Abstract
By using a series of overlapping synthetic peptides covering 98% of the amino acid sequence of the nucleocapsid protein (NP) of severe acute respiratory syndrome coronavirus (SARS-CoV), four helper T-cell (Th) epitopes (NP11, residues 11 to 25; NP51, residues 51 to 65; NP61, residues 61 to 75; and NP111, residues 111 to 125) in C57BL mice (H-2b), four (NP21, residues 21 to 35; NP91, residues 91 to 105; NP331, residues 331 to 345; and NP351, residues 351 to 365) in C3H mice (H-2k), and two (NP81, residues 81 to 95; and NP351, residues 351 to 365) in BALB/c mice (H-2d) have been identified. All of these peptides were able to stimulate the proliferation of NP-specific T-cell lines or freshly isolated lymph node cells from mice immunized with recombinant NP. Immunization of mice with synthetic peptides containing appropriate Th epitopes elicited strong cellular immunity in vivo, as evidenced by delayed-type hypersensitivity. Priming with the helper peptides (e.g., NP111 and NP351) significantly accelerated the immune response induced by recombinant NP, as determined by the production of NP-specific antibodies. When fused with a conserved neutralizing epitope (SP1143-1157) from the spike protein of SARS-CoV, NP111 and NP351 assisted in the production of high-titer neutralizing antibodies in vivo. These data provide useful insights regarding immunity against SARS-CoV and have the potential to help guide the design of peptide-based vaccines.
Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) is a positive-stranded RNA virus of the Coronaviridae family. It is the causative agent of SARS and infected more than 8,000 people around the world in 2002 to 2003 (21, 24). The genome of SARS-CoV encodes several structural proteins, including spike glycoprotein (SP), nucleocapsid protein (NP), membrane protein, and envelope protein (15).
The NP plays an essential role in SARS-CoV genome packaging and virion assembly (6, 9). At the first stage of SARS-CoV infection, it is abundantly expressed in the cells which the virus has entered and induces a strong humoral response in patients (10, 12, 14). Several studies have identified dominant B-cell determinants in human and animals (8, 13). Although the antibodies against NP cannot neutralize viral infectivity, they are useful markers for the early diagnosis of CoV infection (2, 3). Cytotoxic T lymphocytes (CTL) specific for the NP of SARS-CoV could also play an important role in the control of viral infection in vivo. CTL epitopes in the NP of SARS-CoV have also been mapped by several groups (13, 30). Additionally, NP-specific T helper (Th) cells should be able to help the production of neutralizing antibodies against the S protein. For instance, Antón et al. have demonstrated that a synthetic peptide (residues 321 to 335) from the NP of transmissible gastroenteritis virus could assist in the production of neutralizing antibodies against SP in miniswine (1). Zhao and colleagues and Zhu et al. have demonstrated that NP-specific immune responses could be induced by DNA constructs encoding the NP of SARS-CoV (36, 37). By immunizing BALB/c mice with different forms of the NP of SARS-CoV, Gupta and colleagues also identified two H-2d-restricted Th epitopes between amino acid residues 76 to 114 and 352 to 370 (5). However, Th epitopes in C57BL/6 (H-2b) and C3H/He (H-2k) mice have not been investigated. It is also unclear whether the NP-specific Th cells could help in the production of neutralizing antibodies against SARS-CoV in vivo.
The present study was undertaken to identify and characterize murine Th epitopes in the NP of SARS-CoV by using a series of overlapping synthetic peptides that cover 98% of the NP sequence in three strains of mice. The results from this study will not only allow a better understanding of cellular immunity against SARS-CoV infection but also provide useful information for the design of peptide-based anti-SARS-CoV vaccines.
MATERIALS AND METHODS
Synthetic peptides and recombinant proteins.
A series of 41 peptides, covering more than 98% of the SARS-CoV NP sequence (from residues 1 to 415), were synthesized by Hanyu Biotech Co. Ltd. (Shenzhen, China) as 15-mers with five amino acids overlapping the preceding peptide. NP54, from residues 54 to 72 of NP, SP1143, an identified conserved linear neutralizing epitope located in the second heptad repeat region 2 of SARS-CoV SP (residues 1143 to 1157) (11), and two chimeric peptides, NP111-SP1143 and NP351-SP1143 (NP111 and NP351 were chemically linked with SP1143 by two lysine residues [KK] as a spacer [18]), were also synthesized. All the peptides were high-pressure liquid chromatography purified with purities higher than 80% and supplied as lyophilized powder. The peptides maintained high hydrophilicity and were dissolved in phosphate-buffered saline (PBS) at 2 mg/ml, filter sterilized, aliquoted, and stored at −80°C (Table 1).
TABLE 1.
