liter of E. coli BL21(DE3) cells containing wild-type pET15b-NL63-PLP2(1565-1894) or the PLP2-C1678A mutant was grown for 24 h at 25°C. Cells were pelleted by centrifugation and resuspended in 30 ml of buffer A (20 mM Tris [pH 7.5], 500 mM NaCl, 10 mM imidazole) containing lysozyme (0.5 mg/ml). The cells were incubated on ice for 10 min and then lysed via sonication using a 600-watt Model VCX ultrasonicator. The cell debris was pelleted by centrifugation (40,900 × g for 30 min), and the clarified cell lysate was loaded onto a 5-ml Co2+-charged HiTrap column (GE Healthcare) equilibrated with buffer A. Protein was eluted with a 20× column volume gradient from 100% buffer A to 100% buffer B (20 mM Tris [pH 7.5], 500 mM NaCl, 200 mM imidazole). Fractions containing PLP2 were pooled, concentrated, and exchanged into buffer C (20 mM Tris [pH 7.5], 10 mM DTT) and loaded onto a Mono Q 10/10 column (GE Healthcare) equilibrated in buffer C. A 10× column volume gradient from 100% buffer C to 100% buffer D (20 mM Tris [pH 7.5], 250 mM NaCl, 10 mM DTT) was used to elute purified PLP2, which was then exchanged into buffer C containing 20% glycerol, concentrated to approximately 8 mg/ml, flash frozen in dry-ice ethanol, and stored at −80°C.
Reverse-phase HPLC and MALDI-TOF MS.
Twelve-mer peptides representing cleavage site 1 (CS1; FGHGAGSVVFVD), cleavage site 2 (CS2; FTKLAGGKISFS), and cleavage site 3 (CS3; VAKQGAGFKRTY) were synthesized by Sigma Genosys (The Woodlands, TX). To test PLP2 cleavage of the peptides, 2 to 15 μM of purified PLP2 was incubated with 0.2 to 1 mM peptide in 20 mM Tris, pH 7.5, at room temperature for 16 to 48 h. Following the incubation and prior to high-performance liquid chromatography (HPLC) analysis, PLP2 was removed from the reaction using Microcon YM-10 centrifugal filter devices (Millipore). The reaction products were then diluted with an equal volume of 0.1% trifluoroacetic acid and analyzed on an Agilent Technologies 1200 HPLC system with a Zorbax Eclipse XDB-C18 column (4.6 by 150 mm) using a 1 to 40% linear gradient of acetonitrile containing 0.085% trifluoroacetic acid. Eluted peaks, monitored at 215 nm, were collected and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) using an Applied Biosystems Voyager DE-PRO MALDI-TOF MS, with α-cyano-hydroxycinnamic acid as the matrix component, to determine the masses of the cleaved fragments and hence identify the precise cleavage sites.
DUB activity assays.
Proteolytic cleavage of homogeneous K48-Ub6 (Boston Biochem, Cambridge, MA) was carried out under the following conditions. Purified HCoV-NL63 PLP2 (0.01 μg) was incubated with 2.5 μg of K48-Ub6 at 25°C in a 20-μl volume containing 50 mM HEPES (pH 7.5), 0.1 mg/ml bovine serum albumin, 100 mM NaCl, and 2 mM DTT. A control reaction mixture was incubated under identical conditions with the exclusion of PLP2. At the 5- and 60-min time points, the reactions were stopped with the addition of SDS-PAGE sample loading dye to a 1× concentration (25 mM Tris [pH 6.8], 280 mM β-mercaptoethanol, 4% glycerol, 0.8% SDS, 0.02% bromophenol blue) and heat treated at 95°C for 5 min. The samples were analyzed by electrophoresis on a 15% SDS-PAGE gel and stained with Coomassie blue dye.
DUB activity assays were also performed by the suicide substrate probe specific for DUBs as described previously (30). Briefly, 32.5 ng of purified wild-type PLP2 or mutant PLP2-C1678A protein was incubated with 1 μl (0.01 to 0.02 mg/ml) of DUB probe (hemagglutinin [HA]-tagged Ub-VS [HA-Ub-VS]) in a total volume of 20 μl of homogenization buffer (50 mM Tris [pH 7.5], 5 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 2 mM ATP, and 250 mM sucrose). Reaction mixtures were incubated at 37°C for 10 min and then diluted with an equal volume of 2× sample buffer and further incubated at 37°C for 30 min. The samples were separated by SDS-PAGE and then transferred to a polyvinylidene fluoride membrane. The membrane was immunoblotted with the mouse monoclonal anti-HA antibody HA probe (F-7) (SC-7392; Santa Cruz Biotechnology Inc.) at a 1:250 dilution, followed by goat anti-mouse IgG-horseradish peroxidase at a 1:10,000 dilution. Blots were developed using Western Lightning Chemiluminescence Reagent Plus as recommended by the manufacturer (PerkinElmer LAS Inc.).
RESULTS
Identification of HCoV-NL63 replicase products.
