Abstract
The phosphoprotein P of Borna disease virus (BDV) is an essential cofactor of the viral RNA-dependent RNA polymerase. It is preferentially phosphorylated at serine residues 26 and 28 by protein kinase C ɛ (PKCɛ) and, to a lesser extent, at serine residues 70 and 86 by casein kinase II (CKII). To determine whether P phosphorylation is required for viral polymerase activity, we generated P mutants lacking either the PKCɛ or the CKII phosphate acceptor sites by replacing the corresponding serine residues with alanine (A). Alternatively, these sites were replaced by aspartic acid (D) to mimic phosphorylation. Functional characterization of the various mutants in the BDV minireplicon assay revealed that D substitutions at the CKII sites inhibited the polymerase-supporting activity of P, while A substitutions maintained wild-type activity. Likewise, D substitutions at the PKC sites did not impair the cofactor function of BDV-P, whereas A substitutions at these sites led to increased activity. Interestingly, recombinant viruses could be rescued only when P mutants with modified PKCɛ sites were used but not when both CKII sites were altered. PKCɛ mutant viruses showed a reduced capacity to spread in cell culture, while viral RNA and protein expression levels in persistently infected cells were almost normal. Further mutational analyses revealed that substitutions at individual CKII sites were, with the exception of a substitution of A for S86, detrimental for viral rescue. These data demonstrate that, in contrast to other viral P proteins, the cofactor activity of BDV-P is negatively regulated by phosphorylation.
Phosphoproteins (P) of nonsegmented negative-strand RNA viruses are subunits of the viral polymerase complex and known to be extensively phosphorylated by host kinases (15). The impact of P phosphorylation on viral transcription and replication is best described for vesicular stomatitis virus (VSV). VSV-P is phosphorylated in two distinct regions, designated domains I and II (10). Phosphorylation in domain I was shown for both serotype VSV Indiana and New Jersey to be essential for the transcriptional activity of the P protein (2, 3, 10, 12, 32, 43, 45), but it seems to have no effect on replication (32). Domain II phosphorylation affects the viral replication of VSV Indiana (23) and was shown recently to be indispensable for viral growth (13). For other nonsegmented RNA viruses, the role of P phosphorylation in the viral life cycle is less clear. Although the bulk of phosphorylation of the human respiratory syncytial virus P protein is not essential for either RNA transcription or replication (31, 48, 49), it contributes to efficient viral growth in vivo (28). In contrast, for Sendai virus (SV), the major constitutive phosphorylation site of P is dispensable for viral replication in vivo (8, 21, 24). Although the phosphate acceptor sites of human parainfluenza virus type 1 P protein and measles virus have been mapped, their functional significance remains to be shown (7, 14).
Borna disease virus (BDV) causes a persistent infection in the central nervous system of a broad range of warm-blooded animals, resulting in immune-mediated neurological diseases causing behavioral abnormalities (18, 44). In contrast to other Mononegavirales, BDV replicates in the nucleus of infected cells and uses the host cell splicing machinery for maturation of viral transcripts (4, 11, 35). Besides P protein, BDV encodes other proteins, namely, the nucleoprotein (N), protein X, matrix protein (M), glycoprotein (G), and polymerase (L) (5, 51). Whereas M and G are involved in particle formation, P, N, L, and X are components of the polymerase complex (46) in which P acts as a scaffold protein (36). P forms homo-oligomers via a leucine zipper motif located in the C-terminal half of this protein and can interact with all other components of the viral polymerase complex (37, 42). The interaction domain for X is localized to the N-terminal half of P (25), whereas the overlapping L- and N-binding domains are confined to the C-terminal half (37, 42). The function of X in the BDV life cycle is not completely understood. It is a nonstructural protein (40) that is not essential for polymerase activity, as N, P, and L are sufficient to replicate and transcribe artificial minigenomes (33, 38). However, X can negatively modulate the polymerase activity by binding to P (34, 38).
BDV-P is phosphorylated predominantly by protein kinase C ɛ (PKCɛ) and weakly by casein kinase II (CKII) at various serine residues (41). Mapping of the corresponding phosphorylation sites of BDV-P in vitro revealed that replacement of S26 and S28 (S26/28) with alanine (A) residues abrogated PKCɛ phosphorylation completely, whereas substitutions at S70 and S86 (S70/86) prevented CKII phosphorylation (41). Using the BDV minreplicon assay (33, 38, 52) and the recently established technique for generating BDV entirely from cDNA (29, 39), we investigated whether BDV-P phosphorylation supports polymerase activity and viral growth, as in the case of VSV-P, or whether these modifications are dispensable. To this end, we generated P mutants lacking either the PKCɛ or the CKII phosphorylation sites by replacing the corresponding serine residues with A or aspartic acid (D) to abrogate or mimic phosphorylation at these sites (17, 22). Functional characterization of these mutants in the BDV minireplicon assay revealed that D substitutions at the CKII sites inhibited the polymerase activity of P, while mutants with A substitutions showed no reduction. Unimpaired activity was also observed with mutants harboring D substitutions at the PKCɛ sites, whereas A substitutions led to increased activity. Furthermore, initial attempts to generate recombinant viruses were successful only using P mutants with modified PKCɛ sites. These mutant viruses revealed a delayed spread in cell culture. Viruses expressing BDV-P mutants with substitutions at both CKII sites could not be rescued, but substitutions of the individual CKII sites allowed the generation of recombinant BDV when S86 of BDV-P was replaced by A.
MATERIALS AND METHODS
Plasmid construction.
Site-directed mutagenesis on the pCAGGS expression vector encoding the P open reading frame (ORF) (pCA-P) (38) was performed with Turbo Pfu polymerase (Stratagene) employing the assembly PCR technique (27). In the first two separate PCRs, one of the internal overlapping reverse and forward primers containing the desired mutations was combined with an external primer harboring either a NotI site upstream of the translation initiation codon or a BglII site downstream of the stop codon, respectively. The two resulting PCR fragments were linked in a second PCR using only the external primers. The NotI/BglII-restricted product was ligated into a NotI/BglII-linearized pCAGGS vector.
The pCAGGS plasmids encoding the fusion proteins VP16-L, VP16-N, VP16-X, VP16-P-wt (where wt is wild type), and Gal4-P-wt (pCA-VP16-L, pCA-VP16-N, pCA-VP16-X-wt, pCA-VP16-P-wt, and pCA-Gal4-P-wt, respectively) used in the mammalian two-hybrid assay were previously described (37). Expression vectors coding for VP16 or Gal4 fused to the P (17, 22) mutants were obtained by digesting the corresponding pCA-P plasmids with NotI and BglII and subsequent ligation into a NotI/BglII-linearized pCAGGS vector containing the VP16 or Gal4 fusion tag (37). Accordingly, vectors coding for VP16 fused to X mutants were constructed by digesting the corresponding pCA-X plasmids with NotI and BglII and subsequent ligation into a NotI/BglII-linearized pCAGGS vector containing the VP16 fusion tag (37).
To introduce point mutations into the P gene of the full-length BDV genome, assembly PCR using the plasmid pBRPolII-HrBDVc (29) as a template was carried out. The external primers were chosen to anneal upstream or downstream of the unique restriction sites EheI or BbvCI, respectively, in order to allow exchange of the desired DNA fragment in a single cloning step.
For the pCAGGS expression plasmids encoding X or X mutants (pCA-X), the X ORF was amplified by PCR from the respective pBRPolII-HrBDVc plasmid coding for X-wt or an X mutant using primers harboring an NotI site upstream of the translation initiation codon of X or a BglII site downstream of the stop codon. The NotI/BglII-restricted PCR product was ligated into an NotI/BglII-linearized pCAGGS vector.
