Abstract
Mustached monkeys (Cercopithecus cephus), which form a significant component of primate bushmeat in west central Africa, are infected with simian immunodeficiency virus (SIVmus). We identified and genetically characterized five new SIVmus strains infecting wild living mustached monkeys from Cameroon. Phylogenetic analysis of partial pol sequences revealed that SIVmus strains form two distinct groups within the clade comprised of lentiviruses isolated from C. nictitans (SIVgsn), C. mona (SIVmon) and C. cephus (SIVmus). Characterisation of three full-length SIVmus genomes confirmed the presence of two distinct lineages infecting mustached monkeys. These two variants of SIVmus, here designated SIVmus-1 and SIVmus-2, were isolated from animals sharing habitats within the same geographic region. Phylogenetic analyses showed that the diversification of SIVmus, SIVgsn and SIVmon involved inter-lineage recombination, and suggested that one of the SIVmus lineages likely resulted from cross-species transmission and recombination involving SIVmus and an as yet uncharacterized SIV. These results indicate that cross-species transmission and recombination play a major role in the evolution of primate lentiviruses among sympatric primate species.
Keywords: SIV, SIVmus-1, SIVmus-2, lentivirus, primate, cross-species transmission, recombination, polyspecific association
Introduction
It is now well established that the human immunodeficiency viruses, HIV-1 and HIV-2, are the results of cross-species transmissions of lentiviruses naturally infecting non-human primates in sub-Saharan Africa (Hahn et al., 2000). These simian immunodeficiency viruses (SIVs) are generally species-specific, i.e. SIVs cluster as monophyletic lineages according to the species from which they were isolated; the viruses are given a three-letter code reflecting the common name of the host primate, such as SIVmus from mustached monkeys. To date, serological evidence of SIV infection has been reported in 36 different primate species and partial or full-length viral sequences have been characterized from 30 of these (Bibollet-Ruche et al., 2004; Dazza et al., 2005). While the immediate ancestor of HIV-1 was SIVcpz infecting chimpanzees (Pan troglodytes troglodytes) in West Central Africa (Corbet et al., 2000; Gao et al., 1999; Keele et al., 2006) the natural hosts of all other SIVs so far reported are Old World monkey species. Indeed, SIVcpz was derived from a recombination event between two distinct SIV lineages infecting different monkeys; these were the ancestor of SIVrcm from red capped mangabeys (Cercocebus torquatus), and the ancestor of a clade of viruses (SIVgsn, SIVmon, and SIVmus) found infecting three species of Cercopithecus monkeys, greater spot-nosed monkeys (C. nictitans), mona monkeys (C. mona), and mustached monkeys (C. cephus) (Bailes et al., 2003; Courgnaud et al., 2002). A major fragment of the SIVcpz (and hence HIV-1) genome, covering the vpu and env genes, was derived from the SIVgsn/mus/mon lineage (Courgnaud et al., 2003; Sharp et al., 2005) but as yet few of these viruses have been characterized and there is a need to study in more detail the prevalence, genetic diversity and biological characteristics of SIVgsn, SIVmus and SIVmon strains in their natural hosts and environment.
The route of SIV transmission from primates to humans is believed to have been blood exposure resulting from bushmeat hunting (Hahn et al., 2000). SIVcpz and SIVsmm (the ancestor of HIV-2, infecting sooty mangabeys) are each known to have crossed the species barrier to humans on multiple occasions, but the transmission potential of the other primate lentiviruses remains unknown. We have shown that a substantial proportion of wild-living monkeys in Cameroon are infected with SIV, and that humans are exposed to a plethora of genetically diverse viruses through hunting and handling bushmeat (Peeters et al., 2002). Mustached and greater spot-nosed monkeys constitute a large proportion of primate bushmeat, emphasizing the need for further surveillance of these viruses and investigation as to whether they could directly infect humans. Using sensitive and specific serological tools, we recently showed that SIV prevalence in mustached and greater spot-nosed monkeys is very low (3–4%) (Aghokeng et al., 2006) compared to that observed in wild De Brazza monkeys, mantled guerezas, mandrills and sooty mangabeys, where prevalences range from 25% to 50% (Aghokeng et al., 2006; Apetrei et al., 2005; Santiago et al., 2005; Souquiere et al., 2001). However, greater spot-nosed monkey samples and especially mustached monkey samples exhibited much higher rates (up to 38%) of cross-reactivity with HIV-1 and HIV-2 antigens. HIV cross-reactivity usually underestimates the rate of SIV infection, but confirmatory immunoblot analysis (using SIVgsn Env pseudotyped HIV-1 particles as antigen) detected SIV-specific antibodies only in sera that were SIV ELISA positive, indicating false positive HIV cross-reactivity (Aghokeng et al., 2006). Nevertheless, it remains important to determine whether these monkey species harbour other divergent SIVs that are not detected by the current SIVmus/gsn strain specific assays.
Here we focus on SIVmus from mustached monkeys. We have further investigated the SIV infection status of samples that were discordant in SIV-specific and HIV cross-reactive serological tests. Previously, only one full-length genome sequence of SIVmus has been determined (Courgnaud et al., 2003). We have genetically characterized five additional SIVmus strains, and determined complete genome sequences for three of these. Phylogenetic analyses reveal that two distinct variants of SIVmus co-circulate in mustached monkeys in southern Cameroon, and imply that cross-species transmission and recombination has occurred between SIVs infecting sympatric monkey species.
RESULTS
Confirmation of SIV infection by PCR and sequence analysis
A total of 46 samples previously identified with cross-reacting antibodies to HIV antigens and/or SIVmus-specific recombinant gp41 ELISAs were subjected to PCR analyses using a combination of universal and lineage specific primers. DNA quality and species identification by glucose-6-phosphate dehydrogenase (G6PDH) gene amplification and sequencing confirmed that all 46 samples were derived from mustached monkeys (Cercopithecus cephus). Only five of the samples were reactive in the SIVmus-specific recombinant gp41 ELISA, and SIV sequences were amplified only from these samples (Table 1). All five samples were amplified in pol with the lineage-specific primers, but only two out of the five were amplified with the “universal” SIV primers known to amplify sequences from a wide variety of SIV lineages. Sequence analysis revealed mismatches between the three unamplified samples and the inner reverse primer (Uni2), explaining the negative PCR results with the universal SIV primer set for these samples. For env, only three of the five samples were amplified, even though lineage-specific primers were used. All of the other 41 samples, with cross reactive antibodies only in the INNO-LIA HIV assay, remained negative in several PCR attempts, with either the “universal” SIV primers or with the SIVgsn/mus/mon lineage-specific primers.
Table 1.
Antibody profiles in INNO-LIA HIV, of samples positive in the SIVmus-specific ELISAs and results of SIVmus PCR amplifications attempts with different primer sets.
| Sample ID | Serology | PCRa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pol | env | |||||||||||
| gp120 | gp41 | p34 | p24 | p17 | gp105 | gp36 | INNO-LIA Status | SIVMUS rgp41ELISA Status | Universal pol primers650 bp | SIVgsn/mus/mon lineage specific pol primers2050 bp | SIVgsn/mus/mon lineage specific env primers2150 bp | |
| 01CM1239 | (+) | +++ | − | − | − | − | +++ | Pos | Pos | + | + | + |
| 01CM1246 | − | +++ | − | +++ | − | − | +++ | Pos | Pos | + | + | + |
| 01CM2500 | (tr) | ++ | (tr) | +/− | ++ | − | − | Pos | Pos | − | + | + |
| 01CM2573 | − | + | +/− | +/− | (tr) | (tr) | +++ | Pos | Pos | − | + | − |
| 01CM2554 | ++ | + | − | +++ | + | − | +++ | Pos | Pos | − | + | − |
(tr): trace; (+): weakly positive.