HCoV-NL63, a member of the group I CoV with sequence homology to HCoV-229E, is predicted to encode replicase polyproteins that are processed to produce 16 nsp's by three distinct proteases, PLP1, PLP2, and 3CLpro. For a first step toward characterizing these proteases, we focused on the identification of the amino-terminal products and characterization of the activity of the PLP domains.
To identify replicase products processed by PLPs of HCoV-NL63, we generated rabbit polyclonal anti-R3 and anti-R4 antibodies to predicted amino-terminal cleavage products nsp3 and nsp4 (Fig. 1A and Table 1) as described in Materials and Methods and used the antibodies in immunofluorescence, immunoprecipitation, and Western blot assays. HCoV-NL63 replicase products were detected at 24 hpi (Fig. 1B) with both anti-nsp3 and anti-nsp4 sera and localized exclusively in the cytoplasm of infected cells in a perinuclear pattern. These results are consistent with previous studies that show that the amino-terminal replicase products of MHV and SARS-CoV assemble as part of the membrane-associated viral replication complex (10, 20, 22, 43, 53, 54). Future studies will employ these antibodies to further investigate the subcellular localization and assembly of the replicase complex by using colocalization with cellular markers and immunoelectron microscopy.
We also used the anti-R3 and anti-R4 antisera to detect HCoV-NL63 replicase products by immunoprecipitation and Western blot analysis (Fig. 1C and D). We identified nsp3 as an ∼172-kDa protein by immunoprecipitation of radiolabeled, infected cell lysates (Fig. 1C, lane 3). No specific products were immunoprecipitated from mock-infected cells with anti-R3 serum (Fig. 1C, lane 1) or from HCoV-NL63-infected cells with preimmune serum (Fig. 1C, lane 2). Replicase products nsp3 and nsp4 were also detected using Western blotting of HCoV-NL63-infected cell lysates prepared at 96 hpi (Fig. 1D). We noted that nsp4 migrates faster than expected (∼45 kDa) for a predicted mass of 53 kDa. However, the nsp4 protein from MHV-infected cells is predicted to have a molecular mass of 56.5 kDa, including four putative membrane-spanning domains, but it migrates as a 44-kDa protein in SDS-PAGE (29). Overall, we show that the anti-R3 and anti-R4 antibodies recognize specific replicase products generated in HCoV-NL63-infected cells.
HCoV-NL63 PLP1 processes CS1, and PLP2 processes CS2.
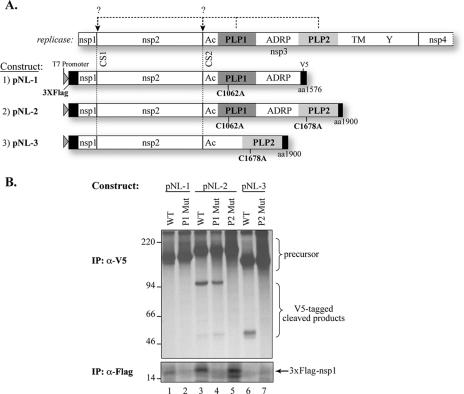
The majority of CoV, including MHV and HCoV-229E, contain two PLPs (termed either PLP1 and PLP2 or PL1pro and PL2pro). Previous studies showed that MHV PLP1 is required for processing CS1 and CS2, whereas PLP2 is responsible for processing CS3 (7, 8, 14, 28, 59). In HCoV-229E, PLP1 is responsible for processing CS1, and both PLP1 and PLP2 are required for efficient processing of CS2 (71). To determine which HCoV-NL63 PLP is responsible for processing CS1 and CS2, three DNA constructs, pNL-1, pNL-2, and pNL-3, were generated as described in Materials and Methods and are diagramed in Fig. 2A. cis-Cleavage assays were performed by transfection of DNA constructs into HeLa cells infected with vaccinia virus expressing T7 polymerase. Newly synthesized proteins radiolabeled with [35S]methionine were subjected to immunoprecipitation with anti-Flag M2 antibody or anti-V5 antibody (Fig. 2B). The results show that when the wild-type PLP1 domain is present, the 15-kDa Flag-tagged nsp1 protein is generated (Fig. 2B, lanes 1, 3, and 5). In contrast, when catalytic cysteine 1062 of PLP1 is changed to alanine or if the PLP1 domain is deleted, no nsp1 is produced (Fig. 2B, lanes 2, 4, 6, and 7). These results show that PLP1 is responsible for processing CS1 to release nsp1. Next, we asked which protease domain is responsible for processing CS2. Cells were transfected with either wild-type construct 2 (pNL-2) or construct 2 encoding a cysteine-to-alanine change at the catalytic site of PLP1 (C1062A) or PLP2 (C1678A). We found that only constructs with wild-type PLP2 were capable of efficiently processing CS2 to generate V5-tagged cleavage products (Fig. 2B, lanes 3, 4, and 6). We also wanted to determine if PLP1 played any assisting role in processing CS2, as has been proposed for PLP2 of HCoV-229E (71). Therefore, we generated pNL-3 with a deletion of the PLP1 domain and found that this construct was sufficient for processing CS2 (Fig. 2B, lane 6), whereas a construct with a mutation in the catalytic cysteine residue of PLP2 was unable to process the polyprotein at CS2 (Fig. 2B, lane 7). Overall, these data show that HCoV-NL63 PLP1 processes CS1 and that PLP2 processes CS2 without assistance from the PLP1 domain.