Synthetic peptides used in this study
| Name | Residues | Sequence |
|---|---|---|
| NP1 | 1-15 | MSDNGPQSNQRSAPR |
| NP11 | 11-25 | RSAPRITFGGPTDST |
| NP21 | 21-35 | PTDSTDNNQNGGRNG |
| NP31 | 31-45 | GGRNGARPKQRRPQG |
| NP41 | 41-55 | RRPQGLPNNTASWFT |
| NP51 | 51-65 | ASWFTALTQHGKEEL |
| NP61 | 61-75 | GKEELRFPRGQGVPI |
| NP71 | 71-85 | QGVPINTNSGPDDQI |
| NP81 | 81-95 | PDDQIGYYRRATRRV |
| NP91 | 91-105 | ATRRVRGGDGKMKEL |
| NP101 | 101-115 | KMKELSPRWYFYYLG |
| NP111 | 111-125 | FYYLGTGPEASLPYG |
| NP121 | 121-135 | SLPYGANKEGIVWVA |
| NP131 | 131-145 | IVWVATEGALNTPKD |
| NP141 | 141-155 | NTPKDHIGTRNPNNN |
| NP151 | 151-165 | NPNNNAATVLQLPQG |
| NP161 | 161-175 | QLPQGTTLPKGFYAE |
| NP171 | 171-185 | GFYAEGSRGGSQASS |
| NP181 | 181-195 | SQASSRSSSRSRGNS |
| NP191 | 191-205 | SRGNSRNSTPGSSRG |
| NP201 | 201-215 | GSSRGNSPARMASGG |
| NP211 | 211-225 | MASGGGETALALLLL |
| NP221 | 221-235 | ALLLLDRLNQLESKV |
| NP231 | 231-245 | LESKVSGKGQQQQGQ |
| NP241 | 241-255 | QQQGQTVTKKSAAEA |
| NP251 | 251-265 | SAAEASKKPRQKRTA |
| NP261 | 261-275 | QKRTATKQYNVTQAF |
| NP271 | 271-285 | VTQAFGRRGPEQTQG |
| NP281 | 281-295 | EQTQGNFGDQDLIRQ |
| NP291 | 291-305 | DLIRQGTDYKHWPQI |
| NP301 | 301-315 | HWPQIAQFAPSASAF |
| NP311 | 311-325 | SASAFFGMSRIGMEV |
| NP321 | 321-335 | IGMEVTPSGTWLTYH |
| NP331 | 331-345 | WLTYHGAIKLDDKDP |
| NP341 | 341-355 | DDKDPQFKDNVILLN |
| NP351 | 351-365 | VILLNKHIDAYKTFP |
| NP361 | 361-375 | YKTFPPTEPKKDKKK |
| NP371 | 371-385 | KDKKKKTDEAQPLPQ |
| NP381 | 381-395 | QPLPQRQKKQPTVTL |
| NP391 | 391-405 | PTVTLLPAADMDDFS |
| NP401 | 401-415 | MDDFSRQLQNSMSGA |
| NP54 | 54-72 | FTALTQHGKEELRFPRGQG |
| SP1143 | S1143-1157 | SPDVDLGDISGINAS |
| NP111-SP1143 | (N111-125)KK(S1143-1157) | FYYLGTGPEASLPYGKKSPDVDLGDISGINAS |
| NP351-SP1143 | (N351-365)KK(S1143-1157) | VILLNKHIDAYKTFPKKSPDVDLGDISGINAS |
Complementary DNA encoding full-length NP of SARS-CoV was a gift from Shenli Bi (China CDC). The DNA was amplified, inserted into a pET28a expression vector (Novagen, Darmstadt, Germany), and verified by sequencing. The His6-tagged recombinant N protein (rNP) was expressed in Escherichia coli BL21(DE3) (Stratagene, La Jolla, CA) and purified using Ni-nitrilotriacetic acid resin (Novagen, Darmstadt, Germany) following the manufacturer's instructions. The protein was desalted by being passed through a PD10 column (Pierce, Rockford, IL) in PBS (pH 7.2). The protein concentration was determined using Coomassie protein assay reagent (Pierce, Rockford, IL) with bovine γ-globulin as the standard protein. A vector containing the gene for green fluorescent protein (GFP) was a gift from L. Barber, Imperial College London, London, United Kingdom. The GFP was expressed and purified as previously described (35).
Patient sera.
Serum samples from a SARS patient (PT32) were collected between July and August 2003, 2 to 3 months after his recovery and subsequent discharge from the hospital. Written consent from the patient was obtained before the blood sample collection.
Animal immunization and serum collection.
Female C57BL/6NCrlBR (H-2b), C3H/HeNCrlBR (H-2k), and BALB/cAnNCrlBR (H-2d) mice, 6 to 8 weeks of age, were obtained from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, and the Department of Laboratory Animal Science of Peking University Health Science Center, Beijing, China, and maintained under specific-pathogen-free conditions. All immunizations were given subcutaneously (s.c.) at the base of the tail.
For the generation of NP-specific T-cell lines or peptide immunization, mice were immunized with rNP or peptides (100 μg/mouse), emulsified in complete Freund's adjuvant (CFA; Sigma, St. Louis, MO). Draining lymph nodes were taken 7 to 9 days later. In all cases, control mice received the adjuvant alone.
For priming of the Th cells in vivo (17), mice were immunized with 100 μg peptides or 50 μg rNP, emulsified in CFA. Booster immunizations were given on day 15 with a suboptimal amount of rNP (25 μg) mixed with incomplete Freund's adjuvant (IFA; Sigma, St. Louis, MO). Sera were collected by tail bleeding every week, aliquoted, and kept at −80°C until further use.
For the generation of neutralizing antibodies, mice were immunized with 150 μg chimeric peptides, a mixture of 75 μg helper peptide and 75 μg SP1143, or 75 μg SP1143 peptide alone, emulsified in CFA. Booster immunizations were given on days 15 and 35 with antigens mixed with IFA. Sera were collected by tail bleeding every week and stored as described above.
Western blot analysis.
Identical quantities of rNP (500 ng) were transferred onto nitrocellulose membranes. The membranes were blocked at room temperature for 2 h with 5% nonfat dried milk and then incubated for 2 h at room temperature with convalescent SARS patient serum (1:200) or rNP-immunized mice antiserum (1:500). After being washed in Tris-buffered saline (pH 7.5) containing 0.05% Tween 20, the membranes were incubated with horseradish peroxidase (HRP)-labeled goat anti-human or anti-mouse immunoglobulin G (IgG). 3,3′-Diaminobenzidine tetrahydrochloride (DAB; Sigma, St. Louis, MO) was used to visualize the reaction.
Generation of NP-specific T-cell lines.
Draining lymph nodes were removed from the mice at 7 to 9 days postimmunization, with rNP and single-cell suspensions made in RPMI 1640 containing 10% fetal calf serum (FCS; HyClone, Logan, UT). Following washing, the cells (4 × 106/well) were seeded in 24-well Costar plates (Nunc, Roskilde, Denmark) supplied with rNP (20 μg/ml). After 5 days of culture at 37°C with 5% CO2, the cells were harvested and viable cells (4 × 105/well) recultured in the presence of rNP and 10 IU/ml recombinant human interleukin-2 (rhIL-2; BRUCP Co., Beijing, China) with 4 × 106/well irradiated (2,000 rads) syngeneic splenocytes as feeder cells. The T-cell lines were maintained on a 7-day refeed cycle and the antigen specificity tested using proliferation assays for every cycle.
Phenotypic analysis of T cells.
NP-specific cell lines (1 × 106) were washed twice and then stained with phycoerythrin- or fluorescein isothiocyanate-labeled anti-mouse CD4 or CD8 monoclonal antibody (BD PharMingen, San Diego, CA) at 4°C for 30 min. The cells were then subjected to flow cytometric analysis using a fluorescence-activated cell sorting machine (FACScan; BD). CellQuest software was used for the analysis of the results.
Cytokine production by NP-specific T-cell lines.