Universal pol primers are NDR1-PolOR (first round) and PolIS4-Uni2 (second round); SIVgsn/mus/mon lineage specific pol primers are CNM.F1-PolOR2 (first round) and CNM.F2-CNM.R2 (second round); and SIVgsn/mus/mon lineage specific env prim ers are CNMenvF1-CNMenvR 1 (first round) and CNMenvF2-CNMenvR2 (second round). Primer sequences are reported in table 2.
Phylogenetic analysis of the 2050 bp fragment in pol showed that these five new SIVmus pol sequences clustered with the previously described sequences from the SIVgsn/mus/mon lineage. However, we observed two distinct SIVmus clusters (Figure 1A). 01CM1239 (i.e., SIVmus-01CM1239) and the previously characterized 01CM1085 (Courgnaud et al., 2003) constituted one group, which we term SIVmus-1. The other four sequences (01CM1246, 01CM2500, 01CM2554 and 01CM2573) formed a second clade, SIVmus-2, which was more closely related to SIVgsn than to SIVmus-1. Within the mus-2 group, sequences from 01CM2500, 01CM2554 and 01CM2573 were very closely related to each other, exhibiting only about 5% nucleotide divergence. These viral sequences were obtained from three animals sampled during the same month in neighbouring villages only 5 km apart, and their sequence similarity is indicative of epidemiological linkage. The 2150 bp env fragment could be amplified for only three samples. Together with the previously characterized sequence, these represented two members of each of the two SIVmus groups identified in the pol tree. However, in the env tree (Figure 1B) all four SIVmus strains formed a monophyletic cluster. This discordance between the relationships derived from pol and env sequences suggests that there was inter-lineage recombination during the divergence of SIVmus and SIVgsn.
Figure 1.
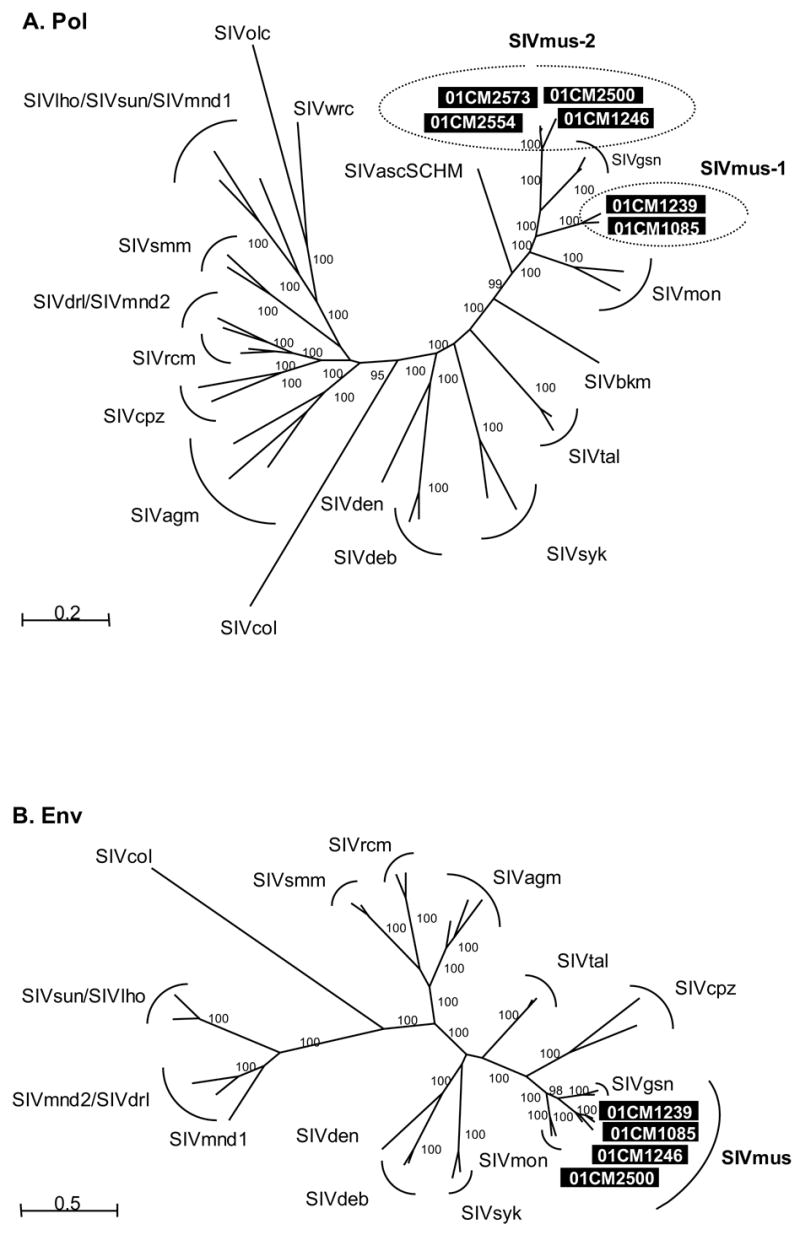
Phylogenetic relationships of the newly derived SIVmus sequences in Pol (A) and Env (B) to representatives of the other SIV lineages. The unrooted trees were inferred from 609 amino acids (Pol) and 536 amino acids (Env). SIVmus sequences are highlighted. The numbers at nodes are posterior probabilities estimated by the Bayesian method. Only those 95% and above are shown. Scale bars represent 0.2 and 0.5 replacements per site.
Full-length genome sequences of SIVmus
To characterise the two SIVmus groups in more detail, we amplified complete genomes of three SIVmus strains: 01CM1239, 01CM1246 and 01CM2500. These viruses came from three wild-caught monkeys from two different geographic locations about 40 km apart in southern Cameroon, 148 km and 190 km east of the capital city Yaoundé (Figure 2). Proviral DNA was amplified from uncultured whole blood. For 01CM1246 circular viral DNA was amplified and overlapping linear PCR fragments were amplified for 01CM1239 and 01CM2500.
Figure 2.