FIG. 2.
Identification of proteases that process the amino-terminal region of the HCoV-NL63 replicase polyprotein. (A) Schematic representation of predicted processing at CS1 and CS2 by PLP1 and PLP2 (top panel) and constructs tested in the cis-cleavage assay. nsp3 conserved domains: Ac, acidic domain; ADRP, ADP-ribose-1′′-phosphatase (49); TM, transmembrane domain; Y, Y domain (70). The proposed catalytic residues (C1602 for PLP1 and C1678 for PLP2) are indicated. (B) cis-Cleavage assay for CS1 and CS2. Constructs were transfected into HeLa-MHVR cells infected with vTF7.3. Newly synthesized proteins were labeled with [35S]methionine from 4.5 to 9.5 hpi. Lysates were prepared and subjected to immunoprecipitation (IP) with anti-V5 (top) or anti-Flag M2 (bottom) antibody. Immunoprecipitated proteins were separated by 10% SDS-PAGE or 8 to 16% Criterion gel and subjected to autoradiography. The precursors and cleaved products are indicated. Molecular mass markers (in kDa) are shown on the left of the gel.
HCoV-NL63 PLP2 is required for processing CS3.
To determine if HCoV-NL63 PLP2 is required for processing CS3, we cloned and expressed constructs pNL-4, pNL-5, and pNL-6 expressing the PLP2-nsp4 region (Fig. 3A). The plasmid DNAs were transfected into HeLa cells and analyzed for proteolytic processing activity as described above. We found that wild-type HCoV-NL63 PLP2 is able to process CS3 (Fig. 3B, lane 1) and release nsp4. We note that nsp4 is sometimes detected as two closely migrating bands, perhaps due to glycosylation of this putative transmembrane protein. To identify the catalytic residues required for PLP2 processing activity, predicted catalytic residues cysteine 1678 and histidine 1836, which were proposed based on sequence alignment analysis of HCoV-NL63 PLP2 with other CoV PLP domains (4), were changed to alanine and tested for activity (Fig. 3B). As expected, introduction of mutations C1678A and H1836A completely abolished PLP2 activity (Fig. 3B, lanes 2 and 4), compared with processing detected with the noncatalytic C1684A mutant. Thus, cysteine 1678 and histidine 1836 are likely two of the three members of the catalytic triad for this PLP. Structural studies will be needed to identify all amino acids involved in the PLP2 catalytic site, particularly the third residue in the catalytic triad of the active site.
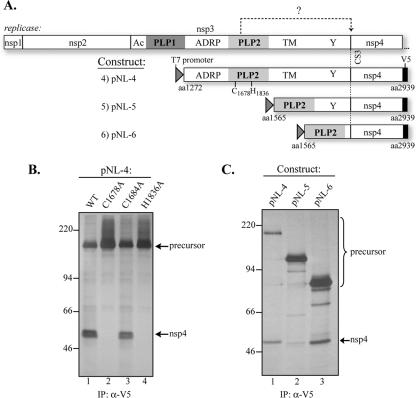
FIG. 3.
Processing of HCoV-NL63 replicase CS3 by PLP2. (A) Schematic diagram of the predicted processing at CS3 by PLP2 and the constructs used in the cis-cleavage assay. The PLP2 catalytic residues (C1678 and H1836) are indicated. (B) Detection of processing at CS3 by PLP2 and identification of PLP2 catalytic sites. cis-Cleavage assays were performed as described for Fig. 2. The processed product (nsp4) and the precursors are indicated. The precursors and processed products were detected with anti-V5 antibody from cells transfected with wild-type PLP2 (WT) (lane 1), the C1678A mutant (lane 2), the C1684A mutant (lane 3), or the H1836A mutant (lane 4). (C) Identification of HCoV-NL63 PLP2 core domain. The processing activities, as detected by immunoprecipitation (IP) of nsp4, from pNL-4 (lane 1), pNL-5 (lane 2), and pNL-6 (lane 3), are shown. Molecular mass markers (in kDa) are shown on the left of each gel.
To determine if a HCoV-NL63 PLP2 core domain is sufficient for processing CS3, we compared the processing activities of three forms of PLP2 (Fig. 3C). We found that a deletion of the transmembrane domain greatly reduced processing activity (Fig. 3C, lane 2) and that removing the Y domain resulted in an increase in PLP2 processing (Fig. 3C, lane 3), indicating that the core form of PLP2 (the PLP2 core domain, aa 1565 to 1900) is sufficient for processing CS3. Overall, these results indicated that PLP2 mediates processing at CS3 and that C1678 and H1836 are the likely catalytic residues required for PLP2 processing activity.
Identification of CS2 and CS3 cleavage sites processed by NL63 PLP2.