The NP-specific T cells (4 × 105/well) were incubated with irradiated feeder cells (4 × 106/well) in 24-well cell culture plates, in the presence of 5 μg/ml rNP, synthetic peptides, or ConA, at 37°C and 5% CO2 for 36 h. The cytokine profiles in the supernatant were then determined by Luminex 100 (Luminex Co., Austin, TX), using a Beadlyte mouse multicytokine flex kit (Upstate, Buffalo, NY).
Proliferation assay.
T cells from NP-specific T-cell lines (2 × 104/well) or lymph nodes (4 × 105/well) from mice at 7 to 9 days postimmunization were incubated with various antigens in 96-well flat-bottomed cell culture plates (Nunc, Roskilde, Denmark) with or without irradiated feeder cells (5 × 105/well). 3H-thymidine (3H-TdR; Atom HighTech Co. Ltd., Beijing, China) (0.2 μCi/well) incorporation into DNA during the last 4 to 8 h of a 3-day culture at 37°C and 5% CO2 was determined as a measure of cell proliferation. Cultures were set up in triplicate and harvested on an automatic cell harvester (Tomtec, San Diego, CA) and the radioactivity counted in a β-counter (EG&G Wallac, Turku, Finland). The results are expressed as either 3H-TdR incorporation (counts per minute [cpm]) or the stimulation index (3H-TdR incorporation [cpm] of cells with antigen stimulation/3H-TdR incorporation [cpm] of cells without antigen stimulation).
ELISAs.
Enzyme-linked immunosorbent assay (ELISA) plates were coated at 4°C overnight with rNP (2 μg/ml) or peptide SP1143 (10 μg/ml) in carbonate buffer (pH 9.6) and subsequently incubated with blocking solution (2% bovine serum albumin in PBS) for 2 h at 37°C. The wells were washed four times with PBS containing 0.05% Tween 20 (PBS-T) and then 100 μl of diluted sera added in triplicate and incubated for 90 min at 37°C. After four washes with PBS-T, the plates were then incubated with HRP-labeled goat anti-human or anti-mouse IgG antibody for 1 h at 37°C. The ortho-phenylenediamine substrate (100 μl/well; Sigma, St. Louis, MO) was added after five washes with PBS-T and the wells incubated for 2 min at room temperature. Fifty microliters of stop solution (2 M H2SO4) was added to each well, and the optical density at 492 nm was read immediately.
DTH induction.
Groups of five mice were primed s.c. at the base of the tail with 100 μg of peptides or 25 μg of rNP emulsified in CFA and challenged in the left hind footpad 6 days later with 15 μg of rNP in 25 μl PBS, with an identical volume of PBS in the right hind footpad. Delayed-type hypersensitivity (DTH) was estimated 24 h later by measuring footpad swelling with a dial caliper (Pocotest, reverse spring-loaded caliper) and the difference between the thickness of the left and right hind footpads expressed as footpad thickness increase in 10−2-mm units (4, 17).
Preparation of pseudovirus.
Pseudovirus expressing the SARS-CoV S protein was prepared as described previously (35). Briefly, 5 × 106 293-T cells, maintained in Dulbecco modified Eagle medium containing 10% FCS, were seeded in 10-cm-diameter tissue culture dishes. One day later, the cells were cotransfected with the defective human immunodeficiency virus type 1 genome pNL4.3-Luc-R−E− (20 μg) and either plasmid pVSV-G (10 μg), encoding vesicular stomatitis virus (VSV) G protein, or plasmid pCMV-S, encoding the S protein of SARS-CoV, using Vigorous transfection reagent (Vigorous Biotech Co., Beijing, China). The defective human immunodeficiency virus type 1 genome contains the gene for luciferase, which can be used as a reporter of successful infection by the pseudovirus. The culture medium was replaced with fresh medium at 24 h posttransfection and the cells cultured for an additional 24 h. The culture supernatant, containing VSV G pseudovirus or SARS S pseudovirus, was then harvested and filtered through a 0.45-μm-pore-sized filter, followed by centrifugation at 90,000 × g for 3 h at 4°C using a Beckman XL-90 ultra centrifuge (Beckman Coulter, Fullerton, CA). The pseudoviral pellets were resuspended in PBS, titrated, aliquoted, and stored at −80°C until use.
Neutralization assays using the SARS-CoV pseudovirus system.
SARS-CoV S pseudovirus and VSV G pseudovirus infection of cells and its neutralization were described previously (35). Briefly, Vero E6 cells maintained in Dulbecco modified Eagle medium with 10% FCS were seeded in 96-well plates, at a density of 8 × 103 cells/well, and cultured overnight. Serially diluted serum samples were mixed with pseudovirus preparations for 30 min at 37°C, and then the mixtures were added to the wells containing monolayers of Vero E6 cells. After incubation at 37°C for 1 h, the mixture in the wells was replaced with fresh medium and the cells were cultured for an additional 48 h. After washes, the cells were lysed using luciferase assay reagent (Promega, Madison, WI) and the luciferase activity in the cell lysate was determined using a Veritas microplate luminometer (Turner Biosystems). The luciferase activity of the reference group (Vero E6 cells treated with pseudovirus alone) was taken as 100% infection. Cells not treated with pseudovirus were included as a specificity control, and their luciferase activity readings were at least 3 logs lower than that of the reference group in all experiments. The luciferase activity of the experimental groups (Vero E6 cells treated with pseudovirus preparations in the presence of serum antibodies) was compared with that of the reference group and the results, expressed as percent infection, calculated as follows: 100 × (luciferase activity of the experimental groups/luciferase activity of the reference group).
Prediction of T-cell epitopes.
H-2b T-cell epitopes of the NP were predicted by the RANKPEP system using motif profiles. Scores were obtained by aligning the position-specific scoring matrices with the protein segments and adding up the appropriate profile coefficients matching the residue type and position in the protein sequence (23). H-2k T-cell epitopes were predicted by the SYFPEITHI system, which was carried out by peptide motifs scoring the anchor residues (22). H-2d T-cell epitopes were predicted by using the PREDBALB/c system class II (I-Ed and I-Ad) models. The PREDBALB/c system scores the binding index of all nanomer sequences to H-2d molecules, based on quantitative matrices (34).