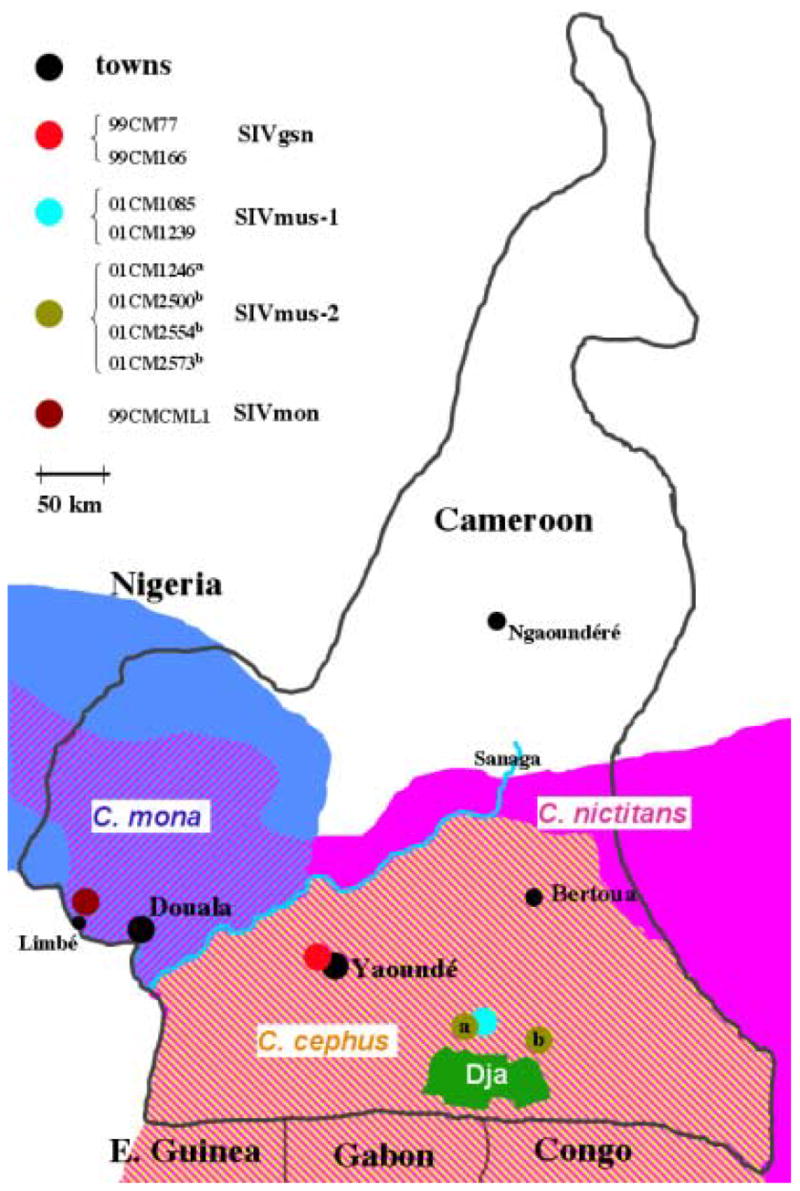
Geographic origins of the SIVmus positive samples and range of C. cephus (in yellow), C. nictitans (in pink) and C. mona (in blue) in Cameroon. Within the area shown, the ranges of C. cephus and C. mona are overlapped by C. nictitans. The previously reported SIVmus-01CM1085 and new SIVmus positive samples from this study were collected in different villages, corresponding to two sampling sites covering a 25 km2 area each, and separated by 40 km around the Dja reserve in south Cameroon. The sites where the previously characterized SIVgsn (99CM71 and 99CM166) and SIVmon (99CMCML1) samples were collected are also indicated on the map.
Previously described degenerate primers were used to amplify a 1984 bp fragment of the pol gene (NDR1/PolOR for the first round and DR4/Uni2 for the second round) for animal 01CM1246. Based on this sequence, specific primers (1246XLS1, 1246XLAS1 and 1246XLAS2) were designed to amplify circular unintegrated viral DNA (Table 2 and Figure 3A). The long PCR product of 8500 bp was purified, then cleaved with EcoRI, and the resulting fragments were cloned and sequenced as previously described (Courgnaud et al., 2003).The full-length genome sequences of 01CM1239 and 01CM2500 were obtained by amplification of overlapping fragments using lineage and strain specific primers as shown in Table 2 and Figure 3B.
Table 2.
Oligonucleotide primers used to amplify full-length genomes of SIVmus according to the schematic representations in Figure 3.
| Fragmenta | Primers | Prime r sequenceb | Region | Size (kb) | ||
|---|---|---|---|---|---|---|
| SIVmus-01CM1246 | ||||||
| Round 1 | NDR1 | TRGAYACAGGRGCWGAYGA | sc | po l | 2.7 | |
| POLOR | ACBACYGCNCCTTCHCCTTTC | as | ||||
| Round 2 | A | PoLIS4 | CCAGCNCACAAAGGNATAGGAGG | s | po l | 0.65 |
| Uni2 | CCCCTATTCCTCCCCTTCTTTTAAAA | as | ||||
| Round 1 | NDR1 | TRGAYACAGGRGCWGAYGA | s | po l | 2.7 | |
| POLOR | ACBACYGCNCCTTCHCCTTTC | as | ||||
| Round 2 | B | DR4 | GGIATWCCICAYCCDGCAGG | s | po l | 1.98 |
| Uni2 | CCCCTATTCCTCCCCTTCTTTTAAAA | as | ||||
| Round 1 | 1246XLS1 | GGAATTCCAAGCTGTTGCATGGTG | s | |||
| 1246XLAS1 | CTCTAGAAGCTTGGCTGTTGTGG | as | circle | 8.8 | ||
| Round 2 | C | 1246XLS1 | GGAATTCCAAGCTGTTGCATGGTG | s | ||
| 1246XLAS2 | AGCTGGTCCCATGTTGTTCACTG | as | circle | 8.5 | ||
| SIVmus-01CM1239 | ||||||
| Round 1 | CNM.F1 | TATCCYTCCYTGTCATCYCTCTTT | s | po l | 2.75 | |
| POLOR2 | ACBACWGCTCCTTCWCCTTTCCA | as | ||||
| Round 2 | A | CNM.F2 | AATGGAGAATGYTMATAGATTTCAG | s | po l | 2.05 |
| CNM.R2 | CCCCYATTCCTCCCTTTTTTTTA | as | ||||
| Round 1 | CNMenvF1 | TGTGTSAAAYTRACHCCNATGTGTGT | s | en v | 2.48 | |
| CNMenvR1 | AACATNNCYTCYAGTCCTCYCTTTTYT | as | ||||
| Round 2 | B | CNMenvF2 | TCCTTYAAYCAGACYACAGARTTYAGRGA | s | en v | 2.14 |
| CNMenvR2 | GGGATAGCCANGAATTNTCNCCAT | as | ||||
| Round 1 | 1239F1 | TGGATTGTACTCACTTAGAAGGAAAAA | s | po l/env | 2.7 | |
| CNMD1 | CCWGTRAAATTRGCATCATTGCATTTTA | as | ||||
| Round 2 | C | 1239F2 | CAGGGAGTAGTAGAAAACAAAAACAAA | s | po l/env | 2.3 |
| CNMD2 | AGTATTGTATNGGRATCGGTTGGAAG | as | ||||
| Round 1 | 1239E1 | GTGGAGCAAGTTGGTGGATAATGA | s | en v/gag | 2.0 | |
| CNM.G1rev | TCAGCATCGCCGAGTGCCTCG | as | ||||
| Round 2 | D | 1239Q1 | GGGAGGACTGGAAGGGATGTTTTA | s | en v/gag | 1.0 |
| SPBSrev | CAAGTCCCTGTTCGGGCGCC | as | ||||
| Round 1 | SPBS | GGCGCCCGAACAGGGACTTG | s | gag/pol | 2.5 | |
| 1239P1 | ATTTTGCTGCCGTGTATTGGAAG | as | ||||
| Round 2 | E | CNM.G1 | CGAGGCACTCGGCGATGCTGA | s | gag/pol | 2.2 |
| 1239P2 | CTTCTTGCTGGTCCCTGGTTATTTA | as | ||||
| SIVmus-01CM2500 | ||||||
| Round 1 | CNM.F1 | TATCCYTCCYTGTCATCYCTCTTT | s | po l | 2.75 | |
| POLOR2 | ACBACWGCTCCTTCWCCTTTCCA | as | ||||
| Round 2 | A | CNM.F2 | AATGGAGAATGYTMATAGATTTCAG | s | po l | 2.05 |
| CNM.R2 | CCCCYATTCCTCCCTTTTTTTTA | as | ||||
| Round 1 | CNMenvF1 | TGTGTSAAAYTRACHCCNATGTGTGT | s | en v | 2.48 | |
| CNMenvR1 | AACATNNCYTCYAGTCCTCYCTTTTYT | as | ||||
| Round 2 | B | CNMenvF2 | TCCTTYAAYCAGACYACAGARTTYAGRGA | s | en v | 2.14 |
| CNMenvR2 | GGGATAGCCANGAATTNTCNCCAT | as | ||||
| Round 1 | 2500F1 | GGACTGCACCCATTTAGAAGGAA | s | po l/env | 2.7 | |
| CNMD1 | CCWGTRAAATTRGCATCATTGCATTTTA | as | ||||
| Round 2 | C | 2500F2 | CACAGAGCCAAGGGGTAGTAGAA | s | po l/env | 2.3 |
| CNMD2 | AGTATTGTATNGGRATCGGTTGGAAG | as | ||||
| Round 1 | 2500L1 | CTATCCCCAAACGCATCCGC | s | en v/gag | 2.0 | |
| CNM.G1rev | TCAGCATCGCCGAGTGCCTCG | as | ||||
| Round 2 | D | 2500L2 | AGAAAAGGGAGGACTGGAAGGGAT | s | en v/gag | 0.8 |
| SPBSrev | CAAGTCCCTGTTCGGGCGCC | as | ||||
| Round 1 | SPBS | GGCGCCCGAACAGGGACTTG | s | gag/pol | 2.5 | |
| 2500P1 | CCTCCTATGTTCCCCTATTTCTCTG | as | ||||
| Round 2 | E | CNM.G1 | CGAGGCACTCGGCGATGCTGA | s | gag/pol | 2.2 |
| 2500P2 | GGAACTGAGAAGGCTGTGTAAGGC | as | ||||
Letters refer to fragments labelled as such in Fig. 3.