Similarly, the P ORF was amplified by PCR from the pBRPolII-HrBDVc plasmid encoding P-wt or P mutants, using a primer containing the NotI site directly upstream of the second codon of the P ORF or a BglII site downstream of the stop codon, respectively. The PCR product was digested with NotI and BglII and ligated into the NotI/BglII-linearized plasmid pCA-Flag-GFP (where GFP is green fluorescent protein) (38). In this way, pCAGGS expression plasmids encoding Flag-P fusion proteins were made (pCA-Flag-P). To generate plasmids encoding hemagglutinin (HA)-P fusion proteins (pCA-HA-P) the pCA-Flag-P plasmids were digested with EcoRI and NotI and ligated with a linker containing the HA tag flanked by an EcoRI and an NotI site. The pCA-Flag-L expression plasmid was previously described (38).
The integrity of all PCR-derived DNA fragments was verified by sequencing. All primers and sequences are available from the authors on request.
Cells and transfection.
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 U of penicillin G per ml, 100 μg of streptomycin per ml, 2 mM glutamine, and 10% fetal bovine serum (FBS) for BSR-T7/5 (6) and HEK 293T cells and with 4% FBS for oligodendroglioma (Oligo), rat astroglia (C6), and Vero cells. For transfection, BSR-T7/5 or HEK 293T cells were seeded in 35-mm or 22-mm dishes and grown to 80% confluence. Plasmids were transfected using Metafectene (Biontex) according to the manufacturer's recommendations.
Virus rescue.
HEK 293T cells in 35-mm dishes were transfected with 4 μg of plasmids encoding the full-length BDV antigenome (pBRPolII-HrBDVc) or the BDV antigenome with mutated P gene together with 0.5 μg of pCA-N, 0.05 μg of pCA-P, and 0.1 μg of pCA-L (29, 38). Three days after transfection, cells were trypsinized and seeded onto 94-mm dishes together with 106 Vero cells. The cocultures were kept in DMEM supplemented with 4% FBS and were split twice a week. Rescue efficiency was monitored by immunofluorescence for at least 8 weeks. A first virus stock was made (see below) when >90% of the Vero cells were infected. To avoid contamination with input DNA from the transfected plasmids, the stock was used to infect Vero cells, and a second virus stock was made when >90% of the cells were infected. This stock was used for further experiments.
Virus stocks.
Recombinant BDV was isolated from persistently infected Vero cells. For this purpose, confluent cells in 94-mm dishes were rinsed with 5 ml of 20 mM HEPES, pH 7.4, and incubated for 1.5 h at 37°C in a 5% CO2-humidified atmosphere with 10 ml of 20 mM HEPES, pH 7.4, 250 mM MgCl2, and 1% FBS. The supernatant was centrifuged at 2,500 × g for 5 min to remove cell debris, followed by centrifugation above a 20% sucrose cushion at 100,000 × g at 20°C for 1 h. Viral pellets were resuspended in phosphate-buffered saline (PBS) and titrated on Vero cells.
Reverse transcription-PCR and sequencing.
RNA of recombinant viruses was prepared from virus stocks with peqGOLD TriFast reagent (PeqLab) according to the manufacturer's instructions. Reverse transcription-PCR was performed with random hexamer primers using a RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas). A total of 2 μl of cDNA was used to amplify the P ORF, and the PCR product was sequenced at GATC Biotech AG.
In vivo labeling of proteins.
To analyze phosphorylation of transiently expressed P mutants, HEK 293T cells in 22-mm dishes were transfected with 0.3 μg of the pCA-P construct coding for P-wt or the various P mutants or with an empty pCAGGS vector as a control. To analyze phosphorylation of P from recombinant BDV, semiconfluent Vero cells persistently infected or uninfected (control) were seeded on 22-mm dishes. Twenty hours after transfection or after seeding, cells were rinsed with 1 ml of DMEM without sodium phosphate and sodium pyruvate (Gibco) and kept for 15 min in DMEM lacking phosphate. Cells were incubated with 100 mCi of [32P]orthophosphate per ml for either 4 or 22 h. After the labeling step, whole-cell extracts were prepared. Therefore, cells were washed with ice-cold PBS, lysed with 120 μl of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris HCl, pH 8, 150 mM NaCL, 1% NP-40, 0.25% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 μM dithiothreitol, and 10 μM phenylmethylsulfonyl fluoride) supplemented with 1% (vol/vol) phosphatase inhibitor cocktail (Calbiochem) and incubated on ice for 15 min, followed by centrifugation for 15 min at 16,000 × g at 4°C. For immunoprecipitation, 100 μl of the supernatant was incubated with 1 μl of P-specific polyclonal rabbit antiserum (38) for 1 h at 4°C. A total of 12.5 μl of packed protein A-Sepharose beads (Amersham) was added, followed by incubation for 1 h at 4°C and four washing steps with RIPA buffer. Finally, beads were taken up in Laemmli buffer (26).
Immunoprecipitation.
For the analysis of the interaction between P and N as well as for P oligomerization, HEK 293T cells seeded in 22-mm dishes were transfected either with 0.25 μg of a pCA-HA-P mutant and 0.25 μg of pCA-N or with 0.25 μg each of a pCA-HA-P construct and the corresponding pCA-Flag-P mutant. For the analysis of the P/L and P/X interactions, HEK 293T cells were seeded into 35-mm dishes and transfected either with 0.2 μg of a pCA-HA-P construct and 2 μg of a pCA-X mutant or with 0.5 μg of a pCA-HA-P construct together with 2.5 μg of pCA-Flag-L, respectively. In the following, the given amounts of solutions correspond to 22-mm dishes, and double amounts were used for 35-mm dishes. Twenty-four hours after transfection, cells were washed with ice-cold PBS and lysed with 200 μl of RIPA buffer or, for the interaction between P and X, with 200 μl of coimmunoprecipitation lysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1% [vol/vol] protease inhibitor G [Serva], and 1 mM dithiothreitol). After incubation for 15 min at 4°C, cell extracts were centrifuged for 15 min at 16,000 × g at 4°C, and 10% (vol/vol) of cell extract was removed to analyze protein expression. The remaining supernatant was used for immunoprecipitation. For the interaction between P mutants, or P and X, 7.5 μl of packed monoclonal anti-HA-agarose (Sigma) was added to the cell extract and incubated for 2 h at 4°C, followed by four washing steps with the corresponding lysis buffer. For the immunoprecipitation of Flag-L or N, 1 μl of polyclonal anti-Flag antibody (Sigma) or 1 μl of N-specific polyclonal rabbit antiserum (39) was added to the cell extract and incubated for 1 h at 4°C. Ten microliters of packed protein A-Sepharose beads (GE Healthcare) was added, followed by incubation for 1 h at 4°C and four washing steps with RIPA buffer. Finally, proteins were eluted by boiling the beads in Laemmli buffer (26).
Immunofluorescence and Western blot analysis.
Immunofluorescence analysis was performed as previously described (38), using N-specific or P-specific polyclonal rabbit antiserum (39) in a 1:1,000 dilution or an anti-X antibody (40) in a 1:200 dilution.
For direct Western blot analysis, BDV-infected or uninfected Vero cells in 35-mm dishes were resuspended in Laemmli buffer (26) and sonicated. Western blot analysis was performed using standard techniques and 15% or 8% SDS-polyacrylamide gel electrophoresis (PAGE) gels. The N-specific and the P-specific polyclonal rabbit antisera (39) were diluted 1:1,000, the M-specific antiserum was diluted 1:800 (kindly provided by K. Tomonaga), and the X/P-specific antiserum (kindly provided by P. Staeheli) was diluted 1:500 in 5% low-fat milk powder in PBS and incubated with membranes overnight at 4°C. After samples were washed three times (each) for 15 min with PBS-0.1% Tween-20, antibodies were detected using horseradish peroxidase-coupled goat anti-rabbit antibodies (Jackson ImmunoResearch) diluted 1:1,000 in 5% low-fat milk powder in PBS, and visualization was performed using ECL Plus Western Blot Detection Reagents (GE Healthcare) according to the manufacturer's instructions.