RESULTS
Expression and purification of rNP.
rNP, expressed in E. coli as a 49-kDa recombinant protein, was purified to near homogeneity by affinity chromatography using a Ni column. The resultant product remained soluble in saline at 650 μg/ml. Immunization of BALB/c mice with the recombinant protein induced specific IgG antibodies against SARS-CoV, as determined by a virus-coated ELISA (data not shown). In subsequent Western blot assays, the recombinant protein was specifically recognized by convalescent-phase serum from a SARS patient (PT32) as well as antiserum from rNP-immunized BALB/c mice (data not shown).
Generation of NP-specific T-cell lines.
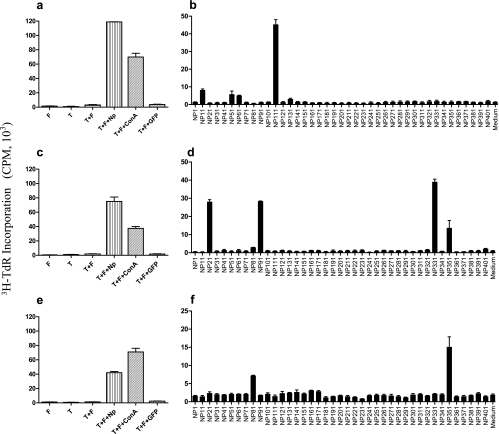
Draining lymph node cells (LNC) isolated from C57BL/6, C3H, and BALB/c mice 7 to 9 days after s.c. immunization with rNP proliferated vigorously in response to stimulation with rNP in vitro (data not shown). NP-specific T-cell lines were established by repeated stimulation, in 7-day intervals, with rNP of these LNC in the presence of irradiated syngeneic splenocytes (as feeder cells) and rhIL-2. All T-cell lines established in this way were CD4+ and CD8−, as determined by flow cytometric analysis (data not shown). They appeared to be Th1 cells, because they secreted mainly gamma interferon and tumor necrosis factor alpha, but very little IL-4 or IL-10, upon stimulation with rNP or the NP-derived synthetic peptides (data not shown). The cell lines were regularly tested for NP specificity by using T-cell proliferation assays (Fig. 1).
FIG. 1.
Screening for stimulatory peptides. To ascertain the specificity of the NP-specific T-cell lines (2 × 104/well) from C57BL/6 (a), C3H (c), or BALB/c (e) mice, resting cell lines were cultured alone or together with irradiated feeder cells (5 × 105/well) in 96-well flat-bottomed plates or stimulated with rNP (20 μg/ml), ConA (5 μg/ml), or GFP (20 μg/ml) in the presence of feeder cells. To screen for stimulatory peptides, the NP-specific cell lines were stimulated with individual synthetic peptides (5 μg/ml) in the presence of feeder cells (b, d, and f). During the last 4 to 8 h of a 3-day culture at 37°C with 5% CO2, the cells were pulsed with 3H-TdR (0.2 μCi/well), followed by counting of the incorporated radioactivity in a β-counter. The results are expressed as 3H-TdR incorporation (cpm).
Screening for stimulatory peptides.
A series of 41 overlapping 15-mer peptides, covering 98% of the SARS-CoV NP sequence, were synthesized and high-pressure liquid chromatography purified to more than 80% homogeneity (Table 1). These peptides were screened for their ability to stimulate the proliferation of NP-specific T cells from BALB/c, C57BL/6, and C3H mice. As illustrated in Fig. 1, four peptides were able to stimulate the proliferation of NP-specific T-cell lines from C57BL/6 mice (NP11, amino acid residues 11 to 25; NP51, residues 51 to 65; NP61, residues 61 to 75; and NP111, residues 111 to 125) and another four peptides stimulated T-cell lines from C3H mice (NP21, residues 21 to 35; NP91, residues 91 to 105; NP331, residues 331 to 345; and NP351, residues 351 to 365). The NP-specific T-cell lines from BALB/c mice recognized two peptides: NP81 (residues 81 to 95) and NP351 (residues 351 to 365). The ability of these peptides to stimulate NP-specific T cells was also confirmed in proliferation assays using freshly isolated LNC from mice after s.c. immunization with rNP (data not shown). Since peptide NP241 was unable to stimulate T cells from any of the three inbred strains of mice, it was selected as a control peptide in subsequent experiments.
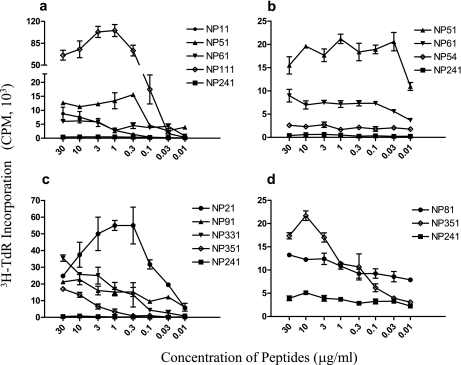
The positive peptides stimulated NP-specific T-cell proliferation in a specific and dose-dependent manner (Fig. 2). It is noteworthy that peptide NP351 was recognized by T cells from both BALB/c (H-2d) and C3H (H-2k) mice. In order to clarify whether the NP51 and NP61 peptides shared the same epitope, a new 19-mer peptide, NP54 (residues 54 to 72), covering the C-terminal 12 residues of NP51 and N-terminal 12 residues of NP61 (5 of which overlap with NP51), was synthesized. Figure 2b indicates that NP54 was unable to stimulate T-cell proliferation, implying that NP51 and NP61 represent two independent epitopes.
FIG. 2.
Dose response of T-cell lines to peptide stimulation. NP-specific T-cell lines (2 × 104/well) from C57BL/6 (a and b), C3H (c), or BALB/c (d) mice were stimulated with various peptides in 96-well flat-bottomed plates in the presence of irradiated feeder cells (5 × 105/well). During the last 4 to 8 h of a 3-day culture at 37°C with 5% CO2, the cells were pulsed with 3H-TdR (0.2 μCi/well), followed by counting of the incorporated radioactivity in a β-counter. The results are expressed as 3H-TdR incorporation (cpm).
Immunogenicity of peptides representing T-cell epitopes.