R = A or G; M = A or C; W = A or T; S = G or C; Y = C or T; B = C, G, or T; H = A, C, or T; and N = A, C, G, or T.
s: forward primer; as: reverse primer.
Figure 3.
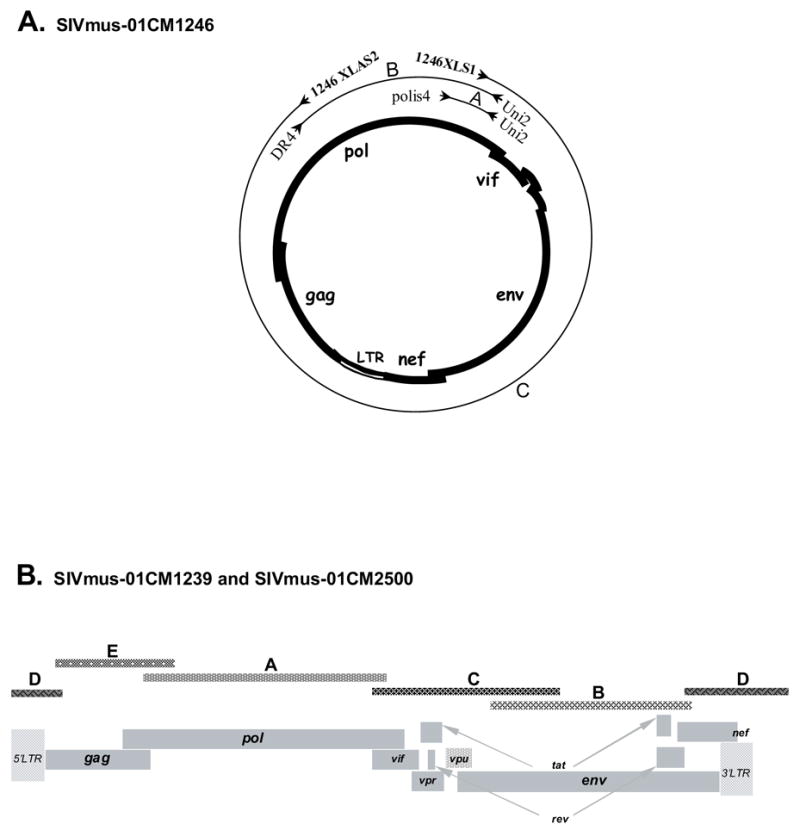
Amplification of full-length genomes of SIVmus: (A) unintegrated circular DNA was targeted for SIVmus-01CM1246 amplification, (B) linear overlapping fragments were amplified to generate the complete genomes for SIVmus-01CM1239 and SIVmus-01CM2500. Primers used to amplify the different PCR fragments are listed in Table 2.
The concatenated 01CM1239, 01CM1246 and 01CM2500 sequences were 9508, 9484 and 9430 nucleotides in length, respectively. The genomic organisation of all three viruses was similar to that of the previously characterized 01CM1085, with reading frames for gag, pol, vif, vpr, tat, rev, env, nef and vpu, characteristic of the SIVgsn/mus/mon lineage as well as SIVcpz/HIV-1.
Genetic distance and phylogenetic analysis of SIVmus genome sequences
Diversity plots
Distances between the newly characterized SIVmus sequences and representatives of the SIVgsn/mus/mon lineage were plotted in windows across the concatenated proteome (Figure 4). 01CM1239 (SIVmus-1) was most similar to 01CM1085 across the entire proteome (Figure 4A). In Gag and Pol, 01CM1239 was as distant from SIVmus-2 (01CM1246 and 01CM2500) as it was from SIVgsn, a virus isolated from a different monkey species; however in Env and Nef SIVmus-2 sequences were only marginally more distant from 01CM1239 than was 01CM1085. In Gag and Pol, 01CM2500 (SIVmus-2) was most similar to 01CM1246 (Figure 4B), while the SIVmus-1 sequences were as distant from 01CM2500 as was SIVgsn; in Env and Nef 01CM2500 was approximately equidistant from each of the other three SIVmus sequences. Thus, in Gag and Pol, there were two distinct and distant SIVmus variants, SIVmus-1 (01CM1085 and 01CM1239) and SIVmus-2 (01CM1246 and 01CM2500). In contrast, in Env and Nef the four SIVmus strains were more similar to each than to SIV from other monkey species, and the two SIVmus-2 strains were no more similar to each other than they were to SIVmus-1 strains.
Figure 4.
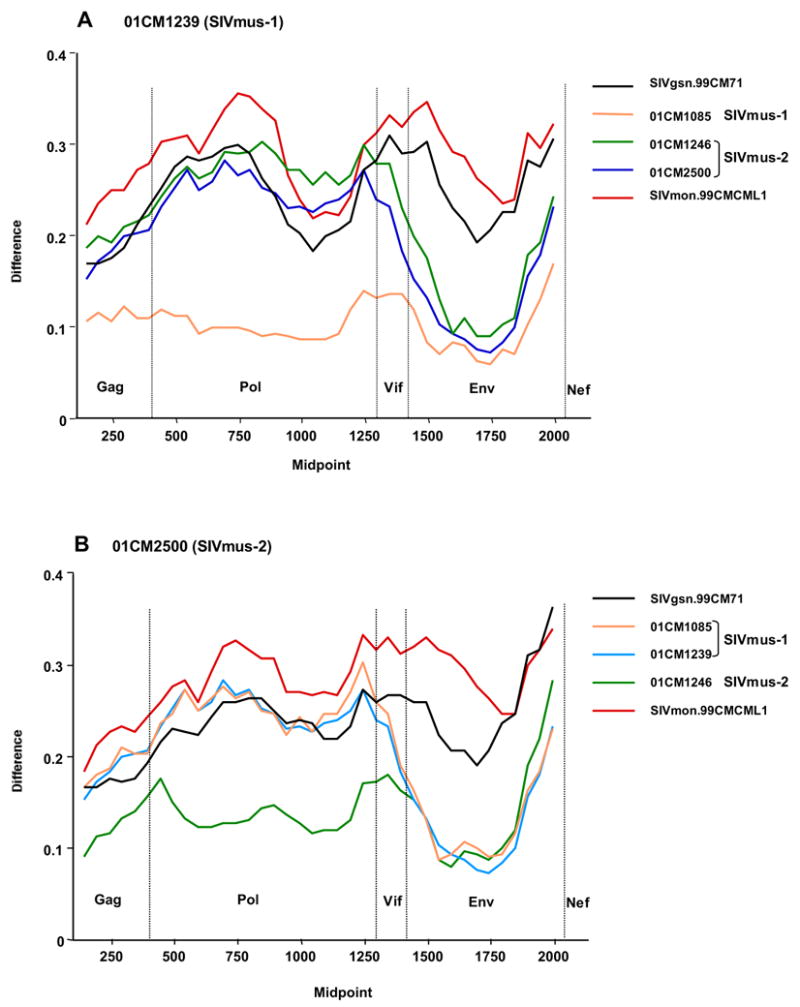
Diversity plots of concatenated Gag, Pol, Vif, Env and Nef protein sequences showing distance from SIVmus-01CM1239 (A) and SIVmus-01CM2500 (B) to other viruses of the SIVgsn/mus/mon lineage. The proportion of amino acid sequence difference per 300 residues window (vertical axis) is plotted against the midpoint of the sequence window (horizontal axis).