RNA preparation and Northern blot analysis.
Total RNA was prepared from BDV-infected or uninfected Vero cells in 25-mm dishes with peqGOLD TriFast reagent (PeqLab). Northern blot analysis with 20 μg of total RNA was performed as previously described (38). For the detection of viral RNA derived from the N and X/P genes or the M, G, and L genes, nucleotides 976 to 1749 or 1014 to 4729, respectively, of the BDV antigenome were amplified by PCR using a plasmid encoding the full-length BDV antigenome (pBRPolII-HrBDVc) as a template. PCR products were radioactively labeled by using a DecaLabel DNA labeling kit (Fermentas).
Minireplicon assays.
Minireplicon assays were carried out according to Schneider et al. (38). Briefly, BSR-T7/5 cells in 22-mm dishes were transfected with 0.4 μg of the BDV minireplicon chloramphenicol acetyltransferase (CAT) reporter construct (pT7-gmgA) together with 0.5 μg of pCA-N, 0.2 μg of pCA-L, 0.05 μg of a pCA-P-construct, and 0.02 to 0.08 μg of a pCA-X-construct. The transfection mixture also contained 0.1 μg of an expression plasmid encoding the firefly luciferase gene under control of a T7 promoter to normalize transfection efficiency. At 60 h posttransfection CAT expression was analyzed, using a CAT enzyme-linked immunosorbent assay kit (Roche). Polymerase activity with P-wt was arbitrarily set to 1. Mean values of at least three independent experiments and standard error of the mean were calculated.
Mammalian two-hybrid assay.
Mammalian two-hybrid assays were performed as previously described (37). Briefly, HEK 293T cells in 22-mm dishes were transfected with 0.3 μg of pCA-VP16-N, -VP16-X, -VP16-P-wt, or -VP16-P-mutant, together with 0.3 μg of pCA-Gal4-P-mutant. To analyze the interaction with L, 0.4 μg of pCA-VP16-L and 0.2 μg of pCA-Gal4-P-wt or pCA-Gal4-P-mutant were transfected. The transfection mixture also contained 0.2 μg of a luciferase reporter construct harboring the firefly luciferase coding sequence downstream of the Gal4-binding site (37) and 0.1 μg of pRL-SV40 (where SV40 is simian virus 40) (Promega). At 24 h posttransfection, firefly and Renilla luciferase expression levels were analyzed by using a Dual Luciferase Kit from Promega and measuring light emission in a Lumat LB9501 luminometer (Berthold). Expression of Renilla luciferase was used to normalize transfection efficiency. Firefly luciferase expression of samples with Gal4-P-wt was arbitrarily set to 100%.
RESULTS
Cofactor activity of BDV-P and its dependency on phosphorylation.
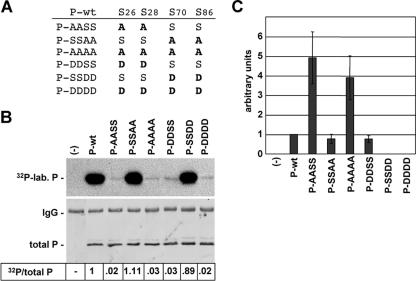
Previous studies have shown that BDV-P is predominantly phosphorylated at S26/28 by PKCɛ and weakly at S70/86 by CKII (41). To study the impact of P phosphorylation on its activity to support the viral polymerase, we mutated the phosphorylation sites (Fig. 1A) by replacing the serine residues at either positions 26 and 28 or positions 70 and 86 with alanine (A) or aspartic acid (D). While substitution with A abolishes phosphorylation, replacement with D can mimic phosphorylated serine residues by its charge distribution (17, 22). We first analyzed the phosphorylation status of the P mutants after transient expression in HEK 293T cells in the presence of 32P-labeled orthophosphate. As expected, P mutants with mutations of the S26/28 residues (P-AASS and P-DDSS) were only weakly phosphorylated (<3%) compared to P-wt, demonstrating that S26/28 are indeed the major phosphorylation sites (Fig. 1B). In contrast, P mutants with substitutions at residues S70/86 (P-SSAA and P-SSDD) were efficiently phosphorylated (Fig. 1B). P-DDDD or P-AAAA were as poorly phosphorylated as P-DDSS or P-AASS (Fig. 1B), indicating that the latter mutants are phosphorylated at very low levels at residues other than S70/86.
FIG. 1.
Phosphorylation and cofactor activity of BDV-P mutants lacking the major or minor phosphate acceptor sites. (A) BDV-P mutants harboring amino acid substitutions at the major (S26/28) and minor (S70/86) phosphorylation sites. (B) To determine the extent of phosphorylation of P-wt and P mutants, these proteins were transiently expressed in HEK 293T cells in the presence of 32P-labeled orthophosphate, immunoprecipitated, separated by SDS-PAGE using a 15% gel, and transferred to a nitrocellulose membrane. The upper panel represents an autoradiography showing the levels of radioactive labeled P (32P-lab. P). The lower panel shows the detection of total P by P-specific antiserum. The immunoglobulin G (IgG) heavy chain detected by the secondary antibody is indicated. Numbers give the ratio of 32P-labeled P proteins (32P) to levels of total P detected by Western blot analysis. The ratio of 32P/total P observed with P-wt was set to 1. −, no transfection of expression plasmids. (C) To determine the activity of the P mutants in supporting viral polymerase, a T7-based BDV minireplicon assay was performed. BSR-T7/5 cells were transfected with the BDV minigenome CAT reporter construct, the expression constructs for N and L, and plasmids encoding P-wt or mutant proteins as well as a plasmid constitutively expressing luciferase, which served to normalize variation in transfection efficiency. Minireplicon activity was analyzed by CAT enzyme-linked immunosorbent assay. Mean values of at least four independent assays are shown. −, complete reaction mixtures without P served as a negative control reaction.
We next tested the ability of the P mutants to support the viral polymerase using the BDV minireplicon system (38). For this purpose, BSR-T7/5 cells were transfected with L, N, and P expression constructs coding for either wt or mutant P proteins and a plasmid expressing a BDV minireplicon CAT reporter construct. Based on the results obtained with VSV-P (17, 32), we assumed that the BDV-P mutants with D substitutions would support the viral polymerase, since they mimic phosphorylated serine residues, whereas the P mutants with A substitutions should be inactive due to the destruction of the phosphorylation sites. As expected, P-DDSS showed activity similar to that of P-wt. However, to our surprise P-SSDD and P-DDDD were completely inactive (Fig. 1C). Furthermore, all mutants with A substitutions were active (Fig. 1C); notably, P-AASS and P-AAAA were fourfold more active than P-wt (Fig. 1C). The differences in the cofactor activity of the P mutants are not due to expression levels, which were comparable to P-wt (data not shown).