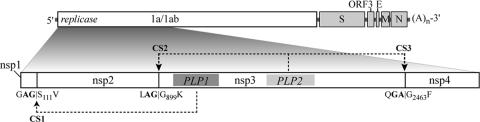
To identify the precise cleavage sites that are processed by HCoV-NL63 PLP2, we overexpressed and purified six-His-tagged PLP2 and analyzed the proteolytic activity of the purified enzyme with synthetic peptides representing the CS2 and CS3 sites. The catalytic core domain of PLP2 (polyprotein residues 1565 to 1894) was expressed in E. coli and purified to homogeneity (Fig. 4A) using a combination of affinity and ion-exchange chromatographic separations. Enzymatic cleavage of two synthetic 12-mer peptides, FTKLAGGKISFS (CS2) and VAKQGAGFKRTY (CS3), was monitored using HPLC separation of the generated peptide fragments (Fig. 4B and C). Under identical assay conditions, cleavage of CS2 was much more efficient than cleavage of CS3, with little or no CS3 product detected in the time required for complete cleavage of CS2. However, increasing the peptide and enzyme concentrations and extending the incubation time afforded detectable products from PLP2 cleavage of CS3. The composition of the cleaved products and the identification of the cleavage sites for both the CS2 and CS3 substrates were determined through MALDI-TOF mass spectral analysis. The cleavage sites were accurately identified as FTKLAG↓GKISFS for CS2 and VAKQGA↓GFKRTY for CS3. These results confirm prior bioinformatics predictions for these cleavage sites (56). Cleavage of the CS1 substrate by PLP2 was not detected under any reaction conditions attempted (data not shown).
FIG. 4.
Identification of CS2 and CS3 cleavage sites. (A) Purified wild-type HCoV-NL63 six-His-PLP2 (lane 2) and mutant PLP2-C1678A (lane 3) are shown resolved on a Coomassie-stained SDS-PAGE gel. Twelve micrograms of purified protein was loaded in each lane. The molecular masses of the marker proteins (lane 1) are shown to the left of the gel in kDa. The expected molecular mass of six-His-PLP2 is 38.6 kDa. HPLC separation of cleavage products from 12-mer peptides representing CS2 (FTKLAGGKISFS) (B) and CS3 (VAKQGAGFKRTY) (C) incubated in the presence (dashed lines) and absence (solid lines) of purified HCoV-NL63 PLP2. The identity of each cleavage product was confirmed by MS (data not shown), as indicated above the peaks.
Previous studies have shown that the P2 and P1 residues are highly conserved in the cleavage sites recognized by CoV PLPs (28) and that specific residues at the P1 and P2 positions were generally required for CoV PLP-mediated processing (4, 29, 36). To identify the critical determinants required for HCoV-NL63 PLP2 recognition and processing of CS2 and CS3, we generated mutants with substitutions of predicted cleavage site residues by site-directed mutagenesis and used a cis-cleavage assay to monitor the effect of each substitution on PLP2 processing. The products of each cis-cleavage assay were immunoprecipitated and resolved by SDS-PAGE as shown in Fig. 5B. For each assay, the presence of the cleavage product indicates that amino acid substitution in the cleavage site had little or no effect on processing, whereas the absence of the cleavage product indicates that amino acid substitution in the cleavage site resulted in a reduction in PLP2 recognition and processing. We found that introduction of mutations L896A (P3) and G899A (P1′) in CS2 had no effect on PLP2 recognition and processing (Fig. 5B, lane 2 and lane 5). However, substitution of alanine for glycine at P1 and substitution of asparagine for alanine at P2 completely abolished PLP2 recognition and processing (Fig. 5B, lane 3 and lane 4). To determine the role of the P1 and P2 residues in recognition and processing at CS3, we performed site-directed mutagenesis to change G2461 to A and A2462 to N and monitored processing activity. We found that the G2461A and A2462N mutants were not processed by PLP2 (Fig. 5C, lane 3 and lane 4), compared to the wild-type control (Fig. 5C, lane 1). These data indicate that the P1 and P2 sites are the critical determinants for recognition and processing by PLP2 at both CS2 and CS3. Further analysis of critical determinants recognized in the context of virus infection will be important to confirm and extend these in vitro studies.
FIG. 5.