Phylogenetic trees
In order to study the relationships of these new SIVmus strains in more detail, we compared multiple trees derived from overlapping windows moved in steps across the proteome. The nature of the results of these analyses is summarized in the four phylogenetic trees in Figure 5. Here the proteome is divided into four regions: Gag, Pol1 (protease and reverse transcriptase), Pol2 (integrase), and Env. In the Gag tree and in both Pol trees the two distinct SIVmus variants described above are evident (Figure 5 A, B and C), whereas in Env all four SIVmus sequences were more closely related to one another and did not separate into two clades (Figure 5D). However, there were additional differences among the trees derived from different regions. In Pol2, SIVmus-2 clustered with SIVgsn (Figure 5C), as for the pol fragment analyzed earlier (Figure 1A). In contrast, in Pol1, while SIVmus-1 and SIVmus-2 formed clearly distinct lineages, they were nevertheless more closely related to each other than to SIVgsn (Figure 5B). In addition, in Pol1 and (albeit without strong support) Pol2, SIVmus and SIVgsn were more closely related to each other than to SIVmon, but in Env SIVgsn and SIVmon formed a clade (Figure 5D). Finally, in Gag, the branching order among the four lineages (SIVmus-1, SIVmus-2, SIVgsn, and SIVmon) was unresolved. Together, the trees provide strong evidence for a complex series of recombination events during the divergence of the four lineages.
Figure 5.
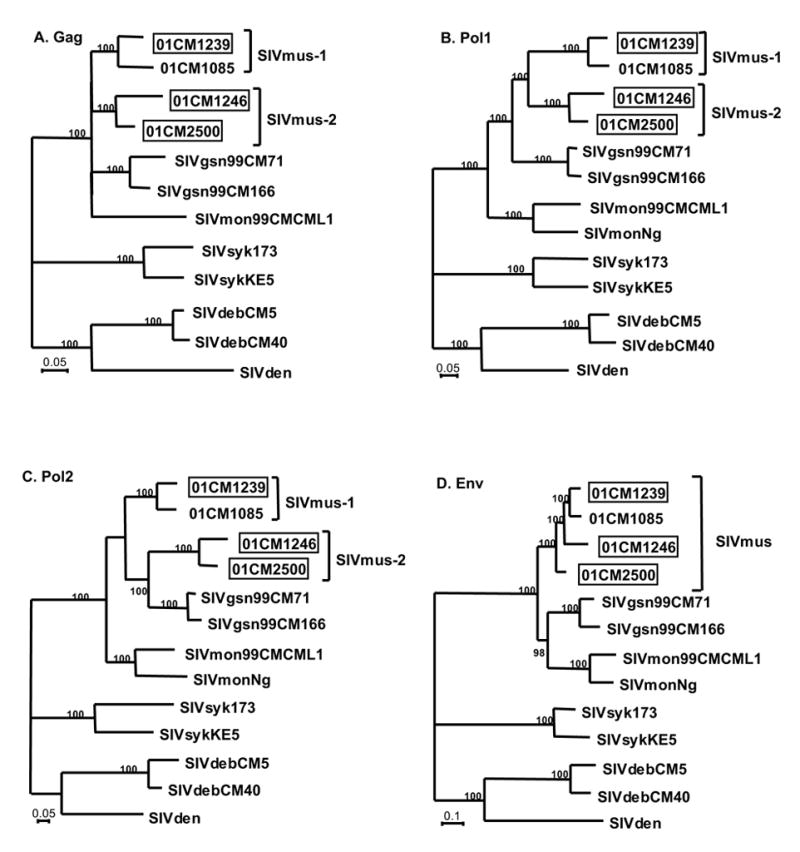
Phylogenetic relationships of SIVmus, SIVgsn and SIVmon. Trees were inferred from protein sequence alignments from different parts of the genome; (A) Gag (sites 1-380 of the proteome alignment), (B) Pol1 (sites 381-896), (C) Pol2 (sites 897-1281) and (D) Env (sites 1415-2057). SIVsyk, SIVdeb and SIVden were used as outgroups to root the trees. Newly characterized full-length genomes are boxed and SIVmus groups are indicated. The numbers on internal branches are the estimated posterior probabilities from the Bayesian method (values of 95% and above are shown). Scale bars indicate 0.05 or 0.1 substitutions per site.
In an attempt to elucidate these events, we calculated the genetic distances within and between the two SIVmus clades and compared them to the intra- and inter-host species distances observed for SIVgsn and SIVmon. A table of pairwise distances was obtained by summing the branch lengths in the trees in Figure 5. We then compared the genetic distances between the two SIVmus variants for the different genomic regions with the other intra- and inter-host genetic distances. The results, summarized in Table 3, show that the genetic distance between SIVmus-1 and SIVmus-2 for Env (Lane A versus B, shaded value) is in the range of within host species comparisons for SIVgsn and SIVmon. In contrast, for Gag and Pol the distances between the two SIVmus variants were more similar to those between viruses from different host species (Lane A versus B, bold values). These analyses suggest that the Env sequences of both SIVmus-1 and SIVmus-2 could represent a “true” SIVmus lineage, whereas the high divergence between SIVmus-1 and SIVmus-2 in Gag and Pol most likely reflects the result of recombination between SIVmus and a more divergent, as yet unidentified, SIV lineage infecting another species. As a result of this cross-species transmission and recombination, the ancestor of either SIVmus-1 or SIVmus-2 in a mustached monkey acquired divergent gag and pol sequences, which then spread further among mustached monkeys. However, the discordance among the trees from different regions with respect to the relationships among SIVmus-1, SIVmus-2, SIVgsn and SIVmon suggests that there were additional recombination events involving their ancestors.
Table 3.
Within and between host species comparisons of genetic distances between the two SIVmus groups, SIVgsn and SIVmon.
| Gag | Pol1 | Pol2 | Env | |
|---|---|---|---|---|
| Aa: SIVmus-1 (1239 vs 1085) | 0.12 | 0.12 | 0.10 | 0.11 |
| B: SIVmus-2 (1246 vs 2500) | 0.14 | 0.15 | 0.13 | 0.18 |
| C: SIVgsn (71 vs 166) | 0.12 | 0.06 | 0.06 | 0.15 |
| D: SIVmon (CML1 vs NG) | d | 0.27 | 0.23 | 0.22 |
| A vs Bb: mus-1 vs mus-2 | 0.24 | 0.43 | 0.38 | 0.19 |
| A vs Cc: mus-1 vs gsn | 0.26 | 0.48 | 0.32 | 0.46 |
| B vs C: mus-2 vs gsn | 0.26 | 0.44 | 0.32 | 0.44 |
| A vs D: mus-1 vs mon | 0.31 | 0.62 | 0.37 | 0.53 |
| B vs D: mus-2 vs mon | 0.32 | 0.59 | 0.49 | 0.51 |
| C vs D: gsn vs mon | 0.33 | 0.53 | 0.43 | 0.51 |
A, B, C, and D are within host species comparisons and are calculated by summing the branch lengths between isolates named in Gag, Pol1, Pol2 and Env trees in Figure 5.