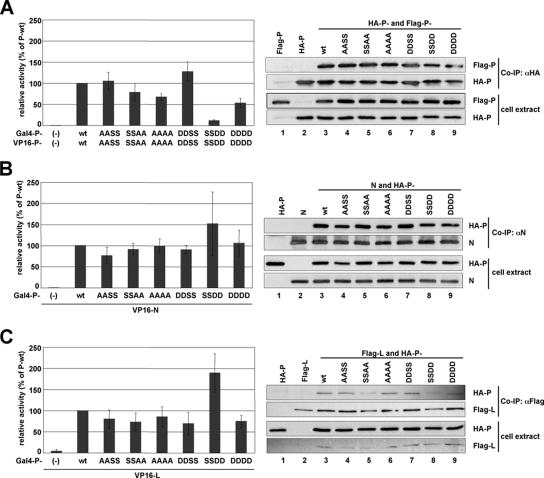
The inactivity of the P-DDDD and P-SSDD mutants in the minireplicon system could be explained by impaired self-oligomerization of these proteins or impaired binding to other nucleocapsid proteins such as L and N. Therefore, we tested all mutant proteins in a mammalian two-hybrid system as well as in coimmunoprecipitation experiments, which were previously shown to be suitable measurements for BDV protein interactions (37, 42). All P mutants, including P-SSDD and P-DDDD, showed no changes in the interaction with N, P, and L in the mammalian two-hybrid system, with the exception of P-SSDD, which interacted more strongly with N and L than P-wt protein (Fig. 2, graphs), whereas the self-oligomerization efficiency of P-SSDD and P-DDDD was reduced compared to P-wt (Fig. 2 A, graph). However, gel filtration analysis with cell extracts of transiently expressed P-SSDD revealed no impairment in formation of homo-oligomers compared to P-wt (data not shown). In addition, coimmunoprecipitation experiments with HA- and Flag-tagged P mutants confirmed the ability of P-SSDD and P-DDDD to form homo-oligomers (Fig. 2 A, right). Furthermore, these studies also revealed that the binding of the P mutants to N and L (Fig. 2 B and C, right panels) was not impaired. In summary, the P mutants P-DDDD and P-SSDD which are inactive in the BDV minireplicon assay still interact with N, L, and itself, excluding the possibility that these mutants are inactive due to a failure in binding.
FIG. 2.
Binding efficiency of BDV-P mutants to themselves, N, and L. Binding efficiencies of BDV-P mutants to themselves (A), N (B), and L (C) were determined by using the mammalian two-hybrid analysis (graphs) or coimmunoprecipitation studies. To study protein-protein interactions in the mammalian two-hybrid system, HEK 293T cells were transfected with a plasmid mix containing the indicated pCA-Gal4 and pCA-VP16 constructs, a luciferase reporter construct harboring the firefly luciferase coding sequence downstream of the Gal4 binding site, and pRL-SV40 expressing constitutively the Renilla luciferase. One day posttransfection the cells were lysed and analyzed for firefly and Renilla luciferase expression. Expression of Renilla luciferase was used to normalize for transfection efficiency. The normalized firefly luciferase expression observed with P-wt was set to 100%. For coimmunoprecipitation (Co-IP) studies, cell extracts were prepared 24 h posttransfection and subjected to immunoprecipitation using anti-HA-agarose (αHA), polyclonal anti-N serum (αN), or anti-Flag antibody (αFlag). Precipitated material was separated on a 15% SDS-PAGE and analyzed by Western blotting (WB) for the presence of the indicated HA- and Flag-tagged proteins and BDV-N using specific antibodies.
Generation of mutant viruses expressing BDV-P variants with mutated phosphorylation sites.
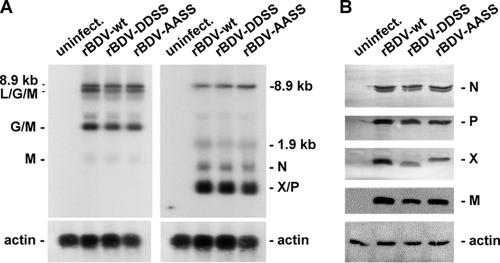
To study the effect of BDV-P phosphorylation on viral replication and growth, we attempted to generate recombinant BDV (rBDV) expressing the various P mutants (Fig. 1 A), using the recently established rescue system (29, 39). As anticipated from the minireplicon results, the rescue of rBDV expressing the inactive P mutants P-DDDD and P-SSDD was not possible. We also failed to generate rBDV expressing P-AAAA or P-SSAA, which did support high-level polymerase or wt-like activity in the BDV mininreplicon system. However, we could successfully rescue rBDV expressing P-AASS (rBDV-AASS) or P-DDSS (rBDV-DDSS). Sequence analysis revealed no other mutations in the P ORF in either mutant virus besides those intentionally introduced. We next compared the viral RNA and protein levels in Vero cells persistently infected with these mutant recombinant viruses or with rBDV-wt. Levels of viral transcripts derived from all three transcription units were similar among these viruses (Fig. 3A), indicating that the viral transcription in Vero cells infected with rBDV-DDSS or rBDV-AASS is not impaired. The same results were obtained with persistently infected Oligo cells (data not shown). In addition, as judged by strand-specific hybridization using an RNA probe complementary to the viral antigenome, no differences in the amount of antigenome between the mutant and wt recombinant viruses were observed (data not shown), indicating that viral replication is also not affected.
FIG. 3.
Viral RNA and protein expression pattern in Vero cells persistently infected with recombinant BDV expressing either wt or the mutant phosphoproteins P-AASS and P-DDSS. (A) Northern blot of uninfected Vero cells or Vero cells persistently infected with rBDV (rBDV-wt) or mutant viruses expressing P-DDSS (rBDV-DDSS) or P-AASS (rBDV-AASS). Blots were hybridized to DNA probes comprising nucleotides 1014 to 4729 (left panel) or 976 to 1749 (right panel) of the BDV antigenome. Rehybridization of the membranes with an actin probe confirmed that similar amounts of RNA were loaded onto the gel. (B) Western blot analysis of lysates of uninfected and persistently infected Vero cells. Proteins were separated by SDS-PAGE using a 15% gel, and Western blot analysis was carried out to detect the viral proteins N, P, X, and M. Detection of actin served as a control reaction to show that equal amounts of proteins were loaded.
The level of viral proteins N, P, and M were comparable in both mutant and wt viruses, whereas the level of X appeared to be reduced in mutant virus-infected cells (Fig. 3B). The X proteins expressed from rBDV-AASS and rBDV-DDSS show a delayed migration in SDS-PAGE gels compared to the wt X (Fig. 3B). Since the P and X are translated from a bicistronic mRNA with overlapping ORFs, multiple mutations were also introduced into the X ORF. Thus, X proteins expressed in rBDV-AASS- or rBDV-DDSS-infected cells contain either two (I42G and V44G) or four (I42G, G43W, V44G, and T45S) amino acid substitutions, respectively, that might have caused the atypical migration in SDS-PAGE.
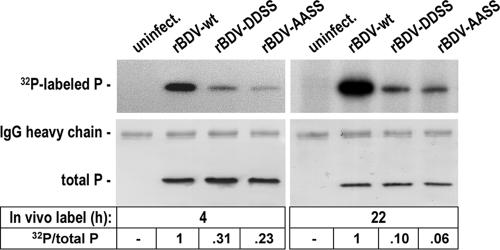
To determine the phosphorylation status of P mutants in the context of virus infection, wt- and mutant-infected Vero cells were incubated with 32P-labeled orthophosphate and analyzed for the incorporation of radioactive 32P. As shown in Fig. 4, P-DDSS and P-AASS were significantly less phosphorylated than P-wt after 4 h of in vivo labeling: compared to P-wt, 31% of P-DDSS and 23% of P-AASS were phosphorylated (left panel). These results indicate that S26/28 represent the major phosphorylation sites of P in persistently infected cells. However, compared to the phosphorylation obtained after transient expression, the residual phosphorylation of the P mutants in Vero cells persistently infected with rBDV-AASS and rBDV-DDSS is very high (compare Fig. 1B and 4). To exclude the possibility that a low turnover in S26/28 phosphorylation caused these discrepancies, we extended in vivo labeling of the infected cells to 22 h (Fig. 4, right panel). With the prolonged incubation time, only 10% of P-DDSS and 6% of P-AASS were phosphorylated compared to P-wt. These results suggest that, in contrast to transiently expressed P, phosphorylation sites other than S26/28 are phosphorylated in persistently infected cells, most likely S70/86 (41).