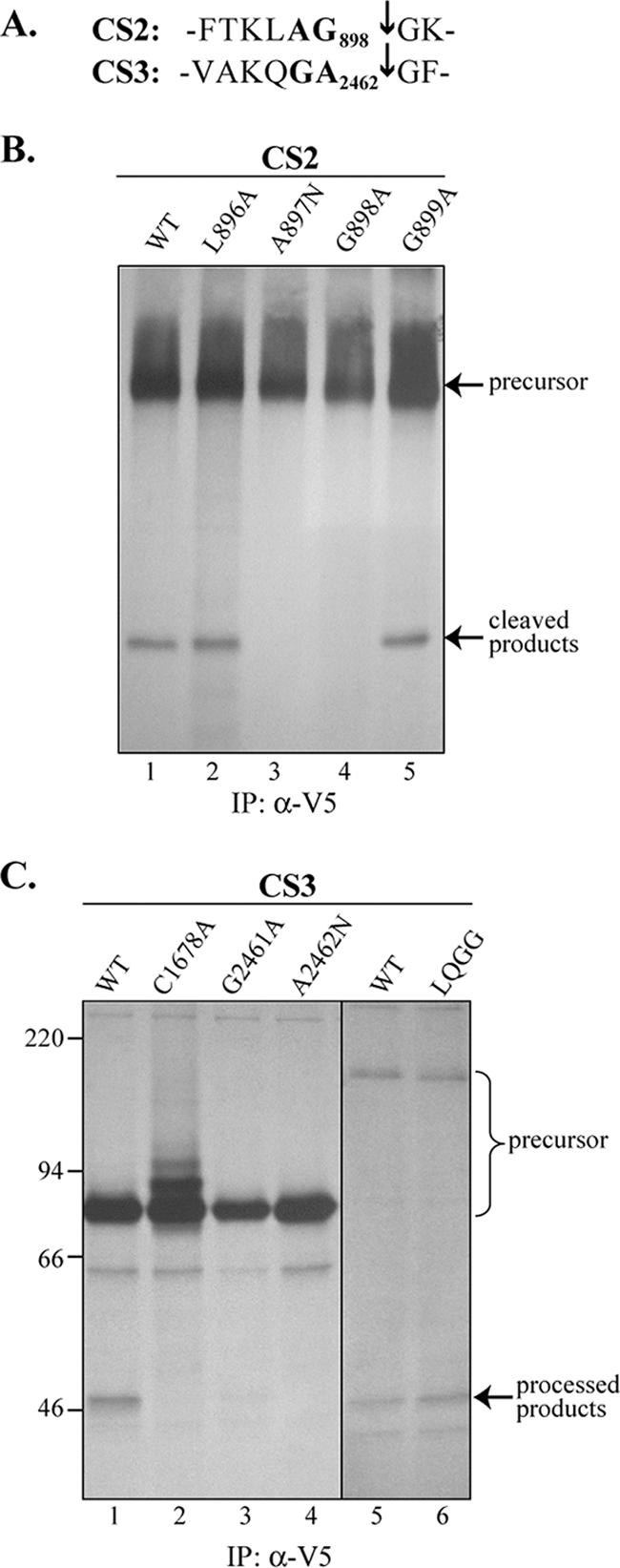
Effect of mutagenesis of critical determinants in CS2 and CS3 on PLP2 recognition and processing. (A) Cleavage site sequences of CS2 and CS3. The arrow indicates the cleavage site identified in the peptide-based cleavage assay. The P1 and P2 residues are highlighted. (B) P1 and P2 residues are critical for processing at CS2. pNL-3 constructs encoding the wild-type (WT) sequence or amino acid substitutions were tested for processing activity as described for Fig. 2. The site of substitution is indicated above each lane. (C) Critical determinants for processing at CS3. pNL-6 constructs (lanes 1 to 4) and pNL-4 constructs (lanes 5 and 6) were analyzed for processing at CS3. Specific substitutions are indicated above each lane. IP, immunoprecipitation.
Previous studies by our group with SARS-CoV PLP indicated that SARS-CoV PLpro recognizes and processes the consensus sequence LXGG at three cleavage sites in the replicase polyprotein to release processed products (22). To determine if HCoV-NL63 PLP2 is also capable of recognizing the LXGG sequence, we introduced LQGG into the CS3 site as VALQGGGF. We found that LQGG at CS3 was processed in the cis-cleavage assay (Fig. 5C, lane 6) to a level similar to that detected in the wild-type control (Fig. 5C, lane 5). This result suggests that besides the protease activity for proteolytic processing of replicase, it is possible that HCoV-NL63 PLP2 is also a viral DUB that can process at conserved LXGG recognition sites (see below).
HCoV-NL63 PLP2 has DUB activity.
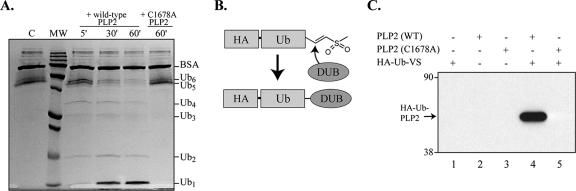
To determine if HCoV-NL63 PLP2 has DUB activity, purified wild-type HCoV-NL63 PLP2 and mutant PLP2-C1678A were tested for DUB activity. First, we tested the ability of PLP2 to disassemble a Ub6 chain. HCoV-NL63 PLP2 was incubated with K48-linked Ub6 for 5, 30, or 60 min, and the products of the reaction were separated by SDS-PAGE and visualized by Coomassie blue staining. We observed visible cleavage of the Ub6 substrate by HCoV-NL63 PLP2 after just 5 min of incubation (Fig. 6A, lane 3), demonstrating that PLP2 has DUB activity. To further confirm that HCoV-NL63 PLP2 recognizes Ub molecules, we used HA-Ub-VS (9) to probe the ability of Ub-VS to covalently modify HCoV-NL63 PLP2. The probe consists of an epitope-tagged, electrophilic Ub-VS derivative that allows covalent adduct formation with DUBs (Fig. 6B). The C-terminal electrophile specifically targets the active-site cysteine residues of DUBs, resulting in a covalent thioether linkage to the protease active site. An N-terminal HA epitope tag facilitates detection of any protease modified in this way. Purified PLP2 was incubated with HA-Ub-VS at 37°C for 10 min, and immunoblot analysis with anti-HA antibody was performed to detect any adduct formation between HA-Ub-VS and PLP2. We found that a prominent protein of approximately 48 kDa was detected using the anti-HA antibody (Fig. 6C, lane 4). These results indicated that HCoV-NL63 PLP2 recognizes Ub-VS and can be covalently modified. The exact function of Ub recognition and associated DUB activity of HCoV-NL63 PLP2 in virus replication and modulation of host cell biological events is currently unknown.