A vs B is the average branch length between SIVmus-1 and SIVmus-2 in each of the trees in Figure 5.
A vs C, B vs C, A vs D, B vs D, and C vs D are between host species comparisons: A versus C is the average branch length between, SIVmus-01CM1085 and SIVgsn.99CM71, SIVmus-01CM1085 and SIVgsn-99CM166, SIVmus-01CM1239 and SIVgsn.99CM71 and SIVmus-01CM1239 and SIVgsn-99CM166 in each of the trees in Figure 5. Comparisons B versus C, A versus D, B versus D and C versus D were similarly obtained.
The distance could not be calculated in Gag for SIVmon since full-length gag sequence is not available for SIVmonNG.
DISCUSSION
Here we report a detailed study of the genetic diversity of SIVmus in mustached monkeys (Cercopithecus cephus) from Cameroon. SIVmus has only recently been identified, together with SIVgsn from greater spot-nosed monkeys (C. nictitans) and SIVmon from mona monkeys (C. mona) from Cameroon and Nigeria (Barlow et al., 2003; Courgnaud et al., 2003; Courgnaud et al., 2002). Within the primate lentivirus tree, these three viruses are each other’s closest relatives, and share a similar genomic organization including the presence of a vpu gene (Courgnaud et al., 2003). These viruses are of particular interest because an ancestor of this lineage is the source of the vpu and env genes of SIVcpz, and hence of HIV-1 (Bailes et al., 2003; Sharp et al., 2005). Furthermore, mustached and greater spot-nosed monkeys represent a large part of the primate bushmeat in west central Africa and thus humans are directly exposed to SIVgsn and SIVmus.
All viruses are not equivalent for transmission and/or infectivity and the factors contributing to successful simian-to-simian or simian-to-human transmissions are not fully determined. Until recently, it was believed that all SIV infections were common and widespread in their hosts, but in a recent study we showed that SIV prevalence can vary significantly from one species to another (Aghokeng et al., 2006). Mustached and greater spot-nosed monkeys were species in which we found a very low SIV prevalence (about 3–4%), approximately ten times lower than prevalence rates observed in African green monkeys, sooty mangabeys, mandrills, and mantled guerezas (Apetrei et al., 2005; Bibollet-Ruche et al., 1997; Courgnaud et al., 2001; Jolly et al., 1996; Santiago et al., 2005; Souquiere et al., 2001; Takehisa et al., 2001). This estimate of low prevalence is reinforced by the work presented here. In particular, we examined whether samples from mustached monkeys that have cross-reactive antibodies in the INNO-LIA HIV assay, but not SIVmus-specific antibodies in the SIVmus rgp41 ELISA, are infected with an SIV. SIV sequences could only be amplified in the SIVmus ELISA positive samples. These results confirm our previous findings that HIV cross-reactive assays such as INNO-LIA overestimate the prevalence of SIV infection among mustached monkeys (Aghokeng et al., 2006). However, we cannot completely exclude the possibility of infection with an SIV that is not detected with the available serological and molecular tools.
To study the diversity of SIVmus, five new strains were genetically characterized and for three of them full length genome sequences were obtained. This revealed that mustached monkeys are infected with two distinct SIVmus variants, here designated SIVmus-1 and SIVmus-2, which differ in gag and pol (but not env) to a similar extent as SIVs from different host species. Mustached monkeys are the second non-human primate species found to be infected with two different SIV variants. Mandrills (Mandrillus sphinx) were the first to be reported to harbour two distinct viruses (Souquiere et al., 2001; Takehisa et al., 2001). However, the mandrills infected with SIVmnd-1 and SIVmnd-2 are geographically separated, to the south and to the north of the Ogooué River in Gabon. In contrast, SIVmus-1 and SIVmus-2 were found in samples collected at sites only 5 km apart, and all the samples in this study were collected from adjacent locations no more than 40 km apart, near the Dja Reserve in southern Cameroon. The habitat range of C.cephus represents 30–90 ha and they can travel up to more than 1 km a day (Gautier-Hion et al., 1999). Samples taken at 40km distance are thus most likeky from geographically separated monkey colonies, but for those taken at 5km distance it can not be excluded that their habitats overlap or that they belong to a same colonie.
SIVs from eight Cercopithecus monkey species have been fully characterized. Six of these, namely SIVgsn, SIVmus and SIVmon together with SIVsyk from Sykes’s monkey (C. albogularis), SIVdeb from De Brazza’s monkey (C. neglectus) and SIVden from Dent’s mona monkey (C. denti), form a single major lineage in the primate lentivirus tree (Bibollet-Ruche et al., 2004; Dazza et al., 2005). These Cercopithecus monkey viruses also share functional motifs in gag and env that distinguish them from other primate lentiviruses (Bibollet-Ruche et al., 2004; Dazza et al., 2005). Within this Cercopithecus monkey SIV radiation SIVgsn, SIVmon and SIVmus form a discrete clade. However, the hosts of these three viruses are not each other’s closest relatives; for example, greater spot-nosed monkeys are classified with Sykes’s monkeys in the C. mitis group, while mona monkeys and Dent’s mona are in the C. mona group (Groves, 2001). Thus, the early radiation of these viruses must have involved cross-species transmission events (Dazza et al., 2005). Phylogenetic analyses including the newly characterized SIVmus genome sequences indicated that there was also recombination during this diversification, since the relationships among SIVmus-1, SIVmus-2, SIVgsn and SIVmon vary depending on the region of the genome analyzed.
Analyses of the diversity between SIVmus-1 and SIVmus-2 (Table 3) suggest that the Env sequences of both could represent a “true” SIVmus lineage, whereas the high divergence in Gag and Pol most likely reflects the result of recombination between SIVmus and a more divergent, as yet unidentified, SIV lineage infecting another species. As a result of this cross-species transmission and recombination, the ancestor of either SIVmus-1 or SIVmus-2 in a mustached monkey acquired divergent gag and pol sequences, which then spread further among mustached monkeys. An alternative explanation of the phylogenetic results (Figure 5) is that mustached monkeys have long been infected by two divergent SIVs, and that the env gene of one form has spread to the other by recombination, replacing the other env gene in at least the samples examined here, and perhaps in all strains. Since there is no other example of sympatric members of a single monkey species harboring two such divergent lineages of SIV, this explanation seems unlikely. Under either scenario, the discordance among the trees from different regions (Figure 5) with respect to the relationships among SIVmus-1, SIVmus-2, SIVgsn and SIVmon suggests that there were additional recombination events involving their ancestors. Thus, cross-species transmission and recombination appear to have played a significant role in the evolution of the SIVgsn/mon/mus lineage. It is well documented that C. cephus and C. nictitans share habitats, and polyspecific associations between them have been reported (Gautier-Hion et al., 1999). These mixed associations are believed to improve foraging efficiency and protection against predators, but can also significantly contribute to inter-species virus exchange through fighting or other risk behaviour. In addition, cross-breeding has been reported within the Cercopithecus genus (Gautier-Hion et al., 1999), and therefore the sexual route could also facilitate virus exchange between different species.