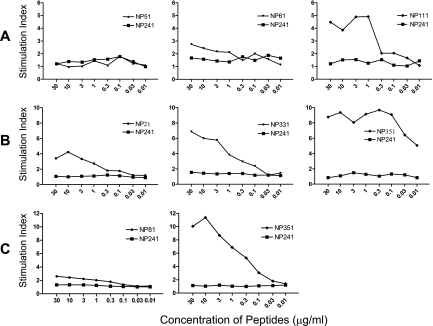
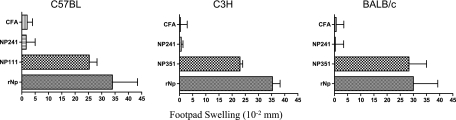
To assess the immunogenicity of the T-cell-stimulating peptides, mice were immunized s.c. with synthetic peptides (emulsified in CFA) representing the appropriate T-cell epitopes. Draining lymph nodes were taken 7 to 9 days later and the LNC stimulated with the immunizing peptides. As illustrated in Fig. 3A, out of the peptides tested in C57BL/6 mice, peptide NP111 was the only one able to induce specific T-cell responses. Peptide NP351 was immunogenic in both C3H and BALB/c mice (Fig. 3B and C), while NP331 was moderately immunogenic in C3H mice. The immunogenicity of peptides NP111 and NP351 was further confirmed by their ability to induce strong DTH responses in C57BL/6 mice and in C3H and BALB/c mice, respectively (Fig. 4).
FIG. 3.
Immunogenicity of peptides. C57BL/6 (A), C3H (B), or BALB/c (C) mice were immunized s.c. at the base of the tail with 100 μg peptides emulsified in CFA. Draining lymph nodes were taken 7 to 9 days later and LNC (4 × 105/well) incubated with the immunizing peptides or the control peptide NP241. During the last 4 to 8 h of a 3-day culture at 37°C with 5% CO2, the cells were pulsed with 3H-TdR (0.2 μCi/well), followed by counting of the incorporated radioactivity in a β-counter. Results are expressed as the stimulation index.
FIG. 4.
DTH induction by peptides P12 and P36. Groups of five mice were primed s.c. at the base of the tail with peptides (100 μg/mouse) or rNP (25 μg/mouse) emulsified in CFA and challenged, 6 days later, with rNP in PBS (15 μg/25 μl) in the left hind footpad and with an identical volume of PBS in the right hind footpad. DTH was estimated 24 h later by measuring footpad swelling, and the difference between the thickness of the left and right hind footpads is expressed as footpad thickness increase in 10−2-mm units.
In mice preimmunized s.c. with peptide NP111 (for BALB/c mice) or NP351 (for C57BL/6 and C3H mice), the humoral response to a booster immunization 2 weeks later with a suboptimal amount of rNP (25 μg/mouse) in IFA was clearly enhanced, as evidenced by the more rapid production of serum IgG against NP in the groups primed with NP111 or NP351 than in those immunized with NP241 or CFA alone (Fig. 5). Thus, these peptides are able to induce Th cells capable of assisting in humoral responses in vivo.
FIG. 5.
Preimmunization with Th peptides accelerated the antibody response to rNP. C57BL/6, C3H, or BALB/c mice were immunized s.c. at the base of the tail with either peptides (100 μg/mouse) emulsified in CFA or, as controls, CFA alone or PBS alone. Booster immunizations were given on day 15, with all the mice (except the PBS control) receiving rNP (25 μg) mixed with IFA. Sera were collected every 7 days and tested using rNP-based ELISAs. HRP-labeled goat anti-mouse IgG served as the secondary antibody. The results are expressed as optical density (OD) readings at 492 nm.
The helper peptides assist in the production of neutralizing antibodies in vivo.
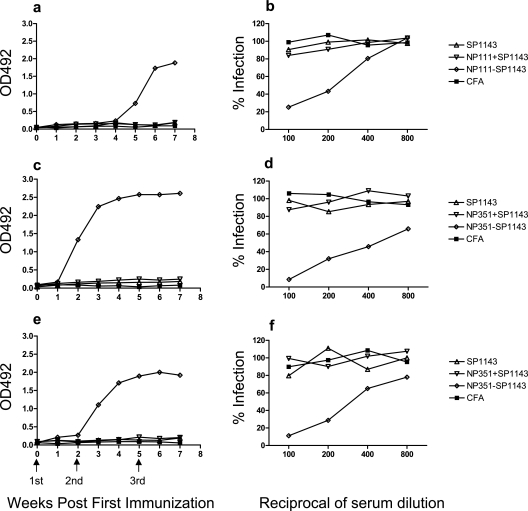
To test whether peptides NP111 and NP351 could assist in the production of neutralizing antibodies in vivo, two fusion peptides, NP111-SP1143 and NP351-SP1143, were synthesized, fusing a neutralizing epitope (SP1143, residues 1143 to 1157 of the S protein of SARS-CoV) with NP111 and NP351, respectively. Mice were immunized with NP111-SP1143, NP351-SP1143, SP1143 peptide alone, or its mixture with NP111 or NP351 emulsified in CFA, followed by two booster immunizations with the same peptides in IFA. The fusion peptides elicited high-titer anti-SP1143 serum IgG, while SP1143 peptide alone, or its mixture with NP111 or NP351, was unable to do so (Fig. 6). The antisera induced by NP111-SP1143 and NP351-SP1143 were employed to inhibit the infection of Vero E6 cells by a pseudovirus expressing the S protein of SARS-CoV together with luciferase as a reporter enzyme (35). Infection by SARS S pseudovirus (Fig. 6b, d, and f), but not VSV G pseudovirus (not shown), was specifically neutralized by antiserum from mice immunized with NP111-SP1143 or NP351-SP1143 peptide. Antiserum from mice immunized with either SP1143 mixed with helper peptide or SP1143 peptide alone was unable to neutralize the SARS-CoV S pseudovirus.
FIG. 6.
Th peptides assisted anti-S protein antibody responses in vivo. C57BL/6 (a), C3H (c), or BALB/c (e) mice were immunized s.c. at the base of the tail with 150 μg of a chimeric peptide (NP111-SP1143 or NP351-SP1143), a mixture of a helper peptide (75 μg) and SP1143 (75 μg), or peptide SP1143 (75 μg) alone. All peptides were emulsified in CFA; mice immunized with CFA alone served as a control. Booster immunizations were given on days 15 and 35 with the immunizing peptides mixed with IFA (or IFA alone for a control). Sera were collected and the anti-SP1143 IgG response determined using SP1143 peptide-based ELISAs. HRP-labeled goat anti-mouse IgG served as the secondary antibody. The results are expressed as optical density (OD) readings at 492 nm. For neutralization assays, serially diluted serum samples from C57BL/6 (b), C3H (d), or BALB/c (f) mice immunized with the chimeric peptides were mixed with medium containing SARS-CoV S pseudovirus and incubated for 30 min at 37°C. The mixtures were then distributed into triplicate wells in 96-well plates containing a monolayer of Vero E6 cells seeded at 8 × 103/well the previous day. Luciferase activity in the infected cells was determined 48 h later and the results expressed as percent infection compared with that of the control group (Vero E6 cells treated with the pseudovirus alone).