FIG. 6.
HCoV-NL63 PLP2 has DUB activity. (A) Processing of K48-Ub6 by PLP2. Lane 1, K48-Ub6 incubated in the absence of PLP2. Lane 2, a molecular-mass (MW) ladder with 250-, 150-, 100-, 75-, 50-, 37-, 25-, 20-, 15-, and 10-kDa markers. Lanes 3 to 5, K48-Ub6 incubated in the presence of wild-type (WT) PLP2 for the indicated time points. Lane 6, K48-Ub6 incubated in the presence of PLP2-C1678A for 60 min. The positions of bovine serum albumin and the various Ub multimers are indicated to the right of the gel. (B) Schematic diagram of the adduct formed between DUBs and HA-Ub-VS. (C) Western blot detection of HA-Ub-PLP2. Purified protein and substrate were incubated for 10 min at 37°C, and the products were separated by SDS-PAGE and analyzed by Western blot analysis using anti-HA antibody.
DISCUSSION
All CoV encode at least one PLP domain that processes the amino-terminal end of the viral replicase polyprotein. Some CoV, such as SARS-CoV, bat-SARS-CoV, and infectious bronchitis virus, encode one PLP domain, termed PLpro, which is responsible for processing cleavage sites upstream and downstream of the protease domain. Other CoV, such as MHV and HCoV-229E, encode two PLP domains, with PLP1 processing CS1, PLP2 processing CS3, and either PLP1 (for MHV) or PLP2 (for HCoV-229E) processing CS2. Here, we provide experimental evidence to show that HCoV-NL63 PLP1 processes CS1 and PLP2 processes CS2 and CS3, summarized in the model presented in Fig. 7. These results are consistent with other studies demonstrating that the P2 and P1 residues are critical determinants for PLP recognition and processing (4, 29, 36) and provide the basis for comparison of PLP activities for other CoV. Proteolytic processing of the replicase polyprotein is proposed to be essential for the proper assembly of the replication complex (20, 31). Furthermore, the kinetics of processing may be important since intermediates, such as nsp2-nsp3 and nsp4-nsp11, have been identified in CoV-infected cells (22, 28) and since the intermediates have been proposed to have an essential role in mediating RNA synthesis (28, 50). Indeed, protease inhibitors have been shown to block replication of CoV (18, 31, 67, 68), and therefore, proteases are attractive targets for development of antiviral drugs. Interestingly, Denison and colleagues, using CoV reverse genetics techniques, found that PLP1-mediated processing at MHV CS1 and CS2 is important for efficient viral replication (13, 21). Future studies will be aimed at determining if PLP2 activity and/or processing at CS3 is essential for CoV replication.
FIG. 7.
Model for processing the HCoV-NL63 replicase polyprotein by PLP1 and PLP2. PLP1 processing at CS1 and PLP2 processing at CS2 and CS3 are indicated by arrows. Critical determinants at the P1 and P2 positions of the cleavage sites are indicated in bold.
Perhaps the most intriguing result from this study is the demonstration of HCoV-NL63 PLP2 DUB activity. This is the second example of a CoV PLP domain with DUB activity. Initially, a bioinformatics study suggested that the SARS-CoV PLpro domain may mimic the folding of cellular DUBs and therefore mediate DUB activity (57). Experiments from two independent laboratories using purified SARS-CoV PLpro showed that PLpro recognized and processed K-48-linked polyubiquitin chains (4) and Ub-like moieties such as ISG15 (37). Furthermore, X-ray crystallographic studies revealed that SARS-CoV PLpro indeed conforms to the structure of known cellular DUBs such as USP14 and HAUSP (46). However, the role of this viral DUB activity during viral replication is not yet clear. The most obvious possibility is that viral DUB activity may target the Ub-proteasome pathway to facilitate virus replication and thwart host defense mechanisms, including innate immunity (52). Another possibility is that DUB activity may modulate posttranslational modification of proteins modified by Ub-like (Ubl) modifiers, such as SUMO, ISG15, or Nedd8. These modifiers have been identified as key players in directing subcellular localization, protein stability, and transcriptional activation (reviewed in references 23 and 65). The modification of proteins with Ub or Ub-like moieties is reversed by DUBs, and the human genome is predicted to encode almost 100 DUBs (40). Thus, the addition or removal of Ub and Ubl for specific proteins is important in many host cell pathways. Interestingly, several virus-encoded DUBs from a variety of virus families have been described recently. These DUBs include UL36 of herpesviruses such as heroes simplex virus type 1, Epstein-Barr virus, and mouse and human cytomegalovirus (30, 51, 64); the adenain protease of adenovirus (3); and now PLP domains from CoV (4, 37) (this report). Currently, the roles of these viral DUBs in viral replication and modification of host innate immunity are not clear. Bioinformatic studies predict that all known CoV will have at least one PLP domain with DUB activity (4, 56, 57). It is possible that CoV DUB activity may target several pathways in host cells. CoV DUBs may deconjugate K-48-linked polyubiquitin chains attached to viral replicase products or structural proteins to protect them from proteasome-mediated degradation. Alternatively, CoV DUBs may deconjugate ISG15 modifications from host proteins, thereby modulating host innate immunity and interferon-mediated antiviral responses (48). While the proteolytic processing activity of HCoV-NL63 PLPs is well established in this study, future work will be focused on determining the functions of DUB activity in CoV replication and pathogenesis.