A candidate for the as yet unidentified monkey host of the SIV which has recombined with SIVmus is C. pogonias, since this monkey is also reported to live in mixed troops with C. cephus and C. nictitans (Gautier-Hion et al., 1999). We have observed some cross-reactions with HIV-1/2 antigens and SIV antigens in samples collected from C. pogonias in Cameroon, but we were unable to confirm SIV infection by PCR using the universal and sensitive primers that have allowed the amplification of all documented major SIV lineages (Aghokeng et al., 2006). C. pogonias may be infected with a highly divergent SIV, or SIV prevalence may be low, as it is in greater spot-nosed and mustached monkeys, such that our sample size was too small to identify a positive animal.
It is interesting to note that the two SIVs found in mandrills also appear to have originated through cross-species transmission and recombination events (Souquiere et al., 2001; Takemura and Hayami, 2004). SIVmnd-1 is most closely related to SIVlho and SIVsun from L’Hoest’s monkeys and sun-tailed monkeys, two species from the Cercopithecus lhoesti group, while SIVmnd-2 is most closely related to SIVmnd-1 in env but to SIVrcm (from redcapped mangabeys) in the 5′ end of its genome. Recombination events are likely to generate a variety of mosaic genomes, with different combinations of regions from the two parental viruses, which vary with respect to their fitness in the host. Notably, in the case of both the two mandrill SIVs and the two mustached monkey SIVs described here, the env genes are similar in the two variants from a single host, perhaps because the Env protein of the invading virus was less well adapted to the new host.
Elucidating the origin and evolution of primate lentiviruses is very complex and our results clearly show a major role of cross-species transmission and recombination. To determine the extent to which cross-species transmission between co-habiting species has shaped the evolution of primate lentiviruses the screening of geographically separated populations of the same species will be necessary. Knowledge of monkey behaviour and past and current geographical distributions will aid the interpretation of the primate lentivirus phylogeny.
MATERIALS AND METHODS
Animals and samples
Whole-blood samples from 46 wild-caught mustached monkeys with cross-reacting antibodies to HIV antigens using the INNO-LIA HIV confirmatory test or the previously described SIVmus specific recombinant gp41 ELISAs were further genetically analyzed to confirm SIV infection (Aghokeng et al., 2006). The majority of these samples (n = 32) were from a previously published bushmeat survey (Peeters et al., 2002), and additional sera were identified more recently during a follow-up survey (n = 14). Samples were obtained from geographically diverse sites throughout southern Cameroon, including bushmeat markets in and around the capital city Yaoundé, logging concessions in southeastern Cameroon, and villages in southwestern Cameroon. The selection criterion for all samples to be further analyzed was that they were positive for HIV cross-reacting antibodies and/or SIV specific antibody assays, were available in sufficient quantities, and contained non-degraded cellular DNA as confirmed by glucose-6-phosphate dehydrogenase (G6PDH) gene amplification (von Dornum and Ruvolo, 1999). As described previously, blood was collected from primate bushmeat by cardiac puncture (Peeters et al., 2002); all samples were obtained with approval from the Cameroonian Ministry of Environment and Forestry.
Confirmation of SIV infection by PCR and sequence analysis in pol and/or env.
Total DNA was isolated from whole blood using the QIAamp blood kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s instructions. For all samples a small fragment of pol (650bp) spanning the RT/integrase 3′ region was PCR amplified with degenerate consensus primers NDR1-PolOR for the first round and Polis4-Uni2 for the second round as previously described (Courgnaud et al., 2001). In order to increase the amplification rates of SIVmus strains, we designed SIVgsn/mus/mon lineage specific primers based on previously published full-length sequences of SIVmus (01CM1085), SIVgsn (99CM71 and 99CM166), and SIVmon (99CMCML1 and SIVmonNG1) (Barlow et al., 2003; Courgnaud et al., 2003; Courgnaud et al., 2002). Primers were designed to amplify a 2050 bp fragment in pol with CNM.F1 and PolOR2 for the first round and CNM.F2 and CNM.R2 for the second round (Table 2). An additional SIVgsn/mus/mon lineage primer set was designed to amplify a 2150 bp fragment in the env gene covering the V3 and gp41 region with CNMenvF1 and CNMenvR1 for the first round and CNMenvF2 and CNMenvR2 for the second round (Table 2).
Amplification of complete SIVmus genomes
For three animals, the full-length SIVmus sequences were obtained either by amplification of unintegrated circular DNA (01CM1246) or by overlapping PCR fragments (01CM1239 and 01CM2500). The amplification strategies and primers used to obtain the different fragments are summarized in Table 2 and Figure 3.
For SIVmus-01CM1246 a 1984 bp fragment was obtained in pol using NDR1 and PolOR in the first round, and DR4 and Uni2 in the second round, as previously described (Courgnaud et al., 2003). To obtain the full-length sequence of SIVmus-01CM1246, two specific primers were generated based on the 1984 bp pol sequence to amplify in the first round unintegrated circular viral forms with 1246XLS1 (GGAATTCCAAGCTGTTGCATGGTG) and 1246XLAS1 (CTCTAGAAGCTTGGCTGTTGTGG) and a semi-nested second round with 1246XLS1 and 1246XLAS2 (AGCTGGTCCCATGTTGTTCACTG). A PCR product of 8500 bp was obtained (Table 2 and Figure 3A). This long fragment was then purified, cleaved with EcoRI and the resulting fragments were cloned and sequenced as previously described (Courgnaud et al., 2003).
Unintegrated circular DNA could not be amplified for SIVmus-01CM1239 and SIVmus-01CM2500, and the full-length sequences were obtained by amplification of overlapping PCR fragments with consensus primers designed based on the SIVgsn/mus/mon sequences available or on strain specific sequences. Primers and amplification strategies used to obtain these two complete genomes are summarized in Table 2 and Figure 3B.
All amplifications were performed by using the Long Expand PCR kit (Roche Molecular Biochemicals, Mannhein, Germany) according to the manufacturer’s instructions. Briefly, approximately 500 ng of genomic DNA was used for first round PCR amplifications. For the second round, 5 μl from the first round were used. Each amplification reaction included a manual hot-start (92°C for 2 min) and 30 cycles of denaturation-annealing-extension. Annealing temperatures varied from 45° to 55° and where determined according to the Tm (melting temperature) of primers used. Extension times varied depending on the size of the expecting fragment and were typically set at 1 min/1.5 kb. Amplified fragments were agarose gel purified, and sequenced by cycle sequencing and dye terminator methods with the Big Dye terminator kit v.3.1 (Applied Biosystems, Foster City, CA) using an automated capillary sequencer (ABI Prism 3130xl Genetic Analyzer; Applied Biosystems, Foster City, CA). To reconstitute the full-length genome sequence, overlapping sequences were assembled into contiguous sequences by using SeqMan II software (DNASTAR, Madison, WI).