DISCUSSION
The presentation of antigenic epitopes by major histocompatibility complex class II molecules and the activation of CD4 Th cells are critical steps in generating adaptive immune responses. In this study, we identified and characterized several murine Th epitopes in the NP of SARS-CoV which dictate the specificity of the cellular immune responses to the molecule. The selection of epitopes for T-cell recognition is governed by the H-2 haplotypes of the responding strains. It is noteworthy that peptide NP351 is recognized by T cells from both C3H (H-2k) and BALB/c (H-2d) mice. This peptide is also able to induce cell-mediated DTH responses in vivo (Fig. 4). NP351 in C3H and BALB/c mice seems more potent than NP111 in C57BL/6 mice for assisting in the production of antibodies against the linked B-cell epitopes in vivo, although variation in responsiveness between different strains of animals could also contribute to this phenomenon (Fig. 6). Screening of the 41 overlapping peptides listed in Table 1 for reactivity with antisera from rNP-immunized mice revealed that NP351 and adjacent peptides were the most strongly recognized of the peptides (data not shown). These data indicate that the sequence around NP351 represents an immunodominant region in the NP in mice.
Inactivated whole virus can be used as a vaccine against SARS-CoV and is able to induce the production of neutralizing antibodies providing protective immunity (7, 26, 29, 33). Synthetic peptides containing appropriate epitopes are equally effective in inducing protective immunity in vivo and can therefore also be used as vaccines (16, 25, 27). Peptide vaccines will have the added advantage that they are easily manufactured, chemically stable, and free of contaminating substances or unwanted sequences capable of inducing pathological responses in vivo.
It is necessary for an effective peptide-based vaccine to include Th epitopes, neutralizing B-cell epitopes, and also CTL epitopes for the efficient production of neutralizing antibodies and virus-specific cytotoxic T cells (25, 27, 28, 31). The inclusion of multiple Th epitopes is of great importance as helper T cells are responsible for the generation of adaptive immunity. The identification of Th epitopes in the NP of SARS-CoV has important implications. Another important consideration is the chemical linkage of neutralizing and Th epitopes, since simply mixing the two together does not result in effective priming against the B-cell epitope (Fig. 6). The ligation of the two epitopes is currently often achieved via m-maleimidobenzoyl succinimide ester covalent linkage to the terminal Cys (32) or by adding several amino acid residues as spacers between the two epitopes (18, 20). Our chimeric peptides contain a Th epitope in the N-terminal half and a neutralizing epitope in the C-terminal half, ligated by two lysine residues. Both chimeric peptides are able to induce high-titer neutralizing antibodies against SARS-CoV S pseudovirus (Fig. 6). In contrast to some other immunogens, multiple copies of the peptide were not necessary (19).
It is possible that fusion of two peptides representing a B-cell epitope and a Th epitope would bring changes to their physical properties, but this is unlikely the main reason for the increased immunogenicity of the fusion peptides (e.g., NP111-SP1143) compared with that of mixtures of unconjugated peptides (e.g., NP111 plus SP1143) in their ability to induce humoral responses in vivo. A more likely explanation for this phenomenon is that physical conjugation between NP111 (T-cell epitope) and SP1143 (B-cell epitope) allowed efficient B-cell receptor-mediated uptake of the Th peptide by SP1143-specific B cells. Subsequently, the SP1143-specific B cells can receive activation signals from the NP111-specific helper T cells. In the case of immunization with an unconjugated mixture of the two peptides, the SP1143-specific B cells are unlikely to be able to endocytose significant amounts of the Th peptide.
It should also be noted that the Th epitopes identified herein are murine specific; they almost certainly cannot be presented by HLA molecules. Consequently, specific features of the cellular immune response in humans cannot be deduced from the mouse data. However, animal models are useful tools for studies on the immunological responses elicited by viruses or their recombinant proteins in vivo. Knowledge gained from this study will no doubt facilitate the development of specific therapies that are designed to minimize pulmonary disease and optimize the anti-SARS-CoV immune response.
Computational algorithms capable of predicting T-cell epitopes have important applications in vaccine design. We employed three systems to predict possible H-2b-, H-2k-, and H-2d-restricted Th epitopes in the NP, and the results, summarized in Table 2, are compared with those obtained in this study. The RANKPEP system (23) predicted only one possible epitope, restricted by major histocompatibility complex class II allele I-Ab, which is consistent with the recognition of NP111 in C57BL/6 mice (Fig. 3). The SYFPEITHI system, available only for H-2k (I-Ak or I-Ek allele) epitope prediction (22), predicted 20 epitopes, including all the 4 epitopes identified herein. The PREDBALB/c system predicts the probability of nanomer sequences binding to the I-Ad and I-Ed molecules (34). Approximately 10 regions with the highest scores are listed in Table 2 for each molecule. The NP81 and NP351 peptides are identified by this algorithm. Interestingly, all three systems predicted some epitopes not identified in this or other studies.
TABLE 2.