In summary, we have (i) identified HCoV-NL63 replicase products nsp3 and nsp4 from virus-infected cells, (ii) showed that PLP1 processes the viral replicase polyprotein at CS1 and that PLP2 processes it at CS2 and CS3, (iii) identified peptide cleavage sites recognized by PLP2 and confirmed that the P2 and P1 sites are critical determinants for PLP2 processing, and (iv) showed that purified PLP2 has DUB activity. This characterization of HCoV-NL63 PLP activity will facilitate comparative studies of CoV protease activity and the development of antivirals targeting CoV PLPs.
Acknowledgments
We thank Oana Ciupuliga for assistance in the preparation of pNL-4 mutants and Linda M. Fox for assistance with the immunofluorescence assays. We also thank John Bechill and Naina Barretto and other members of the Baker laboratory for helpful suggestions for this work.
This research was supported by Public Health Service research grants R01-A1045798 (to S.C.B.) and P01-AI060915 (to S.C.B. and A.D.M.).
Footnotes
Published ahead of print on 28 March 2007.
REFERENCES
- 1.Almeida, J. D., and D. A. Tyrrell. 1967. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J. Gen. Virol. 1:175-178. [DOI] [PubMed] [Google Scholar]
- 2.Arden, K. E., M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2005. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J. Med. Virol. 75:455-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastien, N., K. Anderson, L. Hart, P. Van Caeseele, K. Brandt, D. Milley, T. Hatchette, E. C. Weiss, and Y. Li. 2005. Human coronavirus NL63 infection in Canada. J. Infect. Dis. 191:503-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastien, N., J. L. Robinson, A. Tse, B. E. Lee, L. Hart, and Y. Li. 2005. Human coronavirus NL-63 infections in children: a 1-year study. J. Clin. Microbiol. 43:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla, P. J., S. A. Hughes, J. D. Pinon, and S. R. Weiss. 1995. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology 209:489-497. [DOI] [PubMed] [Google Scholar]
- 8.Bonilla, P. J., S. A. Hughes, and S. R. Weiss. 1997. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 71:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borodovsky, A., H. Ovaa, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, H. L. Ploegh, and B. M. Kessler. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9:1149-1159. [DOI] [PubMed] [Google Scholar]
- 10.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu, S. S., K. H. Chan, K. W. Chu, S. W. Kwan, Y. Guan, L. L. Poon, and J. S. Peiris. 2005. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 40:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, E. H., H. J. Lee, S. J. Kim, B. W. Eun, N. H. Kim, J. A. Lee, J. H. Lee, E. K. Song, S. H. Kim, J. Y. Park, and J. Y. Sung. 2006. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin. Infect. Dis. 43:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denison, M. R., B. Yount, S. M. Brockway, R. L. Graham, A. C. Sims, X. Lu, and R. S. Baric. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denison, M. R., P. W. Zoltick, S. A. Hughes, B. Giangreco, A. L. Olson, S. Perlman, J. L. Leibowitz, and S. R. Weiss. 1992. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drosten, C., S. Gunther, W. Preiser, S. Van Der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 16.Ebihara, T., R. Endo, X. Ma, N. Ishiguro, and H. Kikuta. 2005. Detection of human coronavirus NL63 in young children with bronchiolitis. J. Med. Virol. 75:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh, A. K., K. Xi, K. Ratia, B. D. Santarsiero, W. Fu, B. H. Harcourt, P. A. Rota, S. C. Baker, M. E. Johnson, and A. D. Mesecar. 2005. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 48:6767-6771. [DOI] [PubMed] [Google Scholar]
- 19.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, R. L., and M. R. Denison. 2006. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J. Virol. 80:11610-11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, H., K. Pyrc, L. van der Hoek, M. Geier, B. Berkhout, and S. Pohlmann. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 102:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann, H., G. Simmons, A. J. Rennekamp, C. Chaipan, T. Gramberg, E. Heck, M. Geier, A. Wegele, A. Marzi, P. Bates, and S. Pohlmann. 2006. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80:8639-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes, K. V. 2001. Coronaviruses, vol. 1. Lippincott Williams and Wilkins, Philadelphia, PA.