FIG. 4.
Phosphorylation of the viral phosphoprotein in Vero cells persistently infected with the recombinant BDV expressing either P-AASS or P-DDSS. Uninfected Vero cells or Vero cells persistently infected with either rBDV-wt or rBDV-AASS or rBDV-DDSS were labeled with [32P]orthophosphate for 4 h (left) or 22 h (right). P was subsequently immunoprecipitated, separated by SDS-PAGE using a 15% gel, and transferred to a nitrocellulose membrane. The upper panel represents an autoradiography showing the levels of radioactive labeled P. The lower panel shows the detection of total P by P-specific antibodies. The immunoglobulin G (IgG) heavy chain detected by the secondary antibody is indicated. Numbers at the bottom give the ratio of 32P-labeled P (32P) to levels of total P detected by Western blot analysis. The ratio of 32P/total P observed with P-wt was set to 1.
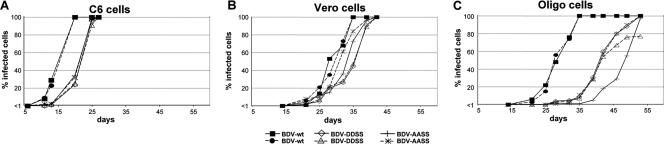
rBDV-DDSS and rBDV-AASS are attenuated in cell culture.
To study the growth properties of the recombinant viruses, we infected C6 cells, Vero cells, and Oligo cells with identical virus doses (1,000 focus-forming units) of rBDV-wt, rBDV-AASS, and rBDV-DDSS. Infection of C6 cells was most efficient, and complete infection of the culture was achieved with rBDV-wt after 20 days, whereas rBDV-AASS and rBDV-DDSS showed a delay in viral spread of approximately 5 days (Fig. 5A). A similar phenotype was observed in Vero cells. While rBDV-wt needed 35 days to infect all cells, rBDV-DDSS showed a delay of about 5 days (Fig. 5B). The rBDV-AASS virus showed an intermediate phenotype, and complete infection was slightly delayed (1 to 3 days) compared to wt virus (Fig. 5B). However, the differences between mutant and wt viruses in the ability to infect Oligo cells were even more pronounced: rBDV-wt completed infection of cells within 35 days, whereas at this time only about 5% of cells were infected by the mutant viruses (Fig. 5C). In one experiment, rBDV-DDSS had not completed infection of Oligo cells after 53 days postinfection. In summary, these results suggest that either the absence of phosphorylation or constitutive phosphorylation at the PKCɛ sites results in attenuated viruses with impaired efficiencies to spread in cell culture.
FIG. 5.
Recombinant BDV expressing P-AASS or P-DDSS is attenuated in cell culture. C6 (A), Vero (B), and Oligo (C) cells were simultaneously infected with the indicated BDV strains using 1,000 focus-forming units. The number of infected cells was determined by immunofluorescence using an N-specific antiserum.
The attenuation of rBDV-DDSS and rBDV-AASS is not due to impaired binding of X to P.
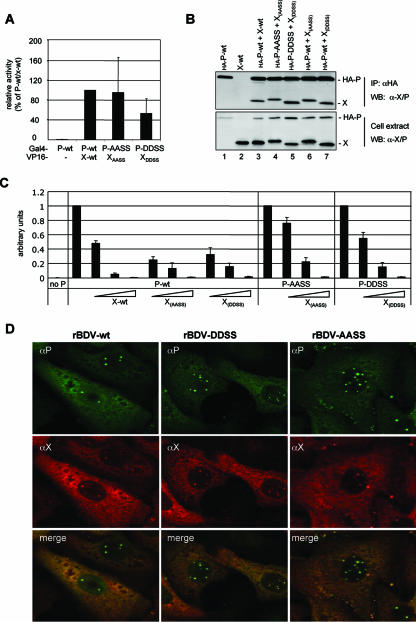
Both the rBDV-DDSS and rBDV-AASS viruses express X mutants, designated X(DDSS) and X(AASS), that harbor either two (I42G and V44G) or four (I42G, G43W, V44G, and T45S) amino acid substitutions. The attenuation of these two rBDV strains might therefore be caused by the amino acid substitutions in X, which may result in an impaired P binding. To test this possibility, we determined the complex formation between the X and P mutants using the mammalian two-hybrid system. These studies revealed efficient binding of X(DDSS) and X(AASS) to P-DDSS and P-AASS (Fig. 6A), although the interaction between X(DDSS) and P-DDSS was slightly impaired. Coimmunoprecipitation experiments confirmed complex formation between the X mutants and the corresponding P mutants (Fig. 6B). These analyses also revealed that X(DDSS) and X(AASS) bind equally well to P-wt (Fig. 6B), suggesting that the mutations introduced into X do not interfere with P binding. We next investigated the ability of the X mutants to block viral polymerase activity in the BDV minireplicon assay. Both X(DDSS) and X(AASS) inhibited the viral polymerase activity with comparable efficacy when P-wt was used in this assay (Fig. 6 C), demonstrating that the introduced mutations in X did not change this function. Similarly, coexpression of X(DDSS) and X(AASS) efficiently reduced the polymerase activity when P-DDSS and P-AASS, respectively, were used (Fig. 6C). Because the X mutants do bind to P and interfere with the polymerase activity in this reporter assay at a level similar to wt X, we next tested whether X(DDSS) and X(AASS) colocalize with P in the nucleus of Vero cells persistently infected with rBDV-DDSS and rBDV-AASS. As shown in Fig. 6D, immunofluorescence analysis using X- and P-specific antibodies revealed that X(DDSS) and X(AASS) do colocalize in the nucleus with P-DDSS and P-AASS, respectively. These results suggest that the mutations introduced into the X mutants abrogated neither their efficiency in binding to P nor their ability to interfere with the polymerase activity.
FIG. 6.
Functional characterization of P-DDSS and P-AASS and their corresponding X mutants. (A) Mammalian two-hybrid analysis with the indicated pCA-Gal4 and pCA-VP16 constructs was carried out as described in the legend of Fig. 2. Expression of Renilla luciferase was used to normalize for transfection efficiency. The normalized firefly luciferase expression observed with P-wt was set to 100%. (B) Cell extracts were prepared for coimmunoprecipitation studies 24 h posttransfection and subjected to immunoprecipitation (IP) using anti-HA-agarose (αHA). Precipitated material was separated by 15% SDS-PAGE and analyzed by Western blotting (WB) for the presence of X and P using an X/P-specific antibody. (C) Cofactor activity of the P mutants in the presence or absence of X mutant proteins was analyzed with the BDV minireplicon assay as described in the legend of Fig. 1C. Increasing levels of X-expressing plasmids correspond to 20, 40, and 80 ng. Mean values of at least three independent assays are shown. Complete reaction mixtures without P served as a negative control reaction (no P). (D) Double immunofluorescence analysis using polyclonal rabbit anti-P-antibodies (αP) and mouse monoclonal anti-X-antibodies (αX) of Vero cells persistently infected with the indicated recombinant viruses.
Effect of single amino acid substitutions at amino acid positions 70 or 86 of BDV-P.