Comparison of actual epitopes and predicted epitopes of SARS-CoV NP
| Actual epitope | Sequence | System used for predicting epitopesa | Sequence(s) for predicted epitope restricted by alleleb:
|
|
|---|---|---|---|---|
| I-A | I-E | |||
| H-2b helper peptides | ||||
| NP11 | 11RSAPRITFGGPTDST25 | |||
| NP51 | 51ASWFTALTQHGKEEL65 | |||
| NP61 | 61GKEELRFPRGQGVPI75 | RANKPEP | ||
| NP111 | 111FYYLGTGPEASLPYG125 | 112YYLGTGPEA (13.82) | ||
| 133WVATEGALN (12.32) | ||||
| 149TRNPNNNAA (12.51) | ||||
| 361YKTFPPTEP (12.86) | ||||
| H-2k helper peptides | ||||
| NP21 | 21PTDSTDNNQNGGRNG35 | 21PTDSTDNNQNGGRNG (16) | ||
| 51ASWFTALTQHGKEEL (24) | ||||
| 80GPDDQIGYYRRATRR (18) | 82DDQIGYYRRATRRVR (18) | |||
| NP91 | 91ATRRVRGGDGKMKEL105 | 96RGGDGKMKELSPRWY (18) | 92TRRVRGGDGKMKELS (22) | |
| 156AATVLQLPQGTTLPK (20) | ||||
| 191SRGNSRNSTPGSSRG (22) | ||||
| SYFPEITHI | 222LLLLDRLNQLESKVS (22) | 222LLLLDRLNQLESKVS (20) | ||
| 286NFGDQDLIRQGTDYK (18) | 289DQDLIRQGTDYKHWP (18) | |||
| NP331 | 309APSASAFFGMSRIGM (18) | |||
| NP351 | 331WLTYHGAIKLDDKDP345 | 338IKLDDKPQFKDNVI (18) | 337AIKLDDKDPQFKDNV (18) | |
| 351VILLNKHIDAYKTFP365 | 351VILLNKHIDAYKTFP (18) | |||
| 369PKKDKKKKTDEAQPL (20) | ||||
| 377TDEAQPLPQRQKKQP (18) | ||||
| 390QPTVTLLPAADMDDF (20) | ||||
| 399ADMDDFSRQLQNSMS (18) | ||||
| H-2d helper peptides | ||||
| 39KQRRPQGLP (9.7) | ||||
| NP81 | 81PDDQIGYYRRATRRV95 | 86GYYRRATRR (9.6) | 86GYYRRATRR (9.7) | |
| 105LSPRWYFYY (9.7) | ||||
| 129EGIVWVATE (9.7) | ||||
| 160LQLPQGTTL (10.0) | ||||
| 207SPARMASGG (9.7) | ||||
| 224LLDRLNQLE (9.7) | ||||
| PREDBALB/c | 248TKKSAAEAS (9.6) | |||
| 260RQKRTATKQ (9.7) | ||||
| 300KHWPQIAQF (9.8) | ||||
| 318MSRIGMEVT (9.5) | ||||
| 331WLTYHGAIK (9.9) | ||||
| 340LDDKDPQFK (9.8) | ||||
| NP351 | 351VILLNKHIDAYKTFP365 | 355NKHIDAYKT (9.5) | 353LLNKHIDAY (9.9) | |
| 405SRQLQNSMS (9.7) | ||||
The RANKPEP system had threshold scores of 9.52 for the I-A allele and 51.80 for the I-E allele, and the PREDBALB/c system had a threshold score of 8 for both alleles.
The number in parentheses is the score obtained with the indicated system. For scoring details, see Materials and Methods.
Taken together, this study has identified nine dominant Th epitopes in the NP of SARS-CoV recognized by T cells from mice of three strains. The peptides containing these epitopes are further characterized for their ability to induce cellular immune responses in vivo and also to assist in the production of antiviral neutralizing antibody. This information is of great value for understanding anti-SARS-CoV immunity and also for designing peptide-based vaccines against the disease.
Acknowledgments
This study was supported by grants from the National Key Basic Research Programs (2001CB510007) and also the National High Technology Research and Development Program (2003AA208412A) of China.
We are indebted to Jingxian Zhao, Institute of Tissue Transplantation and Immunology, Jinan University, Guangzhou, China, for technical assistance in measuring the cytokine profiles. We are grateful to Zhengdong Zhao, Department of Immunology, Peking University Health Science Center, Beijing, China, for the gift of pCMV-S, pVSV-G, and pNL4.3-Luc-R−E− constructs. We thank Peter J. Delves, University College London, London, United Kingdom, for useful comments on the manuscript.
Footnotes
Published ahead of print on 28 March 2007.
REFERENCES
- 1.Antón, I. M., C. Sune, R. H. Meloen, F. Borras-Cuesta, and L. Enjuanes. 1995. A transmissible gastroenteritis coronavirus nucleoprotein epitope elicits T helper cells that collaborate in the in vitro antibody synthesis to the three major structural viral proteins. Virology 212:746-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchholz, U. J., A. Bukreyev, L. Yang, E. W. Lamirande, B. R. Murphy, K. Subbarao, and P. L. Collins. 2004. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. USA 101:9804-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Che, X. Y., W. Hao, Y. Wang, B. Di, K. Yin, Y. C. Xu, C. S. Feng, Z. Y. Wan, V. C. Cheng, and K. Y. Yuen. 2004. Nucleocapsid protein as early diagnostic marker for SARS. Emerg. Infect. Dis. 10:1947-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao, X. M., F. Y. Liew, and J. P. Tite. 1990. A dominant Th epitope in influenza nucleoprotein. Analysis of the fine specificity and functional repertoire of T cells recognizing a single determinant. J. Immunol. 144:2730-2737. [PubMed] [Google Scholar]
- 5.Gupta, V., T. M. Tabiin, K. Sun, A. Chandrasekaran, A. Anwar, K. Yang, P. Chikhlikar, J. Salmon, V. Brusic, E. T. Marques, S. N. Kellathur, and T. J. August. 2006. SARS coronavirus nucleocapsid immunodominant T-cell epitope cluster is common to both exogenous recombinant and endogenous DNA-encoded immunogens. Virology 347:127-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He, R., A. Leeson, M. Ballantine, A. Andonov, L. Baker, F. Dobie, Y. Li, N. Bastien, H. Feldmann, U. Strocher, S. Theriault, T. Cutts, J. Cao, T. F. Booth, F. A. Plummer, S. Tyler, and X. Li. 2004. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 105:121-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He, Y., Y. Zhou, P. Siddiqui, and S. Jiang. 2004. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 325:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, Y., Y. Zhou, H. Wu, Z. Kou, S. Liu, and S. Jiang. 2004. Mapping of antigenic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42:5309-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, Y., P. H. Lin, C. Y. Liu, S. P. Lee, and Y. C. Chao. 2004. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 318:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krokhin, O., Y. Li, A. Andonov, H. Feldmann, R. Flick, S. Jones, U. Stroeher, N. Bastien, K. V. Dasuri, K. Cheng, J. N. Simonsen, H. Perreault, J. Wilkins, W. Ens, F. Plummer, and K. G. Standing. 2003. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics 2:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai, S. C., P. C. Chong, C. T. Yeh, L. S. Liu, J. T. Jan, H. Y. Chi, H. W. Liu, A. Chen, and Y. C. Wang. 2005. Characterization of neutralizing monoclonal antibodies recognizing a 15-residues epitope on the spike protein HR2 region of severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biomed. Sci. 12:711-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung, D. T., F. C. Tam, C. H. Ma, P. K. Chan, J. L. Cheung, H. Niu, J. S. Tam, and P. L. Lim. 2004. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 190:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, S. J., C. H. Leng, S. P. Lien, H. Y. Chi, C. Y. Huang, C. L. Lin, W. C. Lian, C. J. Chen, S. L. Hsieh, and P. Chong. 2006. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24:3100-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, X., Y. Shi, P. Li, L. Li, Y. Yi, Q. Ma, and C. Cao. 2004. Profile of antibodies to the nucleocapsid protein of the severe acute respiratory syndrome (SARS)-associated coronavirus in probable SARS patients. Clin. Diagn. Lab. Immunol. 11:227-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 16.Obeid, O. E., C. D. Partidos, C. R. Howard, and M. W. Steward. 1995. Protection against morbillivirus-induced encephalitis by immunization with a rationally designed synthetic peptide vaccine containing B- and T-cell epitopes from the fusion protein of measles virus. J. Virol. 69:1420-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohishi, K., H. Kabeya, H. Amanuma, and M. Onuma. 1996. Induction of bovine leukaemia virus Env-specific Th-1 type immunity in mice by vaccination with short synthesized peptide-liposome. Vaccine 14:1143-1148. [DOI] [PubMed] [Google Scholar]
- 18.Oishi, Y., A. Onozuka, H. Kato, N. Shimura, S. Imai, and T. Nisizawa. 2001. The effect of amino acid spacers on the antigenicity of dimeric peptide-inducing cross-reacting antibodies to a cell surface protein antigen of Streptococcus mutans. Oral Microbiol. Immunol. 16:40-44. [DOI] [PubMed] [Google Scholar]
- 19.Partidos, C., C. Stanley, and M. Steward. 1992. The influence of orientation and number of copies of T and B cell epitopes on the specificity and affinity of antibodies induced by chimeric peptides. Eur. J. Immunol. 22:2675-2680. [DOI] [PubMed] [Google Scholar]
- 20.Partidos, C. D., C. M. Stanley, and M. W. Steward. 1991. Immune responses in mice following immunization with chimeric synthetic peptides representing B and T cell epitopes of measles virus proteins. J. Gen. Virol. 72:1293-1299. [DOI] [PubMed] [Google Scholar]
- 21.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rammensee, H., J. Bachmann, N. P. Emmerich, O. A. Bachor, and S. Stevanovic. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213-219. [DOI] [PubMed] [Google Scholar]
- 23.Reche, P. A., J. P. Glutting, H. Zhang, and E. L. Reinherz. 2004. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 56:405-419. [DOI] [PubMed] [Google Scholar]
- 24.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 25.Shen, L., R. Schroers, J. Hammer, X. F. Huang, and S. Y. Chen. 2004. Identification of a MHC class-II restricted epitope in carcinoembryonic antigen. Cancer Immunol. Immunother. 53:391-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spruth, M., O. Kistner, H. Savidis-Dacho, E. Hitter, B. Crowe, M. Gerencer, P. Bruhl, L. Grillberger, M. Reiter, C. Tauer, W. Mundt, and P. N. Barrett. 2006. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine 24:652-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, H., R. P. Morrison, N. G. Watkins, and H. D. Caldwell. 1990. Identification and characterization of T helper cell epitopes of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 172:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun, J. C., M. A. Williams, and M. J. Bevan. 2004. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat. Immunol. 5:927-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, L., Q. Zhu, E. Qin, M. Yu, Z. Ding, H. Shi, X. Cheng, C. Wang, G. Chang, Q. Zhu, F. Fang, H. Chang, S. Li, X. Zhang, X. Chen, J. Yu, J. Wang, and Z. Chen. 2004. Inactivated SARS-CoV vaccine prepared from whole virus induces a high level of neutralizing antibodies in BALB/c mice. DNA Cell Biol. 23:391-394. [DOI] [PubMed] [Google Scholar]
- 30.Tsao, Y. P., J. Y. Lin, J. T. Jan, C. H. Leng, C. C. Chu, Y. C. Yang, and S. L. Chen. 2006. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem. Biophys. Res. Commun. 344:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, J. C., and A. M. Livingstone. 2003. Cutting edge: CD4+ T cell help can be essential for primary CD8+ T cell responses in vivo. J. Immunol. 171:6339-6343. [DOI] [PubMed] [Google Scholar]
- 32.Zegers, N. D., C. van Holten, E. Claasen, and W. J. Boersma. 1993. Peptide-induced memory (IgG) response, cross-reactive with native proteins, requires covalent linkage of a specific B cell epitope with a T cell epitope. Eur. J. Immunol. 23:630-634. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, C. H., J. H. Lu, Y. F. Wang, H. Y. Zheng, S. Xiong, M. Y. Zhang, X. J. Liu, J. X. Li, Z. Y. Wan, X. G. Yan, S. Y. Qi, Z. Cui, and B. Zhang. 2005. Immune responses in Balb/c mice induced by a candidate SARS-CoV inactivated vaccine prepared from F69 strain. Vaccine 23:3196-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, G. L., K. N. Srinivasan, A. Veeramani, J. T. August, and V. Brusic. 2005. PREDBALB/c: a system for the prediction of peptide binding to H2d molecules, a haplotype of the BALB/c mouse. Nucleic Acids Res. 33:W180-W183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, J., W. Wang, R. Jia, Z. Yuan, Z. Zhao, X. Xu, P. Lv, Y. Zhang, and X. Gao. 2007. A study on antigenicity and receptor-binding ability of fragment 450-650 of the spike protein of SARS coronavirus. Virology 359:362-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, P., J. Cao, L. J. Zhao, Z. L. Qin, J. S. Ke, W. Pan, H. Ren, J. G. Yu, and Z. T. Qi. 2005. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology 331:128-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, M. S., Y. Pan, H. Q. Chen, Y. Shen, X. C. Wang, Y. J. Sun, and K. H. Tao. 2004. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol. Lett. 92:237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]