- 27.Kaiser, L., N. Regamey, H. Roiha, C. Deffernez, and U. Frey. 2005. Human coronavirus NL63 associated with lower respiratory tract symptoms in early life. Pediatr. Infect. Dis. J. 24:1015-1017. [DOI] [PubMed] [Google Scholar]
- 28.Kanjanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 74:7911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanjanahaluethai, A., D. Jukneliene, and S. C. Baker. 2003. Identification of the murine coronavirus MP1 cleavage site recognized by papain-like proteinase 2. J. Virol. 77:7376-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J. C., R. A. Spence, P. F. Currier, X. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 33.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 35.Li, W., S. K. Wong, F. Li, J. H. Kuhn, I. C. Huang, H. Choe, and M. Farzan. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 80:4211-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim, K. P., L. F. Ng, and D. X. Liu. 2000. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 74:1674-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, D. X., and T. D. Brown. 1995. Characterisation and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology 209:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIntosh, K., J. H. Dees, W. B. Becker, A. Z. Kapikian, and R. M. Chanock. 1967. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA 57:933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773-786. [DOI] [PubMed] [Google Scholar]
- 41.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyrc, K., B. J. Bosch, B. Berkhout, M. F. Jebbink, R. Dijkman, P. Rottier, and L. van der Hoek. 2006. Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother. 50:2000-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyrc, K., M. F. Jebbink, B. Berkhout, and L. van der Hoek. 2004. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 48.Ritchie, K. J., C. S. Hahn, K. I. Kim, M. Yan, D. Rosario, L. Li, J. C. de la Torre, and D. E. Zhang. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10:1374-1378. [DOI] [PubMed] [Google Scholar]
- 49.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathogens 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shackelford, J., and J. S. Pagano. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41:139-156. [DOI] [PubMed] [Google Scholar]
- 53.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song, H. D., C. C. Tu, G. W. Zhang, S. Y. Wang, K. Zheng, L. C. Lei, Q. X. Chen, Y. W. Gao, H. Q. Zhou, H. Xiang, H. J. Zheng, S. W. Chern, F. Cheng, C. M. Pan, H. Xuan, S. J. Chen, H. M. Luo, D. H. Zhou, Y. F. Liu, J. F. He, P. Z. Qin, L. H. Li, Y. Q. Ren, W. J. Liang, Y. D. Yu, L. Anderson, M. Wang, R. H. Xu, X. W. Wu, H. Y. Zheng, J. D. Chen, G. Liang, Y. Gao, M. Liao, L. Fang, L. Y. Jiang, H. Li, F. Chen, B. Di, L. J. He, J. Y. Lin, S. Tong, X. Kong, L. Du, P. Hao, H. Tang, A. Bernini, X. J. Yu, O. Spiga, Z. M. Guo, H. Y. Pan, W. Z. He, J. C. Manuguerra, A. Fontanet, A. Danchin, N. Niccolai, Y. X. Li, C. I. Wu, and G. P. Zhao. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 102:2430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sulea, T., H. A. Lindner, E. O. Purisima, and R. Menard. 2006. Binding site-based classification of coronaviral papain-like proteases. Proteins 62:760-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sulea, T., H. A. Lindner, E. O. Purisima, and R. Menard. 2005. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 79:4550-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki, A., M. Okamoto, A. Ohmi, O. Watanabe, S. Miyabayashi, and H. Nishimura. 2005. Detection of human coronavirus-NL63 in children in Japan. Pediatr. Infect. Dis. J. 24:645-646. [DOI] [PubMed] [Google Scholar]
- 59.Teng, H., J. D. Pinon, and S. R. Weiss. 1999. Expression of murine coronavirus recombinant papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides. J. Virol. 73:2658-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 61.Vabret, A., T. Mourez, J. Dina, L. van der Hoek, S. Gouarin, J. Petitjean, J. Brouard, and F. Freymuth. 2005. Human coronavirus NL63, France. Emerg. Infect. Dis. 11:1225-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welchman, R. L., C. Gordon, and R. J. Mayer. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6:599-609. [DOI] [PubMed] [Google Scholar]
- 66.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, C. Y., J. T. Jan, S. H. Ma, C. J. Kuo, H. F. Juan, Y. S. Cheng, H. H. Hsu, H. C. Huang, D. Wu, A. Brik, F. S. Liang, R. S. Liu, J. M. Fang, S. T. Chen, P. H. Liang, and C. H. Wong. 2004. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 101:10012-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, H., W. Xie, X. Xue, K. Yang, J. Ma, W. Liang, Q. Zhao, Z. Zhou, D. Pei, J. Ziebuhr, R. Hilgenfeld, K. Y. Yuen, L. Wong, G. Gao, S. Chen, Z. Chen, D. Ma, M. Bartlam, and Z. Rao. 2005. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 3:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng, L., U. Baumann, and J. L. Reymond. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]