While substitutions with D residues at amino acid residues 70 and 86 of BDV-P (P-SSDD and P-DDDD) are detrimental, substitutions with A residues (P-SSAA and P-AAAA) did not abrogate the cofactor activity of these proteins in the BDV minireplicon system (Fig. 1C). Nevertheless, we failed to rescue rBDV expressing these active P mutants. To explore the impact of single amino acid substitutions, we analyzed such BDV-P mutants in the BDV minireplicon assay for their polymerase-supporting activity. The BDV-P mutant with an S-to-D change at amino acid residue 86 (P-SSSD) retained only about 10% of wt activity (Fig. 7A). Attempts to rescue rBDV-SSSD were not successful (Fig. 7B), suggesting that S-to-D changes at amino acid position 86 also caused the failure to rescue rBDV expressing P-DDDD or P-SSDD. In contrast, S-to-A changes at this amino acid position (P-SSSA) did not significantly impair its cofactor activity (Fig. 7A), and rBDV expressing this mutant protein could be rescued (Fig. 7B).
FIG. 7.
Impact of single amino acid substitutions at amino acid position 70 or 86 of BDV-P. (A) Cofactor activity of P mutants using the BDV minireplicon assay as described in the legend of Fig. 1C. Mean values of at least three independent assays are shown. Complete reaction mixtures without P served as a negative control reaction (no P). (B) Table showing the ability to rescue BDV mutants. (C) Mammalian two-hybrid analysis. Expression of Renilla luciferase was used to normalize for transfection efficiency. The normalized firefly luciferase expression observed with P-wt was set to 100%. (D) Coimmunoprecipitation studies. Cell extracts were prepared 24 h posttransfection and subjected to immunoprecipitation (IP) using anti-HA-agarose (αHA). Precipitated material was separated by 15% SDS-PAGE and analyzed by Western blotting (WB) for the presence of the indicated HA-tagged and X proteins using an X/P-specific antibody (αX/P). (E) Cofactor activity of the P mutants in the presence or absence of X wt or mutant proteins using the BDV minireplicon system. Mean values of at least three independent assays are shown. Complete reaction mixtures without P served as a negative control reaction (no P). Increasing levels of X-expressing plasmids correspond to 20, 40, and 80 ng.
We next studied P mutants with D or A substitutions at amino acid position 70. These mutations resulted in different cofactor activities: whereas P-SSDS retained 35% of its activity compared to wt P, the P mutant harboring an A substitution at this amino acid position (P-SSAS) showed almost the same activity as wt P (Fig. 7A). We failed to rescue rBDV expressing P-SSDS (Fig. 7B), which might be attributable to the low cofactor activity of this P mutant. However, several attempts to generate rBDV coding for the highly active P-SSAS also failed (Fig. 7B), indicating that other functions of P are disturbed. Since amino acid 70 of BDV-P is located near the X-binding site of P (25), we speculated that the failure to rescue rBDV-SSAS is due to impaired X binding by P-SSAS. We therefore studied the interaction of P-SSAS with wt X and X(SSAS), which harbors a single amino acid substitution (I86G). As shown in Fig. 7C, mammalian two-hybrid analysis revealed that P-SSAS is only poorly associated with both X(SSAS) and wt X (Fig. 7C). Coimmunoprecipitation studies using X and HA-tagged P proteins confirmed that P-SSAS inefficiently binds to X(SSAS) as well as wt X (Fig. 7D, lanes 4 and 5). Thus, the inefficient complex formation of P-SSAS with X(SSAS) is due to the S-to-A change at position 70 of P. Since P-AAAA also harbors an A at position 70, we examined its ability to interact with wt X and X(AAAA), harboring 5 amino acid substitutions (I42S, G43W, V44G, T45S, and I86G). As expected, mammalian two-hybrid and coimmunoprecipitation analyses showed that P-AAAA only inefficiently binds to wt X and X(AAAA) (Fig. 7C and D, lane 7). On the other hand, efficient binding was observed between X(SSSA) and P-SSSA (Fig. 7C and D, lane 6) as well as between X(AASS) and P-AASS (Fig. 6A and B, lane 4). These results strongly suggest that amino acid residue 70 is critical for efficient X binding.
By using the BDV minireplicon system, we next addressed the question of whether the impaired X binding of P-SSAS and P-AAAA also influences the inhibitory activity of X(SSAS) and X(AAAA). As shown in Fig. 7E, both X mutants could block the viral polymerase activity as efficiently as wt X when P-wt was used in the assay. However, when P-SSAS was used for the reconstitution of the viral polymerase complex, efficiency was reduced by up to eightfold at the highest X(SSAS) plasmid concentrations (Fig. 7E). The failure in blocking the polymerase activity was even more pronounced (>20-fold) when X(AAAA) was coexpressed with P-AAAA (Fig. 7E). In summary, these results suggest that the single S-to-A amino acid change at position 70 of BDV-P interferes with X binding and impairs the regulatory activity of X.
DISCUSSION
In this study we addressed the question of whether BDV-P phosphorylation is necessary to support polymerase activity and viral growth. To this end, we analyzed P mutants with modified PKCɛ or the CKII phosphorylation sites by replacing the corresponding phosphate acceptor sites with alanine (A) or aspartic acid (D) to mimic the conditions of no phosphorylation or constitutive phosphorylation, respectively. The functional characterization of these P mutants in the BDV minireplicon assay indicated that phosphorylation at the CKII sites and, to a lesser extent, at the PKCɛ sites negatively regulates its cofactor activity. This is based on the observation that replacement of the CKII sites (S70/86) with D abrogated the polymerase activity completely, whereas substitutions with A had no effect. Furthermore, P mutants with A substitutions at the PKCɛ sites led to a fivefold increase in polymerase activity in the minireplicon assay, whereas substitution with D slightly impaired its activity. The view that phosphorylation of P negatively regulates its activity is also consistent with the results obtained by the P mutants harboring four amino acid substitutions: while P-DDDD was inactive, P-AAAA was highly active. Thus, BDV is the first example among Mononegavirales that does not activate but exclusively downregulates its polymerase activity by phosphorylation of P. Clearly, for VSV, phosphorylation of P is mandatory for efficient transcription and replication (2, 23, 32, 45). However, hyperphosphorylation of VSV-P can also cause the inhibition of replication, thereby most likely promoting efficient progeny maturation during the lytic infection cycle (9, 23). For BDV, which persistently infects cells without causing a cytopathic effect, P phosphorylation might be necessary to prevent uncontrolled polymerase activity and subsequent lysis of cells.
As anticipated, we failed to rescue rBDV expressing P-DDDD or P-SSDD, which lacked detectable cofactor activity in the minireplicon assay. Similarly, the generation of recombinant viruses expressing P mutants with single D substitutions (P-SSDS and P-SSSD) also failed, which could be attributed to the reduced cofactor activity of these mutant proteins. However, it was also not possible to generate mutant viruses expressing P-AAAA and P-SSAA although these proteins supported polymerase activity. In principle, both A substitutions at positions 70 and 86 may have caused this failure. In addition, since P and X are expressed from a bicistronic mRNA, the nucleotide exchanges introduced into the P ORF coding for S70 also affected the X ORF, resulting in a single amino acid substitution (I86G). Analysis of P mutants harboring single A substitutions at amino acid position 70 (P-SSAS) or 86 (P-SSSA) revealed no significant impairment in their cofactor activity. However, only mutant virus expressing P-SSSA could be rescued, whereas several attempts to rescue rBDV coding for P-SSAS failed. We speculate that the failure to rescue mutant virus expressing P-SSAS is caused by an impaired regulatory activity of X since we can provide experimental evidence that the single S-to-A amino acid change at position 70 of BDV-P prevents efficient binding to X. However, we cannot exclude the possibility that the substitution in X might account for the inability to generate the latter recombinant virus by abrogating yet unknown functions of X. Unfortunately, this issue cannot further be clarified, since it is not possible to create a recombinant virus harboring the A mutation at position 70 of P without also changing position 86 of X.