Diversity plots and phylogenetic analyses
Amino acid sequences were aligned using Clustal W (Thompson et al., 1994), with minor manual adjustments. Sites that could not be unambiguously aligned and sites with a gap in any sequence were discarded. The proteome alignment was generated by joining Gag, Pol, Vif, Env, and Nef amino acid sequence alignments; the carboxyl termini of Gag, Pol, and Env that overlap with Pol, Vif and Nef, respectively, were excluded. Diversity plots were made using a sliding window of 300 amino acids, moved in steps of 50 residues. The predicted protein sequences encoded by the new SIVmus strains (SIVmus-01CM1239, SIVmus-01CM1246, SIVmus-01CM2500, SIVmus-01CM2554 and SIVmus-01CM2573) were compared with other SIV strains obtained from GenBank (http://www.ncbi.nlm.nih.gov/): SIVmus-01CM1085 (accession number AY340700), SIVgsn-99CM71 (AF468658), SIVgsn-99CM166 (AF468659), SIVmon-99CMCML1 (AY340701), SIVmonNG1 (AJ549283), SIVsyk173 (L06042), SIVsykKE5 (AY523867), SIVdebCM5 (AY523866), SIVdebCM40 (AY523865), SIVden (AJ580407), SIVtal266 (AF478595), SIVtal-00CM8023 (AM182197), SIVascSCHM (AJ551401), SIVbkm (AY518534), SIVagmVER155 (M29975), SIVagmGRI677 (M58410), SIVagmTAN1 (U58991), SIVsmmSL92B (AF334679), SIVsmmPBJ (M31325), SIVrcmGAB1 (AF382829), SIVrcmNG411 (AF349680), SIVcpzGAB2 (AF382828), SIVcpzTAN1 (AF447763), SIVmndGB1 (M27470), SIVsunL14 (AF131870), SIVlho7 (AF075269), SIVmnd14 (AF328295), SIVdrlFAO (AY159321), SIVwrc (AY138268), SIVolc (AY138269) and SIVcolCGU1 (AF301156). Trees were inferred by the Bayesian method (Yang and Rannala, 1997) implemented in MrBayes (Huelsenbeck and Ronquist, 2001) using the Jones, Taylor and Thornton model of protein evolution (Jones et al., 1992) with gamma distributed rates at sites (Yang, 1994) and 1 million generations. Bayesian likelihoods and parameters were examined with the Tracer program (http://evolve.zoo.ox.ac.uk/software.html?id=tracer) and all estimated sample sizes were above 1000.
Nucleotide sequence accession number
The complete sequences are available in GenBank under the following accessionnumbers: EF070329 (SIVmus01CM1246), EF070330 (SIVmus01CM1239), EF070331 (SIVmus01CM2500), EF070332 (SIVmus01CM2554) and EF070333 (SIVmus01CM2573).
Acknowledgments
This work was supported in part by grants from the National Institute of Health (RO1 AI 50529) and the Agence Nationale de Recherches sur le SIDA (ANRS). Avelin Aghokeng received a postdoctoral fellowship from the Agence Nationale de Recherches sur le SIDA (ANRS).
We thank the Cameroonian Ministries of Health, Research, and Environment and Forestry and Wildlife, for permission to perform this study, and the staff from project PRESICA for logistical support and assistance in the field.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aghokeng AF, Liu W, Bibollet-Ruche F, Loul S, Mpoudi-Ngole E, Laurent C, Mwenda JM, Langat DK, Chege GK, McClure HM, Delaporte E, Shaw GM, Hahn BH, Peeters M. Widely varying SIV prevalence rates in naturally infected primate species from Cameroon. Virology. 2006;345:174–189. doi: 10.1016/j.virol.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Apetrei C, Metzger MJ, Richardson D, Ling B, Telfer PT, Reed P, Robertson DL, Marx PA. Detection and partial characterization of simian immunodeficiency virus SIVsm strains from bush meat samples from rural Sierra Leone. J Virol. 2005;79:2631–2636. doi: 10.1128/JVI.79.4.2631-2636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Barlow KL, Ajao AO, Clewley JP. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona) J Virol. 2003;77:6879–6888. doi: 10.1128/JVI.77.12.6879-6888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibollet-Ruche F, Bailes E, Gao F, Pourrut X, Barlow KL, Clewley JP, Mwenda JM, Langat DK, Chege GK, McClure HM, Mpoudi-Ngole E, Delaporte E, Peeters M, Shaw GM, Sharp PM, Hahn BH. New simian immunodeficiency virus infecting De Brazza’s monkeys (Cercopithecus neglectus): evidence for a Cercopithecus monkey virus clade. J Virol. 2004;78:7748–7762. doi: 10.1128/JVI.78.14.7748-7762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibollet-Ruche F, Brengues C, Galat-Luong A, Galat G, Pourrut X, Vidal N, Veas F, Durand JP, Cuny G. Genetic diversity of simian immunodeficiency viruses from West African green monkeys: evidence of multiple genotypes within populations from the same geographical locale. J Virol. 1997;71:307–313. doi: 10.1128/jvi.71.1.307-313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbet S, Muller-Trutwin MC, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Abela B, Pourrut X, Mpoudi-Ngole E, Loul S, Delaporte E, Peeters M. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different Cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J Virol. 2003;77:12523–12534. doi: 10.1128/JVI.77.23.12523-12534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Pourrut X, Bibollet-Ruche F, Mpoudi-Ngole E, Bourgeois A, Delaporte E, Peeters M. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J Virol. 2001;75:857–866. doi: 10.1128/JVI.75.2.857-866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Salemi M, Pourrut X, Mpoudi-Ngole E, Abela B, Auzel P, Bibollet-Ruche F, Hahn B, Vandamme AM, Delaporte E, Peeters M. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J Virol. 2002;76:8298–8309. doi: 10.1128/JVI.76.16.8298-8309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazza MC, Ekwalanga M, Nende M, Shamamba KB, Bitshi P, Paraskevis D, Saragosti S. Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent’s Mona monkey (Cercopithecus mona denti) J Virol. 2005;79:8560–8571. doi: 10.1128/JVI.79.13.8560-8571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Gautier-Hion A, Colyn M, Gautier JP. Histoire naturelle des primates d’Afrique centrale. Ecofac Multipress; Gabon: 1999. [Google Scholar]
- Groves C. Smithsonian Series in Comparative Evolutionary Biology. Smithsonian Institution Press; 2001. Primate taxonomy. [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jolly C, Phillips-Conroy JE, Turner TR, Broussard S, Allan JS. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops) J Med Primatol. 1996;25:78–83. doi: 10.1111/j.1600-0684.1996.tb00198.x. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Courgnaud V, Abela B, Auzel P, Pourrut X, Bibollet-Ruche F, Loul S, Liegeois F, Butel C, Koulagna D, Mpoudi-Ngole E, Shaw GM, Hahn BH, Delaporte E. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg Infect Dis. 2002;8:451–457. doi: 10.3201/eid0805.01-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Fruteau C, Noe R, Peeters M, Brookfield JF, Shaw GM, Sharp PM, Hahn BH. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d’Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79:12515–12527. doi: 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Shaw GM, Hahn BH. Simian immunodeficiency virus infection of chimpanzees. J Virol. 2005;79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquiere S, Bibollet-Ruche F, Robertson DL, Makuwa M, Apetrei C, Onanga R, Kornfeld C, Plantier JC, Gao F, Abernethy K, White LJ, Karesh W, Telfer P, Wickings EJ, Mauclere P, Marx PA, Barre-Sinoussi F, Hahn BH, Muller-Trutwin MC, Simon F. Wild Mandrillus sphinx are carriers of two types of lentivirus. J Virol. 2001;75:7086–7096. doi: 10.1128/JVI.75.15.7086-7096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa J, Harada Y, Ndembi N, Mboudjeka I, Taniguchi Y, Ngansop C, Kuate S, Zekeng L, Ibuki K, Shimada T, Bikandou B, Yamaguchi-Kabata Y, Miura T, Ikeda M, Ichimura H, Kaptue L, Hayami M. Natural infection of wild-born mandrills (Mandrillus sphinx) with two different types of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 2001;17:1143–1154. doi: 10.1089/088922201316912754. [DOI] [PubMed] [Google Scholar]
- Takemura T, Hayami M. Phylogenetic analysis of SIV derived from mandrill and drill. Front Biosci. 2004;9:513–520. doi: 10.2741/1242. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dornum M, Ruvolo M. Phylogenetic relationships of the New World monkeys (Primates, platyrrhini) based on nuclear G6PD DNA sequences. Mol Phylogenet Evol. 1999;11:459–476. doi: 10.1006/mpev.1998.0582. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo Method. Mol Biol Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]