Compared to rBDV-wt, the mutant viruses rBDV-DDSS and rBDV-AASS were impaired in their ability to spread efficiently in various cell types. The attenuation especially of the latter strain is surprising because P-AASS was fivefold more active than wt P in the BDV minireplicon system. Since the P and X ORFs overlap, multiple mutations were also introduced into the X ORF. As a consequence, rBDV-AASS and rBDV-DDSS expressed X mutants with either two (I42G and V44G) or four (I42G, G43W, V44G, and T45S) amino acid substitutions. We cannot exclude the possibility that these mutations in X may disrupt yet unknown functions. However, functional studies revealed that these X mutants were not impaired in their ability to interact with the corresponding P mutants and to block the viral polymerase activity. Alternatively, if efficient viral spread is dependent on the regulation of the cofactor activity of P by phosphorylation and dephosphorylation, the A or D substitutions in P itself would have caused the attenuated phenotype of both strains. In addition, other functions of P could be affected too. Similar to the human respiratory syncytial virus P, where phosphorylation of S54 regulates the viral budding process by blocking the interaction of P with the viral matrix protein (1), deregulated BDV-P phosphorylation may affect efficient virus assembly.
We can only speculate on the cell culture-dependent differences in viral spread observed between wt and mutant viruses. Viral replication in Vero and C6 cells is not affected by interferon (IFN)-induced antiviral gene products since Vero cells are deficient in producing IFN-α/β (16, 30) and since C6 cells lack antiviral gene products that specifically inhibit BDV replication (19). In contrast, BDV growth in Oligo cells is prevented by exogenous exposure to IFN-α/β. Therefore, the pronounced delay in viral spread in Oligo cells might be due to induction of the IFN system by the mutant viruses. As shown by other investigators, wt BDV-P can interfere with the induction of IFN by binding to the Traf family member-associated NF-κB activator-binding kinase 1 (TBK-1) and thereby blocking phosphorylation and activation of the interferon regulatory factor 3 (IRF3) (47). By using a reporter assay system to measure IRF3 activation through TBK-1 (47), we could show that P mutants with A and D substitutions at the PKCɛ sites showed a similar rate of inhibition of IRF3 activity to wt P (S. Schmid and M. Schwemmle, unpublished data). These results argue against the possibility that the delay in viral spread is due to impaired IRF3 inhibition. However, they do not exclude the possibility that the attenuation of both mutant viruses in Oligo cells is caused by the innate immune system.
Transiently expressed P is mainly phosphorylated at S26/28 and inefficiently if at all at S70/86. The reason for the almost exclusive phosphorylation at S26/28 of P when P is expressed alone is unclear. In the context of viral infection, P is phosphorylated at S70/86 (41). This might explain the 6 to 10% phosphorylation of P mutants in rBDV-infected Vero cells expressing P-DDSS and P-AASS. However, we cannot exclude the possibility that these proteins are also phosphorylated at alternative sites. Alternate phosphorylation was shown for SV-P when the major phosphorylation site (S249) was mutated (20, 21). However, alternate phosphorylation was found not only in cells infected with recombinant SV but also after expression of the corresponding mutant protein alone (8). Since the BDV-P mutants P-DDSS and P-AASS are very inefficiently phosphorylated when expressed alone (less than 3% compared to wt P), we speculate that in the mutant-infected cells, both proteins are not phosphorylated at other phosphate acceptor sites than the described CKII sites. In addition, we speculate that proper complex formation of the P mutants P-DDSS and P-AASS with other viral proteins is a prerequisite for S70/86 phosphorylation.
Recent reports suggest that BDV-P not only is an excellent substrate for PKCɛ in neurons but also serves as a kinase decoy, resulting in reduced phosphorylation of other PKCɛ substrates such as the presynaptic vesicle protein myristylated alanine-rich protein kinase C substrate (50). As a consequence, the BDV-infected neurons show synaptic dysfunctions resulting in learning deficiencies observed in BDV-infected newborn rats (18). With the successful generation of BDV lacking the PKC phosphorylation sites, experiments to test this hypothesis are now possible.
Acknowledgments
We thank Christel Hässler for excellent help and Geoffrey Chase, Otto Haller, Peter Staeheli, and Friedemann Weber for critical reading of the manuscript.
D.M. was supported by a grant from the Schweizerische Stiftung für medizinisch biologische Stipendien through a donation by Novartis AG. This work was supported by grant SCHW 632 from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Asenjo, A., L. Rodriguez, and N. Villanueva. 2005. Determination of phosphorylated residues from human respiratory syncytial virus P protein that are dynamically dephosphorylated by cellular phosphatases: a possible role for serine 54. J. Gen. Virol. 86:1109-1120. [DOI] [PubMed] [Google Scholar]
- 2.Barik, S., and A. K. Banerjee. 1992. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein. Proc. Natl. Acad. Sci., USA 89:6570-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barik, S., and A. K. Banerjee. 1992. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J. Virol. 66:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briese, T., J. C. de la Torre, A. Lewis, H. Ludwig, and W. I. Lipkin. 1992. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc. Natl. Acad. Sci. USA 89:11486-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briese, T., A. Schneemann, A. J. Lewis, Y.-S. Park, S. Kim, H. Ludwig, and W. I. Lipkin. 1994. Genomic organization of Borna disease virus. Proc. Natl. Acad. Sci. USA 91:4362-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrappa, S., and K. C. Gupta. 1999. Human parainfluenza virus type 1 phosphoprotein is constitutively phosphorylated at Ser-120 and Ser-184. J. Gen. Virol. 80:1199-1209. [DOI] [PubMed] [Google Scholar]
- 8.Byrappa, S., Y. B. Pan, and K. C. Gupta. 1996. Sendai virus P protein is constitutively phosphorylated at serine249: high phosphorylation potential of the P protein. Virology 216:228-234. [DOI] [PubMed] [Google Scholar]
- 9.Chang, T. L., C. S. Reiss, and A. S. Huang. 1994. Inhibition of vesicular stomatitis virus RNA synthesis by protein hyperphosphorylation. J. Virol. 68:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. L., T. Das, and A. K. Banerjee. 1997. Phosphorylated states of vesicular stomatitis virus P protein in vitro and in vivo. Virology 228:200-212. [DOI] [PubMed] [Google Scholar]
- 11.Cubitt, B., C. Oldstone, V. Valcarel, and J. C. de la Torre. 1994. RNA splicing contributes to the generation of mature mRNAs of Borna disease virus, a non-segmented negative strand RNA virus. Virus Res. 34:69-79. [DOI] [PubMed] [Google Scholar]
- 12.Das, D., A. K. Gupta, P. W. Sims, C. A. Gelfand, J. E. Jentoft, and A. K. Banerjee. 1995. Role of cellular casein kinase II in the function of the phosphoprotein (P) subunit of the RNA polymerase of vesicular stomatitis virus. J. Biol. Chem. 270:24100-24107. [DOI] [PubMed] [Google Scholar]
- 13.Das, S. C., and A. K. Pattnaik. 2004. Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J. Virol. 78:6420-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, T., A. Schuster, S. Schneider-Schaulies, and A. K. Banerjee. 1995. Involvement of cellular casein kinase II in the phosphorylation of measles virus P protein: identification of phosphorylation sites. Virology 211:218-226. [DOI] [PubMed] [Google Scholar]
- 15.De, B. P., and A. K. Banerjee. 1997. Role of host proteins in gene expression of nonsegmented negative strand RNA viruses. Adv. Virus Res. 48:169-204. [DOI] [PubMed] [Google Scholar]
- 16.Desmyter, J., J. L. Melnick, and W. E. Rawls. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, Y., and J. Lenard. 1995. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 14:1240-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Dunia, D., R. Volmer, D. Mayer, and M. Schwemmle. 2005. Borna disease virus interference with neuronal plasticity. Virus Res. 111:224-234. [DOI] [PubMed] [Google Scholar]
- 19.Hallensleben, W., and P. Staeheli. 1999. Inhibition of Borna disease virus multiplication by interferon: cell line differences in susceptibility. Arch. Virol. 144:1209-1216. [DOI] [PubMed] [Google Scholar]
- 20.Hu, C., and K. C. Gupta. 2000. Functional significance of alternate phosphorylation in Sendai virus P protein. Virology 268:517-532. [DOI] [PubMed] [Google Scholar]
- 21.Hu, C. J., A. Kato, M. C. Bowman, K. Kiyotani, T. Yoshida, S. A. Moyer, Y. Nagai, and K. C. Gupta. 1999. Role of primary constitutive phosphorylation of Sendai virus P and V proteins in viral replication and pathogenesis. Virology 263:195-208. [DOI] [PubMed] [Google Scholar]
- 22.Huang, W., and R. L. Erikson. 1994. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc. Natl. Acad. Sci. USA 91:8960-8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang, L. N., N. Englund, T. Das, A. K. Banerjee, and A. K. Pattnaik. 1999. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J. Virol. 73:5613-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonscher, K. R., and J. R. Yates III. 1997. Matrix-assisted laser desorption ionization/quadrupole ion trap mass spectrometry of peptides. Application to the localization of phosphorylation sites on the P protein from Sendai virus. J. Biol. Chem. 272:1735-1741. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, T., G. Zhang, B. J. Lee, S. Baba, M. Yamashita, W. Kamitani, H. Yanai, K. Tomonaga, and K. Ikuta. 2003. Modulation of Borna disease virus phosphoprotein nuclear localization by the viral protein X encoded in the overlapping open reading frame. J. Virol. 77:8099-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. J. Mol. Biol. 80:575-581. [DOI] [PubMed] [Google Scholar]
- 27.Landt, O., H. P. Grunert, and U. Hahn. 1990. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96:125-128. [DOI] [PubMed] [Google Scholar]
- 28.Lu, B., C. H. Ma, R. Brazas, and H. Jin. 2002. The major phosphorylation sites of the respiratory syncytial virus phosphoprotein are dispensable for virus replication in vitro. J. Virol. 76:10776-10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, A., P. Staeheli, and U. Schneider. 2006. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J. Virol. 80:5708-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarro, J., C. Lopez-Otin, and N. Villanueva. 1991. Location of phosphorylated residues in human respiratory syncytial virus phosphoprotein. J. Gen. Virol. 72:1455-1459. [DOI] [PubMed] [Google Scholar]
- 32.Pattnaik, A. K., L. Hwang, T. Li, N. Englund, M. Mathur, T. Das, and A. K. Banerjee. 1997. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J. Virol. 71:8167-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez, M., A. Sanchez, B. Cubitt, D. Rosario, and J. C. de la Torre. 2003. A reverse genetics system for Borna disease virus. J. Gen. Virol. 84:3099-3104. [DOI] [PubMed] [Google Scholar]
- 34.Poenisch, M., G. Unterstab, T. Wolff, P. Staeheli, and U. Schneider. 2004. The X protein of Borna disease virus regulates viral polymerase activity through interaction with the P protein. J. Gen. Virol. 85:1895-1898. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, P. A., A. Schneemann, and W. I. Lipkin. 1994. RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus. J. Virol. 68:5007-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider, U. 2005. Novel insights into the regulation of the viral polymerase complex of neurotropic Borna disease virus. Virus Res. 111:148-160. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, U., K. Blechschmidt, M. Schwemmle, and P. Staeheli. 2004. Overlap of interaction domains indicates a central role of the P protein in assembly and regulation of the Borna disease virus polymerase complex. J. Biol. Chem. 279:55290-55296. [DOI] [PubMed] [Google Scholar]
- 38.Schneider, U., M. Naegele, P. Staeheli, and M. Schwemmle. 2003. Active Borna disease virus polymerase complex requires a distinct nucleoprotein-to-phosphoprotein ratio but no viral X protein. J. Virol. 77:11781-11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, U., M. Schwemmle, and P. Staeheli. 2005. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc. Natl. Acad. Sci. USA 102:3441-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwardt, M., D. Mayer, R. Frank, U. Schneider, M. Eickmann, O. Planz, T. Wolff, and M. Schwemmle. 2005. The negative regulator of Borna disease virus polymerase is a non-structural protein. J. Gen. Virol. 86:3163-3169. [DOI] [PubMed] [Google Scholar]
- 41.Schwemmle, M., B. De, L. Shi, A. Banerjee, and W. I. Lipkin. 1997. Borna disease virus P-protein is phosphorylated by protein kinase Cɛ and casein kinase II. J. Biol. Chem. 272:21818-21823. [DOI] [PubMed] [Google Scholar]
- 42.Schwemmle, M., M. Salvatore, L. Shi, J. Richt, C. Lee, and W. Lipkin. 1998. Interactions of the Borna disease virus P, N, and X proteins and their functional implications. J. Biol. Chem. 273:9007-9012. [DOI] [PubMed] [Google Scholar]
- 43.Spadafora, D., D. M. Canter, R. L. Jackson, and J. Perrault. 1996. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J. Virol. 70:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staeheli, P., C. Sauder, J. Hausmann, F. Ehrensperger, and M. Schwemmle. 2000. Epidemiology of Borna disease virus. J. Gen. Virol. 81:2123-2135. [DOI] [PubMed] [Google Scholar]
- 45.Takacs, A. M., S. Barik, T. Das, and A. K. Banerjee. 1992. Phosphorylation of specific serine residues within the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J. Virol. 66:5842-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomonaga, K., T. Kobayashi, and K. Ikuta. 2002. Molecular and cellular biology of Borna disease virus infection. Microbes Infect. 4:491-500. [DOI] [PubMed] [Google Scholar]
- 47.Unterstab, G., S. Ludwig, A. Anton, O. Planz, B. Dauber, D. Krappmann, G. Heins, C. Ehrhardt, and T. Wolff. 2005. Viral targeting of the interferon-β-inducing Traf family member-associated NF-κB activator (TANK)-binding kinase-1. Proc. Natl. Acad. Sci. USA 102:13640-13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villanueva, N., R. Hardy, A. Asenjo, Q. Yu, and G. Wertz. 2000. The bulk of the phosphorylation of human respiratory syncytial virus phosphoprotein is not essential but modulates viral RNA transcription and replication. J. Gen. Virol. 81:129-133. [DOI] [PubMed] [Google Scholar]
- 49.Villanueva, N., J. Navarro, E. Mendez, and I. Garcia-Albert. 1994. Identification of a protein kinase involved in the phosphorylation of the C-terminal region of human respiratory syncytial virus P protein. J Gen. Virol. 75:555-565. [DOI] [PubMed] [Google Scholar]
- 50.Volmer, R., C. Monnet, and D. Gonzalez-Dunia. 2006. Borna disease virus blocks potentiation of presynaptic activity through inhibition of protein kinase C signaling. PLOS Pathog. 2:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wehner, T., A. Ruppert, C. Herden, K. Frese, H. Becht, and J. A. Richt. 1997. Detection of a novel Borna disease virus encoded 10 kilodalton protein in infected cells and tissues. J. Gen. Virol. 8:2459-2466. [DOI] [PubMed] [Google Scholar]
- 52.Yanai, H., Y. Hayashi, Y. Watanabe, N. Ohtaki, T. Kobayashi, Y. Nozaki, K. Ikuta, and K. Tomonaga. 2006. Development of a novel Borna disease virus reverse genetics system using RNA polymerase II promoter and SV40 nuclear import signal. Microbes Infect. 8:1522-1529. [DOI] [PubMed] [Google Scholar]